Abstract
A-to-I RNA editing is the conversion of adenosine to inosine in double-stranded cellular and viral RNAs. Recently, abundant hyperediting of human transcripts, affecting thousands of genes, has been reported. Most of these editing sites are confined to intramolecular hairpin double-stranded RNA (dsRNA) structures formed by pairing of neighboring, reversely oriented, primate-specific Alu repeats. The biological implication of this extensive modification is still a mystery. A number of studies have shown that heavily edited transcripts are often retained in the nucleus. A recent study found that the edited region in transcripts of the mouse Slc7a2 gene is post-transcriptionally cleaved upon stress, enabling the release of the mRNA to the cytoplasm, followed by its translation. Here, we aim to test whether this scenario might be relevant for many other hyperedited Alu targets. Bioinformatics analysis of publicly available mRNA and expressed sequence tag data provides evidence showing that neighboring, reversely oriented, Alu elements are often cleaved at both ends of the region harboring the inverted repeats followed by rejoining of the two parts of the transcript on both sides of the inverted repeats, resulting in almost inosine-free mRNA products. Deleted segments vary among transcripts of the same gene and are not flanked by the canonical splicing signal sequences. The tissue distribution of these events seems to correlate with known A-to-I editing patterns, suggesting that it depends on the dsRNA structure being edited. Results are experimentally verified by polymerase chain reaction and cloning data. A database of 566 human and 107 mouse putative cleavage loci is supplied.
Keywords: Alu repeats, A-to-I RNA editing, cleavage
INTRODUCTION
A-to-I RNA hyperediting
Adenosine-to-inosine (A-to-I) RNA editing is a post-transcriptional alteration of RNA sequences, catalyzed by members of the double-stranded RNA-specific adenosine deaminases acting on RNA family (ADAR1, ADAR2, ADAR3) (Bass 2002). In recent years, it was found that thousands of human genes undergo A-to-I hyperediting in their 3′ untranslated regions (UTRs) (Athanasiadis et al. 2004; Blow et al. 2004; Kim et al. 2004; Levanon et al. 2004, 2005). An absolute prerequisite for the editing process is the temporary existence of dsRNA. Virtually all A-to-I substitutions in humans occur within such structures formed by two adjacent, reversely oriented Alu elements within a single transcript. The biological impact and significance of this widespread phenomenon is yet elusive.
A-to-I editing is most abundant in brain tissues and is linked with a number of neurological disorders (Levanon et al. 2005). Its high level seems to be primate-specific, due to the lower divergence of the Alu repeats (Eisenberg et al. 2005; Neeman et al. 2006). Therefore, the question naturally arises whether there is any biological significance to A-to-I hyperediting of the Alu repetitive elements. In particular, did it play a role in primate evolution?
Nuclear retention of hyperedited substrates
A number of possible functional implications for the abundant hyperediting phenomena have been raised. These include a possible role in gene silencing (Wang et al. 2005), in augmenting or counteracting the RNAi mechanism (Knight and Bass 2002), or involvement in an anti-retro-element defense mechanism (Levanon et al. 2005).
It has been suggested that the major fate of heavily edited transcripts is retention in the nucleus by the p54nrb (non-POU domain containing, octamer-binding, NONO) complex (Zhang and Carmichael 2001). It was indeed shown that a single pair of reversely oriented Alu repeats in the 3′ UTR of a reporting gene strongly represses its expression, in conjunction with a significant nuclear retention of the mRNAs. Following this work, the localization properties of structured 3′ UTRs undergoing editing is currently of much interest: nuclear retention was observed for the endogenous Nicolin 1 (NICN11) mRNA harboring inverted Alus in its 3′ UTR (Chen et al. 2008) and for mouse Slc7a2 edited transcripts (Prasanth et al. 2005). Yet, another group (Hundley et al. 2008) has recently reported no effect of editing within the 3′ UTR on mRNA localization and translation of several Caenorhabditis elegans and human transcripts.
The possibility of nuclear retention of hyperedited transcripts was first interpreted as a means of protection against abnormal transcripts (Zhang and Carmichael 2001). This is supported by the abundance of hyperediting clusters in splicing-defective transcripts (Kim et al. 2004). This idea is in line with a similar proposed mechanism, suggesting that an I-specific cleavage of RNAs can lead to the selective destruction of edited RNAs (Scadden and Smith 2001).
Cleavage of hyperedited substrates might release mRNAs to the cytoplasm
However, a recent study by Prasanth et al. (2005) has opened a new perspective on the way transcript localization and I-specific cleavage might contribute to cell function. It was shown that the 3′ UTR of the mouse Slc7a2 gene, which is longer than previously believed, contains inverted SINEs (short interspersed nuclear elements) that form a hairpin dsRNA structure and are highly A-to-I edited. The mRNA is then retained in the nucleus in association with the p54nrb (NONO) complex. Moreover, it has been demonstrated that under stress conditions, the edited part is post-transcriptionally cleaved, removing the edited SINEs from the 3′ UTR. Consequently, the mRNA is exported to the cytoplasm, where it translates into a protein. It thus turns out that A-to-I hyperediting may serve as a powerful means of retaining in the nucleus mRNA molecules that are not immediately needed to produce proteins but whose cytoplasmic presence is rapidly required upon a physiologic stress.
Naturally, one wonders whether this intricate regulation mechanism, exhibited so beautifully in one mouse gene, is relevant to the thousands of hyperedited human genes. Currently, the biological relevance of genome-wide A-to-I editing of 3′ human UTRs is questionable. The possibility of a mechanism for nuclear retention followed by release through regulated cleavage might provide the missing link.
Here, we employ a bioinformatics approach, searching for the footprints of such cleavage events. In particular, we focus on mRNA sequences exhibiting alignment gaps that are not regular introns. We analyze these alignment gaps (termed hereafter noncanonical introns [NCIs]) and show they are strongly associated with the existence of paired Alu repeats at the splice junction. Experimental validation data are supplied, and tissue distribution analysis hints at a possible relation to A-to-I editing.
RESULTS AND DISCUSSION
Search for cleavage loci
Virtually all eukaryotic introns are flanked by a 5′ GT or GC dinucleotide and a 3′ AG dinucleotide (Bhasi et al. 2007). Looking for cleavage sites, we have thus scanned all University of California Santa Cruz (UCSC) alignments of human and mouse mRNA sequences, recording all alignment gaps (henceforth termed, for simplicity, introns) and their flanking dinucleotides (see Materials and Methods). This search resulted in 199,654 human mRNAs, including 1,374,601 canonical introns and 19,702 NCIs, virtually all of which are located in UTRs.
NCIs are enriched in Alu repeats and editing sites
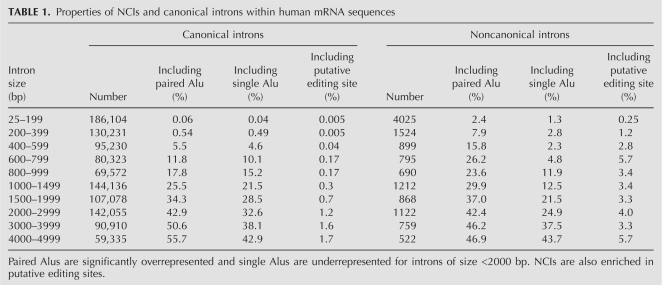
NCIs are distinctively different from canonical introns. They are enriched in putative editing sites (as predicted by the algorithm described by Neeman et al. [2006]): 3.4% of NCIs (662 in number) include a putative editing site, while only 1.0% of canonical introns (13,667) do (P-value < 10−100). They are also slightly enriched in Alu repeats: 54.3% of NCIs (10,694 in number) overlap with an Alu repeat, compared with only 48.1% of canonical introns (661,752) (P-value < 10−60). The tendency strengthens when looking at paired Alus only (see Materials and Methods): 43.7% (8610) of the NCIs overlap with a paired Alu, compared with only 31.4% (431,976) of the canonical introns. Single Alus show the opposite effect and are underrepresented in the NCIs. A detailed comparison for introns of varying sizes is presented in Table 1. It shows that these differences cannot be attributed to different size distributions. We note that similar behavior is observed for mouse NCIs: 52.7% of these (7569) overlap with a B1 repeat and 39.8% (5731) overlap with a B2 repeat, where for canonical introns the fractions are 41.9% (406,581) and 31.1% (301,944), respectively.
TABLE 1.
Properties of NCIs and canonical introns within human mRNA sequences
A potential explanation for the NCIs is the possibility that, occasionally, large parts of the mRNA are somehow skipped during cloning or sequencing, leaving behind gaps that look like introns. In order to exclude this possibility, we compare the properties of the NCIs with those of UTRs. If NCIs are merely a result of cloning or sequencing artifacts, their statistical properties should not differ from those of UTRs. One finds that the density of putative editing sites in UTRs is much higher than in NCIs. Moreover, the density of Alu elements in NCIs is also much higher than in UTRs: 8.3% of the nucleotides within NCIs belong to an Alu repeat, compared with only 3.9% of all RefSeq UTRs (z-test P-value < 10−100). The first difference might be attributed to problems in computational identification of editing sites: the method of detection of these sites relies heavily on the existence of expressed sequences data, and is thus highly ineffective in introns. Similarly, detection of editing sites is difficult in regions that are ineffectively cloned and sequenced. However, the second difference, based on genomic data alone, seems to rule out random cloning or sequencing errors.
The above results suggest that at least some of the NCIs are not merely misaligned introns or mis-sequenced UTRs, but rather are associated with Alu-made dsRNAs (the possibility still exists that there is some sequencing problem that is specifically related to the Alus; see below). The next step is to look for those NCIs flanked by inverted repeats, similarly to the mouse Slc7a2 case (Prasanth et al. 2005).
Identification of human and mouse cleavage loci
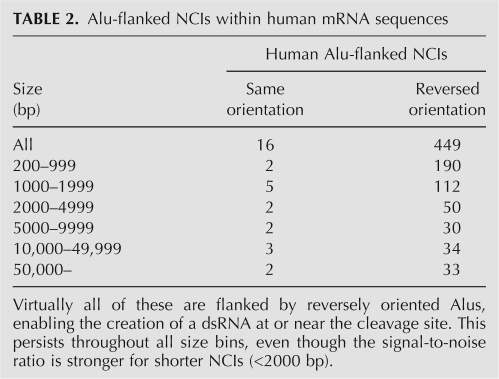
Looking at the resulting list of Alu-flanked NCIs (see Materials and Methods), one is in need of a significance measure: is the number of these NCIs unusually high? In order to provide an answer, one may compare the number of NCIs having their ends overlapping inverted Alus and the number of those with both ends overlapping with same-orientation Alus. Had there been no correlation between the NCIs and the Alu repeats, one would have expected both numbers to be roughly equal (in fact, same-orientation cases should be slightly more prevalent, as there is a weak positive correlation between orientation of successive Alus—the probability of neighboring Alus in the human genome having the same orientation is 57%). Similarly, if the NCIs would have been related to the existence of an Alu at the splice site but not to the formation of a dsRNA by the oppositely orientated Alus, both numbers should have been about the same. Thus, the number of NCIs flanked by same-orientation Alus is used as a measure of the number of random occurrences (or noise) in the set of NCIs flanked by inverted Alus.
We obtained a set of 449 NCIs flanked by inverted repeats, compared with only 16 NCIs flanked by Alus of a similar orientation. It thus appears that most of these 449 NCIs are indeed correlated with the existence of inverted repeats and the dsRNA that follows. The signal to noise ratio is higher for moderate-size NCIs (<2000 base pairs [bp]), but is still very large for the longer NCIs (Table 2). We hereafter use the number of NCIs flanked by same-orientation Alus as a measure of the false-positive detection, or noise, level. A similar scan for human expressed sequence tags (ESTs) resulted in 117 more NCIs (noise level 17). These 566 NCIs are therefore termed putatively cleaved segments (PCSs). One example, including 10 of the PCSs, is presented in Figure 1.
TABLE 2.
Alu-flanked NCIs within human mRNA sequences
FIGURE 1.
UCSC alignment of mRNAs with the 3′ UTR of the apolipoprotein L1 isoform b precursor (APOL1) transcript. (Top panel) Reference sequences for this gene going (left to right) through the end of the last intron (arrowed line), the end of the coding sequence (thick bar), and the 3′ UTR (thin bar). (Middle panel) Different mRNA sequences supporting this region. Most of these sequences (11 out of 15) show signatures of cleavage—a missing part in the middle of the 3′ UTR, looking like an intron (arrowed line). Ten out of the 11 were identified by the present algorithm, while the 11th (CR626091) happened to be cleaved at a canonical splicing signal site. (Bottom panel) Location of the Alu repeats. Unlike splicing, the exact cleavage sites differ between different mRNAs, but they are located within the inverted Alus, or close to their border, in all cases.
The analysis was repeated for 184,554 mouse mRNAs and 1,703,834 mouse ESTs. Seventy-five PCSs with flanking inverted B1 repeats (noise level 2) and 19 PCSs with flanking inverted B2 repeats (noise level 10) were found in mouse mRNAs. The cleavage event reported by Prasanth et al. (2005) was also detected. We note that while the latter publication reported cleavage upstream of the inverted repeats, we found evidence for cleavage downstream from this region, followed by rejoining of the two parts (as demonstrated by mRNA transcript BC127082). Mouse ESTs resulted in only eight B1 and five B2 PCSs (noise levels 3 and 1, respectively). The lower number of mouse PCSs is consistent with the lower number of paired repeats in the mouse genome (Neeman et al. 2006).
A detailed database listing all human and mouse PCSs is provided as Supplemental Material.
Experimental validation
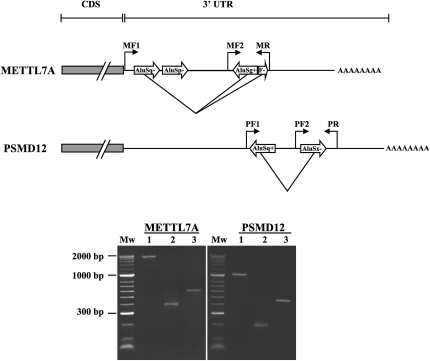
In order to validate the above results, we chose two examples from the above database: the PSMD12 and METTL7A genes. We amplified the region including the inverted Alu structure, and 14 PSMD12 clones and eight METTL7A clones were successfully cloned and sequenced. The electrophoretic analysis of cDNA from both genes clearly shows a band corresponding to transcripts of shorter length, as expected for cleavage products. In contrast, analysis of genomic DNA used as a control yields the expected, full-length amplicon (see Fig. 2). Polymerase chain reaction (PCR) performed using an internal primer demonstrates the existence of full-length transcripts in the cDNA.
FIGURE 2.
Deletion of hyperedited inverted Alu repeat structures within the 3′ UTRs of METTL7A and PSMD12 transcripts. PCR performed on normal human hippocampus genomic DNA using primers flanking the inverted repeat structure (MF1+MR for METTL7A; PF1+PR for PSMD12) results in the expected full-length amplicon (lane 1). However, using the same primers but cDNA from the same tissue as a template, one observes a band of much shorter length, corresponding to completely/partially cleaved isoforms (lane 2). PCR performed on cDNA using an internal primer (MF2+MR for METTL7A; PF2+PR for PSMD12) demonstrates the coexistence of mRNA isoforms with complete 3′ UTRs in which the NCIs were not removed (lane 3). These longer isoforms are not apparent in lane 2 owing to differences in reaction efficiencies.
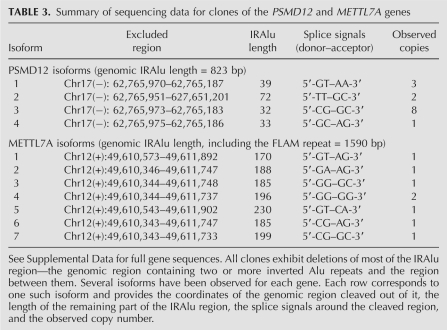
In addition, sequencing of the 14 PSMD clones and eight METTL7A clones reveals that all clones miss most of the inverted Alu region (see Table 3; Supplemental Material). This small-scale study already reveals that the putative cleavage phenomenon is nonspecific: for each gene there are several isoforms of the cleaved product, differing in the exact cleavage site. The splice-site signals also do not show any clear pattern, and they differ from the canonical splicing signals. Some of the clones exhibit A-to-I editing signatures (see Supplemental Material).
TABLE 3.
Summary of sequencing data for clones of the PSMD12 and METTL7A genes
PCSs do not result from editing-assisted normal splicing
Another possible explanation for these PCSs is the possibility of editing-assisted normal splicing, as demonstrated recently for the NARF gene (Lev-Maor et al. 2007). In this process, a canonical splicing site is created in the mRNA modified by A-to-I editing. However, if this was the explanation for the PCSs, one would expect their genomic splice signals to be those that can be edited into the canonical GTAG signal, i.e., ATAG, GTAA, and ATAA. However, only seven of the 449 PCSs are flanked by these three splice signals. Experimental data presented above are also not consistent with this possibility.
PCSs and micro-RNAs
Regardless of the possible relation to nuclear retention, cleavage of parts of the 3′ UTR might affect the transcripts in other ways, including the removal of micro-RNA sequences or targets. Interestingly, one of the PCSs, located within the BX647893 mRNA transcript of the oxysterol binding protein-like 1A gene (OSBPL1A), includes the micro-RNA hsa-mir-320c-2 (Friedlander et al. 2008). This NCI occurs within an intron and seems to be cleaved from a sequence exhibiting intron retention. Similarly, another PCS, located within the DA418713 EST of the zinc finger protein 141 gene (ZNF141), includes the micro-RNA hsa-mir-571 (Cummins et al. 2006). Here too, the PCS occurs within an intron. A few more micro-RNAs occur within very long (>150-kb) PCSs.
Putative micro-RNA targets are found in 48 mRNA PCSs and 10 EST PCSs. As expected, most of these occur within the very large PCSs. However, a significant number occur within shorter PCSs. Notably, one PCS, located within the BX647495 mRNA transcript of the Wolf-Hirschhorn syndrome candidate 1 gene (WHSC1), harbors six micro-RNA targets as predicted by Target Scan (see Materials and Methods).
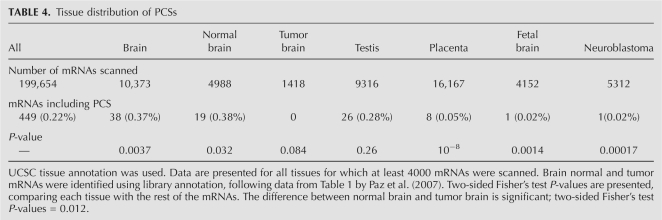
Tissue and histology distribution of the putative cleavage loci suggest an association with RNA editing
Having established the relation between the PCSs and the flanking inverted repeats, two questions arise: (1) Do PCSs represent a biological phenomenon, i.e., cleavage of the dsRNAs as observed in Prasanth et al. (2005), or are they an artifact related to the process of sequencing (reverse transcription, PCR, or sequencing)? In principle, one could have argued that one of these steps is prone to errors that are specifically related to the hairpin dsRNA structure formed by the reversely oriented Alus. (2) If the PCSs do represent a biological phenomenon, do they depend on A-to-I editing of the dsRNA?
Comprehensive and fully convincing answers to both questions would most likely require detailed experimental work. The experimental validation data provided above seem to exclude the possibility of an artifact produced by PCR or cloning procedures, but the possibility of an error resulting from the reverse transcription is still open. However, a closer inspection of the PCSs provides a hint that the answer to both of the above questions might be positive. RNA editing is known to be tissue selective, being mostly pronounced in brain tissues. Cancerous cells have been shown to undergo global hypoediting. If the PCSs are due to a measurement artifact, or even if they depend on the dsRNA secondary structure alone rather than on being edited, one would not expect any tissue or histology preference in the NCI set. Table 4 presents a comparison of PCS prevalence in a number of tissues, as well as a comparison of normal and tumor brain tissues. One finds that PCSs are overrepresented in brain tissues and underrepresented in placenta, fetal brain, and neuroblastoma tissues. Testis tissues exhibit an average PCS level. In addition, normal brain cells are enriched in PCSs compared with brain tumors. These findings are in full agreement with known trends for A-to-I editing (Paz et al. 2007). This provides strong support for the hypothesis that the PCSs do reflect true in vivo, editing-dependent cleavage of dsRNAs.
TABLE 4.
Tissue distribution of PCSs
In addition, we used the database for annotation, visualization, and integrated discovery (DAVID) (Dennis et al. 2003) to analyze the list of genes exhibiting PCSs. The 449 human PCSs supported by mRNAs were mapped to 293 different characterized genes (a number of genes have more than one supporting mRNA, while some PCSs belong to uncharacterized genes). Notably, these genes include a significantly high proportion of genes known to be up-regulated in specific tissues (a gene is said to be up-regulated in a specific tissue if its expression in that tissue, as measured by the GNF gene expression atlas [https://biogps.gnf.org/] is within the top quartile of its expression spectrum across all tissues measured in the GNF gene expression atlas). The list of overrepresented associated tissues is presented in Table 5, and it includes a surprisingly high proportion of brain tissues, suggesting another link to RNA editing. Another observation found by DAVID is the unusually high number of genes in this list that include the zinc finger protein domain. This last finding is in concordance with a similar trend shown for genes harboring inverted Alus in their 3′ UTRs (Chen et al. 2008).
TABLE 5.
Top 10 tissues showing a significant association with the list of genes harboring PCSs
Possible relation to inosine-specific cleavage mechanism
Scadden and Smith (2001) have identified a ribonuclease activity in various cell extracts that specifically cleaves hyperedited dsRNA containing IU pairs. This cleavage involves Tudor Staphylococcal Nuclease, which binds specifically to dsRNAs containing multiple IU pairs, and occurs 5′ of U residues within the sequence 5′-IIUI-3′/3′-UUIU-5′ and leaves a 3′ phosphate (Scadden and O'Connell 2005). It is therefore possible that this previously characterized cleavage scheme is related to the phenomenon described in this work. If this is indeed the case, then the cleavage phenomenon reported here is indeed editing-dependent. However, one should note that in the cases analyzed here, the resulting transcripts include parts coming from both sides of the dsRNA formed by the reversely oriented Alu elements, so the process should presumably involve not only cleavage at both ends but also the reattachments of the two splice sites at both ends together. We should also note that the nucleotides observed at the cleavage site do not conform to the abovementioned IIUI-3′/3′-UUIU-5′ pattern. Yet, the possibility that this disagreement might be an artifact of the alignment algorithm trying to avoid the A-to-I mismatches and force the GT-AG splice signal cannot be ruled out.
Conclusion
Hundreds of alignment gaps flanked by Alu repeats have been identified in human mRNAs and ESTs. They are enriched in A-to-I editing sites, their flanking repeats are reversely oriented, rather than being arranged in the same orientation, and their tissue and histology distribution resembles that of A-to-I RNA editing. These hundreds of gaps seem to be a hallmark of a widespread cleavage mechanism, one example of which was recently demonstrated (Prasanth et al. 2005). Thus, the novel regulatory scheme depicted by Prasanth et al. (2005) might be relevant for thousands of human genes harboring hyperediting substrates, and might shed light on the yet poorly understood functional role of the abundant A-to-I RNA editing of human transcripts.
MATERIALS AND METHODS
Search for noncanonical introns
Alignments of human and mouse mRNAs to the respective genomes have been taken from the mRNA track on the UCSC genome browser (Kent et al. 2002). Sequences aligned to multiple genomic loci were discarded, as well as mRNA sequences having no overlap with any RefSeq sequence. In addition, we discarded mRNA sequences in which at least one intron is flanked by a 5′ CT and 3′ AC (the reverse complement of the canonical GT–AG signal), while no other intron shows the canonical splice signals. These are, most likely, canonically spliced sequences, erroneously aligned to the reverse strand. Introns shorter than 25 bp were removed, as they usually represent sequencing errors and/or deletions. Finally, introns flanked by the rare splicing signals 5′ AT and 3′ AC (Wu and Krainer 1999) were excluded from the analysis.
Single Alus and paired Alus
Alu locations were obtained from the RepeatMasker track on the UCSC genome browser (Kent et al. 2002). An Alu having a neighboring (distance up to 2000 bp) Alu repeat with a reversed orientation is termed a Paired Alu. All other Alu elements are termed Single Alus.
Search for Alu-flanked noncanonical introns
The list of all NCIs was scanned, looking for NCIs harboring an Alu repeat at both the donor and acceptor splice sites. An offset of up to 20 bp was used (i.e., a donor site with an Alu repeat starting <20 bp downstream, or an acceptor site with an Alu repeat starting <20 bp upstream are acceptable). In order to allow for the formation of a significant dsRNA, only Alus overlapping with the NCI in at least half of the length of the typical Alu (150 bp for human; 75 bp for mouse) were considered.
Experimental materials and methods
Human adult hippocampus total RNA and genomic DNA from the same subject were purchased from Biochain. For RT-PCR, each RNA sample was treated with DNase I (Invitrogen) and reverse transcribed using M-MLV RT and random hexamers (Promega). PCR amplification of template cDNA/DNA was performed using the following primers:
METTL7A
MF1, 5′-GAGCTGGCAGTTAAGAGCTGA-3′
MF2, 5′-TTAAGAATCTGAGTCTAAACAGCACAG-3′; and
MR, 5′-TCTGAAAGCCAGACCAGTGA-3′.
PSMD12
PF1, 5′-TGCATTGGTCACACTAATAACATC-3′;
PF2, 5′- CTTGTCAAACCTGGACTATTTGG-3′; and
PR, 5′- CGAGTGCAATTTGGTCACTG-3′.
PCR products were resolved on an agarose gel, or alternatively were purified with a HiYield Gel/PCR DNA fragment extraction kit (RBC Bioscience) and cloned into the pGEM-T Easy vector (Promega). Plasmid DNA was extracted and amplified using an illustra TempliPhi Amplification kit (GE Healthcare). Inserts were sequenced with T7 primer using an ABI PRISM 3100 Genetic Analyzer. The resulting sequences were aligned to the UCSC genomic DNA database by BLAT (UCSC).
Micro-RNAs and micro-RNA targets
The locations of micro-RNA sequences have been taken from the Sanger Center's miRBase database (http://microrna.sanger.ac.uk/). Putative micro-RNA targets were taken from the TargetScan microRNA track on the UCSC genome browser (Kent et al. 2002).
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank E.Y. Levanon for numerous helpful discussions, comments, and suggestions, as well as for a critical reading of the manuscript. This work was supported by The Israel Science Foundation (grant number 365/06). This work was supported in part by grants from the Flight Attendant Medical Research Institute (FAMRI), Bio-Med Morasha ISF (Grant No. 1942/08), and The Israel Ministry for Science and Technology (Scientific Infrastructure Program). We thank the Kahn Family Foundation for their support of our research. G.R. holds the Djerassi Chair in Oncology at the Sackler Faculty of Medicine, Tel-Aviv University. This work was performed in partial fulfillment of the requirements for Ph.D. degrees (to S.O. and D.D.), Sackler Faculty of Medicine, Tel-Aviv University.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1581809.
REFERENCES
- Athanasiadis A, Rich A, Maas S. Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLoS Biol. 2004;2:e391. doi: 10.1371/journal.pbio.0020391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass BL. RNA editing by adenosine deaminases that act on RNA. Annu Rev Biochem. 2002;71:817–846. doi: 10.1146/annurev.biochem.71.110601.135501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhasi A, Pandey RV, Utharasamy SP, Senapathy P. EuSplice: A unified resource for the analysis of splice signals and alternative splicing in eukaryotic genes. Bioinformatics. 2007;23:1815–1823. doi: 10.1093/bioinformatics/btm084. [DOI] [PubMed] [Google Scholar]
- Blow M, Futreal PA, Wooster R, Stratton MR. A survey of RNA editing in human brain. Genome Res. 2004;14:2379–2387. doi: 10.1101/gr.2951204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LL, DeCerbo JN, Carmichael GG. Alu element-mediated gene silencing. EMBO J. 2008;27:1694–1705. doi: 10.1038/emboj.2008.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummins JM, He Y, Leary RJ, Pagliarini R, Diaz LA, Jr, Sjoblom T, Barad O, Bentwich Z, Szafranska AE, Labourier E, et al. The colorectal microRNAome. Proc Natl Acad Sci. 2006;103:3687–3692. doi: 10.1073/pnas.0511155103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:P3. doi: 10.1186/gb-2003-4-5-p3. [DOI] [PubMed] [Google Scholar]
- Eisenberg E, Nemzer S, Kinar Y, Sorek R, Rechavi G, Levanon EY. Is abundant A-to-I RNA editing primate-specific? Trends Genet. 2005;21:77–81. doi: 10.1016/j.tig.2004.12.005. [DOI] [PubMed] [Google Scholar]
- Friedlander MR, Chen W, Adamidi C, Maaskola J, Einspanier R, Knespel S, Rajewsky N. Discovering microRNAs from deep sequencing data using miRDeep. Nat Biotechnol. 2008;26:407–415. doi: 10.1038/nbt1394. [DOI] [PubMed] [Google Scholar]
- Hundley HA, Krauchuk AA, Bass BL. C. elegans and H. sapiens mRNAs with edited 3′ UTRs are present on polysomes. . RNA. 2008;14:2050–2060. doi: 10.1261/rna.1165008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DD, Kim TT, Walsh T, Kobayashi Y, Matise TC, Buyske S, Gabriel A. Widespread RNA editing of embedded alu elements in the human transcriptome. Genome Res. 2004;14:1719–1725. doi: 10.1101/gr.2855504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight SW, Bass BL. The role of RNA editing by ADARs in RNAi. Mol Cell. 2002;10:809–817. doi: 10.1016/s1097-2765(02)00649-4. [DOI] [PubMed] [Google Scholar]
- Lev-Maor G, Sorek R, Levanon EY, Paz N, Eisenberg E, Ast G. RNA-editing-mediated exon evolution. Genome Biol. 2007;8:R29. doi: 10.1186/gb-2007-8-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levanon EY, Eisenberg E, Yelin R, Nemzer S, Hallegger M, Shemesh R, Fligelman ZY, Shoshan A, Pollock SR, Sztybel D, et al. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat Biotechnol. 2004;22:1001–1005. doi: 10.1038/nbt996. [DOI] [PubMed] [Google Scholar]
- Levanon K, Eisenberg E, Rechavi G, Levanon EY. Letter from the editor: Adenosine-to-inosine RNA editing in Alu repeats in the human genome. EMBO Rep. 2005;6:831–835. doi: 10.1038/sj.embor.7400507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neeman Y, Levanon EY, Jantsch MF, Eisenberg E. RNA editing level in the mouse is determined by the genomic repeat repertoire. RNA. 2006;12:1802–1809. doi: 10.1261/rna.165106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paz N, Levanon EY, Amariglio N, Heimberger AB, Ram Z, Constantini S, Barbash ZS, Adamsky K, Safran M, Hirschberg A, et al. Altered adenosine-to-inosine RNA editing in human cancer. Genome Res. 2007;17:1586–1595. doi: 10.1101/gr.6493107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasanth KV, Prasanth SG, Xuan Z, Hearn S, Freier SM, Bennett CF, Zhang MQ, Spector DL. Regulating gene expression through RNA nuclear retention. Cell. 2005;123:249–263. doi: 10.1016/j.cell.2005.08.033. [DOI] [PubMed] [Google Scholar]
- Scadden AD, O'Connell MA. Cleavage of dsRNAs hyperedited by ADARs occurs at preferred editing sites. Nucleic Acids Res. 2005;33:5954–5964. doi: 10.1093/nar/gki909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scadden AD, Smith CW. Specific cleavage of hyperedited dsRNAs. EMBO J. 2001;20:4243–4252. doi: 10.1093/emboj/20.15.4243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Zhang Z, Blackwell K, Carmichael GG. Vigilins bind to promiscuously A-to-I-edited RNAs and are involved in the formation of heterochromatin. Curr Biol. 2005;15:384–391. doi: 10.1016/j.cub.2005.01.046. [DOI] [PubMed] [Google Scholar]
- Wu Q, Krainer AR. AT-AC pre-mRNA splicing mechanisms and conservation of minor introns in voltage-gated ion channel genes. Mol Cell Biol. 1999;19:3225–3236. doi: 10.1128/mcb.19.5.3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Carmichael GG. The fate of dsRNA in the nucleus: a p54(nrb)-containing complex mediates the nuclear retention of promiscuously A-to-I edited RNAs. Cell. 2001;106:465–475. doi: 10.1016/s0092-8674(01)00466-4. [DOI] [PubMed] [Google Scholar]