Abstract
The Prp19-associated complex (NineTeen Complex [NTC]) is required for spliceosome activation by specifying interactions of U5 and U6 with pre-mRNA on the spliceosome after the release of U4. The NTC consists of at least eight protein components, including two tetratricopeptide repeat (TPR)-containing proteins, Ntc90 and Ntc77. Ntc90 has nine copies of the TPR with seven clustered in the carboxy-terminal half of the protein, and interacts with all identified NTC components except for Prp19 and Ntc25. It forms a stable complex with Ntc31, Ntc30, and Ntc20 in the absence of Ntc25, when other interactions between NTC components are disrupted. In this study, we used both biochemical and genetic methods to analyze the structure of Ntc90, and its function in maintaining the integrity of the NTC and in NTC-mediated spliceosome activation. Our results show that Ntc90 interacts with Ntc31, Ntc30, and other NTC components through different regions of the protein, and that its function may be regulated by Ntc31 and Ntc30. Ntc90 is not required for the association of Prp19, Ntc85, Ntc77, Ntc25, and Ntc20, or for their binding to the spliceosome. It is also not required for NTC-mediated spliceosome activation, but is required for the recruitment of Yju2, which is involved in the first catalytic reaction after the function of Prp2. Our results demonstrate a novel role of the NTC in recruiting splicing factors to the spliceosome after its activation.
Keywords: first catalytic reaction, Ntc90, RNA splicing, splicing reaction
INTRODUCTION
Pre-mRNA splicing is an essential process and plays an important role in the regulation of eukaryotic gene expression. The splicing reaction is catalyzed by dynamic macromolecular machinery, called the spliceosome, which is composed of five small nuclear RNAs (snRNAs), U1, U2, U4, U5, and U6 in the forms of small nuclear ribonucleoprotein (snRNP) complexes, and many other protein factors (Will and Lührmann 1997, 2006; Staley and Guthrie 1998; Burge et al. 1999; Wahl et al. 2009). Assembly of the spliceosome is a stepwise process involving sequential binding of snRNPs, in the order of U1, U2, and then U4/U6.U5 as a tri-snRNP, and other protein components. After binding of all the snRNPs, the spliceosome undergoes a major conformational rearrangement, releasing U1 and U4, and is activated to allow catalysis. A protein complex associated with Prp19, named the NineTeen Complex (NTC), is required for the activation process (Tarn et al. 1993, 1994).
The NTC is not associated with any snRNA (Tarn et al. 1993, 1994), and binds to the spliceosome immediately after the dissociation of U4 (Chan et al. 2003). The NTC is required for specific interactions of U5 and U6 with pre-mRNA to stabilize their association with the spliceosome (Chan et al. 2003; Chan and Cheng 2005). Deficiency in NTC function also results in failure in spliceosome recycling due to impaired U4/U6 biogenesis (Chen et al. 2006).
We have previously identified eight NTC components, Prp19, Ntc90/Syf1, Ntc85/Cef1, Ntc77/Syf3, Ntc31/Syf2, Ntc30/Isy1, Ntc25/Snt309, and Ntc20 (Tarn et al. 1994; Chen et al. 1998, 1999, 2001, 2002; Tsai et al. 1999). These proteins are associated with the spliceosome at the same time during spliceosome assembly, suggesting that they might function as an integral complex. All of these proteins were also found in the yeast penta-snRNP complex, which is proposed to be a functional particle of the preassembled spliceosome capable of splicing added pre-mRNA (Stevens et al. 2002). A similar complex has also been identified in human and in the fission yeast Schizosaccharomyces pombe, suggesting evolutionary conservation of the complex (Ohi et al. 2002).
Among NTC components, Ntc31, Ntc30, Ntc25, and Ntc20 are encoded by genes not essential for cellular growth. Ntc25 interacts with Prp19, and plays a role in modulating the interaction of Prp19 with its associated components (Chen et al. 1999). In the absence of Ntc25, the NTC is dissociated into four components with Ntc90, Ntc31, Ntc30, and Ntc20 remaining stably associated (Chen et al. 1999, 2002). Genetic analysis reveals that Ntc31, Ntc30, and Ntc20 might have overlapping functions in regulating the function of Ntc90 since double deletions of each pair of genes cause various degrees of growth impairment, and deletion of all three genes results in cellular lethality. Furthermore, the growth impairment caused by NTC30- and NTC20-deleted (30Δ20Δ) cells can be partially rescued by overexpression of NTC90 (Chen et al. 2002).
NTC90 was also identified as “synthetic lethal with cdc forty” (SYF1) in a genetic screen for genes that interact with CDC40/PRP17 (Ben-Yehuda et al. 2000). The SYF1 gene product was found to be a protein that is highly conserved throughout the evolutionary scale and contains nine semiconserved copies of the tetratricopeptide repeat (TPR) motif, which is implicated in protein–protein interactions (for review, see D'Andrea and Regan 2003). Two-hybrid analysis revealed that Ntc90 interacts with most identified NTC components except for Prp19 and Ntc25 (Chen et al. 2002), suggesting that Ntc90 might serve as a scaffold in maintaining the integrity of NTC. In addition, Ntc90 also interacts with a recently identified splicing factor, Yju2, in two-hybrid assays (Liu et al. 2007). Yju2 interacts with NTC in a dynamic manner, but is not required for NTC-mediated spliceosome activation. Instead, it is required for splicing after the ATP-dependent Prp2 action to promote the first catalytic reaction (Liu et al. 2007). We have previously speculated that dynamic interaction between Yju2 and the NTC might govern the recruitment of Yju2 to the spliceosome via the interaction of Yju2 with Ntc90 and/or with Ntc77, which also interacts with Yju2 in two-hybrid assays (Liu et al. 2007).
Here, to further investigate the function of Ntc90, we generated deletion mutants of NTC90 to examine the function of specific TPR motifs in cellular growth and in the interaction with NTC components. These analyses reveal that Ntc31 and Ntc30 interact with distinct regions of Ntc90 to regulate its function. Furthermore, the proposed TPR superhelix is sufficient for the function of Ntc90. Using extracts prepared from Ntc90-depleted cells, we also found that Ntc90 is neither required for the binding of a subset of NTC components to the spliceosome, nor required for NTC-mediated spliceosome activation, but is required for recruiting Yju2 to the spliceosome. Our studies thus uncover a novel function of NTC components in the recruitment of splicing factors that participate in catalytic steps of splicing.
RESULTS
Deletion analysis of NTC90
Using the Prosite Profiles database, Ben-Yehuda et al. (2000) predicted that the yeast Syf1/Ntc90 contained nine TPR motifs, with TPRs 3–9 forming a TPR superhelix (Fig. 1, first row; Das et al. 1998). Similarly, the human homolog of Syf1, XAB2, was also predicted by Nakatsu et al. (2000) to have 15 TPR motifs (Fig. 1, second row). Ben-Yehuda et al. (2000) predicted TPRs 1–9 correspond to TPRs III, V, VII, VIII, and X–XIV, respectively; Nakatsu et al. (2000) predicted that TPR-containing proteins are involved in different biological pathways; and TPR motifs have been implicated in protein–protein interactions. Indeed, Ntc90 has been shown to interact with several NTC components, including Ntc85, Ntc77, Ntc31, Ntc30, and Ntc20 (Chen et al. 2002), and other putative NTC components Cwc2, Prp46 (Ohi and Gould 2002), and Prp45 (Albers et al. 2003). Another protein, Yju2, associated with NTC, but only required for the first catalytic reaction after the Prp2 step, was also recently shown to interact with Ntc90 (Liu et al. 2007).
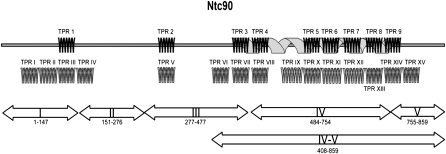
FIGURE 1.
Predicted Ntc90 domain structure. Zigzags on the line denote the TPR motifs 1–9 predicted by Ben-Yehuda et al. (2000) using Prosite Profiles. The spiraling ribbon shown under TPRs 3–9 denotes the TPR superhelix. The second line of zigzags denotes the TPR motifs predicted by Nakatsu et al. (2000) in their study of XAB2. The thick double-headed arrows labeled I, II, III, IV, V, and IV-V mark the regions either deleted or contained in deletion mutants.
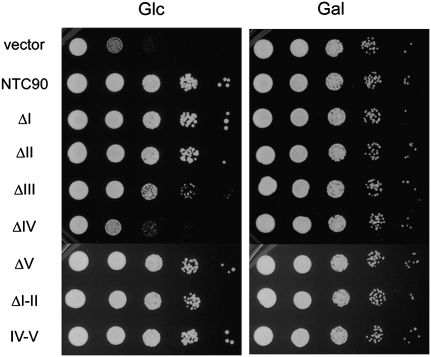
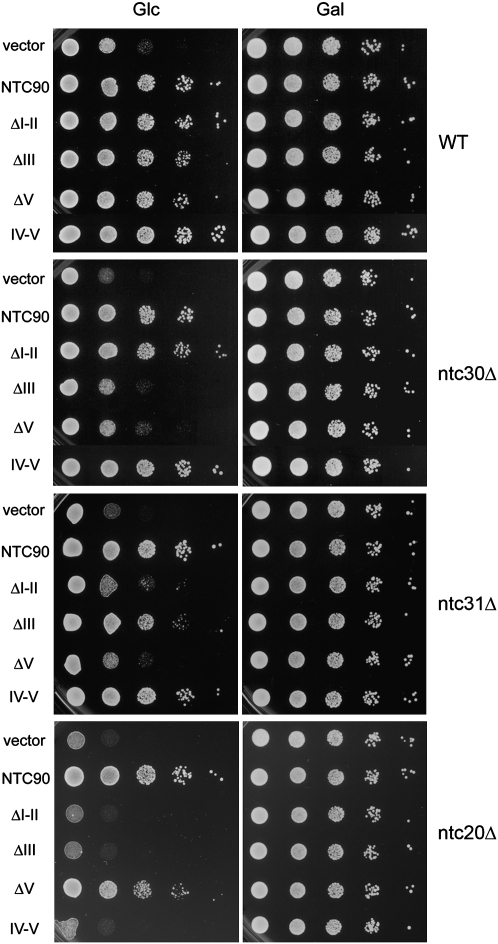
To dissect the function of the TPR motifs on Ntc90, we roughly divided the protein into five regions and generated mutants with deletions from each region (Fig. 1). Region I is the N-terminal fragment of 147 amino acid residues containing TPR1. Region II spans amino acid residues 151–276, containing no TPR. Region III contains TPR2 and TPR3 with amino acid residues 277–477. Region IV contains TPR4–TPR8 and half of TPR9 of amino acid residues 484–754. Region V is the C-terminal fragment of amino acid residues 755–859. The C-terminal half of the protein containing the entire putative TPR superhelix was also constructed and named IV-V as it contained the entire regions IV and V and a fraction of region III. Deletion mutants constructed in plasmid vector pKC414 were transformed into strain YSCC903, in which the expression of NTC90 is under the control of GAL1 promoter. The spot assays shown in Figure 2 reveal that while ΔI, ΔII, ΔV, and IV-V showed no significant impairment in cell growth, ΔIII had a slower growth phenotype, and the growth of ΔIV was severely impaired. These results indicate that region IV is essential for cell growth, and that the proposed TPR superhelix is sufficient for the function of Ntc90.
FIGURE 2.
Growth analysis of NTC90 deletion mutants. YSCC903 cells harboring pRS414-based plasmid vector containing NTC90 deletion mutants were analyzed for growth at 30°C in glucose- and galactose-based media by spot assays. Glc, glucose; Gal, galactose.
Ntc31 and Ntc30 interact with subsidiary regions of Ntc90 to regulate its function
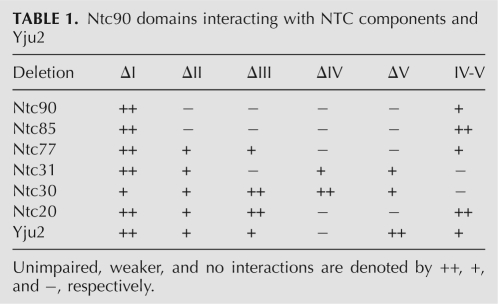
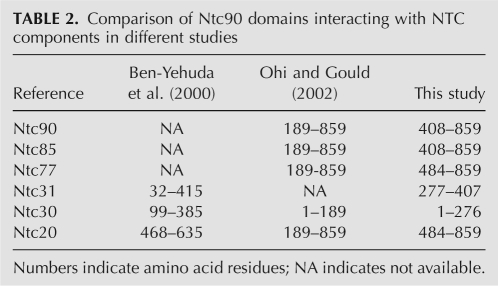
We have previously shown that Ntc90 interacts with all known NTC components except Prp19 and Ntc25. Immunoprecipitation analysis of extracts prepared from an NTC25-deleted strain has further defined a subcomplex formed between Ntc90, Ntc31, Ntc30, and Ntc20 (Chen et al. 2002). Among the genes encoding these proteins, only NTC90 is essential for cell growth, and individual deletion of NTC31, NTC30, or NTC20 does not cause any impairment in growth. While cells with both NTC31 and NTC20 deleted grow normally, cells with NTC31 and NTC30 deleted are temperature sensitive, and show slow-growth phenotypes if both NTC30 and NTC20 are deleted. Deletion of all three genes results in lethality, suggesting an overlapping function of NTC31, NTC30, and NTC20 in regulating the function of NTC90 (Chen et al. 2002). To determine the domains involved in the interactions of Ntc90 with NTC components, two-hybrid assays were conducted with NTC90 deletion mutants. The results summarized in Table 1 show that deletion of region I only affected the interaction of Ntc90 with Ntc30, whereas deletion of region IV abolished interactions of Ntc90 with most of the NTC components except Ntc31 and Ntc30. Domain IV-V appeared to be sufficient for interactions with Ntc85, Ntc77, Ntc20, and itself, but not with Ntc30 and Ntc31. Nevertheless, deletion of region II abolished interactions with Ntc85 and itself, suggesting that an extra fragment containing region I, along with a portion of region III, added to domain IV-V might interfere with the interaction. Our results are largely consistent with results of other studies using different proteins as baits to screen libraries for interacting components (Ben-Yehuda et al. 2000; Ohi and Gould 2002). A comparison of results from three different studies is shown in Table 2.
TABLE 1.
Ntc90 domains interacting with NTC components and Yju2
TABLE 2.
Comparison of Ntc90 domains interacting with NTC components in different studies
Since Ntc31, Ntc30, and Ntc20 interact with different regions of Ntc90 to regulate its function, depletion of any of these three proteins is expected not to exacerbate the defect of mutant Ntc90 when the corresponding interacting region is deleted. The growth of NTC90 deletion mutants in combination with null alleles of NTC31, NTC30, or NTC20, respectively, was analyzed by spot assays. Figure 3 shows that ΔI-II grew normally when NTC30 was deleted, but had a synthetic defect when NTC31 or NTC20 was deleted. ΔIII had a synthetic defect when NTC30 or NTC20 was deleted, but not when NTC31 was deleted. Growth of ΔV was not affected by the deletion of NTC20, but exacerbated when NTC31 or NTC30 was deleted, and IV-V was synthetic lethal only with null NTC20. These results are consistent with the synthetic effects of double deletion mutants of NTC31, NTC30, and NTC20 (Chen et al. 2002), and corroborates the notion of overlapping functions of these three proteins in regulating the function of Ntc90.
FIGURE 3.
Growth analysis of NTC90 deletions in NTC31-, NTC30-, or NTC20-deleted strains. SKC903, SKC311 (ntc31Δ), SKC301 (ntc30Δ), and SKC201 (ntc20Δ) cells harboring pRS414-based plasmid vector containing NTC90 deletion mutants were analyzed for growth at 30°C in glucose- and galactose-based media by spot assays. Glc, glucose; Gal, galactose.
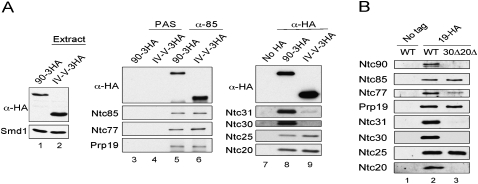
It is worth noting that domain IV-V, which contains the entire putative TPR superhelix, is sufficient for the function of Ntc90, but only in the presence of Ntc20. Splicing extracts prepared from the IV-V-3HA strain contained two to three times more Ntc90 than from the full-length NTC90-3HA strain for unknown reasons (Fig. 4A, lanes 1,2). Nevertheless, immunoprecipitation analysis reveals that IV-V, like th full-length Ntc90, is associated with all identified NTC components except for Ntc31 and Ntc30. When extracts prepared from the NTC90-3HA or IV-V-3HA strain were precipitated with anti-Ntc85 antibody, the amount of Ntc77 and Prp19 co-precipitated from the IV-V-3HA extracts (Fig. 4A, lane 6) was approximately the same as that from the 90-3HA extracts (Fig. 4A, lane 5), despite finding that the amount of Ntc90 co-precipitated from IV-V-3HA was also higher due to higher amounts of the protein present in the extract. When the same amount of the extract was precipitated with anti-HA antibody for Ntc90, the amount of IV-V-3HA precipitated was also ∼2–3 times that of NTC90-3HA (Fig. 4A, lanes 8,9), although similar amounts of Ntc25 and Ntc20 were co-precipitated. Nevertheless, consistent with the growth phenotype and two-hybrid results, only minute amounts of Ntc31 and Ntc30 were co-precipitated with IV-V-3HA despite both also interacting with Ntc85 and Ntc77 in two-hybrid assays (Chen et al. 2002). These results further confirm that domain IV-V is sufficient to maintain the association of all NTC components except for Ntc31 and Ntc30, and that such a complex retains the function of NTC.
FIGURE 4.
Immunoprecipitation of NTC components. (A) Extracts prepared from untagged (lane 7), NTC90-3HA or IV-V-3HA (lanes 1,2) were immunoprecipitated with no antibody (lanes 3,4) purified anti-Ntc85 (lanes 5,6) or anti-HA antibody (lanes 8,9), and probed with antibodies against NTC components. Full-length Ntc90 was in vivo depleted when preparing the IV-V-3HA extracts. (B) Extracts prepared from untagged (lane 1), PRP19-HA (lane 2), or PRP19-HA with deletions of NTC30 and NTC20 (lane 3), were immunoprecipitated with anti-HA antibody, and probed with antibodies against NTC components. PAS, protein A-Sepharose; 30Δ20Δ, deletions of NTC30 and NTC20.
We have previously shown that deletion of NTC30 and NTC20 (30Δ20Δ) resulted in the most severely impaired growth phenotype among the double deletion mutants of NTC31, NTC30, and NTC20 (Chen et al. 2002; Dahan and Kupiec 2002). It was conceivable that the impairment in cell growth in 30Δ20Δ reflected down-regulation of Ntc90 function. To confirm whether this was the case, we first examined whether the association of Ntc90 with the NTC was affected in 30Δ20Δ cells. Splicing extracts were prepared from the 30Δ20Δ strain, in which Prp19 was tagged with HA, and precipitated with anti-HA antibody. As shown in Figure 4B, in the absence of Ntc30 and Ntc20—while Ntc85 and Ntc25 remained stably associated with Prp19—Ntc90 and Ntc31 were no longer associated (Fig. 4B, lane 3), indicating that Ntc30 and/or Ntc20 are required for stable association of Ntc90 with Prp19 through interaction with Ntc85. Since the association of Ntc77 was also affected, Ntc30 and/or Ntc20 might also regulate the interaction between Ntc77 and Ntc85. Thus, the lack of Ntc30 and Ntc20 greatly affects the integrity of the NTC, and consequently, might affect the function of NTC in spliceosome activation.
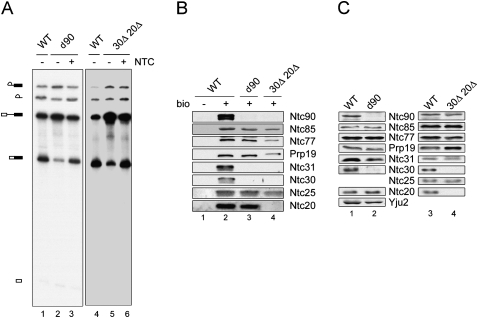
To see whether the function of the NTC was affected in 30Δ20Δ cells, we first examined whether such impaired NTC could efficiently bind to the spliceosome. Extracts prepared from 30Δ20Δ cells, as well as from the Ntc90-depleted cells, were defective in splicing but could be complemented by affinity-purified NTC (Fig. 5A). Spliceosomes formed with biotinylated pre-mRNA in 30Δ20Δ extracts were precipitated with streptavidin Sepharose and the associated proteins probed with antibodies against NTC components. While the binding of Prp19, Ntc85, Ntc77, and Ntc25 was not affected by the absence of Ntc30 and Ntc20, Ntc90 and Ntc31 could no longer bind to the spliceosome (Fig. 5B, lane 4). This implies that the NTC can be dissected into two parts, one containing Prp19, Ntc85, Ntc77, and Ntc25, and the other containing Ntc90, Ntc31, Ntc30, and Ntc20. Prp19, Ntc85, Ntc77, and Ntc25 can bind to the spliceosome in the absence of the other four proteins. Ntc77, although becoming more loosely associated with Prp19, retains its ability to bind to the spliceosome. Consistent with this result, these four proteins were able to bind to the spliceosome formed in Ntc90-depleted extracts (Fig. 5B, lane 3). We have previously demonstrated that in ntc25Δ extracts, the interaction between Prp19 and Ntc85 is severely affected, resulting in weakening of most of the other interactions except for that between Ntc90, Ntc31, Ntc30, and Ntc20, which remain stably associated with each other. This suggests that Ntc90, Ntc31, Ntc30, and Ntc20 form a subcomplex in the NTC, independent of interactions with others (Chen et al. 2002). The result in Figure 4B shows that in 30Δ20Δ extracts, Ntc85, Ntc77, and Ntc25 remained associated with Prp19, further suggesting that the association of these four proteins were independent of the other four. It is likely that the NTC complex consisted of two four-subunit complexes. The result in Figure 5B is consistent with the notion that these four proteins act in concert. Interestingly, although Ntc31 and Ntc30 failed to bind to the spliceosome in the absence of Ntc90, binding of Ntc20 was not affected, indicating that Ntc20 acts more independently of Ntc90. Western blotting of total proteins in splicing extracts (Fig. 5C) revealed that although the level of NTC components was not affected in 30Δ20Δ extracts (Fig. 5C, lane 4), the amount of Ntc30 was greatly reduced in Ntc90-depleted extracts (Fig. 5C, lane 2). In fact, the amount of Ntc90 was also reduced about twofold in 30Δ extracts (data not shown) despite there being no obvious change in 30Δ20Δ extracts (Fig. 5C, lane 4). This suggests that Ntc90 and Ntc30 might be more closely associated with each other both physically and functionally. This is supported by the observation that the association of Ntc90 with Ntc30, but not with other NTC components, was resistant to high-salt wash during immunoprecipitation (data not shown).
FIGURE 5.
Analysis of NTC components in the spliceosome precipitated with streptavidin Sepharose. (A) Splicing reactions were carried out in wild-type (lanes 1,4), Ntc90-depleted (lanes 2,3), and 30Δ20Δ extracts (lanes 5,6), in the absence (lanes 2,5) or presence (lanes 3,6) of added NTC. (B) The spliceosome formed with nonbiotinylated (lane 1) or biotinylated pre-mRNA was precipitated with streptavidin Sepharose using extracts prepared from wild-type (lane 2), Ntc90-depleted (lane 3) or NTC30- and NTC20-deleted cells (lane 4), and probed with antibodies against NTC components. (C) Western blotting of total proteins from extracts prepared from wild-type (lanes 1,3), Ntc90-depleted (lane 2) or NTC30- and NTC20-deleted cells (lane 4).
Ntc90 is not required for NTC-mediated spliceosome activation
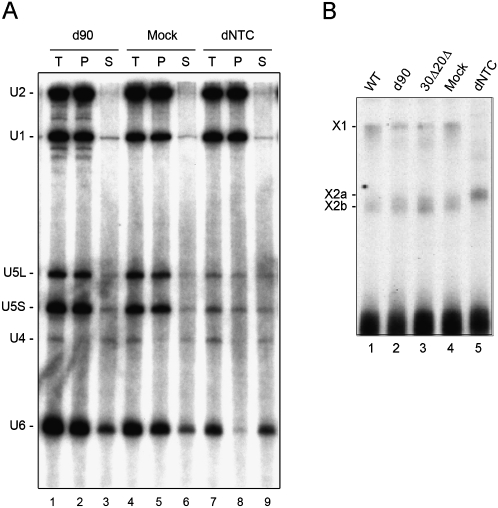
The fact that Prp19, Ntc85, Ntc77, Ntc25, and Ntc20 can bind to the spliceosome in the absence of Ntc90 raises the question of whether the function of the NTC in mediating spliceosome activation requires Ntc90. We have previously demonstrated that the NTC is required for stable association of U5 and U6 with the spliceosome after U4 is released (Chan et al. 2003). The spliceosome formed in NTC-depleted extracts contains lower amounts of U5 and U6, and approximately half of U5 and most of U6 are dissociated after reincubation at 25°C in the presence of ATP (Fig. 6A, lanes 8,9; Chan et al. 2003). The recruitment of tri-snRNP is not affected as evidenced by normal binding U4, U5, and U6 when splicing reactions were carried out at low ATP concentrations to block spliceosome activation. When splicing was performed in Ntc90-depleted extracts, U5 and U6 remained stably associated with the spliceosome (Fig. 6A, lanes 2,3) as in mock-treated extracts (Fig. 6A, lanes 5,6). This suggests that Ntc90 is not required for stable association of U5 and U6 during spliceosome activation. UV-cross-linking analysis also revealed cross-linked products of U6 to pre-mRNA from Ntc90-depleted (Fig. 6A, lane 2) or 30Δ20Δ extracts (Fig. 6B, lane 3), identical to those from wild-type (Fig. 6B, lane 1) or mock-depleted extracts (Fig. 6B, lane 4), but distinct from those from in vitro NTC-depleted extracts (Fig. 6B, lane 5). This provides further evidence to suggest that Ntc90 does not have a role in NTC-mediated spliceosome activation.
FIGURE 6.
Ntc90 is not required for spliceosome activation. (A) The U5, U6 stability assay. Splicing reactions were carried out in mock-depleted (lanes 4–6), NTC-depleted (lanes 7–9), or Ntc90-depleted extracts (lanes 1–3) using biotinylated Ac/Cla pre-mRNA as the substrate, and the spliceosome was precipitated with streptavidin Sepharose. After washing off unbound materials, the pellet was separated into two fractions: one was for total precipitate (T, lanes 1,4,7), and the other had splicing buffer added and was incubated at room temperature for 20 min. After separating supernatant (S) and pellet (P) fractions, RNA was extracted and analyzed by Northern blotting. (B) UV cross-linking of the U6 snRNA with pre-mRNA. Splicing reactions were carried out in wild-type (lane 1), Ntc90-depleted (lane 2), 30Δ20Δ (lane 3), mock-depleted (lane 4), or NTC-depleted extracts (lane 5), using Ac/Cla pre-mRNA as the substrate. The reaction mixtures were precipitated with the anti-Smd1 antibody followed by UV irradiation. The cross-linked products X1, X2a, and X2b are as described by Chan et al. (2003) and Chan and Cheng (2005).
Ntc90 is required for the recruitment of Yju2 to the spliceosomes but not for functioning of Prp2
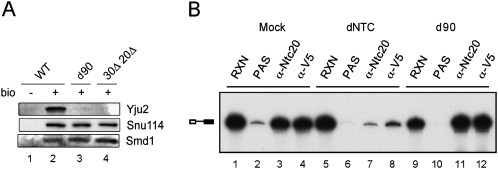
As Ntc90 is not involved in spliceosome activation, we speculated that Ntc90 must play some roles in the post-activation steps considering that it is required for growth and for splicing in vivo (Russell et al. 2000; Chen et al. 2006). We have recently identified a novel splicing factor, Yju2, required for the first catalytic reaction after Prp2 action (Liu et al. 2007). Yju2 interacts with the NTC in a dynamic manner, but is not required for NTC-mediated spliceosome activation. Two-hybrid assays have revealed that Yju2 interacts with Ntc90 and Ntc77 (Liu et al. 2007), and region IV of Ntc90 is essential for the interaction between Ntc90 and Yju2, as its deletion completely abolished the interaction (Table 1). We have previously speculated that the interaction between Yju2 and NTC might play a role in the recruitment of Yju2 to the spliceosome through the interaction of Yju2 with Ntc90 and/or Ntc77 (Liu et al. 2007). We therefore tested whether Ntc90 functions in recruiting Yju2. As shown in Figure 7A, the spliceosome formed in Ntc90-depleted or 30Δ20Δ extracts contained no Yju2, despite the amount of Yju2 not being greatly affected by depletion of Ntc90 (Fig. 5C), indicating that Ntc90 is required for recruiting Yju2 to the spliceosome.
FIGURE 7.
Analysis of Yju2 and Prp2 in the spliceosome precipitated with streptavidin Sepharose. (A) Spliceosomes formed with nonbiotinylated (lane 1) or biotinylated pre-mRNA were precipitated with streptavidin Sepharose using extracts prepared from wild-type (lane 2), Ntc90-depleted (lane 3) or NTC30- and NTC20-deleted cells (lane 4), and probed with antibodies against Yju2, Snu114 and Smd1. (B) Spliceosomes formed in the presence of recombinant V5-tagged prp2S387L were precipitated with anti-Ntc20 or anti-V5 antibody in mock-depleted (lanes 1–4), NTC-depleted (lanes 5–8) extracts, or extracts prepared from in vivo Ntc90-depleted cells (lanes 9–12).
Since Prp2 acts before Yju2 in the first catalytic step, we further investigated whether Ntc90 is also required for the functional step of Prp2 by examining the binding of Prp2 to the spliceosome in Ntc90-depleted extracts. To arrest Prp2 on the spliceosome, we used a dominant negative mutant of Prp2, prp2S378L, which carries a mutation in the SAT-motif of the helicase and could neither function nor dissociate from the spliceosome after binding (Plumpton et al. 1994). The recombinant prp2S378L protein tagged with V5 was added to wild-type extracts or Ntc90-depleted extracts for the splicing reaction. The spliceosome formed was examined for the presence of prp2S378L by immunoprecipitation with anti-V5 antibody. While depletion of NTC largely prevented binding of prp2S378L (Fig.7B, lane 8), depletion of Ntc90 still allowed its binding (Fig.7B, lane 12), indicating that Ntc90 is not required for the functional step of Prp2. We therefore conclude that the function of Ntc90 is to recruit Yju2 for the first catalytic reaction, and such recruitment could occur either before or after the action of Prp2.
DISCUSSION
To study the structure and function of Ntc90, we first evaluated the importance of different TPR motifs in supporting cell growth by dividing the protein into five regions for generation of deletion mutants. Region IV, which forms the main body of the proposed TPR superhelix (Ben-Yehuda et al. 2000), was most important since its deletion resulted in severe growth impairment. While cell growth was slightly impaired when region III was deleted, deletion of region I, II, or V, or both regions I and II, did not result in any growth impairment. As region III contains the N-terminal region of the proposed TPR superhelix, we concluded that the integrity of the superhelix is important for normal cellular growth. This is confirmed by the fact that mutant IV-V, which comprises the C-terminal half of the protein with intact superhelix, shows a normal growth phenotype. Furthermore, deletion of one, two or three TPRs from either the N-terminus or C-terminus of IV-V showed different degrees of growth impairment (data not shown).
Ntc90 is presumed to function as a scaffold because of the presence of multiple TPRs. Two-hybrid analysis has revealed its interaction with six known NTC components and a step-one factor Yju2 (Liu et al. 2007). The region comprising the TPR superhelix interacts with three large subunits of the NTC, Ntc77, Ntc85, and Ntc90 itself, and also with Ntc20. The three large proteins are encoded by genes essential for cell growth. By contrast, the N-terminal half of Ntc90 interacts with Ntc30 and Ntc31, which are smaller in size and dispensable. This suggests that the putative TPR superhelix is the core of Ntc90, and the N-terminus is the auxiliary domain that modulates the structure and function of Ntc90. This pattern of domain interactions coincides with the results of the synthetic effects of NTC31, NTC30, or NTC20 null alleles with different NTC90 deletions. Deletion of NTC30 had no synthetic effect with mutant ΔI-II or mutant IV-V since the region that interacts with Ntc30 was deleted, and the presence of Ntc30 was no longer important for Ntc90 function. Similarly, deletion of NTC31 had hardly any synthetic effect with mutant IV-V or mutant ΔIII when the region that interacts with Ntc31 was deleted, and deletion of NTC20 had no synthetic effect with mutant ΔV.
Like Ntc90, Ntc77 also contains multiple TPRs, and interacts with the same set of proteins in the NTC and with Yju2. However, Ntc31, Ntc30, and Ntc20 appear to interact with Ntc90 more tightly than with Ntc77. While stable association of Ntc77 with these three proteins is regulated by Prp19 via its interaction with Ntc85, Ntc90 forms a stable complex with Ntc31, Ntc30, and Ntc20, regardless of Prp19–Ntc85 interaction (Chen et al. 2002). Furthermore, overproduction of Ntc90 could partially complement the growth defect of cells with both NTC30 and NTC20 deleted, suggesting that Ntc31, Ntc30, and Ntc20 might play roles in modulating the function of Ntc90 and its interaction with other NTC components (Chen et al. 2002). This is corroborated by our biochemical analyses reported here, which showed that in 30Δ20Δ extracts, neither Ntc90 nor Ntc31 is closely associated with the NTC, and neither protein could be efficiently recruited to the spliceosome, despite the recruitment of other NTC components (Fig. 5B). Ntc30 appears to be more tightly linked to Ntc90 as in vivo depletion of Ntc90 results in deprivation of Ntc30 from cell extracts, suggesting that either the expression of NTC30 or the stability of Ntc30 protein is affected by Ntc90. Although the protein level of Ntc31 is not severely affected in 30Δ20Δ extracts, or extracts prepared from in vivo Ntc90-depleted cells, Ntc31 could not be recruited to the spliceosome. Neither could Ntc31 associate with NTC in 30Δ20Δ extracts. These results suggest that Ntc31 and Ntc30 may coordinate to regulate the function of Ntc90. By contrast, the association of Ntc20 with the spliceosome is not severely affected by the absence of Ntc90. Thus, Ntc20 behaves more independently of Ntc90, Ntc31, and Ntc30. In the absence of Ntc25, the integrity of the NTC is disrupted as interactions between Ntc85 and other NTC components are weakened, but Ntc20 remains associated with Ntc90, Ntc31, and Ntc30 to form a stable complex via its interaction with Ntc90 (Chen et al. 2002). In the absence of Ntc90, Ntc20 becomes associated with Ntc85, Prp19, Ntc25, and Ntc77 via interactions with Ntc85 and Ntc77, although the association is weakened (data not shown).
Despite being a part of the NTC, Ntc90 is not required for spliceosome activation. Instead, it is required for the recruitment of Yju2, which functions in the first catalytic reaction after the action of Prp2 in an ATP-independent manner (Liu et al. 2007). This indicates that the NTC has dual functions in the splicing reaction, both in spliceosome activation and in recruiting factors required for the later steps of the spliceosome pathway. The interaction of Yju2 with Ntc90 has been demonstrated by two-hybrid assays (Liu et al. 2007). Analysis of deletion mutants revealed that such interaction does not require region I or II of Ntc90. Thus, the putative superhelix of Ntc90 is not only involved in the interaction with core NTC components to maintain the architecture of the NTC complex, but also in the interaction with Yju2 for its recruitment. Another TPR-containing NTC component, Ntc77, also interacts with Yju2 by two-hybrid assays (Liu et al. 2007). Distinct from Ntc90, Ntc77 is required for spliceosome activation (our unpublished data), but whether it also plays roles in post-activation steps is not known. It remains possible that Ntc77 has an auxiliary role in recruiting Yju2 through its interaction with Yju2.
The recruitment of Prp2 does not require Ntc90, but requires the NTC, or at least Prp19, suggesting that Prp2 binds to the spliceosome only after its activation. This is also consistent with our previous observation that Yju2 and Prp2 are associated with the spliceosome independently of each other. The function of Prp2 requires a co-factor Spp2, which interacts with Prp2 and presumably modulates the function of Prp2 in the splicing reaction or the binding of Prp2 to the spliceosome in the ATP-dependent step (Roy et al. 1995; Silverman et al. 2004). The ATP-independent step requires a heat-resistant factor HP in addition to Yju2, but the identity of HP remains unknown (Kim and Lin 1996). How Yju2 coordinates with HP in mediating the catalytic reaction also remains unexplored. It will be of interest to see whether HP recruitment also requires Ntc90.
MATERIALS AND METHODS
Yeast strains
BJ2168: MATa prc1 prb1 pep4 leu2 trp1 ura3;
EGY48: MATα ura3 his3 trp1 LexAop6-LEU2;
SEY6210: MATα leu2 ura3 his3 trp1 lys2 suc2;
SKC201: MATα leu2 ura3 his3 trp1 lys2 suc2 ntc20Δ∷LEU2 URA3∷GAL1-NTC90;
SKC301: MATα leu2 ura3 his3 trp1 lys2 suc2 ntc30Δ∷HIS3 URA3∷GAL1-NTC90;
SKC311: MATα leu2 ura3 his3 trp1 lys2 suc2 ntc31Δ∷TRP1 URA3∷GAL1-NTC90;
SKC903: MATα leu2 ura3 his3 trp1 lys2 suc2 URA3∷GAL1-NTC90;
YSCC1: MATa prc1 prb1 pep4 leu2 trp1 ura3 PRP19-HA;
YSCC14: MATa prc1 prb1 pep4 leu2 trp1 ura3 PRP19-HA ntc20Δ∷LEU2 ntc30Δ∷URA3;
YSCC20: MATa prc1 prb1 pep4 leu2 trp1 ura3 NTC20-HA;
YSCC93: MATa prc1 prb1 pep4 leu2 trp1 ura3 NTC90-3HA;
YSCC201: MATa prc1 prb1 pep4 leu2 trp1 ura3 NTC20-HA ntc25Δ∷LEU2; and
YSCC903: MATa prc1 prb1 pep4 leu2 trp1 ura3 URA3∷GAL1-NTC90.
Oligonucleotides
The following oligonucleotides were used:
HA1: CGTATGGGTAAGCGTAGTCTGGGACGTCGTATGGGTAAGCTACCCATACGACGT;
HA2: CGTATGGGTAAGCGTAGTCTGGGACGTCGTATGGGTAAGCGTAGTCTGGGACGT;
P2-6: CTTTAAAATTGCTTATATCATTAGCAACAATGAACGCAAAAAA; and
P2-7: TTTTTTGCGTTCATTGTTGCTAATGATATAAGCAATTTTAAAG.
Antibodies
The anti-HA monoclonal antibody 8G5F was produced by immunizing mice with a KLH-conjugated HA-peptide (TY Tsao and S-C Chang, unpubl.), and 12CA5 was purchased from Berkeley Antibody. Anti-V5 antibody was from Serotec Inc. Antibodies against Prp19, Ntc90, Ntc85, Ntc77, Ntc31, Ntc30, Ntc25, Ntc20, Yju2, and Smd1 were produced by immunizing rabbits with corresponding recombinant proteins, and against Snu114 with GST fusion of the N-terminal fragment of 129 amino acid residues, all expressed in Escherichia coli.
Plasmids
pRS406GAL
A 0.8-kb EcoRI-BamHI fragment containing the GAL1-GAL10 promoter region was inserted into the EcoRI-BamHI sites of pRS406.
pKC90
A 445-bp BamHI fragment containing the N-terminus of the NTC90 ORF was inserted into the BamHI site of pRS406GAL.
pKC414
Plasmid vector pRS414 was deleted from the KpnI and SacI sites.
pKC900
A 4.7-kb Eco47III-SpeI fragment containing the ORF of NTC90, 1.6 kb of the upstream region sequence and 0.5 kb of the downstream sequence, was inserted into the SmaI-SpeI sites of pKC414.
pKC901
A sequence containing EcoRI-NdeI-BamHI restriction sites immediately upstream of the initiation codon of NTC90 was inserted into pKC900 followed by deletion of a 445-bp BamHI fragment of the N-terminal region.
pKC902
A 380-bp BamHI-MscI fragment was deleted from pKC900.
pKC903
A 600-bp KpnI-MscI fragment was deleted from pKC900.
pKC904
An 810-bp BglII-SacI fragment was deleted from pKC900.
pKC905
A 322-bp fragment from the SacI site to the stop codon of NTC90 was deleted from pKC900.
pKC906
A 824-bp fragment from the initiation codon to the MscI site of NTC90 was deleted from pKC900.
pKC907
A 1.22-kb N-terminal fragment starting from the second codon of NTC90 was deleted from pKC900.
pKC9003
A 4.7-kb EcoRI-SpeI fragment from pKC900 was cloned into the EcoRI-SpeI sites of pRS413.
pKC9004
The 27-base sequence encoding the HA-epitope was inserted immediately upstream of the termination codon of the NTC90 in pKC900.
pKC9005
Primers HA1 and HA2 were annealed and inserted into the AatII site of pKC9004.
pKC9033
A 4.1-kb EcoRI-SpeI fragment from pKC903 was cloned into the EcoRI-SpeI sites of pRS413.
pKC9053
A 4.4-kb EcoRI-SpeI fragment from pKC905 was cloned into the EcoRI-SpeI sites of pRS413.
pKC9063
A 3.9-kb XhoI-SpeI fragment from pKC906 was cloned into the XhoI-SpeI sites of pRS413.
pKC9073
A 3.5-kb XhoI-SpeI fragment from pKC907 was cloned into the XhoI-SpeI sites of pRS413.
pKC9001
A 3.1-kb StuI-SpeI fragment from pKC900 was blunt ended and cloned into the blunt-ended NcoI-XhoI sites of pACT2.
pKC9011
A 2.6-kb StuI-SpeI fragment from pKC901 was blunt ended and cloned into the blunt-ended NcoI-XhoI sites of pACT2.
pKC9021
A 2.7-kb StuI-SpeI fragment from pKC902 was blunt ended and cloned into the blunt-ended NcoI-XhoI sites of pACT2.
pKC9031
A 2.5-kb StuI-SpeI fragment from pKC903 was blunt ended and cloned into the blunt-ended NcoI-XhoI sites of pACT2.
pKC9041
A 2.3-kb StuI-SpeI fragment from pKC904 was blunt ended and cloned into the blunt-ended NcoI-XhoI sites of pACT2.
pKC9051
A 2.8-kb StuI-SpeI fragment from pKC905 was blunt ended and cloned into the blunt-ended NcoI-XhoI sites of pACT2.
pKC9071
A 1.0-kb SacI-BamHI fragment from pKC907 was cloned into the SacI-BamHI sites of PkC9011.
pKC9002
A 3.1-kb StuI-NotI fragment form pKC900 was cloned into the BamHI (blunt-ended)-NotI sites of pEG202.
pKC9012
A 2.6-kb StuI-NotI fragment from pKC901 was cloned into the blunt-ended BamHI (blunt-ended)-NotI sites of pEG202.
pKC9022
A 2.7-kb StuI-NotI fragment from pKC902 was cloned into the BamHI (blunt-ended)-NotI sites of pEG202.
pKC9032
A 2.5-kb StuI-NotI fragment from pKC903 was cloned into the BamHI (blunt-ended)-NotI sites of pEG202.
pKC9042
A 2.3-kb StuI-NotI fragment from pKC904 was cloned into the BamHI (blunt-ended)-NotI sites of pEG202.
pKC9052
A 2.8-kb StuI-NotI fragment from pKC905 was cloned into the BamHI (blunt-ended)-NotI sites of pEG202.
pKC9072
A 1.9-kb StuI-NotI fragment from pKC907 was cloned into the BamHI (blunt-ended)-NotI sites of pEG202.
pKC9075
A 1.7-kb BglII-SpeI fragment from pKC9005 was cloned into the BglII-SpeI sites of pKC907.
Preparation of splicing extracts
Splicing extracts were prepared according to Cheng et al. (1990). For preparation of Ntc90-depleted extracts, YSCC903 cells were cultured in uracil-dropout synthetic complete (SC) medium supplemented with galactose at 30°C overnight. Yeast cells were collected, washed 3×in 2 mL of sterile water, resuspended in 1 mL of sterile water, inoculated into YPD, and cultured at 30°C for 16 h. For preparation of the 90-3HA and IV-V-3HA extracts, YSCC903 cells bearing pKC9005 or pKC9075 were cultured in uracil and tryptophan-dropout SC medium supplemented with glucose at 30°C overnight, inoculated into YPD, and cultured at 30°C for 16–18 h.
Splicing assays, immunoprecipitation, precipitation of the spliceosome with streptavidin Sepharose, spliceosome stability assays, and UV-cross-linking analysis
Splicing assays and immunoprecipitation were performed as described by Tarn et al. (1993) with an anti-Ntc85, anti-Ntc20, or 8G5F antibody. Precipitation of the spliceosome with streptavidin Sepharose, spliceosome stability assays, and UV-cross-linking analysis were carried out according to Chan et al. (2003).
Purification of Prp2 and prp2S378L
The PRP2 gene tagged with four copies of V5-epitope at the amino terminus (Liu et al. 2007) and two copies of the HA-epitope at the carboxyl terminus were cloned into pET15b. A leucine to serine mutation at position 378 was introduced by site-directed mutagenesis using primers P2-6 and P2-7. Both wild-type and prp2S378L mutant proteins were purified by consecutive chromatography on a nickel affinity column and anti-HA antibody conjugated protein A-Sepharose beads.
ACKNOWLEDGMENTS
We thank P. Lin and members of the Cheng (S.-C.C.) laboratory for helpful discussions and M. Loney for editing the manuscript. This work was supported by a grant from the Academia Sinica and National Science Council (Taiwan), NSC96-2321-B-001-006.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1625309.
REFERENCES
- Albers M, Diment A, Muraru M, Russell CS, Beggs JD. Identification and characterization of Prp45p and Prp46p, essential pre-mRNA splicing factors. RNA. 2003;9:138–150. doi: 10.1261/rna.2119903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Yehuda S, Dix I, Russell CS, McGarvey M, Beggs JD, Kupiec M. Genetic and physical interactions between factors involved in both cell cycle progression and pre-mRNA splicing in Saccharomyces cerevisiae. Genetics. 2000;156:1503–1517. doi: 10.1093/genetics/156.4.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge CB, Tuschl TH, Sharp PA. Splicing of precursors to mRNAs by the spliceosome. In: Gesteland RF, et al., editors. RNA world II. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1999. [Google Scholar]
- Chan SP, Cheng SC. The Prp19-associated complex is required for specifying interactions of U5 and U6 with pre-mRNA during spliceosome activation. J Biol Chem. 2005;280:31190–31199. doi: 10.1074/jbc.M505060200. [DOI] [PubMed] [Google Scholar]
- Chan S-P, Kao D-I, Tsai W-Y, Cheng S-C. The Prp19p-associated complex in spliceosome activation. Science. 2003;302:279–282. doi: 10.1126/science.1086602. [DOI] [PubMed] [Google Scholar]
- Chen HR, Jan SP, Tsao TY, Sheu YJ, Banroques J, Cheng SC. Snt309p, a component of the Prp19p-associated complex that interacts with Prp19p and associates with the spliceosome simultaneously with or immediately after dissociation of U4 in the same manner as Prp19p. Mol Cell Biol. 1998;18:2196–2204. doi: 10.1128/mcb.18.4.2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HR, Tsao TY, Chen CH, Tsai WY, Her LS, Hsu MM, Cheng SC. Snt309p modulates interactions of Prp19p with its associated components to stabilize the Prp19p-associated complex essential for pre-mRNA splicing. Proc Natl Acad Sci. 1999;96:5406–5411. doi: 10.1073/pnas.96.10.5406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CH, Tsai WY, Chen HR, Wang CH, Cheng SC. Identification and characterization of two novel components of the Prp19p-associated complex, Ntc30p and Ntc20p. J Biol Chem. 2001;276:488–494. doi: 10.1074/jbc.M006958200. [DOI] [PubMed] [Google Scholar]
- Chen C-H, Yu W-C, Tsao TY, Wang L-Y, Chen H-R, Lin J-Y, Tsai W-Y, Cheng S-C. Functional and physical interactions between components of the Prp19p-associated complex. Nucleic Acids Res. 2002;30:1029–1037. doi: 10.1093/nar/30.4.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C-H, Kao D-I, Chan S-P, Kao T-C, Lin J-Y, Cheng S-C. Functional links between the Prp19-associated complex, U4/U6 biogenesis and spliceosome recycling. RNA. 2006;12:765–774. doi: 10.1261/rna.2292106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S-C, Newman A, Lin R-J, McFarland GD, Abelson JN. Preparation and fractionation of yeast splicing extract. Methods Enzymol. 1990;181:89–96. doi: 10.1016/0076-6879(90)81114-a. [DOI] [PubMed] [Google Scholar]
- Dahan O, Kupiec M. Mutations in genes of Saccharomyces cerevisiae encoding pre-mRNA splicing factors cause cell cycle arrest through activation of the spindle checkpoint. Nucleic Acids Res. 2002;30:4361–4370. doi: 10.1093/nar/gkf563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Andrea LD, Regan L. TPR proteins: The versatile helix. Trends Biochem Sci. 2003;28:655–662. doi: 10.1016/j.tibs.2003.10.007. [DOI] [PubMed] [Google Scholar]
- Das AK, Cohen PW, Barford D. The structure of the tetratricopeptide repeats of protein phosphatase 5: Implications for TPR-mediated protein-protein interactions. EMBO J. 1998;17:1192–1199. doi: 10.1093/emboj/17.5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S-H, Lin R-J. Spliceosome activation by PRP2 ATPase prior to the first transesterification reaction of pre-mRNA splicing. Mol Cell Biol. 1996;16:6810–6819. doi: 10.1128/mcb.16.12.6810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y-C, Chen H-C, Wu N-Y, Cheng S-C. A novel splicing factor Yju2 is associated with NTC and acts after Prp2 in promoting the first catalytic reaction of pre-mRNA splicing. Mol Cell Biol. 2007;27:5403–5413. doi: 10.1128/MCB.00346-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsu Y, Asahina H, Citterio E, Rademakers S, Vermeulen W, Kamiuchi S, Yeo JP, Khaw MC, Saijo M, Kodo N, et al. XAB2, a novel tetratricopeptide repeat protein involved in transcription-coupled DNA repair and transcription. J Biol Chem. 2000;275:34931–34937. doi: 10.1074/jbc.M004936200. [DOI] [PubMed] [Google Scholar]
- Ohi MD, Gould KL. Characterization of interactions among the Cef1p-Prp19p-associated splicing complex. RNA. 2002;8:798–815. doi: 10.1017/s1355838202025050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohi MD, Link AJ, Ren L, Jennings JL, McDonald WH, Gould KL. Proteomics analysis reveals stable multiprotein complexes in both fission and budding yeasts containing Myb-related Cdc5p/Cef1p, novel pre-mRNA splicing factors, and snRNAs. Mol Cell Biol. 2002;22:2011–2024. doi: 10.1128/MCB.22.7.2011-2024.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plumpton M, McGarvey M, Beggs JD. A dominant negative mutation in the conserved RNA helicase motif ‘SAT’ causes splicing factor PRP2 to stall in spliceosomes. EMBO J. 1994;13:879–887. doi: 10.1002/j.1460-2075.1994.tb06331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy J, Kim K, Maddock JR, Anthony JG, Woolford JL. The final stages of spliceosome maturation require Spp2p that can interact with the DEAH box protein Prp2p and promote step 1 of splicing. RNA. 1995;1:375–390. [PMC free article] [PubMed] [Google Scholar]
- Russell CS, Ben-Yehuda S, Dix I, Kupiec M, Beggs JD. Functional analysis of interacting factors involved in both pre-mRNA splicing and cell cycle progression in Saccharomyces cerevisiae. RNA. 2000;6:1565–1572. doi: 10.1017/s1355838200000984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman EJ, Maeda A, Wei J, Smith P, Beggs JD, Lin R-J. Interaction between a G-patch protein and a spliceosome DEXD/H-box ATPase that is critical for splicing. Mol Cell Biol. 2004;24:10101–10110. doi: 10.1128/MCB.24.23.10101-10110.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley JP, Guthrie C. Mechanical devices of the spliceosome: Motors, clocks, springs, and things. Cell. 1998;92:315–326. doi: 10.1016/s0092-8674(00)80925-3. [DOI] [PubMed] [Google Scholar]
- Stevens SW, Ryan DE, Ge HY, Moore RE, Young MK, Lee TD, Abelson J. Composition and functional characterization of the yeast spliceosomal penta-snRNP. Mol Cell. 2002;9:31–44. doi: 10.1016/s1097-2765(02)00436-7. [DOI] [PubMed] [Google Scholar]
- Tarn W-Y, Lee K-R, Cheng S-C. The yeast PRP19 protein is not tightly associated with small nuclear RNAs, but appears to associate with the spliceosome after binding of U2 to the pre-mRNA and prior to formation of the functional spliceosome. Mol Cell Biol. 1993;13:1883–1891. doi: 10.1128/mcb.13.3.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarn WY, Hsu CH, Huang KT, Chen HR, Kao HY, Lee KR, Cheng SC. Functional association of essential splicing factor(s) with PRP19 in a protein complex. EMBO J. 1994;13:2421–2431. doi: 10.1002/j.1460-2075.1994.tb06527.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai WY, Chow YT, Chen HR, Huang KT, Hong RI, Jan SP, Kuo NY, Tsao TY, Chen CH, Cheng SC. Cef1p is a component of the Prp19p-associated complex and essential for pre-mRNA splicing. J Biol Chem. 1999;274:9455–9462. doi: 10.1074/jbc.274.14.9455. [DOI] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Lührmann RL. The spliceosome: Design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- Will CL, Lührmann R. Protein functions in pre-mRNA splicing. Curr Opin Cell Biol. 1997;9:320–328. doi: 10.1016/s0955-0674(97)80003-8. [DOI] [PubMed] [Google Scholar]
- Will CL, Lührmann R. Spliceosome structure and function. In: Gesteland RF, et al., editors. The RNA world. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2006. pp. 369–400. [Google Scholar]