Abstract
Metazoan replication-dependent histone mRNAs are not polyadenylated, and instead terminate in a conserved stem–loop structure generated by an endonucleolytic cleavage of the pre-mRNA involving U7 snRNP. U7 snRNP contains two like-Sm proteins, Lsm10 and Lsm11, which replace SmD1 and SmD2 in the canonical heptameric Sm protein ring that binds spliceosomal snRNAs. Here we show that mutations in either the Drosophila Lsm10 or the Lsm11 gene disrupt normal histone pre-mRNA processing, resulting in production of poly(A)+ histone mRNA as a result of transcriptional read-through to cryptic polyadenylation sites present downstream from each histone gene. This molecular phenotype is indistinguishable from that which we previously described for mutations in U7 snRNA. Lsm10 protein fails to accumulate in Lsm11 mutants, suggesting that a pool of Lsm10–Lsm11 dimers provides precursors for U7 snRNP assembly. Unexpectedly, U7 snRNA was detected in Lsm11 and Lsm1 mutants and could be precipitated with anti-trimethylguanosine antibodies, suggesting that it assembles into a snRNP particle in the absence of Lsm10 and Lsm11. However, this U7 snRNA could not be detected at the histone locus body, suggesting that Lsm10 and Lsm11 are necessary for U7 snRNP localization. In contrast to U7 snRNA null mutants, which are viable, Lsm10 and Lsm11 mutants do not survive to adulthood. Because we cannot detect differences in the histone mRNA phenotype between Lsm10 or Lsm11 and U7 mutants, we propose that the different terminal developmental phenotypes result from the participation of Lsm10 and Lsm11 in an essential function that is distinct from histone pre-mRNA processing and that is independent of U7 snRNA.
Keywords: U7, snRNP, Lsm10, Lsm11, RNA processing, histone
INTRODUCTION
Replication-coupled histone biosynthesis is an essential aspect of genome duplication during cell proliferation, and is controlled primarily by regulating histone mRNA abundance. The 3′ end of histone mRNA is required for this cell cycle regulation (Luscher and Schumperli 1987; Stauber and Schumperli 1988; Harris et al. 1991). However, unlike all other metazoan mRNAs, the histone mRNA 3′ end contains a conserved terminal stem–loop structure rather than a poly(A) tail (Marzluff et al. 2008). Specialized machinery is needed to generate this unique mRNA 3′ end, and determining how this machinery functions is necessary for fully understanding replication-coupled histone mRNA biosynthesis.
Histone pre-mRNA utilizes two cis-acting elements for proper 3′ end processing: the stem–loop and a purine rich region downstream from the cleavage site termed the histone downstream element (HDE) (Mowry and Steitz 1987a). The stem–loop is recognized by a protein called the stem–loop binding protein (SLBP) (Wang et al. 1996), or hairpin binding protein (HBP) (Martin et al. 1997), and U7 snRNP interacts with histone pre-mRNA by base pairing between the 5′ end of U7 snRNA and the HDE (Galli et al. 1983; Schaufele et al. 1986; Mowry and Steitz 1987a; Bond et al. 1991). These factors recruit a complex that triggers endonucleolytic cleavage via the CPSF73 endonuclease between the stem–loop and the HDE forming the mature mRNA (Dominski et al. 2005; Kolev and Steitz 2005; Wagner et al. 2007). After processing, the mature histone mRNA bound to SLBP exits the nucleus and enters the cytoplasm where SLBP stimulates translation (Sanchez and Marzluff 2002).
Because U7 snRNP functions to recognize histone pre-mRNA and to recruit the appropriate cleavage factors, it is essential for histone pre-mRNA processing in vitro and in vivo (Galli et al. 1983; Gick et al. 1986; Mowry and Steitz 1987b; Godfrey et al. 2006). U7 snRNP is related to spliceosomal snRNPs, all of which are composed of a core particle containing a short, noncoding, nonpolyadenylated snRNA bound by seven structurally similar Sm proteins: SmD1, SmD2, SmE, SmF, SmG, SmB/B′, and SmD3 (Lührmann et al. 1990; Matera et al. 2007). These proteins bind to a conserved uridine-rich sequence in the snRNA termed the Sm binding site (Branlant et al. 1982). Sm proteins consist of two conserved motifs, SM1 and SM2, separated by a linker region of variable length (Hermann et al. 1995; Seraphin 1995). Structural analysis of the SmD3/SmB and SmD1/SmD2 complexes reveals that the SM1 and SM2 domains together form a common fold, termed the Sm fold, containing an NH2-terminal α-helix followed by a strongly bent five-stranded anti-parallel beta sheet (Kambach et al. 1999). These data, along with structural information from two related bacterial proteins—Lsmα from the thermophilic archaeon Methanobacterium thermoautotrophicum and AF-Sm1 and AF-Sm2 from the hyperthermophilic euryarchaeon Archaeoglobus fulgidus—suggest that the seven Sm proteins assemble into a donut-shaped, heptameric ring and that the snRNA may thread through the center hole (Collins et al. 2001; Toro et al. 2001, 2002).
U7 snRNP differs from spliceosomal snRNPs in an important way. U7 snRNP contains two unique Sm-like proteins, Lsm10 and Lsm11, which replace SmD1 and SmD2 in the canonical heptameric Sm ring. Lsm10 and Lsm11 contain the SM1 and SM2 motifs and are very similar in structure to Sm proteins. Lsm10 is closely related to SmD1, whereas Lsm11 is related to SmD2 (Pillai et al. 2001, 2003). U7 snRNA contains a noncanonical Sm binding site that recruits Lsm10 and Lsm11 specifically to the U7 particle. Mutation of the U7 Sm binding site to a spliceosomal consensus Sm binding site disrupts U7 snRNA function (Stefanovic et al. 1995), and results in replacement of Lsm10 and Lsm11 with SmD1 and SmD2 in the U7 snRNP (Pillai et al. 2003). This finding demonstrates that Lsm10 and Lsm11 confer functional properties to U7 snRNP that are essential for histone pre-mRNA processing. Previous work suggests that one role for Lsm11 in histone 3′ end processing is to bind directly to ZFP100 (Pillai et al. 2003; Azzouz et al. 2005), a 100 kDa zinc finger protein that is part of U7 snRNP and helps to stabilize the cleavage complex on histone pre-mRNA (Dominski et al. 2002). A specific binding site for ZFP100 was mapped to amino acids 63–82 in the NH2-terminal domain of human Lsm11 (Wagner et al. 2006). Lsm11 likely plays additional roles in the processing reaction, since mutation of other conserved amino acids in the NH2-terminal domain of Lsm11 impairs processing without affecting binding to ZFP100 (Azzouz et al. 2005).
How Lsm10 and Lsm11 contribute to histone pre-mRNA processing in vivo, or which aspects of their structure are necessary in vivo, has not been determined. For instance, ZFP100 has not been identified in invertebrate species like Drosophila, which contain a single U7 snRNA (Dominski et al. 2003) and single genes encoding Lsm10 and Lsm11 (Azzouz and Schumperli 2003; Dominski et al. 2003). In particular, it is not known whether Lsm10 or Lsm11 participate in other aspects of RNA metabolism as do other Lsm proteins. For instance, a complex of Lsm2–8 binds to U6 snRNA and is required for the U4/U6 formation during splicing (Achsel et al. 1999), while a complex of Lsm1–7 functions in cytoplasmic mRNA decay (Bouveret et al. 2000). We hypothesized that if Lsm10 and Lsm11 only function in histone pre-mRNA processing as part of U7 snRNP, then mutations in Drosophila Lsm10, Lsm11, and U7 would cause identical phenotypes. We previously demonstrated that U7 null mutants fail to properly process histone mRNA beginning at the third larval instar stage, but nonetheless develop into fully formed but sterile adults. Here we identify mutations of Lsm10 and Lsm11 and demonstrate that, like U7 snRNA mutations, disruption of Lsm10 and Lsm11 function results in the production of misprocessed histone mRNAs beginning at the third larval instar stage. However, unlike U7 null mutants, Lsm10 and Lsm11 mutants do not survive to adulthood, dying as non-pharate pupae. This strongly suggests that Lsm10 and Lsm11 are required for an essential process distinct from histone pre-mRNA processing.
RESULTS
Identification of Lsm10 and Lsm11 mutations
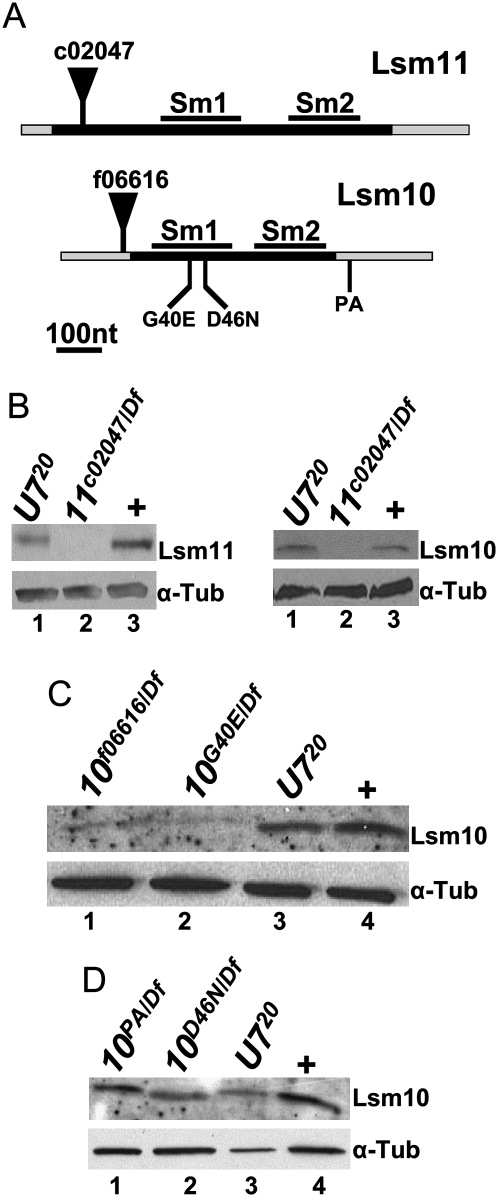
To examine the contribution of Lsm10 and Lsm11 proteins to histone pre-mRNA processing during development, we isolated mutations in each gene. We first identified from the Exelixis collection (Thibault et al. 2004) a single piggyBac (pBac) insertion allele of both Lsm10 and Lsm11. pBac transposons have a higher incidence of inserting into the coding sequence of genes relative to P-element transposons, which more often insert into the 5′ UTR (Thibault et al. 2004). Accordingly, Lsm11c02047 contains a pBac insertion near the 5′ end of the coding sequence (Fig. 1A), and animals containing this mutation in trans with a deficiency (Df) do not contain detectable amounts of Lsm11 protein by Western analysis (Fig. 1B, left panel). This suggests that Lsm11c02047 is a null allele. Lsm10f06616 contains a pBac insertion in the 5′ UTR (Fig. 1A), and this mutant is a hypomorph that expresses small amounts of Lsm10 protein (Fig. 1C, lane 1). In an effort to identify additional Lsm10 alleles, including a null, we employed the targeting-induced local lesions in genomes (TILLING) strategy. TILLING is a high-throughput method to molecularly identify EMS-induced mutations in specific regions of the genome (Till et al. 2003). The strategy was adapted for use with a collection of Drosophila strains that carry an EMS-mutagenized chromosome 2 (Koundakjian et al. 2004). We screened a ∼800 nucleotide region containing the entire Lsm10 gene from ∼3000 of these mutagenized lines. Three mutations were recovered that we suspected might affect Lsm10 function: a Glu substitution for an evolutionarily invariant Gly (Lsm10G40E) needed for a sharp bend in β-strand 3 of the SM1 domain (Lührmann et al. 1990), an Asn substitution for an evolutionarily invariant Asp that, based on structural analysis of related Sm proteins (Toro et al. 2001, 2002), is predicted to contact the snRNA (Lsm10D46N), and a single nucleotide change in the 3′ UTR that alters a canonical poly(A) signal sequence (AATAAA→TATAAA; Lsm10PA) (Fig. 1A). The Lsm10G40E/Df mutant expressed a small amount of Lsm10 protein (Fig. 1C), suggesting that the G40E substitution disrupts folding of the SM1 domain resulting in reduced protein accumulation. The Lsm10PA/Df and Lsm10D46N/Df mutants did not show any reproducible reduction in Lsm10 protein compared to wild type (Fig. 1D, lanes 1,2).
FIGURE 1.
Identification of Lsm10 and Lsm11 mutations. (A) Schematic diagram of the Lsm11 and Lsm10 genes. The black bars represent the coding sequence and the gray bars represent the 5′ and 3′ UTRs. Note there are no introns in these two genes. The position of the Sm1 and Sm2 domains are marked with black bars above the coding sequence. The positions of the pBac insertions are indicated by black triangles. The positions of the three Lsm10 EMS alleles are indicated below the gene. (B–D) Protein extracts of brain and salivary gland tissue from third instar larvae or adult flies (D) of the indicated genotypes were probed with anti-Lsm11 or anti Lsm10-antibodies by Western blotting. w1118 was used as a normal control here and in subsequent figures (+). α-Tubulin is used as a loading control.
To test whether the accumulation of Lsm10 and Lsm11 are interdependent, we examined Lsm10 and Lsm11 accumulation in U7 mutants, and Lsm10 accumulation in Lsm11 mutants. Both Lsm10 and Lsm11 protein were present at wild-type levels in the U720 null mutant (Fig. 1B–D), indicating that a pool of these proteins can exist independent of U7 snRNA. In contrast, Lsm10 protein was undetectable in the Lsm11c02047/Df null mutant (Fig. 1B, right panel). We previously obtained a similar result when Lsm11 was depleted by RNAi in cultured Drosophila S2 cells (Wagner et al. 2007). These data indicate that Lsm10 accumulation depends on Lsm11, and suggest that a free pool of Lsm10 and Lsm11 dimers exist in cells. This is consistent with current models of snRNP assembly, which suggest that SmD1/SmD2 dimers are precursors for snRNP assembly in the cytoplasm and that both SmD1/SmD2 and Lsm10/Lsm11 are located adjacent to one another in the Sm ring of their respective snRNP particles (Schumperli and Pillai 2004).
Lsm10 and Lsm11 mutants fail to properly process histone pre-mRNA
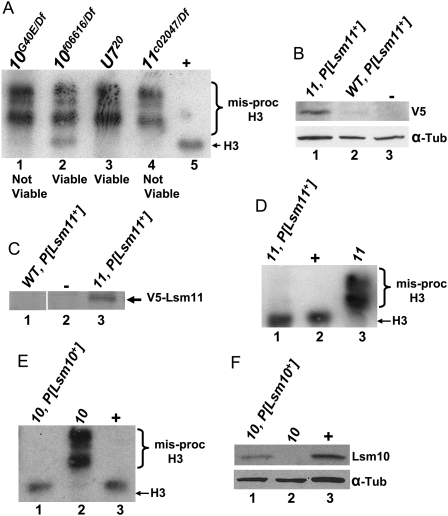
We have previously shown that mutations in U7 snRNA prevent normal histone pre-mRNA processing, resulting in aberrantly long histone mRNAs that are polyadenylated due to the use of cryptic polyadenylation signals downstream from the cleavage site in each histone gene (Godfrey et al. 2006). To test whether a similar phenotype occurs after loss of Lsm10 and Lsm11, we used Northern blotting to hybridize total RNA samples prepared from Lsm10 or Lsm11 mutant third instar larvae with a histone H3 probe, and compared the results to a U720 null mutant (Fig. 2A). As with U720, we found that the strong Lsm10G40E and Lsm11c02047 alleles caused nearly complete misprocessing of H3 mRNA, with very little wild-type H3 mRNA present in these mutants (Fig. 2A, cf. lanes 1,4 and lanes 3,5). In the Lsm10f06616 mutant, which expresses a small amount of Lsm10 protein, we observed misprocessed H3 mRNA, but unlike the other mutants, we could also detect a small amount of correctly processed wild-type H3 mRNA (Fig. 2A, lane 2). These data indicate that Lsm10 and Lsm11 are required for normal histone pre-mRNA processing in vivo.
FIGURE 2.
Lsm10 and Lsm11 mutants fail to properly process histone pre-mRNA and are necessary for development. (A) Total RNA isolated from whole third instar larvae of the indicated genotypes was subjected to Northern analysis with a 32P-labeled H3 probe. Note that the severity of the misprocessed H3 phenotype is similar in U720, Lsm10G40E, and Lsm11c02047 mutants, while the terminal developmental phenotype is different: viable for U720 and not viable for the Lsm10G40E and Lsm11c02047 null mutants. The Lsm10f06616 hypomorph is viable. (B) Protein extracts prepared from embryos of the indicated genotypes were probed with anti-V5 antibodies by Western blotting. P[Lsm11+] is a transgene expressing a V5-Lsm11 with the endogenous Lsm11 promoter. “11” Refers to the homozygous Lsm11c02047 genotype. Lane 3 contains protein from a nontransgenic control. Note that in a wild-type Lsm11 background there is very little accumulation of V5-Lsm11 protein. α–Tubulin is used as a loading control. (C) Protein extracts isolated from whole third instar larvae of the indicated genotypes were subjected to immunoprecipitation then Western blot analysis with anti-V5 antibody. Lane 2 contains protein from nontransgenic control. (D,E) RNA isolated from whole third instar larvae of the indicated genotypes was subjected to Northern analysis with 32P-labeled H3 probe. “10” Refers to the Lsm10G40E/Df mutant genotype. Note that there is very little misprocessed H3 in both Lsm11c02047, P[Lsm11+] and Lsm10G40E/Df, P[Lsm10+] genotypes. (F) Protein extracts prepared from whole third instar larvae of the indicated genotypes were probed with anti-Lsm10 antibodies. Note that the lane 1 genotype contains a single copy of P[Lsm10+], accounting for the reduction in Lsm10 accumulation relative to wild type (+). All other P[Lsm] transgenes are present in two copies.
Lsm10 and Lsm11 are necessary for development
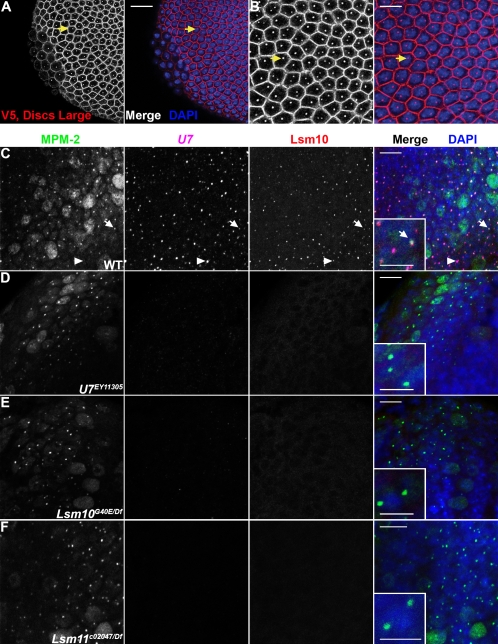
The strong Lsm10G40E and Lsm11c02047 alleles cause lethality: In crosses segregating Lsm10 or Lsm11 mutant progeny, none of the adults that eclosed were of the mutant genotype (n = 935 for Lsm11 and n = 994 for Lsm10; P < 0.005). The Lsm mutants progress through larval stages and die as nonpharate pupae. This result was surprising since U7 null mutants are fully viable yet display the same H3 mRNA misprocessing phenotype (Godfrey et al. 2006). We therefore tested if the Lsm10 and Lsm11 mutant phenotypes could be complemented with P element transgenes containing a functional copy of the respective genes. We first engineered a transgene expressing an NH2-terminal V5 epitope-tagged Lsm11 with the endogenous Lsm11 promoter. Curiously, we could detect little, if any, transgenic V5-Lsm11 in a wild-type background when using anti-V5 on a Western blot (Fig. 2B, lane 2), immunoprecipitation (Fig. 2C, lane 1), or immunofluorescent staining of fixed tissue (data not shown). In contrast, we readily detected V5-Lsm11 using all of these methods when the V5-Lsm11 transgene was present in the Lsm11c02047 mutant background (Fig. 2B, lane 1; Fig. 2C, lane 3; Fig. 7A,B, see below). One explanation for this result is that V5-Lsm11 is a poor substrate for U7 snRNP assembly relative to normal Lsm11, and the resulting free V5-Lsm11 is degraded. In spite of this, V5-Lsm11 completely rescued the H3 misprocessing defect and the lethality of Lsm11c02047 mutants, indicating that V5-Lsm11 is fully functional (Fig. 2D) and that Lsm11 function is necessary for Drosophila development. Overexpression may overcome any slight U7 snRNP assembly defects that Lsm11 proteins containing an NH2-terminal epitope tag may have, as Liu et al. (2006) previously demonstrated that YFP-Lsm11 expressed using the strong da promoter behaves similarly to wild-type Lsm11.
FIGURE 7.
The U7 snRNP formed in an Lsm11 mutant does not localize to the histone locus body. (A) Stage 5 Lsm11c02047, P[V5-Lsm11+] homozygous embryos were stained with anti-Discs Large antibodies, to visualize cell boundaries, and anti-V5 antibodies (left panels, both red in merge). Anti-mouse secondary antibodies were used to simultaneously detect V5-Lsm11 and Discs Large. Embryos were also stained with DAPI (blue in merge). Note that V5-Lsm11 localizes to one or two nuclear foci just like endogenous Lsm11. Arrows indicate the same cell in (A) (20 μm scale bar) and (B) (10 μm scale bar). (C–F) Brains dissected from w1118, U7EY11305, Lsm10G40E/Df, and Lsm11c02047/Df third instar larvae were stained with MPM-2 (first column; green in merge), hybridized with a fluorescent probe recognizing U7 snRNA (second column; magenta in merge), anti-Lsm10 antibodies (third column; red in merge), and DAPI (blue in merge). Arrows indicate a histone locus body that contains MPM-2 antigen(s), U7, and Lsm10. (Insets) A higher magnification view. Arrowheads indicate a histone locus body lacking MPM-2 staining. This nucleus is likely not within S phase, and therefore lacks the Cyclin E/Cdk2 activity necessary to produce the MPM-2 epitope. Note that both U7 and Lsm10 are undetectable in histone locus bodies marked by MPM-2 staining in both Lsm10 (E) and Lsm11 (F) mutant brains. Bar, 10 μm (main panels) and bar, 5 μm (insets).
Because of theV5-Lsm11 results, we elected not to epitope tag Lsm10, and instead generated a transgene carrying a genomic DNA fragment containing the wild-type Lsm10 gene. This transgene completely rescued both the H3 misprocessing defect and lethality of Lsm10G40E mutants (Fig. 2E). Note that we did not achieve wild-type expression of Lsm10 protein in this experiment because the genotype we constructed only contained a single copy of the wild-type Lsm10 transgene (Fig. 2F). This suggests that Lsm10 is normally present in functional excess. These data show that reintroducing a functional copy of Lsm10 and Lsm11 can rescue both the lethality and histone mRNA misprocessing phenotypes of Lsm10 and Lsm11 mutants, confirming that these phenotypes are a direct consequence of the loss of each gene.
Lsm11 mutant lethality is independent of histone mRNA misprocessing
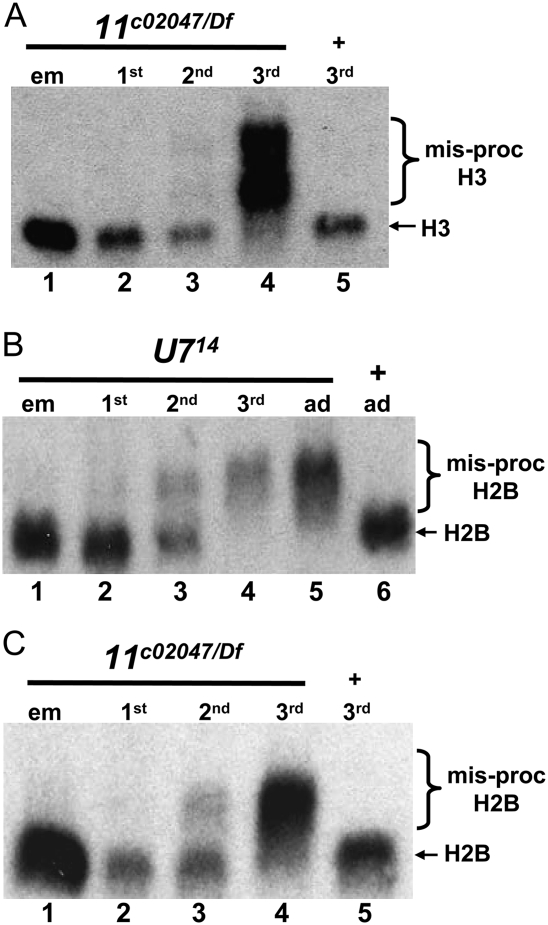
Because U7 null mutants are viable, we were surprised to find that our strongest Lsm10 and Lsm11 alleles caused lethality. We previously showed that a maternal supply of U7 snRNA is sufficient for normal histone pre-mRNA processing through the first larval instar stage of development, such that in U7 mutants misprocessed histone mRNAs first appear in second instar larvae and wild-type histone mRNA is undetectable by the third larval instar stage (Fig. 3B; Godfrey et al. 2006). In contrast to U7, Slbp null mutations cause lethality in late larval or pupal stages, and this is likely due to an earlier onset of the histone mRNA misprocessing mutant phenotype: there is no functional maternal supply of SLBP and misprocessed histone mRNA can be detected as soon as zygotic transcription begins in Slbp mutant embryos (Lanzotti et al. 2002). Based on these observations, we hypothesized that an earlier onset of the histone mRNA misprocessing phenotype in Lsm10 and Lsm11 mutants relative to U7 mutants might account for the Lsm10 and Lsm11 mutant lethality. To test this, we examined histone mRNA processing at different stages of development in U7 and Lsm11c02047 mutants (which also lack Lsm10 protein) (Fig. 1B). Total RNA was extracted from mutant embryos as well as first, second, and third instar larvae and hybridized with a histone H3 mRNA probe. As we previously reported for histone H3 (Godfrey et al. 2006), in U714 null mutants a small amount of misprocessed histone H2B mRNA was expressed in second instar larvae and only misprocessed histone mRNA was present in third instar larvae (Fig. 3B). In Lsm11 mutants, the longer, misprocessed H3 mRNA was also not detected until the second larval instar stage of development, and the correctly processed, wild-type H3 mRNA was barely detectable by the third instar stage (Fig. 3A). Similar results were obtained with histone H2B mRNA (Fig. 3C), as well as histone H2A, H4, and H1 mRNAs (Supplemental Fig. S1). For all of these genes we never detected an onset of misprocessing in Lsm11c02047 mutants any earlier than the 2nd larval instar stage of development. Consistent with Lsm10 acting together with Lsm11 in histone pre-mRNA processing, we first detected longer, misprocessed H3 mRNA at the third larval instar stage in Lsm10G40E mutants (Supplemental Fig. S2A).
FIGURE 3.
Lsm11 mutant lethality is independent of histone mRNA misprocessing. (A–C) Total RNA was extracted from animals at different stages of development of the indicated genotypes and subjected to Northern analysis with a 32P-labeled probe to H2B (B,C) or H3 (A). Note that the misprocessed histone mRNA is first detectable in small amounts at the second larval instar stage and that wild-type histone mRNA is absent in the third larval instar stage.
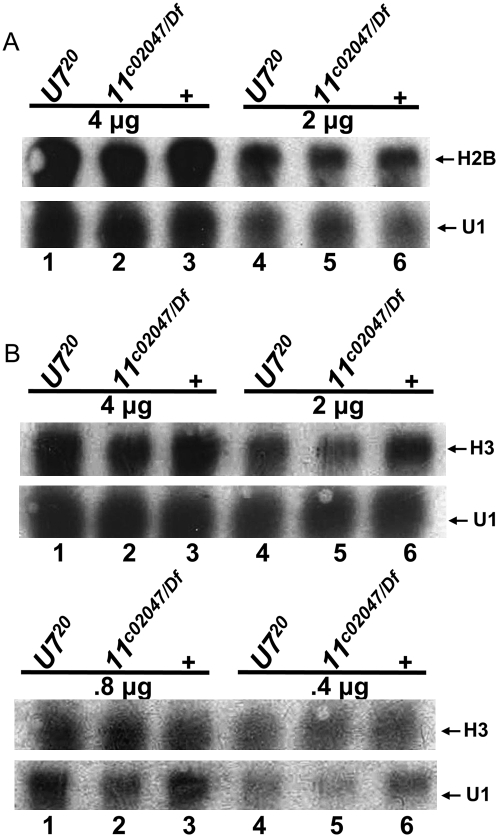
To determine if there was any quantitative difference in histone expression between U7 and Lsm11 mutants prior to the onset of histone pre-mRNA misprocessing, we performed Northern analysis for H2B (Fig. 4A) and H3 (Fig. 4B) using RNA extracted from wild-type, U7, and Lsm11 null mutant first instar larvae. Relative to a U1 snRNA loading control, we did not detect any reproducible difference in the accumulation of H3 or H2B mRNA between the two mutants and control (Fig. 4). Thus, Lsm11 mutants do not begin to accumulate misprocessed histone mRNA earlier in development than U7 mutants and there is no difference in histone mRNA expression. We conclude from these data that the difference in terminal developmental phenotype of Lsm11 and U7 mutants (i.e., lethal versus viable, respectively) does not result from differences in histone mRNA biosynthesis. Because strong Lsm10 mutants are also not viable, these data suggest that Lsm10 and Lsm11 perform an essential function during development independent of histone mRNA metabolism.
FIGURE 4.
Quantitative analysis of histone mRNA levels between U7 and Lsm11 mutants. (A,B) Total RNA was extracted from first instar larvae of the indicated genotypes and different quantities subjected to Northern analysis with a 32P-labeled probe to H2B (A) or H3 (B). U1 is used as a loading control.
Hypomorphic Lsm10 alleles are viable with compromised fertility
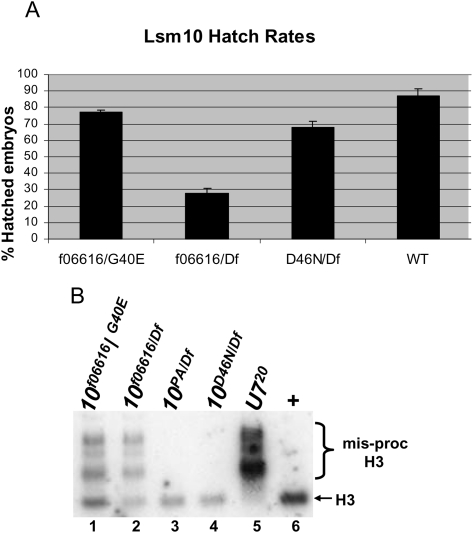
Although U7 null mutants are viable, both males and females are completely sterile, and defects during late stages of oogenesis prevent U7 mutant females from laying eggs (Godfrey et al. 2006). Since the Lsm10f06616 and Lsm10D46N mutant alleles support development to adulthood when in trans to a deficiency, we assessed whether the adults were fully fertile. Lsm10f06616/Df and Lsm10D46N/Df trans-heterozygous males and females are fertile. We then self-crossed Lsm10f06616/Df and Lsm10D46N/Df mutant males to the corresponding Lsm10 mutant virgin females, and determined the fraction of eggs that would hatch into larvae as an assessment of the relative fecundity. We found that 30% of the eggs from the Lsm10f06616 cross hatched, which was significantly (P < 0.00001) reduced relative to the 85% hatching of wild-type controls and significantly (P < 0.005) less than the 75% expected to hatch if Lsm10 was not required for embryogenesis (the Df/Df genotype is embryonic lethal and expected to be 25% of progeny) (Fig. 5A). Eggs from the Lsm10D46N/Df self-cross also had reduced hatching relative to wild type (70%, P < 0.02), and this was slightly less than the 75% expected (P < 0.05). DAPI staining of mutant embryos revealed that these decreases in hatching were not due to a failure of fertilization (Supplemental Fig. S3). These results are consistent with our Western data, which indicate that the Lsm10f06616 mutant has a much larger reduction in Lsm10 protein levels compared to the Lsm10D46N mutant (Fig. 1C,D). We also found that placing the Lsm10f06616 hypomorphic allele in trans to our strongest Lsm10 mutant allele, Lsm10G40E, results in viable adults, and that eggs from Lsm10f06616/G40E males and females hatched less than wild-type controls (75% hatching; P < 0.05). Because this hatching rate is significantly greater than that obtained from Lsm10f06616/Df males and females (P < 0.00005), we conclude that the Lsm10G40E mutation is not null and can be characterized as a strong hypomorph (i.e., Lsm10G40E is not equivalent to a Df in this assay). This is consistent with our detection of a small amount of Lsm10 protein in the Lsm10G40E mutant (Fig. 1C).
FIGURE 5.
Hypomorphic Lsm10 alleles are viable, with some fertility defects. (A) The average and standard deviation of the percent of hatched embryos for the indicated genotypes. Measurements were made on six collections of 100 eggs. (B) RNA isolated from 1–2-d-old adult females of the indicated genotypes was subjected to Northern analysis with a 32P-labeled H3 probe. Note misprocessed H3 mRNA is detected in only two of the Lsm10 mutant genotypes, and that these hypomorphic mutants also contain wild-type, processed mRNA.
To determine whether these changes in fertility correlate with defects in histone pre-mRNA processing, we extracted total RNA from adult females of each mutant genotype and hybridized it with a histone H3 mRNA probe. The mutants of genotype Lsm10f06616/Df and Lsm10f06616/G40E contain the longer, misprocessed histone H3 mRNA that we detect in U7 null mutants (Fig. 5B, cf. lanes 1,2 and lane 5). These mutants also contain correctly processed H3 mRNA, consistent with a hypomorphic condition that is not completely defective in histone pre-mRNA processing. Interestingly, we detected only normal H3 mRNA and no misprocessed H3 mRNA in the Lsm10D46N/Df allele combination (Fig. 5B, cf. lanes 3,4 and lane 6). This indicates that Asn substitution of the highly conserved Asp46 does not affect U7 snRNP function. In addition, the slight fertility defects in this mutant are likely not caused by aberrant histone pre-mRNA processing.
U7 snRNA is present in a snRNP particle in Lsm11 mutants
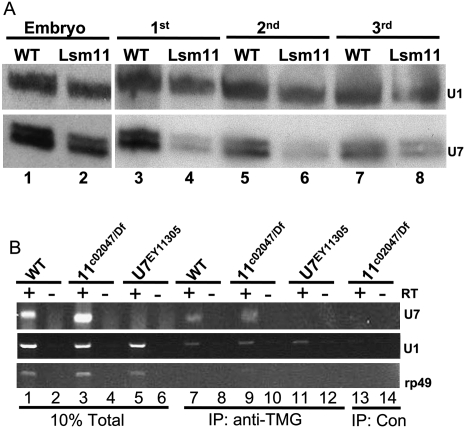
Previous studies suggest that U snRNAs, including U7, that cannot bind Sm proteins are unstable and do not accumulate in the cell (Jones and Guthrie 1990; Grimm et al. 1993). We therefore hypothesized that a U7 snRNP would not form in the absence of Lsm10 and Lsm11, and that we consequently would not detect any U7 snRNA in an Lsm11 mutant. To test this we used Northern blotting to measure the accumulation of U7 snRNA in Lsm11c02047 mutants, compared to wild type, throughout development. Total RNA was extracted from embryos, as well as first, second, and third instar larvae, and hybridized with a U7 probe. We found that in Lsm11c02047 mutants the U7 snRNA levels begin to drop compared to wild type at the first instar stage (Fig. 6A, cf. lane 3 and lane 4). Surprisingly, we repeatedly detected U7 snRNA in Lsm11c02047 mutant 3rd instar larvae (Fig. 6A, lane 8), a stage at which all histone mRNA are misprocessed in this mutant (Fig. 3B,C, lane 4) and U7 snRNA is not detected in mutants (Figs. 6B, 7D). Using densitometry we determined that Lsm11c02047 mutants have 60% the amount of U7 snRNA compared to wild type at this developmental stage. We also detected U7 snRNA in Lsm10G40E mutant third instar larvae (Supplemental Fig. S2B). We considered two interpretations of these results: either the U7 snRNA is stable in the absence of a bound Sm protein ring, or the U7 snRNA assembles into a snRNP particle lacking Lsm10 and Lsm11.
FIGURE 6.
U7 snRNA can form a snRNP particle in Lsm11 mutants. (A) U7 Northern analysis of RNA isolated from Lsm11c02047/Df mutant (Lsm11) or w1118 control (WT) embryos and first to third instar larvae. Note that in the Lsm11 mutant U7 snRNA is detected in third instar larvae when all the histone mRNA is misprocessed. A U1 probe was used as a loading control. U7 snRNA migrates as a doublet as described previously (Dominski et al. 2003). (B) Reverse transcriptase (RT)-PCR analysis of RNA extracted from anti-TMG immunoprecipitates of whole third instar larvae RNA samples of the indicated genotypes. (Lanes 1–6) 10% of total input RNA; (lanes 7–12) anti-TMG IP; and (lanes 13,14) mock IP negative control. (Top panel) U7 snRNA primer pair. Note that there is no U7 present in the U7EY11305 mutant or in the control IP lane, but U7 is detected in both WT and Lsm11c02047/Df TMG IP samples. (Middle panel) U1 snRNA primer pair. Note that U1 is present in all three TMG IP samples, but not in the IP control. (Bottom panel) Ribosomal protein 49 (rp49) primer pair. Note that rp49 mRNA is not precipitated by anti-TMG antibodies because the mRNA lacks a trimethylguanosine cap.
To distinguish between these two possibilities, we determined whether the U7 snRNA observed in the Lsm11 mutant contains a 5′ trimethylguanosine (TMG) cap. During snRNP assembly, the canonical m7G cap of newly transcribed snRNAs is hypermethylated to a TMG cap, and this requires Sm protein binding to the snRNA (Mattaj 1986). Thus, if the U7 snRNA present in the Lsm11 mutant has a TMG cap, we would infer that it had assembled into a snRNP particle. To test this, we developed an assay that couples immunoprecipitation with reverse transcription (RT)-PCR. Total RNA was prepared from wild type, Lsm11 mutant, or U7 mutant third instar larvae and incubated with anti-TMG antibodies that were coupled to agarose beads. Precipitated RNA was extracted from the beads and U7 snRNA was detected by RT-PCR. We found that anti-TMG antibodies could precipitate U7 snRNA from wild-type control samples, but not from U7 mutant samples, which lack U7 snRNA (Fig. 6B, top panel, lanes 7,11). The anti-TMG antibodies precipitated U1 snRNA from all samples (Fig. 6B, middle panel), and did not precipitate rp49 mRNA, which contains an m7G cap rather than a TMG cap, indicating that the antibody was specific (Fig. 6B bottom panel). No U snRNA was precipitated when a nonspecific antibody was used (Fig. 6B, lanes 13,14). Thus, our assay specifically detects U7 snRNA. In Lsm11 mutant samples we reproducibly detected U7 snRNA with the anti-TMG antibodies (Fig. 6B, lane 9). These data suggest that even in the absence of Lsm10 and Lsm11, U7 snRNA can assemble into a snRNP particle.
We considered the possibility that this aberrant U7 snRNP particle, which is not functional in histone pre-mRNA processing, could be toxic and thus account for the lethality of Lsm10 and Lsm11 mutants. This hypothesis predicts that removing U7 snRNA from an Lsm11 mutant will suppress lethality. To test this we constructed a U720 Lsm11c02047double mutant strain. We found that this double mutant was still lethal, indicating that eliminating the U7 snRNP formed in an Lsm11 mutant could not rescue the lethal phenotype, and that the nonfunctional U7 snRNP in Lsm11 mutants was not the basis of Lsm11 mutant lethality.
The U7 snRNP formed in an Lsm11 mutant does not localize to the histone locus body
In mammalian cells, U7 snRNP localizes to Cajal bodies (CBs), which are nuclear structures involved in the assembly and modification of the machinery needed for pre-mRNA splicing, pre-ribosomal RNA processing, and histone pre-mRNA processing (for reviews, see Gall 2000; Carmo-Fonseca 2002; Gall 2003; Matera 2003; Cioce and Lamond 2005). Some of these Cajal bodies are associated with histone genes, and may represent sites of histone mRNA biosynthesis. Drosophila cells also contain a Cajal body, but the Cajal body lacks U7 snRNP (Liu et al. 2006). Drosophila cells have a distinct body, termed the histone locus body (HLB), which is invariably associated with the histone gene locus and where the U7 snRNP localizes (Fig. 7A–C; Liu et al. 2006; White et al. 2007). HLBs likely contain all of the factors necessary for histone mRNA transcription and pre-mRNA processing (Marzluff et al. 2008). Our observation that a nonfunctional U7 snRNP particle forms in Lsm11 mutants provided an opportunity to determine if U7 snRNA localization to the HLB depends on Lsm11 and Lsm10. We used fluorescent in situ hybridization (FISH) to detect U7 snRNA, and MPM-2 antibodies to detect HLBs independently from U7 snRNP. The MPM-2 monoclonal antibody recognizes a cell cycle-regulated, Cyclin E/Cdk2-dependent phospho-epitope that localizes to HLBs during S phase (White et al. 2007). We stained third instar larvae brains, because HLBs are easy to identify in this tissue and the Lsm11 mutant phenotype is fully expressed at this developmental stage. In wild-type cells, HLBs were clearly detected by the co-localization of MPM-2 staining, anti-Lsm10 staining, and U7 snRNA FISH (Fig. 7C). The lack of U7 FISH signal in MPM-2 positive HLBs in U7 null mutant cells confirmed the specificity of our U7 probe (Fig. 7D). As we previously observed for Lsm11 (White et al. 2007), we could not detect Lsm10 signal in the HLBs of U7 null mutants, indicating that neither Lsm10 nor Lsm11 accumulate in HLBs in the absence of U7 snRNA (Fig. 7D). In Lsm10 and Lsm11 mutants we failed to detect U7 snRNA in the HLBs, and with respect to FISH signal for the U7 snRNA these preparations were indistinguishable from the U7 null mutant (Fig. 7E,F). This indicates that in the Lsm10 and Lsm11 mutants, the aberrant U7 snRNP does not concentrate in a particular location within the nucleus. We know that in these mutants other proteins can correctly localize to the HLB, because MPM-2 staining is similar to that in wild-type controls (Fig. 7E,F). Finally, we did not detect Lsm10 protein in HLBs of Lsm11 mutants, consistent with our Western blot results showing a lack of Lsm10 protein accumulation in Lsm11 null mutants (Fig. 7F). Together these results suggest that Lsm10 and Lsm11 are required for U7 snRNA localization to the HLB.
DISCUSSION
Recent studies of snRNAs and their associated Sm proteins have revealed new snRNP particles and novel functions for existing particles (Beggs 2005). Here we report the first genetic analysis of Lsm10 and Lsm11, which are both components of the Sm protein ring of the U7 snRNP particle. Our data indicate that, like U7 snRNA, Lsm10 and Lsm11 are essential for histone pre-mRNA processing in vivo. Surprisingly, our data also suggest that these Lsm proteins may play an essential role in development that is independent of U7 snRNA and histone mRNA metabolism.
Genetic evidence for a novel function for Lsm10 and Lsm11
As predicted from previous studies of U7 snRNP (Pillai et al. 2001; Azzouz and Schumperli 2003; Pillai et al. 2003; Azzouz et al. 2005; Wagner et al. 2006), including our phenotypic analysis of U7 snRNA mutations in Drosophila (Godfrey et al. 2006), we find that Drosophila Lsm10 and Lsm11 are both essential for normal histone mRNA biosynthesis during development. The longer, aberrant histone mRNAs produced in Lsm10 and Lsm11 mutants are identical to those we previously described in mutants of U7 snRNA and other components of the histone pre-mRNA processing machinery, and arise from the use of cryptic downstream polyadenylation signals located within each histone gene (Sullivan et al. 2001; Lanzotti et al. 2002; Godfrey et al. 2006; Wagner et al. 2007). Thus, as expected, loss of Lsm10 and Lsm11 results in the same molecular phenotype as loss of U7 snRNA. However, unlike viable U7 snRNA mutants, both Lsm10 and Lsm11 mutants die as nonpharate pupae. This is reminiscent of Slbp mutants, which also die as larvae or pupae (Lanzotti et al. 2002). We attribute the lethality of Slbp mutants to an earlier onset of histone pre-mRNA misprocessing during development (i.e., embryonic stage in Slbp mutants versus the third instar larval stage in U7 mutants) resulting from the lack of maternal SLBP and a large store of maternal U7 snRNA. However, this model cannot explain the lethality of Lsm11 and Lsm10 mutants, since the timing of onset of histone misprocessing during development in Lsm11 and U7 mutants is identical for the replication-dependent histone mRNAs.
What might cause the Lsm10 and Lsm11 mutants to die? Surprisingly, there is still U7 snRNA present in Lsm10 and Lsm11 mutant third instar larvae. One possibility is that this U7, or an aberrant U7 snRNP particle that assembles in the absence of Lsm10 and Lsm11, may be detrimental and result in a dominant negative effect on some essential process. However, this model is not supported by our observation that Lsm11 U7 double mutants are not viable, because it predicts that removing U7 snRNA should suppress the lethality of Lsm11 mutants. Therefore, we interpret our data as an indication that Lsm10 and Lsm11 are involved in a U7-independent function that is required for viability.
Since Lsm10 and Lsm11 are binding partners in the U7 Sm ring, it is possible that they could both be part of another Sm or Lsm ring. Lsm proteins participate in many aspects of RNA metabolism, and the full repertoire of Lsm complexes that exists in vivo is not known (Beggs 2005). The best understood Lsm complexes are the heptameric Lsm2–8 complex, which binds U6 and functions in the nucleus during pre-mRNA splicing, and the heptameric Lsm1–7 complex, which functions in cytoplasmic mRNA decay (Beggs 2005). There is evidence for other novel Lsm complexes whose composition remains incompletely understood. For instance, in Saccharomyces cerevisiae a complex containing Lsm2–7 that resides in nucleoli associates with the small nucleolar RNA (snoRNA) snR5, which is a member of the box H/ACA class of snoRNAs that function in pseudouridylation of rRNA (Fernandez et al. 2004). This Lsm complex is likely distinct from the Lsm2–8 complex (Fernandez et al. 2004). In Xenopus a complex containing Lsm2, Lsm3, Lsm4, Lsm6, Lsm7, and Lsm8, and apparently lacking Lsm5, associates with the U8 snoRNA, which is a member of the box C/D class of snoRNAs (Tomasevic and Peculis 2002). It is unclear whether this is the same Lsm2–8 complex that binds the U6 snRNA or whether an unidentified Lsm protein (perhaps in place of Lsm5) binds only to U8 (Tomasevic and Peculis 2002). In neither the yeast nor the frog complex have all the Lsm components been identified. Thus, in principle, Lsm10 and/or Lsm11 could be part of an uncharacterized Lsm ring that can transiently bind to any of the many snRNAs or snoRNAs.
There is recent evidence that Lsm10 and Lsm11 could function in aspects of RNA metabolism other than histone pre-mRNA processing. RNAi-mediated depletion of Lsm11 in Drosophila S2 cells causes a shift from one alternatively spliced variant of the Drosophila paralytic gene to another (Park et al. 2004). D. paralytic encodes a neuronal sodium channel that is essential for development (Loughney et al. 1989). Therefore, in Lsm11 mutants a shift in paralytic splice variants may disrupt expression of the sodium channel in a way that causes lethality. However, we were unable to detect any significant difference in accumulation of paralytic splice variants between wild-type and Lsm11 mutants (Supplemental Fig. S4). Nevertheless, there are many alternatively spliced, essential genes in Drosophila. Disruption of normal expression of even a single one of these by loss of Lsm10 or Lsm11 may be sufficient to cause the lethality of Lsm10 and Lsm11 mutants. Our future work will involve testing whether we can identify a novel function for Lsm10 and Lsm11.
U7 snRNA assembles into a snRNP without Lsm10 and Lsm11
During the biogenesis of snRNP particles, newly transcribed snRNA is exported to the cytoplasm where it is bound by the survival motor neuron (SMN) complex, which then assembles the Sm ring onto the snRNA (Matera et al. 2007). Based on previous work indicating snRNAs that are incapable of binding Sm proteins do not accumulate (Jones and Guthrie 1990; Grimm et al. 1993), we did not expect to detect U7 snRNA in Lsm11 mutants (which also lack Lsm10). Instead, we readily detected trimethylguanosine cap-modified U7 snRNA in Lsm11 mutant larvae when histone pre-mRNA processing was completely defective. Because the 5′ cap of snRNA is hypermethylated only after assembly into a snRNP particle (Mattaj 1986), we infer that the U7 snRNA present in an Lsm11 mutant is part of an intact snRNP. The composition of such an aberrant U7 snRNP is not known. We speculate that because of their sequence similarities, perhaps SmD1 and SmD2 replace Lsm10 and Lsm11 in the Sm protein ring that binds U7 snRNA. There is some precedence for this possibility. Changing the U7 Sm binding site to the canonical site found in spliceosomal snRNPs results in U7 snRNP particles that cannot function in histone 3′ end processing and that contain SmD1 and SmD2 instead of Lsm10 and Lsm11 (Stefanovic et al. 1995; Pillai et al. 2003). In the protozoan parasite Trypanosoma brucei two novel Sm proteins, Sm15K and Sm16.5K, replace SmB and SmD3 in the U2 snRNP. The Sm site in T. brucei U2 snRNA differs by one base pair from the consensus Sm site in other T. brucei U snRNAs, and a single base pair change in the U2 Sm site can convert the special Sm ring containing Sm15K and Sm16.5K to the canonical one containing SmB and SmD3 (Wang et al. 2006). Since the SmD1/SmD2 heterodimer is very similar in structure to the Lsm10/Lsm11 heterodimer, perhaps, in the absence of competing Lsm10 and Lsm11 protein, SmD1 and SmD2 bind to the nonconsensus Sm site in wild-type U7 snRNA. Further studies are needed to test this hypothesis directly. An alternative possibility is that in the absence of Lsm10 and Lsm11 an intermediate in the U7 snRNP assembly pathway accumulates sufficiently to be detected.
The aberrant U7 snRNP we detect in Lsm11 mutants is nonfunctional in histone pre-mRNA processing, presumably because it lacks both Lsm10 and Lsm11 proteins, which contribute to U7 snRNP function in at least two ways. First, we cannot detect U7 snRNA, and by inference the aberrant U7 snRNP particle, at the histone locus in Lsm11 or Lsm10 mutants. This suggests that Lsm10 and/or Lsm11 are required for the proper localization of U7 snRNP to the sites of histone mRNA biosynthesis. Second, Lsm11 plays a direct role in histone pre-mRNA processing by interacting with other components of the processing machinery. For instance, in human cells Lsm11 interacts with ZFP100, a zinc finger protein that helps coordinate the processing machinery on nascent histone mRNA (Dominski et al. 2002; Pillai et al. 2003; Wagner et al. 2006). This interaction requires a unique NH2-terminal domain of Lsm11 that is not found in other members of the Sm/Lsm protein family. Consequently, even if a small amount of the aberrant U7 snRNP localized correctly to the HLB, histone pre-mRNA processing would likely remain defective.
In summary, our genetic analysis of Lsm10 and Lsm11 provide the groundwork for exploring novel roles for these two proteins in both histone pre-mRNA processing and other aspects of RNA metabolism.
MATERIAL AND METHODS
Western blots
Tissue extracts from third instar larval brains plus salivary glands or adult flies were prepared in NET buffer (0.05 M Tris at pH 7.5, 0.4 M NaCl, 0.005 M EDTA, and 1% NP40) with 100 mM PMSF, 1 mg/mL Leupeptin, and 0.5 mg/mL Pepstatin and cleared by centrifugation at 10,000g for 10 min at 4°C. GFP-negative, homozygous U7 mutant larvae were collected from U720/TM3 P[act-GFP] parents. Lsm11 mutant larvae were collected from Lsm11c02047/CyO P[twist-GFP] × Df(2R)M073/CyO P[twist-GFP] parents. Lsm10 mutant larvae were collected from Lsm10f06616/CyO P[twist-GFP] or Lsm10G40E/CyO P[twist-GFP] × Df(2R)17/CyO P[twist-GFP] parents. Proteins were separated through either a 12% (Lsm11) or a 15% (Lsm10) acrylamide gel (Bio-Rad) and transferred to a 0.45 μm Pure Nitrocellulose Membrane (Bio-Rad). Membranes were probed with anti-Lsm11, anti-Lsm10 (1:1,000; Liu et al. 2006), or anti-α-tubulin (1:1,000; Sigma). Horseradish peroxidase linked secondary (Amersham Biosciences) was used at 1:5,000 for Lsm10 and 1:1,000 for Lsm11 and α-Tubulin and visualized with ECL (Amersham Biosciences).
Northern analysis
For the detection of U1 or U7 snRNA with an α[32P]-UTP anti-sense RNA probe, 15 μg/lane of total cellular RNA isolated using TRIzol Reagent (Gibco) was denatured with 8 M urea and subjected to electrophoresis through an 8% polyacrylamide gel containing 7 M urea. Separated RNA were transferred to a N+ nitrocellulose membrane (Amersham) using a Genie Blotter (Idea Scientific). For the analysis of histone mRNA, 2 μg/lane of RNA were subjected to electrophoresis through a 1% agarose gel containing 0.01 M MOPS (pH 7.0) and 6.75% formaldehyde and transferred via the wick method to N+ nitrocellulose membrane. DNA containing histone coding regions were random primer labeled with α[32P]-dCTP (Stratagene). Hybridizations were performed at 58°C for snRNA probes and at 60°C for histone probes.
Drosophila genetics
The U714, U720, and U7EY11305 null alleles are described by Godfrey et al. (2006); w1118 was used throughout as a wild-type control. To assess relative fertility, eggs were collected overnight from broods of Lsm10 mutant males and virgin females. One hundred eggs from each genotype were transferred to a fresh grape juice plate and the total number that hatched within three days was determined. The data are reported as the average and standard deviation of six independent measurements. P values for mutant–wild-type comparisons were determined using a paired student t-test. A χ2 test was used to determine significance of deviations from expected Mendelian ratios.
P element transgenes were constructed for rescue of Lsm10 and Lsm11 mutant phenotypes. DNA containing either the Lsm10 or Lsm11 locus (Fig. 1A) was amplified by PCR from adult female w1118 genomic DNA and subcloned into the pCaSpeR 4 transformation vector and confirmed by sequencing. The V5 epitope was added to the NH2-terminus of Lsm11 by using a 5′ primer with the V5 sequence immediately downstream from the ATG start codon. All phenotypic rescue experiments employed strains containing a second chromosome recombinant of genotype Lsm11c02047, P[V5-Lsm11+] or Lsm10G40E, P[Lsm10+]. Each recombinant chromosome was confirmed molecularly by PCR (Lsm11c02047allele) or sequencing (Lsm10G40E allele).
Immunoprecipitations and RT-PCR
Twenty micrograms of total cellular RNA were added to anti-TMG antibody coated beads (EMD Biosciences, Inc.) in 1 mL of buffer A (20 mM HEPES-KOH at pH 7.6, 0.2 mM EDTA, 0.5 mM DTT, 0.2 M KCl, 5% glycerol) plus 80 units of RiboLock RNase Inhibitor (Fermentas). Samples were incubated at 4°C for 2 h and the beads were recovered by centrifugation at 2300 rpm for 1 min., and then washed 3X with buffer A at 0.3 M KCl. Bound RNA was eluted by incubation in 50 mM Tris-HCl at pH 8.0, 100 mM NaCl, 10 mM EDTA, 1% SDS for 10 min at 60°C and subsequently precipitated with PCI (Invitrogen). Recovered RNA was treated with DNase (Promega) prior to generating cDNA by using an N6 random Hexamer (IDT) and SuperScriptII Reverse Transcriptase (Invitrogen). Primers specific for U7, U1, and RP49 were used in separate PCR reactions to amplify each gene. For each separate PCR the cycle number was optimized to ensure that the PCR reaction was in the linear range.
Immunostaining, in situ hybridization, and microscopy
To detect transgenic Lsm11, Lsm11c02047, P[V5-Lsm11+] homozygous embryos were dechorionated, fixed in a 1:1 mixture of 7% formaldehyde/heptane for 25 min, and incubated with primary and secondary antibodies overnight at 4°C and for 1 h at 25°C, respectively. Primary antibodies used were monoclonal mouse anti-V5 (1:1000; Invitrogen), monoclonal mouse anti-Discs Large (1:1000, Developmental Studies Hybridoma Bank). Secondary antibodies used were goat anti-mouse IgG-Cy3 (Jackson Immuno Research Laboratories). DNA was detected by staining embryos with 4,6-diamidino-2-phenylindole (DAPI) (1:1000 of 1 mg/mL stock, Dako North America) for 1 min.
Brains were dissected from third instar larvae in PBS, fixed in 4% formaldehyde for 20 min, permeabilized in 1% Triton X-100 for 1 h, and incubated with primary antibodies at 37°C for 1 h or overnight and secondary antibodies at 25°C for 1 h or overnight at 4°C. Brains were fixed again in 4% formaldehyde for 20 min prior to in situ hybridization. The following primary and secondary antibodies were used: monoclonal mouse anti-Ser/Thr-Pro MPM-2 (1:1000; Millipore), affinity-purified polyclonal rabbit anti-Lsm10 (1:1000; Liu et al. 2006), goat anti-mouse Cy5 and goat anti-rabbit Cy2 (both from Jackson Immuno Research Laboratories). Brains were hybridized with anti-sense U7 digoxigenin-labeled RNA probes as described previously (Lanzotti et al. 2002; White et al. 2007). Probes to U7 were derived from a clone containing a cDNA for U7 (Dominski et al. 2003). Hybrids were detected using the Cyanine 3 tyramide signal amplification system (PerkinElmer Life and Analytical Sciences).
Confocal images were taken at a zoom of 1.0–5.0 with a 40X (numerical aperture 1.30) Plan Neofluor objective on a Zeiss 510 laser scanning confocal microscope using the LSM data acquisition software (Carl Zeiss). Image false coloring and contrast was adjusted using Photoshop (Adobe Systems).
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank Joe Gall for donating the Lsm10 and Lsm11 antibodies, Graydon Gonsalvez for help with anti-TMG precipitations, Ashmita Chatterjee for assistance with Lsm genetics, and Greg Matera for critical reading of the manuscript. This work was supported by NIH grant GM58921 to W.F.M. and NSF grant MCB-9984068 to R.J.D.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1518009.
REFERENCES
- Achsel T, Brahms H, Kastner B, Bachi A, Wilm M, Lührmann R. A doughnut-shaped heteromer of human Sm-like proteins binds to the 3′-end of U6 snRNA, thereby facilitating U4/U6 duplex formation in vitro. EMBO J. 1999;18:5789–5802. doi: 10.1093/emboj/18.20.5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzouz TN, Schumperli D. Evolutionary conservation of the U7 small nuclear ribonucleoprotein in Drosophila melanogaster. RNA. 2003;9:1532–1541. doi: 10.1261/rna.5143303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzouz TN, Gruber A, Schumperli D. U7 snRNP-specific Lsm11 protein: Dual binding contacts with the 100 kDa zinc finger processing factor (ZFP100) and a ZFP100-independent function in histone RNA 3′ end processing. Nucleic Acids Res. 2005;33:2106–2117. doi: 10.1093/nar/gki516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beggs JD. Lsm proteins and RNA processing. Biochem Soc Trans. 2005;33:433–438. doi: 10.1042/BST0330433. [DOI] [PubMed] [Google Scholar]
- Bond UM, Yario TA, Steitz JA. Multiple processing-defective mutations in a mammalian histone pre-mRNA are suppressed by compensatory changes in U7 RNA both in vivo and in vitro. Genes & Dev. 1991;5:1709–1722. doi: 10.1101/gad.5.9.1709. [DOI] [PubMed] [Google Scholar]
- Bouveret E, Rigaut G, Shevchenko A, Wilm M, Seraphin B. A Sm-like protein complex that participates in mRNA degradation. EMBO J. 2000;19:1661–1671. doi: 10.1093/emboj/19.7.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branlant C, Krol A, Ebel JP, Lazar E, Haendler B, Jacob M. U2 RNA shares a structural domain with U1, U4, and U5 RNAs. EMBO J. 1982;1:1259–1265. doi: 10.1002/j.1460-2075.1982.tb00022.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmo-Fonseca M. New clues to the function of the Cajal body. EMBO Rep. 2002;3:726–727. doi: 10.1093/embo-reports/kvf154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cioce M, Lamond AI. Cajal bodies: A long history of discovery. Annu Rev Cell Dev Biol. 2005;21:105–131. doi: 10.1146/annurev.cellbio.20.010403.103738. [DOI] [PubMed] [Google Scholar]
- Collins BM, Harrop SJ, Kornfeld GD, Dawes IW, Curmi PM, Mabbutt BC. Crystal structure of a heptameric Sm-like protein complex from archaea: Implications for the structure and evolution of snRNPs. J Mol Biol. 2001;309:915–923. doi: 10.1006/jmbi.2001.4693. [DOI] [PubMed] [Google Scholar]
- Dominski Z, Erkmann JA, Yang X, Sanchez R, Marzluff WF. A novel zinc finger protein is associated with U7 snRNP and interacts with the stem–loop binding protein in the histone pre-mRNP to stimulate 3′-end processing. Genes & Dev. 2002;16:58–71. doi: 10.1101/gad.932302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski Z, Yang XC, Purdy M, Marzluff WF. Cloning and characterization of the Drosophila U7 small nuclear RNA. Proc Natl Acad Sci. 2003;100:9422–9427. doi: 10.1073/pnas.1533509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski Z, Yang XC, Marzluff WF. The polyadenylation factor CPSF-73 is involved in histone-pre-mRNA processing. Cell. 2005;123:37–48. doi: 10.1016/j.cell.2005.08.002. [DOI] [PubMed] [Google Scholar]
- Fernandez CF, Pannone BK, Chen X, Fuchs G, Wolin SL. An Lsm2–Lsm7 complex in Saccharomyces cerevisiae associates with the small nucleolar RNA snR5. Mol Biol Cell. 2004;15:2842–2852. doi: 10.1091/mbc.E04-02-0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall JG. Cajal bodies: The first 100 years. Annu Rev Cell Dev Biol. 2000;16:273–300. doi: 10.1146/annurev.cellbio.16.1.273. [DOI] [PubMed] [Google Scholar]
- Gall JG. The centennial of the Cajal body. Nat Rev Mol Cell Biol. 2003;4:975–980. doi: 10.1038/nrm1262. [DOI] [PubMed] [Google Scholar]
- Galli G, Hofstetter H, Stunnenberg HG, Birnstiel ML. Biochemical complementation with RNA in the Xenopus oocyte: A small RNA is required for the generation of 3′ histone mRNA termini. Cell. 1983;34:823–828. doi: 10.1016/0092-8674(83)90539-1. [DOI] [PubMed] [Google Scholar]
- Gick O, Kramer A, Keller W, Birnstiel ML. Generation of histone mRNA 3′ ends by endonucleolytic cleavage of the pre-mRNA in a snRNP-dependent in vitro reaction. EMBO J. 1986;5:1319–1326. doi: 10.1002/j.1460-2075.1986.tb04362.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godfrey AC, Kupsco JM, Burch BD, Zimmerman RM, Dominski Z, Marzluff WF, Duronio RJ. U7 snRNA mutations in Drosophila block histone pre-mRNA processing and disrupt oogenesis. RNA. 2006;12:396–409. doi: 10.1261/rna.2270406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm C, Stefanovic B, Schumperli D. The low abundance of U7 snRNA is partly determined by its Sm binding site. EMBO J. 1993;12:1229–1238. doi: 10.1002/j.1460-2075.1993.tb05764.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris ME, Bohni R, Schneiderman MH, Ramamurthy L, Schumperli D, Marzluff WF. Regulation of histone mRNA in the unperturbed cell cycle: Evidence suggesting control at two post-transcriptional steps. Mol Cell Biol. 1991;11:2416–2424. doi: 10.1128/mcb.11.5.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann H, Fabrizio P, Raker VA, Foulaki K, Hornig H, Brahms H, Lührmann R. snRNP Sm proteins share two evolutionarily conserved sequence motifs which are involved in Sm protein–protein interactions. EMBO J. 1995;14:2076–2088. doi: 10.1002/j.1460-2075.1995.tb07199.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MH, Guthrie C. Unexpected flexibility in an evolutionarily conserved protein–RNA interaction: Genetic analysis of the Sm binding site. EMBO J. 1990;9:2555–2561. doi: 10.1002/j.1460-2075.1990.tb07436.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kambach C, Walke S, Young R, Avis JM, de la Fortelle E, Raker VA, Lührmann R, Li J, Nagai K. Crystal structures of two Sm protein complexes and their implications for the assembly of the spliceosomal snRNPs. Cell. 1999;96:375–387. doi: 10.1016/s0092-8674(00)80550-4. [DOI] [PubMed] [Google Scholar]
- Kolev NG, Steitz JA. Symplekin and multiple other polyadenylation factors participate in 3′-end maturation of histone mRNAs. Genes & Dev. 2005;19:2583–2592. doi: 10.1101/gad.1371105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koundakjian EJ, Cowan DM, Hardy RW, Becker AH. The Zuker collection: A resource for the analysis of autosomal gene function in Drosophila melanogaster. Genetics. 2004;167:203–206. doi: 10.1534/genetics.167.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanzotti DJ, Kaygun H, Yang X, Duronio RJ, Marzluff WF. Developmental control of histone mRNA and dSLBP synthesis during Drosophila embryogenesis and the role of dSLBP in histone mRNA 3′ end processing in vivo. Mol Cell Biol. 2002;22:2267–2282. doi: 10.1128/MCB.22.7.2267-2282.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JL, Murphy C, Buszczak M, Clatterbuck S, Goodman R, Gall JG. The Drosophila melanogaster Cajal body. J Cell Biol. 2006;172:875–884. doi: 10.1083/jcb.200511038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loughney K, Kreber R, Ganetzky B. Molecular analysis of the para locus, a sodium channel gene in Drosophila. Cell. 1989;58:1143–1154. doi: 10.1016/0092-8674(89)90512-6. [DOI] [PubMed] [Google Scholar]
- Lührmann R, Kastner B, Bach M. Structure of spliceosomal snRNPs and their role in pre-mRNA splicing. Biochim Biophys Acta. 1990;1087:265–292. doi: 10.1016/0167-4781(90)90001-i. [DOI] [PubMed] [Google Scholar]
- Luscher B, Schumperli D. RNA 3′ processing regulates histone mRNA levels in a mammalian cell cycle mutant. A processing factor becomes limiting in G1-arrested cells. EMBO J. 1987;6:1721–1726. doi: 10.1002/j.1460-2075.1987.tb02423.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin F, Schaller A, Eglite S, Schumperli D, Muller B. The gene for histone RNA hairpin binding protein is located on human chromosome 4 and encodes a novel type of RNA binding protein. EMBO J. 1997;16:769–778. doi: 10.1093/emboj/16.4.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluff WF, Wagner EJ, Duronio RJ. Metabolism and regulation of canonical histone mRNAs: Life without a poly(A) tail. Nat Rev Genet. 2008;9:843–854. doi: 10.1038/nrg2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matera AG. Cajal bodies. Curr Biol. 2003;13:R503. doi: 10.1016/S0960-9822(03)00438-X. [DOI] [PubMed] [Google Scholar]
- Matera AG, Terns RM, Terns MP. Noncoding RNAs: Lessons from the small nuclear and small nucleolar RNAs. Nat Rev Mol Cell Biol. 2007;8:209–220. doi: 10.1038/nrm2124. [DOI] [PubMed] [Google Scholar]
- Mattaj IW. Cap trimethylation of U snRNA is cytoplasmic and dependent on U snRNP protein binding. Cell. 1986;46:905–911. doi: 10.1016/0092-8674(86)90072-3. [DOI] [PubMed] [Google Scholar]
- Mowry KL, Steitz JA. Both conserved signals on mammalian histone pre-mRNAs associate with small nuclear ribonucleoproteins during 3′ end formation in vitro. Mol Cell Biol. 1987a;7:1663–1672. doi: 10.1128/mcb.7.5.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowry KL, Steitz JA. Identification of the human U7 snRNP as one of several factors involved in the 3′ end maturation of histone premessenger RNA's. Science. 1987b;238:1682–1687. doi: 10.1126/science.2825355. [DOI] [PubMed] [Google Scholar]
- Park JW, Parisky K, Celotto AM, Reenan RA, Graveley BR. Identification of alternative splicing regulators by RNA interference in Drosophila. Proc Natl Acad Sci. 2004;101:15974–15979. doi: 10.1073/pnas.0407004101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai RS, Will CL, Lührmann R, Schumperli D, Muller B. Purified U7 snRNPs lack the Sm proteins D1 and D2 but contain Lsm10, a new 14 kDa Sm D1-like protein. EMBO J. 2001;20:5470–5479. doi: 10.1093/emboj/20.19.5470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai RS, Grimmler M, Meister G, Will CL, Lührmann R, Fischer U, Schumperli D. Unique Sm core structure of U7 snRNPs: Assembly by a specialized SMN complex and the role of a new component, Lsm11, in histone RNA processing. Genes & Dev. 2003;17:2321–2333. doi: 10.1101/gad.274403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez R, Marzluff WF. The stem–loop binding protein is required for efficient translation of histone mRNA in vivo and in vitro. Mol Cell Biol. 2002;22:7093–7104. doi: 10.1128/MCB.22.20.7093-7104.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaufele F, Gilmartin GM, Bannwarth W, Birnstiel ML. Compensatory mutations suggest that base-pairing with a small nuclear RNA is required to form the 3′ end of H3 messenger RNA. Nature. 1986;323:777–781. doi: 10.1038/323777a0. [DOI] [PubMed] [Google Scholar]
- Schumperli D, Pillai RS. The special Sm core structure of the U7 snRNP: Far-reaching significance of a small nuclear ribonucleoprotein. Cell Mol Life Sci. 2004;61:2560–2570. doi: 10.1007/s00018-004-4190-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seraphin B. Sm and Sm-like proteins belong to a large family: Identification of proteins of the U6 as well as the U1, U2, U4, and U5 snRNPs. EMBO J. 1995;14:2089–2098. doi: 10.1002/j.1460-2075.1995.tb07200.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stauber C, Schumperli D. 3′ Processing of pre-mRNA plays a major role in proliferation-dependent regulation of histone gene expression. Nucleic Acids Res. 1988;16:9399–9414. doi: 10.1093/nar/16.20.9399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanovic B, Hackl W, Lührmann R, Schumperli D. Assembly, nuclear import and function of U7 snRNPs studied by microinjection of synthetic U7 RNA into Xenopus oocytes. Nucleic Acids Res. 1995;23:3141–3151. doi: 10.1093/nar/23.16.3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan E, Santiago C, Parker ED, Dominski Z, Yang X, Lanzotti DJ, Ingledue TC, Marzluff WF, Duronio RJ. Drosophila stem–loop binding protein coordinates accumulation of mature histone mRNA with cell cycle progression. Genes & Dev. 2001;15:173–187. doi: 10.1101/gad.862801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibault ST, Singer MA, Miyazaki WY, Milash B, Dompe NA, Singh CM, Buchholz R, Demsky M, Fawcett R, Francis-Lang HL, et al. A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat Genet. 2004;36:283–287. doi: 10.1038/ng1314. [DOI] [PubMed] [Google Scholar]
- Till BJ, Colbert T, Tompa R, Enns LC, Codomo CA, Johnson JE, Reynolds SH, Henikoff JG, Greene EA, Steine MN, et al. High-throughput TILLING for functional genomics. Methods Mol Biol. 2003;236:205–220. doi: 10.1385/1-59259-413-1:205. [DOI] [PubMed] [Google Scholar]
- Tomasevic N, Peculis BA. Xenopus LSm proteins bind U8 snoRNA via an internal evolutionarily conserved octamer sequence. Mol Cell Biol. 2002;22:4101–4112. doi: 10.1128/MCB.22.12.4101-4112.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toro I, Thore S, Mayer C, Basquin J, Seraphin B, Suck D. RNA binding in an Sm core domain: X-ray structure and functional analysis of an archaeal Sm protein complex. EMBO J. 2001;20:2293–2303. doi: 10.1093/emboj/20.9.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toro I, Basquin J, Teo-Dreher H, Suck D. Archaeal Sm proteins form heptameric and hexameric complexes: Crystal structures of the Sm1 and Sm2 proteins from the hyperthermophile Archaeoglobus fulgidus. J Mol Biol. 2002;320:129–142. doi: 10.1016/S0022-2836(02)00406-0. [DOI] [PubMed] [Google Scholar]
- Wagner EJ, Ospina JK, Hu Y, Dundr M, Matera AG, Marzluff WF. Conserved zinc fingers mediate multiple functions of ZFP100, a U7snRNP associated protein. RNA. 2006;12:1206–1218. doi: 10.1261/rna.2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner EJ, Burch BD, Godfrey AC, Salzler HR, Duronio RJ, Marzluff WF. A genome-wide RNA interference screen reveals that variant histones are necessary for replication-dependent histone pre-mRNA processing. Mol Cell. 2007;28:692–699. doi: 10.1016/j.molcel.2007.10.009. [DOI] [PubMed] [Google Scholar]
- Wang ZF, Whitfield ML, Ingledue TC, 3rd, Dominski Z, Marzluff WF. The protein that binds the 3′ end of histone mRNA: A novel RNA-binding protein required for histone pre-mRNA processing. Genes & Dev. 1996;10:3028–3040. doi: 10.1101/gad.10.23.3028. [DOI] [PubMed] [Google Scholar]
- Wang P, Palfi Z, Preusser C, Lucke S, Lane WS, Kambach C, Bindereif A. Sm core variation in spliceosomal small nuclear ribonucleoproteins from Trypanosoma brucei. EMBO J. 2006;25:4513–4523. doi: 10.1038/sj.emboj.7601328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White AE, Leslie ME, Calvi BR, Marzluff WF, Duronio RJ. Developmental and cell cycle regulation of the Drosophila histone locus body. Mol Biol Cell. 2007;18:2491–2502. doi: 10.1091/mbc.E06-11-1033. [DOI] [PMC free article] [PubMed] [Google Scholar]