Abstract
To date, ∼700 microRNA (miRNA) molecules have been identified in humans. Accurate and sensitive quantification of miRNA levels will help unveil their biological functions. Here, we extend the isothermal ramification amplification (RAM) approach to a sensitive and specific real-time assay for quantitative analysis of miRNA. This RAM miRNA assay is based on the threshold cycle (CT) principle similar to that of real-time PCR. It has a dynamic range of at least seven orders of magnitude, allowing for the quantification of miRNA input from 103 to 1010 copies per reaction (10 nM to 1 fM). The capabilities of discriminating single-base mismatch and distinguishing mature miRNAs from their precursors are achieved by coupling the reverse-transcription of miRNA to the generation of a closed C-probe, rather than using expensive detection probes like in real-time PCR. Quantitative measurement of 5 miRNAs (mir-1, miR-122, mir-150, mir-143, and let-7a) across 12 mouse tissues is validated in total RNA samples without further purification. U6 snRNA, snoRNA 135, and miRNA-191 could be simultaneously quantified as endogenous controls. These results suggest that our RAM miRNA assay might provide a universal tool for miRNA detection and functional studies to meet the needs for bench examination, clinical diagnosis, and on-site detection.
Keywords: microRNA, quantification, RAM, rolling cycle amplification
INTRODUCTION
MicroRNAs (miRNAs) are a group of endogenous noncoding single-strand RNAs, typically, 21–23 nucleotides (nt) in length, which have been convincingly shown to play a critical role in a wide variety of physiological processes such as cell proliferation, differentiation, apoptosis, tumor metastasis, etc. (Reinhart et al. 2000; Lagos-Quintana et al. 2001; Hutvagner and Zamore 2002; Johnson et al. 2005; Ma et al. 2007; van Rooij et al. 2007; Yu et al. 2007). The accurate and quantitative analysis of miRNA expression has become imperative for the understanding of miRNA's functions, especially when the notion that miRNAs fine-tune the expression of most of their target mRNAs is becoming widely accepted (Giraldez et al. 2005; Flynt and Lai 2008).
However, given the fact that mature miRNAs are short in length and some miRNAs contain a highly homologous sequence (i.e., the let-7 family), the characterization of miRNA expression pattern remains a challenging task. A great deal of effort has been made on developing miRNA detection methods, including modified Northern blot (Lee et al. 1993; Lagos-Quintana et al. 2002), invader assay (Allawi et al. 2004), splinted ligation (Maroney et al. 2007), and LNA probes-based assay (Weinholds et al. 2005; Kloosterman et al. 2006). Compared with these aforementioned methods, several real-time RT-PCR-based approaches are becoming a prominent tool for miRNA quantitative analysis due to their high sensitivity and specificity (Chen et al. 2005; Raymond et al. 2005; Shi and Chiang 2005; Li et al. 2009).
Besides the RT-PCR technique, rolling circle amplification (RCA) is a novel isothermal DNA amplification technology based on the rolling replication of short, single-stranded DNA circles by certain DNA polymerases (Lizardi et al. 1998; Zhang et al. 1998, 2001). RCA has proven to be promising for target DNA, mRNA, and even protein detection in various assays and revealed advantages over traditional PCR. In this work, we present a real-time quantitative assay for mature miRNA based on the modified ramification amplification (RAM) approach. Through coupling the reverse transcription of miRNA to the ligation of a C-probe as well as isothermal amplification, we demonstrate that the new assay could detect miRNA molecules expression in concentration ranging from 10 nM down to 1 fM, and has remarkable capability of single-base mismatch and precursor discrimination. In use of common DNA probes and fluorescent dyes, this assay could accurately quantify miRNAs in total RNA samples without further enrichment.
RESULTS
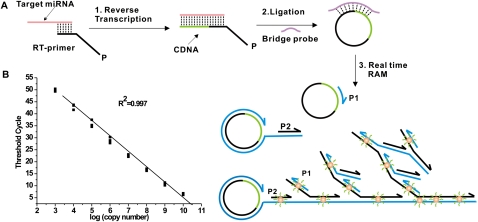
This RAM miRNA assay consisted of three steps: reverse transcription of miRNA, C-Probe ligation, and ramification amplification (Fig. 1A). First, an oligonucleotide RT-primer bound to the 3′ end of the miRNA and was reverse transcribed. Secondly, the reverse-transcribed product was circularized in the presence of a bridge probe and then ligated to form a closed C-probe by Taq DNA ligase. Finally, ramification amplification was initiated upon the addition of Bst DNA polymerase, primer 1 (P1) and primer 2 (P2) following a previous description (Zhang et al. 2001; Li et al. 2005; Yi et al. 2006). Since RAM followed a similar exponential nature to that of conventional real-time PCR (Supplemental Fig. S1), data processing methodology commonly used in quantitative PCR was employed herein (see more details in Materials and Methods).
FIGURE 1.
(A) Schematic description of the real-time RAM miRNA assay. This assay consisted of three steps: miRNA reverse transcription, C-Probe ligation, and ramification amplification. Those steps coordinated each other to ensure the detection specificity. SYBR Green I dye was employed here. (B) Dynamic range and sensitivity of let-7a. Synthetic let-7a input ranged from 103 to 1010 copies per reaction. Each experiment was repeated three times.
Real-time observation of each concentration of let-7a in triplicate is presented in Figure 1B. The amplification plot showed excellent linearity between the log of initial miRNA copy number input versus its CT values. This RAM assay was capable of detecting as few as 103 copies (zeptomole) of let-7a and had a dynamic range of at least seven orders of magnitude for each reverse-transcriptase reaction. We observed the same sensitivity and dynamic ranges when characterizing other miRNA molecules such as miR-1, miR-122 (Supplemental Fig. S2).
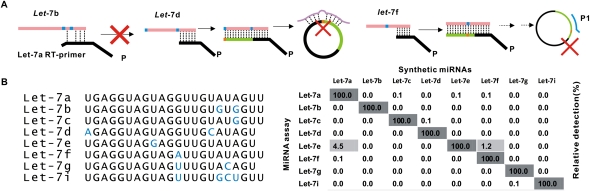
Next, we investigated the specificity of the RAM miRNA assay by using eight closely related sequences of let-7 family. Figure 2 showed that the let-7 family assays produced <0.1% nonspecific signal only with a few of exceptions. We assumed that three steps coordinated each other to ensure the detection specificity (Fig. 2A). In brief, a RT-primer was able to discriminate a mismatch at 3′ end of miRNA, while the following C-probe ligation of transcribed cDNAs could distinguish a 5′ end mismatch. A pair of amplification primers (mainly P1) could further help discriminate a mismatch site in the middle of the miRNA sequence. Thus, to tailor the sequences of those RT-primers, bridge probes or amplification primers would enable us to create a RAM assay, which was capable of discriminating a single mutation along miRNA sequence, for each miRNA molecule.
FIGURE 2.
Discrimination power of let-7 family assays. (A) Schematic description of the discrimination capability of let-7a assay from other let-7 members such as let-7b, let-7d, and let-7f. In brief, the reverse transcription step allowed us to discriminate let-7b from 7a by employing well-designed RT-primers, and let-7d could be successfully eliminated by a bridge probe in the following ligation step. Finally, let-7f was excluded by a pair of amplification primers hybridizing to the closed C-probe. (B) Eight closely related sequences of the let-7 family. Bases that differ from let-7a are highlighted in blue. (C) Relative detection (%) by RAM reaction was calculated on CT difference between the perfect match and mismatched target; 1 × 108 copies of miRNAs per reaction were used as template in real-time RAM detection assays. The concentration was estimated based on the A260 values. Each experiment was repeated three times.
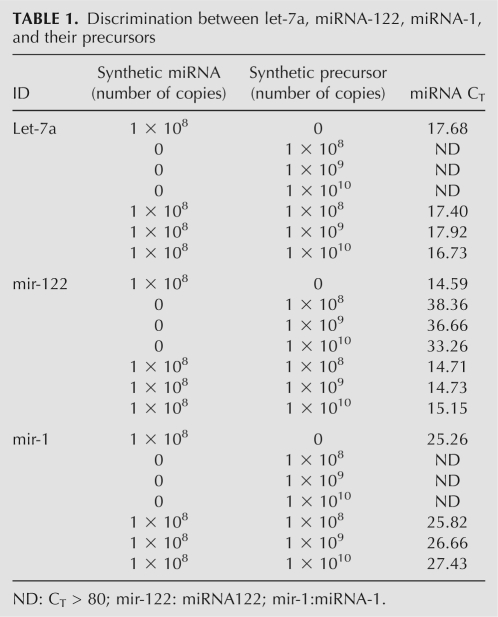
Precursor miRNA (pre-miRNA) is a stable hairpin containing an entire miRNA sequence, which may interfere with mature miRNA detection. Our assay was able to distinguish mature miRNAs such as let-7a, mir-122, and mir-1 from their precursors (Table 1). When the assays designed to detect mature miRNAs were tested with in vitro transcript precursors, we observed that even if precursor miRNAs were of 100 times higher concentration than mature miRNAs in reaction, they would not contribute any higher than background signal to the assay for mature targets. Thus, we claimed that this RAM assay was superior to TaqMan miRNA assay in aspect of their capability of precursor discrimination (Chen et al. 2005).
TABLE 1.
Discrimination between let-7a, miRNA-122, miRNA-1, and their precursors
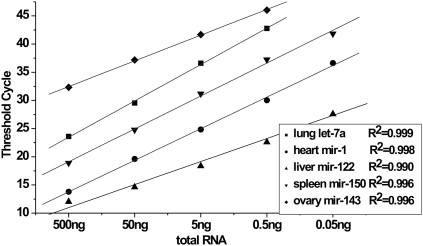
Prior to investigate the expression profile of miRNAs across different tissues, we evaluated the anti-interference capability of this RAM assay. Since miR-122 was renowned as a liver-specific miRNA, we quantitively examined this miRNA molecule in the absence or presence of nonliver total RNAs. The results demonstrated that the anti-inference capability of our assay was excellent when no more than 200 ng of total RNA was included per RT reaction (Supplemental Fig. S3). In the meanwhile, we investigated the dynamic range of the RAM assay and determined the appropriate loading amount when in use of total RNA sample. Figure 3 showed that when RNA input ranged from 0.05 ng to 500 ng, the CT values correlated to the RNA input (R2 > 0.990) over four orders of magnitude. To guarantee good experimental repeatability, 100 ng of total RNA was preferable in this assay.
FIGURE 3.
Correlation of total RNA input to the CT values for five miRNAs in total RNA samples. Mouse tissue total RNA input ranged from 0.05 ng to 500 ng per reaction.
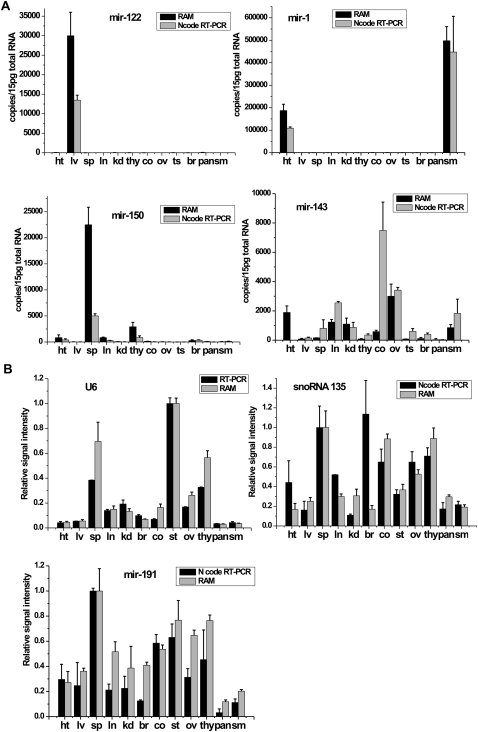
The expression profile of five miRNAs (miR-1, miR-122, miR-143, miR-150, and let-7a) was quantitively analyzed across 12 different mouse tissues (Fig. 4A; Supplemental Fig. S3). We observed that miR-122 abundantly expressed in liver (mir-122, ∼30,000 copies per cell), miR-1 in heart or skeletal muscle (∼190,000 or 500,000 copies per cell, respectively), and miR-150 in spleen (∼22,000 copies per cell) if assuming average 15 pg of total RNA per cell (Fig. 4A). By comparison, miR-143 and let-7a exhibited less tissue-specific patterns and existed in most of these tissues ranging from 50 to 3000 copies per cell. These results were further confirmed by the use of the Ncode RT-PCR assay, a commercially available miRNA detection kit, with exception of let-7a.
FIGURE 4.
(A) The expression profiles of miRNA-122, miRNA-1, miRNA-150, and miRNA-143 per cell were examined by the RAM assay (black bars) and Ncode RT-PCR assay (gray bars) across 12 mouse tissues. We assumed average 15 pg of total RNA per cell. ht: heart, lv: liver, sp: spleen, ln: lung, kd: kidney, thy: thymus, co: colon, ov: ovary, ts: testicle, br: brain, pan: pancreas, sm: skeletal muscle. (B) The relative signal intensity of endogenous controls including U6 snRNA, snoRNA 135, or miRNA-191. The black bars represented the relative signal intensities of U6, snoRNA 135 or miRNA-191 detected by the RAM assay. The gray bars represented their intensities quantified by conventional real-time RT-PCR or Ncode RT-PCR assay. Expression amount of U6 in testicle, or snoRNA 135 and miRNA-191in spleen was normalized to 1.
Finally, we adapted the RAM assay to the direct detection of U6 snRNA, a universal endogenous control, to normalize the amount of total RNA sample. A specially designed RT-primer for U6 snRNA was reverse transcribed into 72 nt of cDNA, further ligated into a closed C-probe, and exposed to RAM amplification. We compared this RAM method with conventional RT-PCR for the characterization of U6 expression across 12 tissues. As presented in Figure 4B, both methods agreed well with each other in all tested tissues. Small RNAs such as snoRNA 135 and miRNA-191 were recently showed as more stable normalizers in miRNA qRT-PCR experiments (Applied Biosystems) (Peltier and Latham 2008). Thus, we examined the expression levels of both snoRNA 135 and miRNA-191 across 12 tissues by use of either the RAM assay or Ncode RT-PCR assay. We found that both assays presented almost the same expression patterns of both small RNAs across all tested tissues. Therefore, all those results strongly supported the assertion that our assay can be used for the quantitative characterization of miRNAs among or across tissues.
DISCUSSION
Jonstrup et al. (2006) recently demonstrated a miRNA detection system based on padlock probes and rolling circle amplification (RCA), in which a target miRNA functioned as both the mediator for the circularization of padlock probes and the primer for RCA. Cheng et al. (2009) more recently improved the sensitivity of this method by using branched-RCA. As we know, ordinarily a single nucleotide mismatch at the middle position could be easily distinguished by C-probe ligation, but a terminal mismatch would lead to poor discrimination. Furthermore, simultaneously discriminating a group of highly analogical miRNAs (such as the let-7 family) is still a challenging task for ligase itself. In this work, we incorporated an additional step, reverse transcription, prior to C-probe ligation and isothermal amplification. Although such incorporation compromised a little operating convenience, the coupling of these steps endowed this miRNA assay with the remarkable features such as high sensitivity, single mutation discrimination, as well as wide dynamic range. Our assay demonstrated the increase in sensitivity by at least five orders of magnitude over padlock-based miRNA assay. The reverse transcription step allowed us to discriminate let-7b, 7c, 7g, and 7i from 7a by employing well-designed RT-primers (Supplemental Table S3). Then, let-7d, with a 5′ end mismatch, could be successfully eliminated by a bridge probe in the following ligation step (Fig. 2 A). Finally, let-7e and 7f were excluded by a pair of amplification primers hybridizing to the closed C-probe.
Our assay realized the quantitative analysis of miRNA expression level by use of total RNA samples. Four miRNAs, including mir-122, mir-1, mir-150, mir-143, and let-7a, examined by this assay showed same expression patterns to ones quantified with Ncode RT-PCR assay across a dozen of the mouse tissues. The expression level of those miRNAs also agreed very well with the previous reports. For example, Chang et al. (2004) quantified ∼66,000 copies of miR-122 per cell in mouse liver tissue by Northern analysis, while Zhou et al. (2007) also found that miRNA-150 was highly expressed in the spleen, >20 times than in the thymus and heart. However, our assay showed lower let-7a expression levels than the Ncode RT-PCR assay did in almost all tested tissues (Supplemental Fig. S4). We presumed that the variation might result from the discrimination capability of both assays to let-7a family.
The characterization of relative expression of a miRNA among or across tissues required the normalization of the amount of loading samples according to a universal endogenous control. Our assay was demonstrated to have the capability of quantifying several well-know endogenous controls including U6 snRNA, snoRNA-135, and miRNA-191 across different tissues, which were confirmed by conventional real-time RT-PCR or Ncode RT-PCR assay, respectively.
The detection throughput was another major concern in the quantitative analysis of miRNA profiling. Although microarray-based techniques were particularly appealing since they could examine thousands of miRNAs in parallel, and their accuracy and sensitivity were still arguable. We believed that once incorporated with a multiplexed detection system, our assay also held the possibility of being an assay of quantitative analysis of hundreds of miRNA molecules simultaneously, which should be satisfied with general needs given no more than 1000 of the miRNA molecules identified in a cell so far.
In summary, RAM was adapted to quantitively examine miRNA for the first time through the coupling of reverse transcription, C-probe ligation, and isothermal amplification. Like the TaqMan assay, this RAM miRNA assay demonstrated the remarkable capabilities of sensitive and specific quantification of mature miRNAs. However, instead of demanding steps of thermal cycling and expensive fluorescent probes akin to the TaqMan assay, this assay took advantage of isothermal reaction steps and ordinary DNA probes as well as fluorescent dyes, thus enabling researchers to perform practical and prevalent detection of miRNAs with lower cost and greater convenience. Those features are becoming more and more important, since there are many lines of evidence showing that miRNAs might be a novel class of biomarkers for cancer diagnosis, the study of cancer origin (Lu et al. 2005; Shell et al. 2007; Gilad et al. 2008), and even on-site viral detection until the recent identification of miRNAs in viruses (Pfeffer et al. 2005; Gupta et al. 2006). Further studies will utilize this assay to quantitively characterize other small RNAs such as piwiRNAs.
MATERIALS AND METHODS
Mature miRNA, precursor miRNAs, probes, and primers
Sequences of mature and precursor miRNAs including the let-7 family, mir-122, mir-1, mir-143, mir-150, and mir-191 were selected from the Sanger Center miRbase (http://microrna.sanger.ac.uk/sequences), and the sequences of snoRNA 135 and U6 were obtained from NCBI database (http://www.ncbi.nlm.nih.gow). Synthetic mature miRNAs were purchased from Shanghai GenePharma, and all precursor miRNAs were produced through in vitro transcription (Supplemental Table S1).
All of the DNA probes and primers were purchased from Invitrogen. Oligonucleotides were purified by polyacrylamide gel electrophoresis, and sequences are listed in Supplemental Tables S2 and S3. Nucleic acid concentrations were determined by absorption readings at 260 nm on a NanoDrop Spectrophotometer (ND-100, ThermoFisher).
Tissue preparation and total RNA extraction
Dissected organs including the heart, intestine, liver, spleen, lung, kidney, thymus, colon, ovary, testicle, brain, pancreas, and skeletal muscle from 8-wk-old mice were utilized for the preparation of total RNA samples using TRNzol reagent (Catalog No. DP405-02, Tiangen) following the manufacturer's procedure. In brief, tissue samples were thoroughly homogenized in 1 mL of TRIZol reagent per 50–100 mg of tissue sample. After that, 0.2 mL of chloroform was added per 1 mL of TRIZol reagent, and then the samples were mixed vigorously for 15 sec and incubated at room temperature for 2 to 3 min. The samples were centrifuged at 12,000g for 15 min at 4°C and the upper aqueous phase were carefully removed without disturbing the lower phase into a fresh tube. The same volumes of isopropyl alcohol were added and mixed well. The samples were incubated at 15°C–30°C for 20 min and centrifuged at 12,000g at 4°C for 10 min. Following centrifugation, the supernatants were removed completely and RNA pellet was washed with 75% ethanol twice. Finally, RNA pellet was air dried and eluted in nuclease-free water. All animal procedures were in accordance with Institutional Animal Care and Use Committee (IACUC) and OECD guidelines.
Preparation of dsDNA templates
A total of 50 pmol of forward and reverse DNA oligomers was annealed by incubation at 75°C for 5 min, then slowly cooled to room temperature (∼30 min). The fill-in reaction to form dsDNA templates was performed in a 20 μL volume containing 10 mM Tris-HCl, 50 mM NaCl, 10 mM MgCl2, 1 mM DTT, 0.25 mM dNTPs, and 5 U Klenow Fragment (3′–5′ exo) (New England BioLabs) at 37°C for 1 h. Then the reaction mixture was heated at 75°C for 20 min to inactivate the enzyme and slowly cooled to room temperature for dsDNA annealing.
In vitro transcription reaction
A total of 20 μL of the aforementioned dsDNA template mixture was added into 30 μL of in vitro transcription buffer containing 0.5 mM NTPs, 40 mM Tris-HCl, 6 mM MgCl2, 10 mM DTT, 2 mM spermidine, 40–200 U Ribonuclease Inhibitor (Takara), and 50 U T7 RNA Polymerase (New England BioLabs). The reaction ran at 37°C for 4 h. After that, 1 U of RNase-Free DNase I (Fermentas) was added to digest DNA templates. Finally, the precursor miRNAs in the transcription mixture were purified with phenol/chloroform extraction by disposing of extra salt and proteins. The transcripted precursor miRNAs were examined by 2% Agarose gel, and the concentration was determined from the absorption at 260 nm with NanoDrop spectrophotometer.
Phosphorylation and reverse transcriptase reaction
Before miRNA was reverse transcribed, the RT primer was chemically 5′-phosphorylated. A total of 0.5 nmol RT primer was heated to 75°C for 5 min, and then chilled on ice prior to the treatment with kinase. The 50 μL reaction volume containing 50 mM Tris-HCl (pH 8.0), 10 mM MgCl2, 5 mM Dithiothreitol (DTT), 1 mM ATP, 20 U T4 polynucleotide kinase (Takara), and DNA probes was performed at 37°C for 2 h. Finally, T4 polynucleotide kinase was inactivated by incubating at 65°C for 20 min.
Reverse transcriptase reaction for the formation of cDNA from miRNA was performed in a 10 μL volume, which contained 1 μL of total RNA sample, 500 nM 5′-phosphorylated RT primers, 20 U PrimeScript reverse transcriptase (Takara), 50 μM dNTPs and 1× reaction buffer (pH 8.3, 50 mM Tris-HCl, 75 mM KCl, and 3 mM MgCl2) at 41°C for 30 min. Then, 2.5 U Ribonuclease H (Fermentas) were added at 37°C for 20 min for the degradation of miRNA strand in the RNA–DNA hybrid.
Ligation of C-probe
The mixture of 10 μL of the reverse transcribed product and 1 μL of the bridge probe (10 μM) was boiled at 95°C for 2 min, which was then slowly cooled to room temperature to form an open C-probe hybrid. The nick in the C-probe was sealed by 10 U thermophilic Taq DNA ligase (New England BioLabs) in reaction buffer containing 20 mM Tris-HCl (pH 7.6), 25 mM KAc, 10 mM Mg(Ac)2, 10 mM DTT, 1 mM NAD, and 0.1% Triton X-100 (total volume of 20 μL), at 55°C for 45 min.
Real-time RAM assay
The isothermal amplification was performed in a 25-μL volume containing 1 μL of ligation product mixture, 1.2 μM of each primer-1 and -2, 100 μM dNTPs, 20 mM Tris-HCl (pH 8.8), 10 mM KCl, 10 mM (NH4)2SO4, 2 mM MgSO4, 0.1% Triton X-100, 6% dimethyl sulfoxide, 6 μL of 1× SYBR Green I dye (Bio-Vision), and 6.4 U Bst DNA polymerase large fragment (New England BioLabs). The reactions were incubated at 65°C in a 96-well plate and were real-time monitored for 100 cycles with a 25-sec period by an Opticon2 DNA Engine (Bio-Rad). All reactions were run in triplicate. The threshold cycle (CT) was defined as the fractional cycle number at which the fluorescence passed the fixed threshold.
Ncode RT-PCR and conventional real-time PCR assay
To quantify the miRNA expression level by the real-time PCR method, the cDNA was synthesized using the NCode VILO miRNA cDNA Synthesis Kit (Invitrogen). Briefly, all the miRNAs in total RNA samples or snoRNA 135 were polyadenlyated and reverse-transcribed using poly A polymerase, ATP, SuperScript III RT, and a specially designed universal RT primer in a single reaction. The resulting cDNA was subjected to real-time PCR using SYBR Green ER qPCR Mix (Invitrogen). The Universal qPCR Primer was provided in the VILO kit and the forward primer was designed according to NCode miRNA Database at http://escience.invitrogen.com/ncode. Quantitative Real-time PCR was used under the following thermocycler conditions: 50°C for 2 min, 95°C for 2 min, 40 cycles of 95°C for 15 sec, and 60°C for 50 sec.
U6 small nuclear RNA was quantified by the traditional real-time PCR method. Total RNA were reversed-transcribed using random primers. A total of 20 μL of the reaction volume, including total RNA samples, 25 nM random primers, 100 U PrimeScript reverse transcriptase (Takara), 0.25 mM dNTPs, and 1× reaction buffer (pH 8.3, 50 mM Tris-HCl, 75 mM KCl, and 3 mM MgCl2) were incubated at 42°C for 60 min. The cDNA was subjected to real-time PCR using SYBR Green ER qPCR Mix and the cycling parameters were the same as the procedures of miRNAs.
Data processing methodology
A 10-fold dilution series of synthetic miRNAs were used as standard template to generate a plot of log numbers of miRNAs (x-axis) at different dilutions versus the corresponding CT Value (y-axis). A linear regression line or standard curve was constructed and followed using the equation y = ax + b or CT = a(log quantity) + b. Thus, the unknown samples assayed in the same run could be determined according to the standard curve.
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank Yanyi Huang, Cheng Zhu, Hanjoong Jo, and Seth Brodie for reading the manuscript and the National Natural Science Foundation of China (Nos. 30600142 and 90607004), Ministry of Education (No. 708002), and the Ministry of Science and Technology of the People's Republic of China (No. 2007AA04Z313) for support. We also thank the Shanghai GenePharma Co., Ltd., for kindly offering synthetic RNA and related technical consultations.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1555209.
REFERENCES
- Allawi HT, Dahlberg JE, Olson S, Lund E, Olson M, Ma WP. Quantitation of microRNAs using a modified invader assay. RNA. 2004;10:1153–1161. doi: 10.1261/rna.5250604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J, Nicolas E, Marks D, Sander C, Lerro A, Buendia MA, Xu C, Mason WS, Molosho T, Bort R, et al. mir-122, a mammalian liver-specific microRNA, is processed from hcr mRNA and may down-regulate the high affinity cationic amino acid transporter CAT-1. RNA Biol. 2004;1:106–113. doi: 10.4161/rna.1.2.1066. [DOI] [PubMed] [Google Scholar]
- Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, et al. Real-time quantification of microRNAs by stem–loop RT–PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Zhang X, Li Z, Jiao X, Wang Y, Zhang Y. Highly sensitive determination of microRNA using target-primed and branched rolling-circle amplification. Angew Chem Int Ed Engl. 2009;48:3268–3272. doi: 10.1002/anie.200805665. [DOI] [PubMed] [Google Scholar]
- Flynt AS, Lai EC. Biological principles of microRNA-mediated regulation: Shared themes amid diversity. Nat Rev Genet. 2008;9:831–842. doi: 10.1038/nrg2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilad S, Meiri E, Yogev Y, Benjamin S, Lebanony D, Yerushalmi N, Benjamin H, Kushnir M, Cholakh H, Melamed N, et al. Serum microRNAs are promising novel biomarkers. PLoS One. 2008;3:e3148. doi: 10.1371/journal.pone.0003148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldez AJ, Cinalli RM, Glasner ME, Enright AJ, Thomson JM, Baskerville S, Hammond SM, Bartel DP, Schier AF. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 2005;308:833–838. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- Gupta A, Gartner JJ, Sethupathy P, Hatzigeorgiou AG, Fraser NW. Anti-apoptotic function of a microRNA encoded by the HSV-1 latency-associated transcript. Nature. 2006;442:82–85. doi: 10.1038/nature04836. [DOI] [PubMed] [Google Scholar]
- Hutvagner G, Zamore PD. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297:2056–2060. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert K, Brown D, Slack F. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- Jonstrup SP, Koch J, Kjems J. A microRNA detection system based on padlock probes and rolling circle amplification. RNA. 2006;12:1747–1752. doi: 10.1261/rna.110706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloosterman W.P., Weinholds E., de Bruijn E., Kauppinen S., Plasterk R.H.A. In situ detection of microRNAs in animal embryos using LNA modified oligonucleotide probes. Nat Methods. 2006;3:27–29. doi: 10.1038/nmeth843. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Li F, Zhao C, Zhang W, Cui S, Meng J, Wu J, Zhang DY. Use of ramification amplification assay for detection of Escherichia coli 0157:H7 and other E. coli shiga toxin-producing strains. J Clin Microbiol. 2005;43:6086–6090. doi: 10.1128/JCM.43.12.6086-6090.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Yao B, Huang H, Wang Z, Sun C, Fan Y, Chang Q, Li S, Wang X, Xi J. Real-time polymerase chain reaction microRNA detection based on enzymatic stem–loop probes ligation. Anal Chem. 2009;81:5446–5451. doi: 10.1021/ac900598d. [DOI] [PubMed] [Google Scholar]
- Lizardi PM, Huang X, Zhu Z, Brayward P, Thomas DC, Ward DC. Mutation detection and single-molecule counting using isothermal rolling circle amplification. Nat Genet. 1998;19:225–232. doi: 10.1038/898. [DOI] [PubMed] [Google Scholar]
- Lu L, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- Ma L, Teruya-Feldstein J, Weinberg RA. Tumor invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–689. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- Maroney PA, Chamnongpol S, Souret F, Nilsen TW. A rapid, quantitative assay for direct detection of microRNAs and other small RNAs using splinted ligation. RNA. 2007;13:930–936. doi: 10.1261/rna.518107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltier HJ, Latham GJ. Normalization of microRNA expression levels in quantitative RT-PCR assays: Identification of suitable reference RNA targets in normal and cancerous human solid tissues. RNA. 2008;14:844–852. doi: 10.1261/rna.939908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer S, Sewer A, Lagos-Quintana M, Sheridan R, Sander C, Grasser FA, van Dyk L, Ho C, Shuman S, Chien M, et al. Identification of microRNAs of the herpesvirus family. Nat Methods. 2005;2:269–276. doi: 10.1038/nmeth746. [DOI] [PubMed] [Google Scholar]
- Raymond CK, Roberts BS, Garrett-Engele P, Lim LP, Johnson JM. Simple, quantitative primer-extension PCR assay for direct monitoring of microRNAs and short-interfering RNAs. RNA. 2005;11:1737–1744. doi: 10.1261/rna.2148705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- van Rooij E, Sutherland LB, Qi X, Richardson JA, Hill J, Olson EN. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science. 2007;316:575–579. doi: 10.1126/science.1139089. [DOI] [PubMed] [Google Scholar]
- Shell S, Park SM, Radjabi AR, Schickel R, Kistner EO, Jewell DA, Feig C, Lengyel E, Peter ME. Let-7 expression defines two differentiation stages of cancer. Proc Natl Acad Sci. 2007;104:11400–11405. doi: 10.1073/pnas.0704372104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi R, Chiang VL. Facile means for quantifying microRNA expression by real-time PCR. Biotechniques. 2005;39:519–525. doi: 10.2144/000112010. [DOI] [PubMed] [Google Scholar]
- Weinholds E, Kloosterman WP, Miska E, Alvarez-Saavedra E, Berezikov E, de Bruijn E, Horvitz HR, Kauppinen S, Plasterk RHA. MicroRNA expression in zebrafish embryonic development. Science. 2005;309:310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- Yi J, Zhang W, Zhang DY. Molecular zipper: A fluorescent probe for real-time isothermal DNA amplification. Nucleic Acids Res. 2006;34:e81. doi: 10.1093/nar/gkl1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong C, Huang Y, Hu X, Su F, Lieberman J, et al. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 2007;131:1109–1123. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]
- Zhang DY, Brandwein M, Hsuih TC, Li H. Amplification of target-specific, ligation-dependent circular probe. Gene. 1998;211:277–285. doi: 10.1016/s0378-1119(98)00113-9. [DOI] [PubMed] [Google Scholar]
- Zhang D, Zhang W, Li X, Konomi Y. Detection of rare DNA targets by isothermal ramification amplification. Gene. 2001;274:209–216. doi: 10.1016/s0378-1119(01)00607-2. [DOI] [PubMed] [Google Scholar]
- Zhou B, Wang S, Mayr C, Bartel DP, Lodish HF. miR-150, a microRNA expressed in mature B and T cells, blocks early B cell development when expressed prematurely. Proc Natl Acad Sci. 2007;104:7080–7085. doi: 10.1073/pnas.0702409104. [DOI] [PMC free article] [PubMed] [Google Scholar]