Abstract
The RNA genome of the hepatitis C virus (HCV) contains multiple conserved structural cis domains that direct protein synthesis, replication, and infectivity. The untranslatable regions (UTRs) play essential roles in the HCV cycle. Uncapped viral RNAs are translated via an internal ribosome entry site (IRES) located at the 5′ UTR, which acts as a scaffold for recruiting multiple protein factors. Replication of the viral genome is initiated at the 3′ UTR. Bioinformatics methods have identified other structural RNA elements thought to be involved in the HCV cycle. The 5BSL3.2 motif, which is embedded in a cruciform structure at the 3′ end of the NS5B coding sequence, contributes to the three-dimensional folding of the entire 3′ end of the genome. It is essential in the initiation of replication. This paper reports the identification of a novel, strand-specific, long-range RNA–RNA interaction between the 5′ and 3′ ends of the genome, which involves 5BSL3.2 and IRES motifs. Mutants harboring substitutions in the apical loop of domain IIId or in the internal loop of 5BSL3.2 disrupt the complex, indicating these regions are essential in initiating the kissing interaction. No complex was formed when the UTRs of the related foot and mouth disease virus were used in binding assays, suggesting this interaction is specific for HCV sequences. The present data firmly suggest the existence of a higher-order structure that may mediate a protein-independent circularization of the HCV genome. The 5′–3′ end bridge may have a role in viral translation modulation and in the switch from protein synthesis to RNA replication.
Keywords: HCV, IRES, 3′ UTR, HCV CRE, long-range RNA–RNA interactions
INTRODUCTION
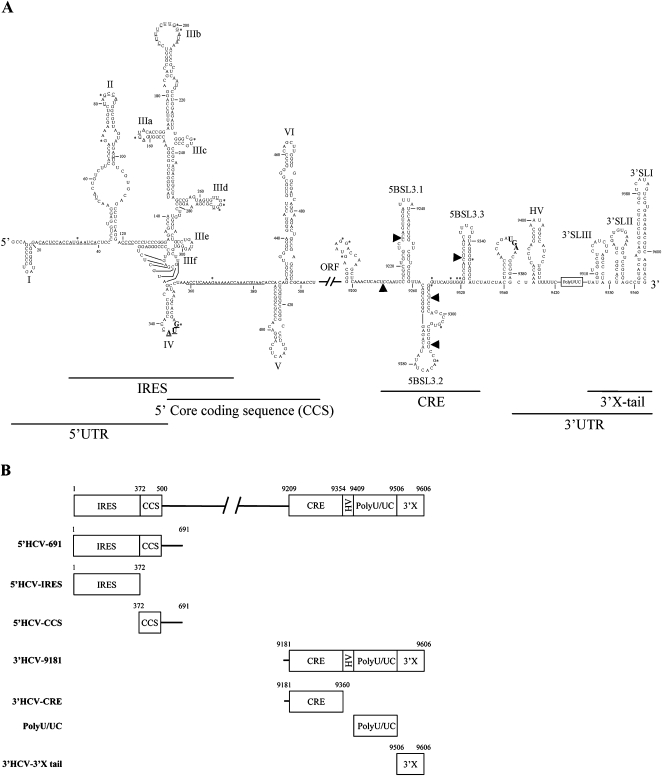
The hepatitis C virus (HCV) genome is a 9.6 kb long, (+) polarity, single-stranded RNA molecule (Choo et al. 1989; Kato et al. 1990; Takamizawa et al. 1991). The single open reading frame (ORF) is flanked by highly structured 5′ and 3′ untranslatable regions (UTRs), which contain essential domains for viral translation, replication, and infectivity (Fig. 1A; Tsukiyama-Kohara et al. 1992; Wang et al. 1993; Kolykhalov et al. 2000; Friebe et al. 2001; Friebe and Bartenschlager 2002; Yi and Lemon 2003a,b). The genomic RNA serves as mRNA for the production of a single polyprotein product, which is co- and post-translationally processed by viral and cellular proteases to yield structural and nonstructural proteins. Viral protein synthesis is initiated in a cap-independent manner by a highly conserved domain that acts as an internal ribosome entry site (IRES) (Fig. 1A), largely located at the 5′ UTR (Tsukiyama-Kohara et al. 1992; Wang et al. 1993). This represents an alternative mechanism to that employed by cellular mRNAs, in which 40S ribosomal subunits are directly recruited in the absence of any other canonical initiation factor, directly positioning the start codon at the ribosome P site (Lytle et al. 2002; Ji et al. 2004; Otto and Puglisi 2004). The HCV IRES spans a region of 340 nucleotides (nt) that includes a short stretch of the 5′ core coding sequence (Fig. 1A; Reynolds et al. 1995; Wang et al. 2000), and has a complex organization that must be preserved for it to be active (Lukavsky et al. 2000; Odreman-Macchioli et al. 2000; Collier et al. 2002; Kieft et al. 2002). Under physiological magnesium concentrations, the IRES is folded into four stem–loop motifs (designated I–IV) that define different functional domains (Fig. 1A), each with essential roles in ribosome recruitment and viral RNA synthesis. Domain IIId, which is particularly important in IRES function (Barria et al. 2008), is composed of two short helices separated by a loop E and capped by an apical loop folded into a U-turn motif (Fig. 1A; Jubin et al. 2000; Klinck et al. 2000). Domain IIId acts as the main anchoring site for the 40S ribosomal subunit (Kolupaeva et al. 2000; Lukavsky et al. 2000; Babaylova et al. 2009). The high sequence and structural conservation rate among the hepaciviruses and even related RNA viruses firmly supports its requirement for viral persistence (Jubin et al. 2000; Barria et al. 2008).
FIGURE 1.
(A) Sequence and secondary structure for the 5′ and the 3′ ends of the HCV genome. The 5′ UTR plus domains V and VI located at the core coding sequence are included. The minimum region for IRES activity is shown. The 3′ end of the viral genomic RNA is organized into two structural elements: the CRE region and the 3′X-tail, separated by a hypervariable sequence and the polyU/UC stretch. Numbers refer to the nucleotide positions of the HCV Con1 isolate. Residues accessible to RNase T1, RNase V1, or lead processing under nondenaturing conditions are indicated by an asterisk, an arrow, or underlined, respectively. Start and stop translation codons placed at positions 342 and 9371, respectively, are shown in bold. (B) Diagram of the transcripts encompassing different functional domains of both the 5′ and the 3′ ends of the HCV genome used in this study.
The 3′ UTR is an evolutionarily conserved structural element around 200 nt long located at the 3′ end of the HCV RNA genome (Fig. 1A; Kolykhalov et al. 1996), and has a critical function in HCV replication (Kolykhalov et al. 2000; Friebe and Bartenschlager 2002; Yi and Lemon 2003a, b) and virion infectivity (Yanagi et al. 1999). Three different domains can be identified in this UTR: the highly variable region (VR), a polyU/UC tract of variable length and composition, and the 3′X tail, composed of three stem-loops (Fig. 1A, 3′SLI, 3′SLII, 3′SLIII; Tanaka et al. 1996; Blight and Rice 1997; Ito and Lai 1997).
Other functional and evolutionarily conserved RNA motifs distinct from those present in the UTRs have been recently described by analyzing the variability of synonymous sites, through structural alignment studies, and through classical phylogenetic comparisons. These motifs may act as cis signals that modulate essential steps of the viral cycle (Tuplin et al. 2002, 2004; Lee et al. 2004; You et al. 2004). The 5′ core coding sequence shows a high sequence conservation rate, which was initially thought to be related to the existence of alternative reading frames (Walewski et al. 2001; Xu et al. 2001; Choi et al. 2003; Branch et al. 2005); however, its importance has recently been shown in the preservation of structures important for IRES activity and replication (domains V and VI) (Fig. 1A; Wang et al. 2000; Kim et al. 2003; Beguiristain et al. 2005; McMullan et al. 2007; Vassilaki et al. 2008). Within the 3′ end of the NS5B coding sequence, the stem–loop 5BSL3.2 is embedded in a cruciform structure that has been identified as a cis-essential element for viral RNA synthesis (cis-acting replication element [CRE]) (Fig. 1A; Lee et al. 2004; You et al. 2004). The 5BSL3.2 stem–loop consists of two G-C rich helices connected by an eight-base bulge, and an apical loop (Fig. 1A; Friebe et al. 2005). Disruptions in this folding lead to replication-incompetent HCV genomes (You et al. 2004; Friebe et al. 2005). Moreover, subtle changes in the loop region prevent RNA replication, indicating that sequence specificity is required for interaction with protein factors and/or RNA functional elements. Indeed, a kissing complex important for viral RNA synthesis is established between the apical loop of 5BSL3.2 and 3′SL2 in the 3′HCV-3′X tail (Friebe et al. 2005). More recently, the internal loop of 5BSL3.2 has been involved in the formation of an apical loop–internal loop (ALIL) interaction with structural elements located upstream in the NS5B coding sequence, suggesting the importance of long-range RNA–RNA interactions in the modulation of multiple steps of the viral cycle (Diviney et al. 2008).
Most cellular mRNAs and many viral RNAs adopt a circular conformation that promotes efficient protein synthesis (Sachs et al. 1997; Edgil and Harris 2006). The acquisition of this conformation is mainly mediated by the recruitment of protein factors that simultaneously interact with both ends of the mRNA, facilitating the bridging between the 5′ and the 3′ ends (Sachs et al. 1997). Alternatively, some viruses have developed RNA motifs located at both ends of the genome with complementary sequences that promote the protein-independent circularization of the mRNA. This is the case of flavivirus (Harris et al. 2006), certain picornaviruses, such as FMDV (Serrano et al. 2006), retroviruses (Ooms et al. 2007; Kenyon et al. 2008), or Dengue virus (Polacek et al. 2009), among others. With respect to HCV, conflicting results regarding the role of the 3′ UTR in viral protein synthesis have been reported (Ito et al. 1998; Ito and Lai 1999; Fang and Moyer 2000; Michel et al. 2001; Murakami et al. 2001; Kong and Sarnow 2002; McCaffrey et al. 2002; Imbert et al. 2003; Bradrick et al. 2006; Song et al. 2006; Lourenco et al. 2008). Today it is commonly accepted that the 3′ end exerts an enhancer translational effect through the recruitment of viral and cellular protein factors that stimulate IRES activity (McCaffrey et al. 2002; Bradrick et al. 2006; Song et al. 2006; Lourenco et al. 2008). However, little is known about the possibility of establishing direct, long-range RNA–RNA interactions between the ends of the HCV genome that might modulate not only translation, but other viral processes as well.
The present work describes a novel long-range RNA–RNA interaction between the apical loop of subdomain IIId in the IRES and the internal loop of 5BSL3.2. To our knowledge, this is the first reported connection based on sequence complementarity between the ends of the HCV genome. The functional importance of the interacting RNA motifs suggests the formation of this complex to have a modulating role in viral protein synthesis and in the switch from translation to replication during the viral cycle.
RESULTS
Identification of a long-distance RNA–RNA interaction in the HCV genome
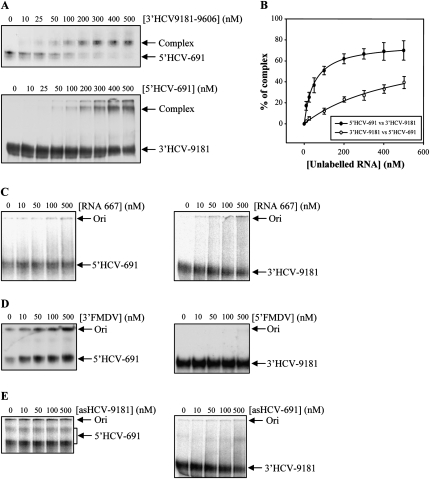
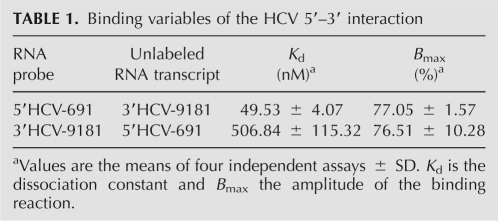
To look for functional links between the ends of the HCV genome mediated by direct RNA–RNA interactions, two RNA transcripts resembling the natural 5′ and 3′ ends of the HCV were constructed (Fig. 1B). The RNA molecule representing the 5′ end (5′HCV-691) contained the first 691 nt of the HCV genome, therefore encompassing the whole 5′ UTR and a significant portion of the core-coding sequence. This should resemble the functional folding of the IRES to be examined (Wang et al. 2000; Kim et al. 2003; Beguiristain et al. 2005). For the 3′ construct, the 3′ UTR was extended at its 5′ end with the three last hairpins of the NS5B coding sequence (3′HCV-9181), thus enabling the correct interplay between these regions and favoring functional folding (You et al. 2004; Friebe et al. 2005). 32P-internally labeled 5′HCV-691 RNA was incubated with a molar excess of the nonlabeled 3′ end construct under low magnesium concentrations and resolved in native polyacrylamide gels (Fig. 2A). A dose-dependent complex with lower electrophoresis mobility was detected. This provides evidences of the existence of an interaction between the ends of the HCV genome. Binding assays were repeated four times and the data fitted to a nonlinear equation to obtain the dissociation constant, Kd. The 3′ fragment bound at high affinity with a Kd value of 49.5 nM; the amplitude of the reaction reached up to 77% (Fig. 2A; Table 1). This led us to ask whether the inverse reaction was also possible. To answer this question, the 3′ end was internally labeled and incubated with increasing concentrations of the nonlabeled 5′HCV-691 RNA. A retarded complex was observed, indicating that the interaction occurs irrespective of the RNA probe used (Fig. 2B). Surprisingly, a 10-fold increase in the Kd value was observed with respect to that obtained for the 5′ probe (Fig. 2B; Table 1). This asymmetry in the reaction might be related to the specific reaction conditions.
FIGURE 2.
RNA–RNA interactions established between the 5′ and the 3′ ends of the HCV genome. (A,B) 5′–3′ ends binding is concentration dependent. Transcripts 5′HCV-691 and 3′HCV-9181 were alternatively used as probes and challenged with increasing concentrations (10–500 nM) of their corresponding interacting partners in binding buffer. Complexes were fractionated in 5% native polyacrylamide gels with TBM. These assays were repeated four times and data were fitted to a non/linear equation for the calculation of the Kd value. This interaction is sequence and strand specific, since no complex was detected under similar conditions when the probes were incubated with a nonrelated RNA sequence, RNA 667 (C); the UTRs for the FMDV, 5′FMDV, and 3′FMDV (D); or the corresponding antisense transcripts asHCV-691 and asHCV-9181 (E).
TABLE 1.
Binding variables of the HCV 5′–3′ interaction
To further study the specificity of the interaction, the 5′ and the 3′ transcripts were challenged with a nonrelated RNA molecule, RNA 667 (Fig. 2C). This was unable to interact with either of the HCV genome ends. Similar results were obtained when the 5′ and the 3′ probes were incubated with the corresponding 3′- and 5′-end transcripts of FMDV (Fig. 2D). A lack of interaction was also detected when the 5′- and 3′-end constructs were incubated with the respective antisense transcripts (Fig. 2E), verifying the strand specificity of the complex.
Taken together, these results demonstrate the occurrence of an interaction between functional domains at the 5′ and 3′ ends of the HCV genome. This complex is specific for HCV and occurs in a dose-dependent manner, in the absence of proteins, and at low magnesium concentrations.
The specific RNA–RNA interaction between the 5′ and 3′ ends of the HCV RNA genome is restricted to the IRES domain and the CRE region
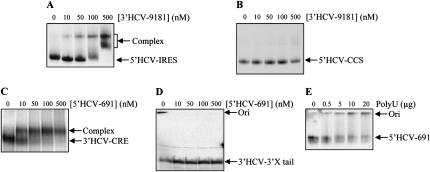
Truncated mutants encompassing different viral RNA regions were constructed, aimed at experimentally identifying the functional domains essential for 5′–3′ contact. The 5′ partner was the IRES and the 5′core-coding sequence (CCS), while the 3′ end was divided into the CRE region, the polyU/UC stretch, and the 3′HCV-3′X tail (Fig. 1B). Each of these molecules was challenged with increasing concentrations of the corresponding full-length nonlabeled RNA to test their ability to interact. For the 5′ end, only the transcript containing the IRES efficiently associated with the 3′ end (Fig. 3A,B), excluding the core-coding sequence as a participant in complex formation. The essential interacting region at the 3′ end was restricted to the CRE domain (Fig. 3C). Neither the 3′HCV-3′X tail nor the polyU stretch complexed with the 5′ fragment (Fig. 3D,E). The IRES and the CRE region therefore appear to interact in a dose-dependent manner to yield the complex.
FIGURE 3.
The essential interacting domains reside in the IRES and the CRE regions. Different truncated variants for the functional elements of the 5′ and the 3′ ends of the HCV genome were 32P-unifomly labeled and incubated with a range of concentrations (10–500 nM) of their corresponding interacting partners. (A,B) Unlabeled 3′HCV-9181 transcripts were challenged with the RNA probes 5′HCV-IRES and 5′HCV-CCS, respectively. (C,D) Unlabeled 5′-end transcript, 5′HCV-691, with the probes corresponding to the CRE region, 3′HCV-CRE, and the 3′HCV-3′X tail, respectively. (E) PolyU RNA was incubated with internally labeled 5′HCV-691.
Theoretical model for the interaction between the 5′ and 3′ ends of the HCV genome
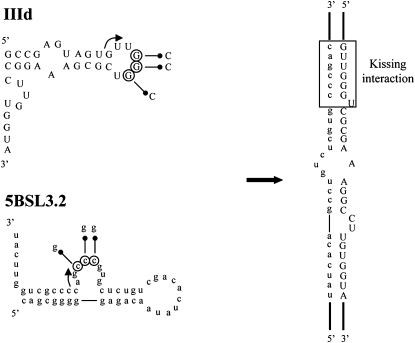
Folding softwares based on dynamic programming algorithms—RNAcofold and RNAup—were used to decipher the most plausible motifs involved in the establishment of the complex formed between the 5′ and 3′ ends of the HCV genome. RNAcofold allows the input of two RNA sequences that are concatenated and folded into a dimer (Hofacker et al. 1994; Bernhart et al. 2006). This program computes the minimum free energy of all possible dimers and reports the most stable structure. RNAup contemplates the thermodynamics of an RNA–RNA interaction as the sum of two contributing energies, one required to alter the secondary structure of the interacting residues, the other gained as a result of dimer formation (Hofacker et al. 1994; Mueckstein et al. 2006). Both tools suggested the same theoretical model in which domain IIId of the IRES associates with 5BSL3.2 in the CRE region (Fig. 4). The essential nucleotide G263 located at the base of the apical loop of domain IIId may initiate contact with C9301 adjacent to the internal loop of 5BSL3.2. The interaction might extend up to A288 for the 5′ end and A9275 for the 3′ end. The resulting dimer, with a ΔG value of −6.48 kcal/mol, was stable enough to be detected experimentally under the present assay conditions. Though the formation of the whole duplex is plausible in a thermodynamic context, the structure of the interacting domains makes the progression of the duplex beyond the loops unlikely. One might assume the formation of a kissing complex involving ALIL interactions between the IIId and 5BSL3.2 domains of the HCV genome (Fig. 4).
FIGURE 4.
Theoretical model for the interacting domains. RNAcofold and RNAup softwares were used to predict the residues involved in the binding between the 5′ and the 3′ ends of the HCV genome. Complementary sequences were identified in the IIId domain in the IRES and in the 5BSL3.2 hairpin in the CRE region. The interaction is proposed to be initiated at the nucleotides indicated by arrows. The kissing interaction between the apical loop of domain IIId and the internal loop of 5BSL3.2 region is boxed. The encircled residues were mutated as noted to generate the respective inactive variants.
In summary, in silico predictions suggest that the sequences involved in the 5′-3′ HCV ends map within the IIId domain at the 5′ end and the 5BSL3.2 domain at the 3′ end.
Domains IIId and 5BSL3.2 are key elements in the interaction between the 5′ and 3′ ends of the HCV genome
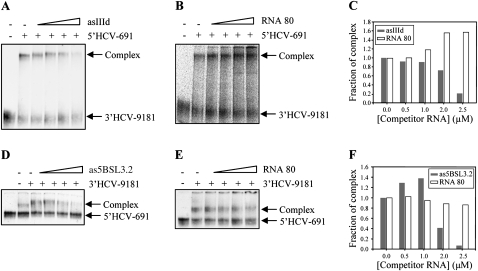
In vitro binding competition assays were performed to experimentally validate the role of the IIId and 5BSL3.2 domains in complex formation. 3′HCV-9181 was incubated with a molar excess of nonlabeled 5′HCV-691 to yield a complex with retarded mobility in native polyacrylamide gels (Fig. 5A). A reduction in the proportion of this product up to 70% was detected in the presence of increasing amounts of an antisense transcript for domain IIId, asIIId (Fig. 5A,C), whereas the use of a nonrelated RNA, RNA 80, as a competitor induced no reduction in complex formation (Fig. 5B,C). This confirms the specificity of the interaction and suggests domain IIId to be an essential component in the interaction between the 5′ and 3′ ends of the HCV genome. The inverse assay was then performed to analyze the role of 5BSL3.2 in the formation of the complex. The 5′ probe was incubated with a molar excess of the nonlabeled 3′-end construct, either in the presence or absence of as5BSL3.2, an antisense transcript for 5BSL3.2. This molecule effectively competed with the complex formation (Fig. 5D,F), confirming its importance in the establishment of the interaction. Again, RNA 80 induced no changes in complex formation (Fig. 5E,F).
FIGURE 5.
Binding competition assays. Antisense RNA molecules for the putative interacting domains IIId and 5BSL3.2 were used in competition assays. 32P-labeled 3′HCV-9181 transcripts were incubated with a molar excess of their interacting partners, 5′HCV-691, and increasing concentrations (0.5–2.5 μM) of an antisense RNA for domain IIId, asIIId (A) or a nonrelated RNA, RNA 80 (B). Similar assays were performed for the reciprocal interaction (D,E). Changes in complex formation were quantified and data are represented in graphs (C,F).
Taken together, these results confirm the importance of domains IIId and 5BSL3.2, and identify them as essential partners in the establishment of the long-range RNA–RNA interaction between the 5′ and 3′ ends of the HCV genome.
RNA structure probing of the interacting nucleotides
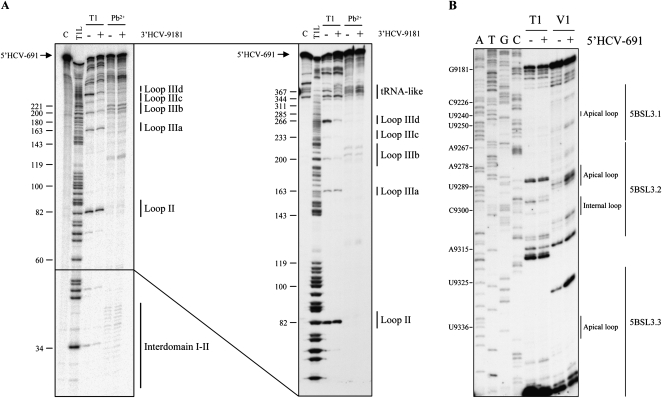
RNA structure probing of the residues involved in the association was performed by partial digestion with RNases and lead. To analyze the 5′ end, the 5′HCV-691 transcript was 5′-end labeled and used as a substrate for partial cleavage with the single-stranded endonucleolytic reagents RNase T1 (G residues) and lead (any nucleotide). The magnesium concentrations were the same as those used in the binding assays. The degradation pattern resembled that previously described (Figs. 1A, 6A; Kieft et al. 1999), in which unpaired nucleotides in loops and single-stranded regions appeared sensitive to cleavage. Changes in this map were detected when the reactions proceeded in the presence of the 3′ construct 3′HCV-9181 (Fig. 6A). Residues in the apical loop of domain IIId were now clearly resistant to degradation by RNase T1, suggesting their involvement in the establishment of an interaction with the 3′ end of the HCV genome. Slight protection around nucleotide 360 was also noticeable, as well as in the apical loop IIIc (Fig. 6A). The in silico modeling predicted no interaction at these sites (data not shown).
FIGURE 6.
Secondary structure analysis of the 5′ and the 3′ ends of the HCV genome and identification of the interacting residues. (A) 32P-5′-end-labeled 5′HCV-691 was partially digested with RNase T1 or Pb2+, either in the absence (−) or presence (+) of the 3′HCV-9181. The right panel shows a different run length aimed to resolve the higher molecular weight cleavage products. The functional subdomains of the IRES region are indicated. C, 5′HCV-691 incubated in binding buffer. T1L, T1 cleavage ladder. (B) Primer extension analysis of the 3′HCV-9181 transcript treated with RNase T1 or RNase V1 in the absence (−) or presence (+) of the 5′-end RNA. cDNA products were analyzed in 6% denaturing polyacrylamide gels in parallel with a sequence ladder obtained with the same labeled primer. The autoradiograph shows the results obtained for the CRE region.
The 3′ end of the NS5B coding sequence adopts a well-defined cruciform structure previously detected by endonuclease cleavage and subsequent primer extension of the digestion products (You et al. 2004). Similar experimental conditions were used to map the interacting residues in 3′ end. The 3′HCV-9181 RNA was treated with RNase T1, which is specific for single-stranded G residues, and RNase V1, which cleaves base-paired nucleotides or stacked helical single-stranded regions. This assay confirmed many of the structural features previously detected for this region, particularly around 5BSL3.2 (Figs. 1A, 6B; You et al. 2004; data not shown). Clear signs of cleavage by RNase V1 were noticeable in the theoretical single-stranded region upstream of 5BSL3.1. This is likely due to the involvement of these residues in the establishment of the interaction with the 5′ segment of the 3′ UTR that generates the cruciform structure in which the CRE region is embedded (You et al. 2004). The presence of the 5′HCV-691 RNA rendered residues in the internal loop of the 5BSL3.2 domain clearly resistant to cleavage by RNase T1 and sensitive to RNase V1 processing, suggesting their involvement in the formation of a duplex with the 5′-end construct (Fig. 6B). In agreement with these results, a clear increase in RNase V1-mediated processing was detected at the flanking stems of the internal loop, C9306 and U9289. This was likely due to stabilization of these stems or to the formation of the heteroduplex formed between the interacting molecules. No changes in the degradation pattern were detected in other regions of the 3′-end RNA (data not shown), which supports the idea of an interacting site being located in 5BSL3.2.
These results are in good agreement with the theoretical model by which interaction between the 5′ and 3′ ends of the viral genome is mediated by the essential domains IIId and 5BSL3.2.
5′–3′ complex formation is mediated by an apical loop–internal loop (ALIL) interaction
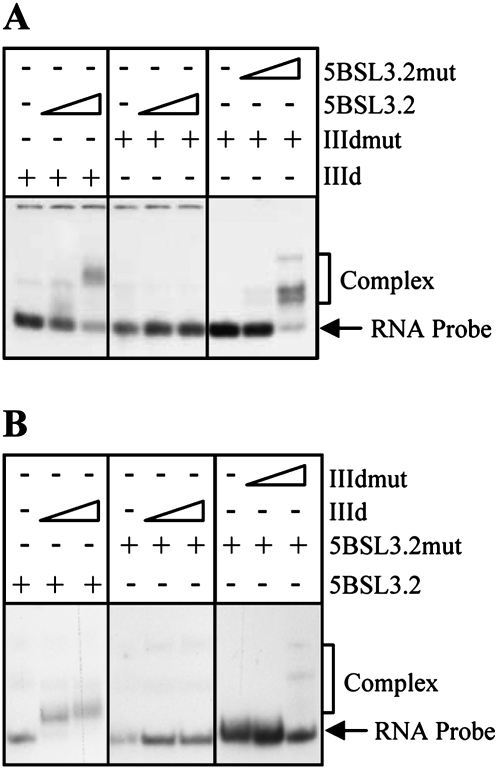
The key determinants in the interaction between the HCV IRES and the CRE region were then studied in more detail. Complementary sequences map to the apical loop of domain IIId and the internal loop of 5BSL3.2. Therefore, it is tempting to assume that these motifs are involved in the initiation of the complex formation via an ALIL interaction. To test this hypothesis, RNA molecules encompassing the wild-type domain IIId and 5BSL3.2, as well as mutant versions, were generated (Fig. 4). Mutants for domain IIId (IIIdmut) contained a three nucleotide substitution in the apical loop (GGG to CCC); similarly, 5BSL3.2mut had three modified residues in its internal loop (CCC to GGG). These changes did not alter the in silico predicted secondary structure of the interacting domains (data not shown). In vitro binding assays were performed to study the occurrence of interactions between these molecules. As expected, complex formation was detected for the wild-type versions of the independent stem-loops irrespective of the RNA probe used (Fig. 7A,B, lanes 1–3), consistent with the proposed interaction between the IIId and 5BSL3.2 domains. Mutant variants for both domain IIId and 5BSL3.2 were unable to generate efficient and/or stable complexes with their corresponding wild-type targets under the assay conditions (Fig. 7A,B, lanes 4–6). Binding was restored when both mutants were simultaneously incubated (Fig. 7A,B, lanes 7–9), though to a lesser extent than for the wild-type versions, supporting the idea of an ALIL interaction being necessary for efficient complex formation.
FIGURE 7.
Mutations in the loops of the interacting domains interfere with the association between IIId and 5BSL3.2. Binding assays were accomplished with the probes IIId or 5BSL3.2 and their nonlabeled interacting partners. Variations in nucleotide composition were introduced as indicated in Figure 4 to generate the respective mutants. (A) The internally labeled IIId or IIIdmut RNAs were challenged with the 5BSL3.2 or 5BSL3.2mut transcripts. Complexes were resolved in 8% native polyacrylamide gels with TBM. (B) Reciprocal experiments where the RNA probes 5BSL3.2 or 5BSL3.2mut were incubated with the nonlabeled IIId or IIIdmut transcripts.
DISCUSSION
General models of translation initiation for eukaryotic mRNAs propose a closed loop conformation for these molecules mediated by RNA–protein and/or protein–protein interactions (for review, see Komarova et al. 2006). Prokaryotic mRNAs and many viruses, however, have simpler systems based on direct end-to-end RNA communication via base-pairing (Edgil and Harris 2006; Komarova et al. 2006). With respect to positive single-stranded RNA viruses, such us luteovirus and tombusvirus, these are promoted by stem–loop structures that interact to generate efficient and specific kissing complexes (Komarova et al. 2006). In hepacivirus genomes, circularization has been proposed to be mediated by protein factors. Their interaction with each of the ends has been proved (Edgil and Harris 2006), however, to date, no evidence of circularization has been reported. Neither has direct contact between both RNA ends been demonstrated.
The HCV genome contains well-defined RNA structural elements in its UTRs that have been studied in depth. More recently, other phylogenetically conserved secondary structures in the coding sequence have been identified by bioinformatic methods (Tuplin et al. 2002; 2004; You et al. 2004; McMullan et al. 2007; Diviney et al. 2008). Our knowledge of these regions, however, is limited, and both their three-dimensional folding and function are now subjects of much research. One of the most studied domains is the short stem loop 5BSL3.2, which is located at the 3′ end of the NS5B coding sequence. Its structural integrity is essential for replication (You et al. 2004; Friebe et al. 2005), probably due to its participation in distant interactions with structural elements of the 3′ UTR (Friebe et al. 2005). The results of the present study show that 5BSL3.2 also associates with the essential domain IIId of the IRES. To our knowledge, this is the first report describing a long-range RNA–RNA interaction between functional domains of the 5′ and 3′ ends for the HCV genome.
The present results show that the 5′–3′ contact is stable at low magnesium concentrations and that this contact is sequence and strand specific (Fig. 2).
The complex is efficiently generated in vitro in the absence of protein factors, though a further stabilization of the interaction by auxiliary elements is likely in a biological context. In fact, a role for the NFAR protein family in the preservation of the 5′–3′ UTR contacts of the pestivirus genome has been suggested (Isken et al. 2004). For HCV, numerous cellular proteins, including polypyrimidine tract-binding protein (PTB) (Tsuchihara et al. 1997; Ito and Lai 1999), La autoantigen (Spangberg et al. 1999), and ribosomal proteins (Wood et al. 2001), among others, have been proposed to bind to the 3′ UTR and stimulate IRES-dependent translation. The well-documented interaction of the HCV NS5B protein to the SL3.2 domain postulates the implication of the viral polymerase in the genome circularization (Zhang et al. 2005). This supports the notion that protein factors might act as molecular bridges that reinforce the interaction shown here.
The RNase probing and binding assays show that the main interacting regions reside in domain IIId of the IRES and 5BSL3.2 of the NS5B coding sequence, as predicted in silico by the RNAcofold and RNAup computations. These regions are highly conserved among hepacivirus and related flaviviruses (Jubin et al. 2000; Tuplin et al. 2002; You et al. 2004; Barria et al. 2008), supporting the idea that this contact may also occur in other closely related viral systems. Interestingly, small changes in the degradation pattern of the apical loop IIIc and the region surrounding nucleotide 360 were observed in 5′ end probing in the presence of the 3′ construct. These positions were not predicted in the in silico experiments to participate in the association between the ends of the RNA molecules. However, they might act as secondary interacting sites, which might further stabilize the complex. They may be also the result of structural rearrangements in the IRES arising as a consequence of the initial interaction.
The present results also provide evidence that the association process is initiated by an ALIL interaction. ALIL complexes are a type of kissing complex widely distributed in nature, and play important roles in many biological processes (Brunel et al. 2002). In vitro-selected RNA aptamers also exploit this strategy to efficiently associate with their targets (Aldaz-Carroll et al. 2002; Da Rocha Gomes et al. 2004). In all cases, the presence of a short stretch of sequence complementarity located in the apical and internal loops of the interacting domains is sufficient to stabilize the resulting complex. A peculiarity shown by all these systems is that the G-C rich stems flank the internal interacting loop (Aldaz-Carroll et al. 2002; Brunel et al. 2002; Da Rocha Gomes et al. 2004). Interestingly, this feature is also shown by 5BSL3.2, further supporting the ALIL interaction model. Thermodynamic predictions propose the extension of the duplex beyond the interacting loops based on sequence complementarity (Fig. 4). However, though the extension of the base-pairing regions is plausible in a thermodynamic context, the structure of the interacting domains, particularly the presence of a loop E in domain IIId, makes the progression of the duplex beyond the loops unlikely. A more detailed structural analysis is needed to confirm this.
Unlike cellular mRNAs, which only contain translational regulatory elements, viral RNA genomes encompass differentially evolved functional regions that act in nonsimultaneous processes of the infective cycle, such as protein synthesis and replication. Consequently, direct communication between these motifs might act as a functional link for the correct switch from translation to replication, defining a riboswitch-like motif. During early HCV infection, uncapped RNAs initiate the IRES-dependent protein synthesis, which is proposed to be stimulated by host factors bound to the 3′ UTR (Bradrick et al. 2006; Song et al. 2006). It seems plausible that the 5BSL3.2 domain should exert an additional modulatory activity over viral translation through its interaction with domain IIId. Once viral protein accumulation is guaranteed, nonstructural factors are gathered within the endoplasmic reticulum to generate the membranous web required for replication. Transition between these processes is complex and requires strict control that needs to integrate multiple functional elements. The interaction involving domains IIId and 5BSL3.2, together with other viral factors previously proposed (Domitrovich et al. 2005; Lourenco et al. 2008), might be involved in this regulation.
In summary, the present results firmly support the existence of a protein-independent 5′–3′ communication in the HCV genome that involves RNA elements essential for viral translation and replication—domain IIId of the IRES and 5BSL3.2. The resulting complex shows sequence and strand specificity and represents a new cis-element involved in the regulation of multiple processes during HCV infection.
MATERIALS AND METHODS
DNA templates and RNA synthesis
All RNAs were synthesized by in vitro transcription and purified as previously described (Barroso-delJesus et al. 1999).
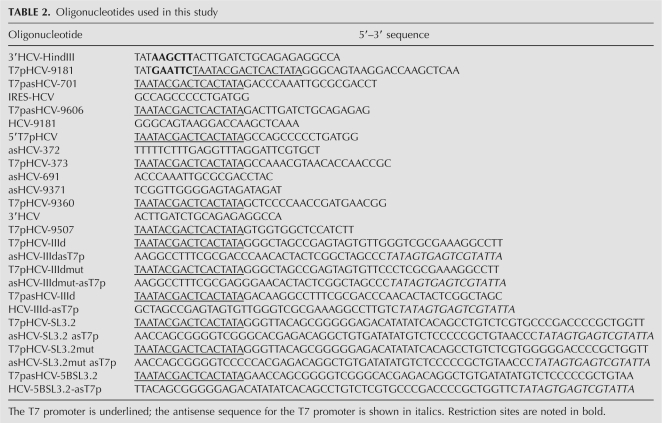
DNA templates for the generation of 5′HCV-691 RNA, RNA 667, and RNA 80 were obtained as previously reported (Romero-López et al. 2005, 2007). A plasmid containing the 3′HCV-9181 was constructed as follows. DNA encoding the CRE domain plus the 3′ UTR of HCV was obtained by reverse transcription and amplification of total RNA from Huh-7 cells harboring a stable subgenomic replicon (Huh-7 NS3-18) (Larrea et al. 2006), a kind gift of Dr. Rafael Aldabe (University of Navarra, Spain). Briefly, cells were cultured in DMEM supplemented with 10% heat-inactivated fetal bovine serum (FBS) (Invitrogen), 2 mM L-glutamine (Sigma), and 0.5 mg/mL of G-418 (Sigma) at 37°C in a 5% CO2 atmosphere. A total of 500,000 cells were lysed with Trizol reagent (Invitrogen) following the manufacturer's instructions to obtain intracellular RNA. cDNA synthesis was accomplished using 100 ng of total RNA and employing the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). A fraction of the resulting cDNA was used for amplification with primers 3′HCV-HindIII and T7pHCV-9181 to yield the construct T7p3′HCV-9181. This was cloned in the EcoRI and HindIII restriction sites in the pUC19 vector to generate the plasmid pU3′HCV-9181, which was then digested with HindIII to obtain the DNA template for the synthesis of 3′HCV-918.
DNA templates encompassing the antisense sequences for the 5′ and 3′ ends of HCV, asHCV-691 and asHCV-9181, respectively, were obtained by PCR. For the asHCV-691 construct, the pUCHCV-691 plasmid was used as a template for amplification with primers T7pasHCV-701 and IRES-HCV. The antisense sequence for the 3′ end was amplified from the plasmid pU3′HCV-9181 using oligonucleotides T7pasHCV-9606 and HCV-9181.
DNAs encoding the truncated forms of the 5′ and 3′ ends of the HCV genome were generated by amplification as follows. Plasmid pUCHCV-691 was used as a template for amplification with the primers 5′T7pHCV and asHCV-372 to yield 5′HCV-IRES; alternatively, hybridization and amplification with the oligonucleotides T7pHCV-373 and asHCV-691 produced 5′HCV-CCS. Several specific primer pairs were used with the plasmid pU3′HCV-9181 to produce amplicons with modified 3′ ends: oligonucleotides T7pHCV-9181 and asHCV-9371 were used to generate 3′HCV-CRE containing the CRE region and T7pHCV-9507 and 3′HCV to generate an amplicon with the 3′HCV-3′X tail.
Plasmids containing the 5′ and 3′ UTRs of FMDV, pGEM-IRES-8 and psubTAG, respectively (Ramos and Martinez-Salas 1999; Lopez de Quinto et al. 2002; Serrano et al. 2006) were the generous gift of Dr. Encarnación Martínez-Salas (Consejo Superior de Investigaciones Cientificas).
The annealing of specific oligonucleotides was used to generate the following dsDNA templates:
Domain IIId: oligonucleotides T7pHCV-IIId and asHCV-IIIdasT7p;
Domain IIIdmut: T7pHCV-IIIdmut and asHCV-IIIdmut-asT7p;
AsIIId: T7pasHCV-IIId and HCV-IIId-asT7p;
Domain 5BSL3.2: T7pHCV-SL3.2 and asHCV-SL3.2 asT7p;
Domain 5BSL3.2: T7pHCV-SL3.2mut and asHCV-SL3.2mut asT7p; and
As5BSL3.2: T7pasHCV-5BSL3.2 and HCV-5BSL3.2 asT7p.
See Table 2 for the oligonucleotide sequences.
TABLE 2.
Oligonucleotides used in this study
Binding assays
The analysis of RNA–RNA interactions was performed essentially as previously described (Serrano et al. 2006). Briefly, 0.5 nM of 32P-labeled RNA was challenged with increasing concentrations of the target molecule. Prior to complex formation, RNA molecules were independently denatured for 3 min at 95°C in binding buffer (50 mM sodium cacodylate at pH 7.5, 300 mM KCl, and 1 mM MgCl2) (Ferrandon et al. 1997) and subsequently transferred to ice for 15 min. Reactions were initiated by mixing both molecules and proceeded at 37°C for 30 min. The resulting complexes were immediately resolved in 6%–8% native polyacrylamide gels supplemented with 2.5 mM MgCl2. Electrophoresis was performed at 4°C over 5 h at 15 mA in TBM buffer (45 mM Tris-HCl at pH 8.3, 43 mM boric acid, and 0.1 mM MgCl2). Gels were dried, scanned, and analyzed as previously described (Romero-López et al. 2007). Kd values were calculated using Sigma Plot 8.02 software according to the equation y = (Bmaxx)/(Kd + x) (Puerta-Fernández et al. 2005), where y is the percentage of complexed inhibitory RNA, Bmax is the amplitude of the reaction, x is the concentration of the target RNA, and Kd the dissociation constant.
Competition binding assays were performed under similar conditions. Briefly, 0.5 nM of internally 32P-labeled RNA was complexed with a constant amount of the target molecule. The complex was challenged with increasing concentrations (0.5–2.5 μM) of the competitor RNA. All RNA molecules were denatured and renatured separately, as noted above and subsequently mixed. Complexes proceeded for 30 min and the reaction products resolved, analyzed, and quantified as indicated.
In silico predictions of the interacting sequences
RNAcofold (Hofacker et al. 1994; Bernhart et al. 2006) and RNAup (Hofacker et al. 1994; Mueckstein et al. 2006) softwares were employed to predict the interacting residues between the 5′ and 3′ ends of the HCV genome using the Web interface at http://rna.tbi.univie.ac.at/ (Vienna RNA web servers). Sequences 5′HCV-691 and 3′HCV-9181, as well as several truncated forms, were subjected to both analyses with the aim of examining all possible interacting residues.
RNA–RNA interaction probing assays
Probing assays of the complex between the 5′ and 3′ end were carried out with 32P 5′-end-labeled 5′HCV-691 RNA. Complexes were constructed as described above by incubating 50 fmol of the 32P 5′-end-labeled 5′HCV-691 RNA (∼200 CPS) with 10 pmol of the nonlabeled 3′ end, 3′HCV-9181. Control reactions were performed in the presence of an equal amount of tRNA. Digestions were initiated by the addition of RNase T1 (0.1 units; Industrial Research) or Pb2+ acetate (30 mM; Merck), and incubated at 30°C for 5 and 20 min, respectively. Reactions were stopped by the addition of EDTA 100 mM and by phenol extraction. RNAs were precipitated with ethanol and analyzed in high resolution denaturing polyacrylamide gels (6% acrylamide, 7 M urea). These were dried and scanned as above.
Alternatively, the identification of the interacting residues at the 3′ end was essentially performed as previously reported (You et al. 2004). Briefly, complex formation was accomplished as noted above and subjected to partial digestion with 0.1 units of cobra venom RNase V1 (Pierce Biotechnology) or 0.1 units of RNase T1 (Industrial Research) at 4°C for 10 and 5 min, respectively. Cleavage reactions were stopped as above and the RNA extracted using phenol-chloroform followed by ethanol precipitation. The degradation pattern was mapped by primer extension with the 32P 5′-end-labeled primer (You et al. 2004). Total processed RNA was mixed with 2 pmol of primer oligonucleotide and denatured for 2 min at 95°C. Extension was then performed at 42°C in a 20 μL reaction volume with RT buffer, dNTPs 0.5 mM and 100 U SuperScript II RT (Invitrogen). cDNA products were analyzed in high resolution denaturing polyacrylamide gels (8% acrylamide, 7 M urea), which were dried and scanned as above. Dideoxy sequencing reactions were prepared using the Sequenase 7-deaza-dGTP DNA Sequencing Kit (USB) with the same oligonucleotide employed for the primer extension reactions and the plasmid pU3′HCV-9181. Reactions were run in parallel to identify the RT extension products.
ACKNOWLEDGMENTS
We thank Dr. Encarnación Martínez-Salas for her kind gift of the plasmid constructs pSUBTAG and pGEM-IRES-8, and Drs. Carlos Briones and Michael Stitch for their help with the two-dimensional modeling in the prediction of the interacting sequences. We also thank Dr. Alicia Barroso-delJesus for helpful discussions and Vicente Augustin-Vacas for excellent technical assistance. This work was supported by grants BFU2006-02568 from the Spanish Ministerio de Educación y Ciencia and CTS-233 from the Junta de Andalucía to A.B.-H. C.R.-L. was funded by grant BFU2006-02568.
Footnotes
Abbreviations: HCV, hepatitis C virus; FMDV; foot and mouth disease virus; IRES, internal ribosome entry site; UTR, untranslatable region; CRE, cis-acting replication element.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1680809.
REFERENCES
- Aldaz-Carroll L, Tallet B, Dausse E, Yurchenko L, Toulme JJ. Apical loop–internal loop interactions: A new RNA–RNA recognition motif identified through in vitro selection against RNA hairpins of the hepatitis C virus mRNA. Biochemistry. 2002;41:5883–5893. doi: 10.1021/bi0121508. [DOI] [PubMed] [Google Scholar]
- Babaylova E, Graifer D, Malygin A, Stahl J, Shatsky I, Karpova G. Positioning of subdomain IIId and apical loop of domain II of the hepatitis C IRES on the human 40S ribosome. Nucleic Acids Res. 2009;37:1141–1151. doi: 10.1093/nar/gkn1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barria MI, Gonzalez A, Vera-Otarola J, Leon U, Vollrath V, Marsac D, Monasterio O, Perez-Acle T, Soza A, Lopez-Lastra M. Analysis of natural variants of the hepatitis C virus internal ribosome entry site reveals that primary sequence plays a key role on cap-independent translation. Nucleic Acids Res. 2008;37:957–971. doi: 10.1093/nar/gkn1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barroso-delJesus A, Tabler M, Berzal-Herranz A. Comparative kinetic analysis of structural variants of the hairpin ribozyme reveals further potential to optimize its catalytic performance. Antisense Nucleic Acid Drug Dev. 1999;9:433–440. doi: 10.1089/oli.1.1999.9.433. [DOI] [PubMed] [Google Scholar]
- Beguiristain N, Robertson HD, Gomez J. RNase III cleavage demonstrates a long range RNA: RNA duplex element flanking the hepatitis C virus internal ribosome entry site. Nucleic Acids Res. 2005;33:5250–5261. doi: 10.1093/nar/gki822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhart SH, Tafer H, Mückstein U, Flamm C, Stadler PF, Hofacker IL. Partition function and base-pairing probabilities of RNA heterodimers. Algorithms Mol Biol. 2006;1:3. doi: 10.1186/1748-7188-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blight KJ, Rice CM. Secondary structure determination of the conserved 98-base sequence at the 3′ terminus of hepatitis C virus genome RNA. J Virol. 1997;71:7345–7352. doi: 10.1128/jvi.71.10.7345-7352.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradrick SS, Walters RW, Gromeier M. The hepatitis C virus 3′-untranslated region or a poly(A) tract promote efficient translation subsequent to the initiation phase. Nucleic Acids Res. 2006;34:1293–1303. doi: 10.1093/nar/gkl019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branch AD, Stump DD, Gutierrez JA, Eng F, Walewski JL. The hepatitis C virus alternate reading frame (ARF) and its family of novel products: The alternate reading frame protein/F-protein, the double-frameshift protein, and others. Semin Liver Dis. 2005;25:105–117. doi: 10.1055/s-2005-864786. [DOI] [PubMed] [Google Scholar]
- Brunel C, Marquet R, Romby P, Ehresmann C. RNA loop–loop interactions as dynamic functional motifs. Biochimie. 2002;84:925–944. doi: 10.1016/s0300-9084(02)01401-3. [DOI] [PubMed] [Google Scholar]
- Choi J, Xu Z, Ou JH. Triple decoding of hepatitis C virus RNA by programmed translational frameshifting. Mol Cell Biol. 2003;23:1489–1497. doi: 10.1128/MCB.23.5.1489-1497.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo QL, Kuo G, Weiner AJ, Overby LR, Bradley DW, Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244:359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- Collier AJ, Gallego J, Klinck R, Cole PT, Harris SJ, Harrison GP, Aboul-Ela F, Varani G, Walker S. A conserved RNA structure within the HCV IRES eIF3-binding site. Nat Struct Biol. 2002;9:375–380. doi: 10.1038/nsb785. [DOI] [PubMed] [Google Scholar]
- Da Rocha Gomes S, Dausse E, Toulme JJ. Determinants of apical loop–internal loop RNA–RNA interactions involving the HCV IRES. Biochem Biophys Res Commun. 2004;322:820–826. doi: 10.1016/j.bbrc.2004.07.185. [DOI] [PubMed] [Google Scholar]
- Diviney S, Tuplin A, Struthers M, Armstrong V, Elliott RM, Simmonds P, Evans DJ. A hepatitis C virus cis-acting replication element forms a long-range RNA–RNA interaction with upstream RNA sequences in NS5B. J Virol. 2008;82:9008–9022. doi: 10.1128/JVI.02326-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domitrovich AM, Diebel KW, Ali N, Sarker S, Siddiqui A. Role of La autoantigen and polypyrimidine tract-binding protein in HCV replication. Virology. 2005;335:72–86. doi: 10.1016/j.virol.2005.02.009. [DOI] [PubMed] [Google Scholar]
- Edgil D, Harris E. End-to-end communication in the modulation of translation by mammalian RNA viruses. Virus Res. 2006;119:43–51. doi: 10.1016/j.virusres.2005.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang JW, Moyer RW. The effects of the conserved extreme 3′ end sequence of hepatitis C virus (HCV) RNA on the in vitro stabilization and translation of the HCV RNA genome. J Hepatol. 2000;33:632–639. doi: 10.1034/j.1600-0641.2000.033004632.x. [DOI] [PubMed] [Google Scholar]
- Ferrandon D, Koch I, Westhof E, Nusslein-Volhard C. RNA–RNA interaction is required for the formation of specific bicoid mRNA 3′ UTR-STAUFEN ribonucleoprotein particles. EMBO J. 1997;16:1751–1758. doi: 10.1093/emboj/16.7.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friebe P, Bartenschlager R. Genetic analysis of sequences in the 3′ nontranslated region of hepatitis C virus that are important for RNA replication. J Virol. 2002;76:5326–5338. doi: 10.1128/JVI.76.11.5326-5338.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friebe P, Lohmann V, Krieger N, Bartenschlager R. Sequences in the 5′ nontranslated region of hepatitis C virus required for RNA replication. J Virol. 2001;75:12047–12057. doi: 10.1128/JVI.75.24.12047-12057.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friebe P, Boudet J, Simorre JP, Bartenschlager R. Kissing-loop interaction in the 3′ end of the hepatitis C virus genome essential for RNA replication. J Virol. 2005;79:380–392. doi: 10.1128/JVI.79.1.380-392.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris E, Holden KL, Edgil D, Polacek C, Clyde K. Molecular biology of flaviviruses. Novartis Found Symp. 2006;277:23–39. discussion 40, 71-23, 251-253. [PubMed] [Google Scholar]
- Hofacker IL, Fontana W, Stadler PF, Bonhoeffer LS, Tacker M, Schuster P. Fast folding and comparison of RNA secondary structures. Monatsh Chem. 1994;125:167–188. [Google Scholar]
- Imbert I, Dimitrova M, Kien F, Kieny MP, Schuster C. Hepatitis C virus IRES efficiency is unaffected by the genomic RNA 3′NTR even in the presence of viral structural or nonstructural proteins. J Gen Virol. 2003;84:1549–1557. doi: 10.1099/vir.0.18907-0. [DOI] [PubMed] [Google Scholar]
- Isken O, Grassmann CW, Yu H, Behrens SE. Complex signals in the genomic 3′ nontranslated region of bovine viral diarrhea virus coordinate translation and replication of the viral RNA. RNA. 2004;10:1637–1652. doi: 10.1261/rna.7290904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Lai MM. Determination of the secondary structure of and cellular protein binding to the 3′-untranslated region of the hepatitis C virus RNA genome. J Virol. 1997;71:8698–8706. doi: 10.1128/jvi.71.11.8698-8706.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Lai MM. An internal polypyrimidine-tract-binding protein-binding site in the hepatitis C virus RNA attenuates translation, which is relieved by the 3′-untranslated sequence. Virology. 1999;254:288–296. doi: 10.1006/viro.1998.9541. [DOI] [PubMed] [Google Scholar]
- Ito T, Tahara SM, Lai MM. The 3′-untranslated region of hepatitis C virus RNA enhances translation from an internal ribosomal entry site. J Virol. 1998;72:8789–8796. doi: 10.1128/jvi.72.11.8789-8796.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H, Fraser CS, Yu Y, Leary J, Doudna JA. Coordinated assembly of human translation initiation complexes by the hepatitis C virus internal ribosome entry site RNA. Proc Natl Acad Sci. 2004;101:16990–16995. doi: 10.1073/pnas.0407402101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jubin R, Vantuno NE, Kieft JS, Murray MG, Doudna JA, Lau JY, Baroudy BM. Hepatitis C virus internal ribosome entry site (IRES) stem–loop IIId contains a phylogenetically conserved GGG triplet essential for translation and IRES folding. J Virol. 2000;74:10430–10437. doi: 10.1128/jvi.74.22.10430-10437.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato N, Hijikata M, Ootsuyama Y, Nakagawa M, Ohkoshi S, Sugimura T, Shimotohno K. Molecular cloning of the human hepatitis C virus genome from Japanese patients with non-A, non-B hepatitis. Proc Natl Acad Sci. 1990;87:9524–9528. doi: 10.1073/pnas.87.24.9524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon JC, Ghazawi A, Cheung WK, Phillip PS, Rizvi TA, Lever AM. The secondary structure of the 5′ end of the FIV genome reveals a long-range interaction between R/U5 and gag sequences, and a large, stable stem–loop. RNA. 2008;14:2597–2608. doi: 10.1261/rna.1284908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieft JS, Zhou K, Jubin R, Murray MG, Lau JY, Doudna JA. The hepatitis C virus internal ribosome entry site adopts an ion-dependent tertiary fold. J Mol Biol. 1999;292:513–529. doi: 10.1006/jmbi.1999.3095. [DOI] [PubMed] [Google Scholar]
- Kieft JS, Zhou K, Grech A, Jubin R, Doudna JA. Crystal structure of an RNA tertiary domain essential to HCV IRES-mediated translation initiation. Nat Struct Biol. 2002;9:370–374. doi: 10.1038/nsb781. [DOI] [PubMed] [Google Scholar]
- Kim YK, Lee SH, Kim CS, Seol SK, Jang SK. Long-range RNA–RNA interaction between the 5′ nontranslated region and the core-coding sequences of hepatitis C virus modulates the IRES-dependent translation. RNA. 2003;9:599–606. doi: 10.1261/rna.2185603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinck R, Westhof E, Walker S, Afshar M, Collier A, Aboul-Ela F. A potential RNA drug target in the hepatitis C virus internal ribosomal entry site. RNA. 2000;6:1423–1431. doi: 10.1017/s1355838200000935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolupaeva VG, Pestova TV, Hellen CU. An enzymatic footprinting analysis of the interaction of 40S ribosomal subunits with the internal ribosomal entry site of hepatitis C virus. J Virol. 2000;74:6242–6250. doi: 10.1128/jvi.74.14.6242-6250.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolykhalov AA, Feinstone SM, Rice CM. Identification of a highly conserved sequence element at the 3′ terminus of hepatitis C virus genome RNA. J Virol. 1996;70:3363–3371. doi: 10.1128/jvi.70.6.3363-3371.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolykhalov AA, Mihalik K, Feinstone SM, Rice CM. Hepatitis C virus-encoded enzymatic activities and conserved RNA elements in the 3′ nontranslated region are essential for virus replication in vivo. J Virol. 2000;74:2046–2051. doi: 10.1128/jvi.74.4.2046-2051.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarova AV, Brocard M, Kean KM. The case for mRNA 5′ and 3′ end cross talk during translation in a eukaryotic cell. Prog Nucleic Acid Res Mol Biol. 2006;81:331–367. doi: 10.1016/S0079-6603(06)81009-3. [DOI] [PubMed] [Google Scholar]
- Kong LK, Sarnow P. Cytoplasmic expression of mRNAs containing the internal ribosome entry site and 3′ noncoding region of hepatitis C virus: Effects of the 3′ leader on mRNA translation and mRNA stability. J Virol. 2002;76:12457–12462. doi: 10.1128/JVI.76.24.12457-12462.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larrea E, Aldabe R, Molano E, Fernandez-Rodriguez CM, Ametzazurra A, Civeira MP, Prieto J. Altered expression and activation of signal transducers and activators of transcription (STATs) in hepatitis C virus infection: In vivo and in vitro studies. Gut. 2006;55:1188–1196. doi: 10.1136/gut.2005.070060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Shin H, Wimmer E, Paul AV. cis-acting RNA signals in the NS5B C-terminal coding sequence of the hepatitis C virus genome. J Virol. 2004;78:10865–10877. doi: 10.1128/JVI.78.20.10865-10877.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez de Quinto S, Saiz M, de la Morena D, Sobrino F, Martinez-Salas E. IRES-driven translation is stimulated separately by the FMDV 3′-NCR and poly(A) sequences. Nucleic Acids Res. 2002;30:4398–4405. doi: 10.1093/nar/gkf569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lourenco S, Costa F, Debarges B, Andrieu T, Cahour A. Hepatitis C virus internal ribosome entry site-mediated translation is stimulated by cis-acting RNA elements and trans-acting viral factors. FEBS J. 2008;275:4179–4197. doi: 10.1111/j.1742-4658.2008.06566.x. [DOI] [PubMed] [Google Scholar]
- Lukavsky PJ, Otto GA, Lancaster AM, Sarnow P, Puglisi JD. Structures of two RNA domains essential for hepatitis C virus internal ribosome entry site function. Nat Struct Biol. 2000;7:1105–1110. doi: 10.1038/81951. [DOI] [PubMed] [Google Scholar]
- Lytle JR, Wu L, Robertson HD. Domains on the hepatitis C virus internal ribosome entry site for 40s subunit binding. RNA. 2002;8:1045–1055. doi: 10.1017/s1355838202029965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaffrey AP, Ohashi K, Meuse L, Shen S, Lancaster AM, Lukavsky PJ, Sarnow P, Kay MA. Determinants of hepatitis C translational initiation in vitro, in cultured cells and mice. Mol Ther. 2002;5:676–684. doi: 10.1006/mthe.2002.0600. [DOI] [PubMed] [Google Scholar]
- McMullan LK, Grakoui A, Evans MJ, Mihalik K, Puig M, Branch AD, Feinstone SM, Rice CM. Evidence for a functional RNA element in the hepatitis C virus core gene. Proc Natl Acad Sci. 2007;104:2879–2884. doi: 10.1073/pnas.0611267104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel YM, Borman AM, Paulous S, Kean KM. Eukaryotic initiation factor 4G-poly(A) binding protein interaction is required for poly(A) tail-mediated stimulation of picornavirus internal ribosome entry segment-driven translation but not for X-mediated stimulation of hepatitis C virus translation. Mol Cell Biol. 2001;21:4097–4109. doi: 10.1128/MCB.21.13.4097-4109.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mückstein U, Tafer H, Hackermüller J, Bernhart SH, Stadler PF, Hofacker IL. Thermodynamics of RNA–RNA binding. Bioinformatics. 2006;22:1177–1182. doi: 10.1093/bioinformatics/btl024. [DOI] [PubMed] [Google Scholar]
- Murakami K, Abe M, Kageyama T, Kamoshita N, Nomoto A. Down-regulation of translation driven by hepatitis C virus internal ribosomal entry site by the 3′ untranslated region of RNA. Arch Virol. 2001;146:729–741. doi: 10.1007/s007050170142. [DOI] [PubMed] [Google Scholar]
- Odreman-Macchioli FE, Tisminetzky SG, Zotti M, Baralle FE, Buratti E. Influence of correct secondary and tertiary RNA folding on the binding of cellular factors to the HCV IRES. Nucleic Acids Res. 2000;28:875–885. doi: 10.1093/nar/28.4.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooms M, Abbink TE, Pham C, Berkhout B. Circularization of the HIV-1 RNA genome. Nucleic Acids Res. 2007;35:5253–5261. doi: 10.1093/nar/gkm564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto GA, Puglisi JD. The pathway of HCV IRES-mediated translation initiation. Cell. 2004;119:369–380. doi: 10.1016/j.cell.2004.09.038. [DOI] [PubMed] [Google Scholar]
- Polacek C, Foley JE, Harris E. Conformational changes in the solution structure of the dengue virus 5′ end in the presence and absence of the 3′ untranslated region. J Virol. 2009;83:1161–1166. doi: 10.1128/JVI.01362-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puerta-Fernández E, Barroso-delJesus A, Romero-López C, Tapia N, Martinez MA, Berzal-Herranz A. Inhibition of HIV-1 replication by RNA targeted against the LTR region. AIDS. 2005;19:863–870. doi: 10.1097/01.aids.0000171399.77500.e0. [DOI] [PubMed] [Google Scholar]
- Ramos R, Martinez-Salas E. Long-range RNA interactions between structural domains of the aphthovirus internal ribosome entry site (IRES) RNA. 1999;5:1374–1383. doi: 10.1017/s1355838299991240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds JE, Kaminski A, Kettinen HJ, Grace K, Clarke BE, Carroll AR, Rowlands DJ, Jackson RJ. Unique features of internal initiation of hepatitis C virus RNA translation. EMBO J. 1995;14:6010–6020. doi: 10.1002/j.1460-2075.1995.tb00289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-López C, Barroso-delJesus A, Puerta-Fernandez E, Berzal-Herranz A. Interfering with hepatitis C virus IRES activity using RNA molecules identified by a novel in vitro selection method. Biol Chem. 2005;386:183–190. doi: 10.1515/BC.2005.023. [DOI] [PubMed] [Google Scholar]
- Romero-López C, Diaz-Gonzalez R, Berzal-Herranz A. Inhibition of hepatitis C virus internal ribosome entry site-mediated translation by an RNA targeting the conserved IIIf domain. Cell Mol Life Sci. 2007;64:2994–3006. doi: 10.1007/s00018-007-7345-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs AB, Sarnow P, Hentze MW. Starting at the beginning, middle, and end: Translation initiation in eukaryotes. Cell. 1997;89:831–838. doi: 10.1016/s0092-8674(00)80268-8. [DOI] [PubMed] [Google Scholar]
- Serrano P, Pulido MR, Saiz M, Martinez-Salas E. The 3′ end of the foot-and-mouth disease virus genome establishes two distinct long-range RNA–RNA interactions with the 5′ end region. J Gen Virol. 2006;87:3013–3022. doi: 10.1099/vir.0.82059-0. [DOI] [PubMed] [Google Scholar]
- Song Y, Friebe P, Tzima E, Junemann C, Bartenschlager R, Niepmann M. The hepatitis C virus RNA 3′-untranslated region strongly enhances translation directed by the internal ribosome entry site. J Virol. 2006;80:11579–11588. doi: 10.1128/JVI.00675-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spangberg K, Goobar-Larsson L, Wahren-Herlenius M, Schwartz S. The La protein from human liver cells interacts specifically with the U-rich region in the hepatitis C virus 3′ untranslated region. J Hum Virol. 1999;2:296–307. [PubMed] [Google Scholar]
- Takamizawa A, Mori C, Fuke I, Manabe S, Murakami S, Fujita J, Onishi E, Andoh T, Yoshida I, Okayama H. Structure and organization of the hepatitis C virus genome isolated from human carriers. J Virol. 1991;65:1105–1113. doi: 10.1128/jvi.65.3.1105-1113.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Kato N, Cho MJ, Sugiyama K, Shimotohno K. Structure of the 3′ terminus of the hepatitis C virus genome. J Virol. 1996;70:3307–3312. doi: 10.1128/jvi.70.5.3307-3312.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchihara K, Tanaka T, Hijikata M, Kuge S, Toyoda H, Nomoto A, Yamamoto N, Shimotohno K. Specific interaction of polypyrimidine tract-binding protein with the extreme 3′-terminal structure of the hepatitis C virus genome, the 3′X. J Virol. 1997;71:6720–6726. doi: 10.1128/jvi.71.9.6720-6726.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukiyama-Kohara K, Iizuka N, Kohara M, Nomoto A. Internal ribosome entry site within hepatitis C virus RNA. J Virol. 1992;66:1476–1483. doi: 10.1128/jvi.66.3.1476-1483.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuplin A, Wood J, Evans DJ, Patel AH, Simmonds P. Thermodynamic and phylogenetic prediction of RNA secondary structures in the coding region of hepatitis C virus. RNA. 2002;8:824–841. doi: 10.1017/s1355838202554066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuplin A, Evans DJ, Simmonds P. Detailed mapping of RNA secondary structures in core and NS5B-encoding region sequences of hepatitis C virus by RNase cleavage and novel bioinformatic prediction methods. J Gen Virol. 2004;85:3037–3047. doi: 10.1099/vir.0.80141-0. [DOI] [PubMed] [Google Scholar]
- Vassilaki N, Friebe P, Meuleman P, Kallis S, Kaul A, Paranhos-Baccala G, Leroux-Roels G, Mavromara P, Bartenschlager R. Role of the hepatitis C virus core+1 open reading frame and core cis-acting RNA elements in viral RNA translation and replication. J Virol. 2008;82:11503–11515. doi: 10.1128/JVI.01640-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walewski JL, Keller TR, Stump DD, Branch AD. Evidence for a new hepatitis C virus antigen encoded in an overlapping reading frame. RNA. 2001;7:710–721. doi: 10.1017/s1355838201010111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Sarnow P, Siddiqui A. Translation of human hepatitis C virus RNA in cultured cells is mediated by an internal ribosome-binding mechanism. J Virol. 1993;67:3338–3344. doi: 10.1128/jvi.67.6.3338-3344.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang TH, Rijnbrand RC, Lemon SM. Core protein-coding sequence, but not core protein, modulates the efficiency of cap-independent translation directed by the internal ribosome entry site of hepatitis C virus. J Virol. 2000;74:11347–11358. doi: 10.1128/jvi.74.23.11347-11358.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood J, Frederickson RM, Fields S, Patel AH. Hepatitis C virus 3′X region interacts with human ribosomal proteins. J Virol. 2001;75:1348–1358. doi: 10.1128/JVI.75.3.1348-1358.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Choi J, Yen TS, Lu W, Strohecker A, Govindarajan S, Chien D, Selby MJ, Ou J. Synthesis of a novel hepatitis C virus protein by ribosomal frameshift. EMBO J. 2001;20:3840–3848. doi: 10.1093/emboj/20.14.3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagi M, St Claire M, Emerson SU, Purcell RH, Bukh J. In vivo analysis of the 3′ untranslated region of the hepatitis C virus after in vitro mutagenesis of an infectious cDNA clone. Proc Natl Acad Sci. 1999;96:2291–2295. doi: 10.1073/pnas.96.5.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi M, Lemon SM. 3′ Nontranslated RNA signals required for replication of hepatitis C virus RNA. J Virol. 2003a;77:3557–3568. doi: 10.1128/JVI.77.6.3557-3568.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi M, Lemon SM. Structure–function analysis of the 3′ stem–loop of hepatitis C virus genomic RNA and its role in viral RNA replication. RNA. 2003b;9:331–345. doi: 10.1261/rna.2144203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You S, Stump DD, Branch AD, Rice CM. A cis-acting replication element in the sequence encoding the NS5B RNA-dependent RNA polymerase is required for hepatitis C virus RNA replication. J Virol. 2004;78:1352–1366. doi: 10.1128/JVI.78.3.1352-1366.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Yamada O, Sakamoto T, Yoshida H, Araki H, Murata T, Shimotohno K. Inhibition of hepatitis C virus replication by pol III-directed overexpression of RNA decoys corresponding to stem–loop structures in the NS5B coding region. Virology. 2005;342:276–285. doi: 10.1016/j.virol.2005.08.003. [DOI] [PubMed] [Google Scholar]