Abstract
Alternative splicing of the metabotropic glutamate receptor 1 (mGluR1) receptor gene generates two major receptor isoforms, mGluR1a and mGluR1b, differing in intracellular function and distribution. However, little is known on the expression profiles of these variants during development. We examined the mRNA expression profile of mGluR1a/b in microdissected layers and acutely isolated mitral cells in the developing mouse olfactory bulb. This analysis showed that the two mGluR1 variants are differentially regulated within each bulb layer. During the first postnatal week, the mGluR1a isoform replaces GluR1b in the microdissected mitral cell layer (MCL) and in isolated identified mitral cells, coinciding with a developmental epoch of mitral cell dendritic reorganization. Although mGluR1a mRNA is expressed at high levels in both the adult external plexiform layer (EPL) and MCL, Western blotting analysis reveals a marked reduction of the mGluR1a protein in the MCL, where mitral cell bodies are located, and strong labeling in the EPL, which contains mitral cell dendrites. This suggests that there is increased dendritic trafficking efficiency of the receptor in adult. The temporal and spatial shift in mGluR1b/a expression suggests distinct roles of the mGluR1 isoforms, with mGluR1b potentially involved in the early mitral cell maturation and mGluR1a in dendritic and synapse function.
Keywords: RNA splicing, GAD transgenic mice, RT-PCR, Western blot, mGluR5, microdissection, single cell
The adult olfactory bulb (OB) is a cortical structure consisting of alternating cellular and neuropil layers. Axons of the olfactory receptor neurons (ORN) project from the olfactory epithelium and surround the olfactory bulb, forming the nerve fiber layer. Individual axons of the olfactory nerve leave the nerve fiber layer to enter the glomeruli, where they synaptically terminate on the dendrites of mitral cells, tufted cells, and periglomerular neurons. ORN axons utilize glutamate as their primary neurotransmitter (Sassoe-Pognetto et al., 1993; Ennis et al., 1996). Glutamate released from the olfactory nerve acts on glutamate receptors located on the dendrites of the OB target cells (Berkowicz et al., 1994; Ennis et al., 1996). The major outputs neurons of the bulb, mitral and tufted cells, are also glutamatergic, and their lateral dendrites form reciprocal synapses with GABAergic granule cells in the external plexiform layer. Fast synaptic responses in mitral and tufted cells are mediated through AMPA/kainate receptors and NMDA receptors (Ennis et al., 1996; Aroniadou-Anderjaska et al., 1997). These cells also express high levels of the group I metabotropic glutamate receptor (mGluR) subtype, mGluR1 (Masu et al., 1991; Martin et al., 1992; Shigemoto et al., 1992; van den Pol, 1995; Sahara et al., 2001). The functional role of this receptor in mitral and tufted cells is still only partially understood (Heinbockle et al., 2004; Ennis et al., 2006). Group I mGluRs (mGluR1 and mGluR5) are coupled to PI turnover and Ca2+ mobilization from intracellular stores. At least four alternatively spliced variants of mGluR1 exist (mGluR1a–d), which diverge in their cytoplasmic C-terminal tails (Conn and Pin, 1997). All mGluR1 splice variants activate phospholipase C, but the intracellular calcium signals generated by the shorter proteins mGluR1b, mGluR1c, and mGluR1d have slower kinetics (Pin and Duvoisin, 1995) and differ from mGluR1a in their subcellular localization (Ferraguti et al., 1998). The functional impact of the different kinetics, and localization, of specific mGluR1 variants in the brain is not known.
The expression and subcellular distribution of group I mGluRs are developmentally regulated in different brain regions (Nicoletti et al., 1986; Minakami et al., 1995; Casabona et al., 1997; Liu et al., 1998). Group I mGluRs have a role in cerebellar development (Catania et al., 2001; Hashimoto et al., 2001), in the early processing of sensory information (Munoz et al., 1999), and in the ocular dominance plasticity in the visual cortex (Bear and Rittenhouse, 1999). mGluR1 is expressed prenatally by rat mitral and tufted cells (Shigemoto et al., 1992), and high expression is maintained by these cells into adulthood (Masu et al., 1991; Shigemoto et al., 1992; Martin et al., 1992; Baude et al., 1993; Fotuhi et al., 1993; van den Pol, 1995; Petralia et al., 1997; Berthele et al., 1998). However, the existing developmental data do not distinguish among the various mGluR1 splicing variants.
This study examined the developmental expression profile of group I mGluR splice variants in different microdissected OB layers and in the major second-order output neurons, mitral cells, at different developmental times. The results showed that before birth mitral cell express mainly mGluR1b. However, there is a rapid shift such that, by 1 week of age, mGluR1a is the primary mRNA transcript in both the mitral cell (MCL) and the external plexiform (EPL) layers. This temporal regulation of mGluR1 splice variants suggests that the mGluR1b isoform could have an early role in mitral cell maturation, and, conversely, mGluR1a could be more important in late events such as activity-dependent remodeling of mitral cell dendrites and synaptic refinement.
MATERIALS AND METHODS
Isolation of OB Layers
Microdissections of individual OB layers were performed on fresh 300-μm vibratome slices harvested from the OBs of E18, P0, P4, P8, and adult mice (the day of positive dam was designated E0 and the day of birth designated P0). Under a high-magnification dissecting microscope, each layer was carefully dissected: the olfactory nerve fiber layer (ONL), glomerular layer (GL), external plexiform layer (EPL), mitral cell layer (MCL), and granule cell layer (GCL). Microdissection performed approximately 40 μm superficial to the GL separated the ONL, and dissection approximately 40 μm deep to the GL separated this layer. Dissection approximately 40 μm either side of the MCL separated the EPL, MCL, and GCL. Therefore, the ONL and EPL samples do not contain any material from the GL, whereas the GL layer contains small amounts of ONL and EPL. Also, the EPL and GCL samples do not contain any MCL material, but the MCL contains small amounts of EPL and GCL.
Isolation of Identified Mitral Cells
To isolate individual mitral cells, fresh vibratome slices of 300 μm thickness were harvested from the OBs of postnatal GAD-LacZ-9B mice (P0, P4, P8, and adult) in ice-cold L15 media (Gibco BRL, Grand Island, NY). The GAD-LacZ-9B line of mice was generated through random insertion of a GAD promoter-LacZ construct driving expression of β-galactosidase (Sekerkova et al., 1997a, b; Katarova et al., 1998). The construct contains an 8-kb segment of the 5′-flanking region of the mouse GAD67 gene rather than 1 kb. The LacZ reporter gene was fused in frame with the GAD67 open reading frame (or coding region) in exon 2. However, because of random insertion of the construct into the genome, rather than observing β-galactosidase expression in GAD-positive cells of the OB, the enzyme was expressed specifically but ectopically in mitral cells (see Fig. 6; Sekerkova et al., 1997a, b; Katarova et al., 1998). Nonolfactory regions of the brain exhibited normal GABAergic interneuron expression of the transgene (Sekerkova et al., 1997a, b; Katarova et al., 1998). Cells in the developing MCL were also selectively expressing β-galactosidase (Fig. 6C). The MCL of these slices was microdissected by cutting through the EPL approximately 40 μm superficial to the MCL and through the GCL approximately 40 μm deep to the MCL. The MCL microdissection separates β-galactosidase-positive tufted cells present in the GL and most of the EPL from positive mitral cells. Additionally, microdissection of this layer removes the bulk of β-galactosidase-“negative” cells, i.e., the GCL, from the dissociation. The microdissected MCL “strip” was enzymatically dissociated via modifications to our previously published methods (Puche and Shipley, 1999). Briefly, the cell layer was gently agitated in a papain enzyme mix containing 100 U cysteine-activated papain (Boehringer-Mannheim, Indianapolis, IN), 0.25% glucose (Sigma, St. Louis, MO), and 1 mM kyurenic acid (Sigma) in minimal Eagle’s medium (MEM; Invitrogen, Carrlsbad, CA), for 5 min (P0 and P4 pups) or 15 min (P8 and adult mice) at 37°C. The tissue was briefly triturated through 23–25-G needles, and dissociated cells were separated from residual tissue pieces by filtration through a 70-μm cell strainer. The dissociated cells were pelleted by centrifugation and resuspended in L15 buffer (minus phenol red) containing 1 mM kynurenic acid. These cells were plated at low density in sterile tissue culture dishes containing L15 buffer (minus phenol red) with 1 mM kynurenic acid. The fluorescent β-galactosidase substrate 5-chloromethylfluorescein-di-o-galactopyranoside (FDG) was used to label living cells. FDG (20 mM) was added and the cells reacted at 37°C for 15 min. After reaction, the dishes were placed at 4°C during cell collection (less than 90 min). Single fluorescent cells were aspirated into a micropipette on an inverted fluorescent microscope and expelled immediately into Trizol reagent. Only fluorescent cells that were not in contact with any other cell and exhibited birefringence under phase-contrast optics were selected. The specificity of FDG labeling was tested in live 300-μm slices through the OB. Upon hydrolysis by β-galactosidase, FDG generates a green fluorescent product that is clearly visible in mitral cells. There is excellent concordance between the distribution of β-galactosidase expression by immunohistochemistry in cryostat sections and FDG fluorescence in fresh brain slices (see Fig. 6D, G). Cell types not expressing β-galactosidase in immunostained sections show negligible nonspecific background hydrolysis of FDG. These findings indicate that FDG is a specific substrate for the detection of LacZ transgene expression in mitral cells. The fluorescent product generated by hydrolysis of FDG by β-galactosidase is retained only in cells with intact membranes. Damaged or dying cells lose membrane integrity and are incapable of retaining the fluorescent reaction product; therefore, we preferentially select live cells with this approach. Pools of 25 cells were collected from each age and from at least two different preparations.
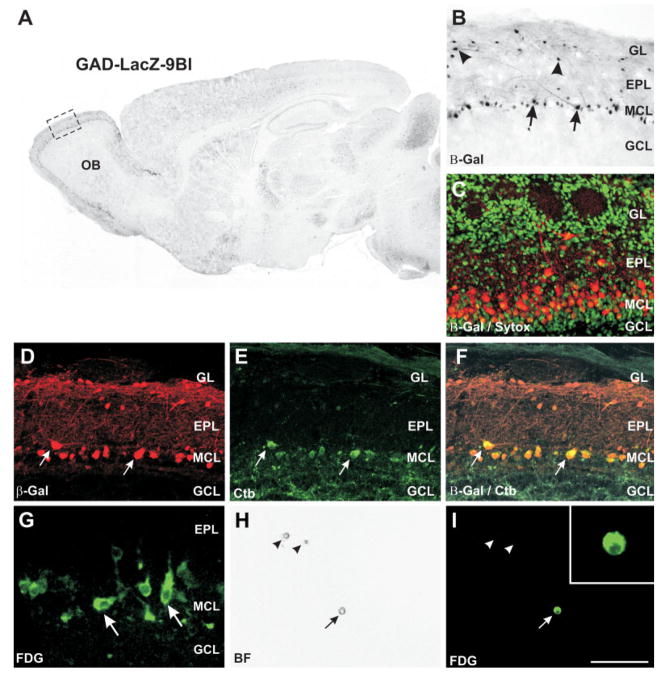
Fig. 6.
Expression of β-galactosidase in GAD-LacZ-9B transgenic mice. A: Immunohistochemistry for β-galactosidase in GAD-LacZ-9B mice. The transgene is expressed in the olfactory bulb (OB) as well as several other brain regions. B: Higher magnification of the region demarcated in A, showing that both mitral (arrows) and tufted (arrowheads) cells express β-galactosidase. C: Immunofluorescence for β-galactosidase (red) in P2 GAD-LacZ-9B mice. Strong expression is present in mitral cells and scattered tufted cells, but not in granule or periglomerular interneurons. Nuclei counterstained with Sytox green. The mitral cells at this age form a layer several cells thick. D–F: Retrograde labeling of mitral cells in GAD-LacZ-9B mice by cholera toxin β(CTb) injections into the piriform cortex. Immunofluorescence for β-galactosidase is shown in D, CTb in E, and overlay in F. All CTb-stained cells expressed β-galactosidase (arrows). A small number of β-galactosidase-positive cells in the mitral cell layer did not label with CTb, as expected, insofar as it is unlikely that a CTb injection into the piriform cortex will label all mitral cells. G: Hydrolysis of the fluorescent β-galactosidase substrate 5-chloromethylfluorescein-di-o-galactopyranoside (FDG) in 300-μm slices through fresh olfactory bulbs yield a strong green fluorescent signal in mitral cells (arrows). H: Dissociated cells from the mitral cell layer viewed with brightfield optics on a confocal microscope. Three dissociated cells are visible in this field (arrows). I: The same field as in H viewed under fluorescence, showing that only one of the three cells expresses β-galactosidase and labels with FDG (arrow). Scale bar = 1 mm for A; 200 μm for B; 25 μm for G; 100 μm in I (applies to C–F, H, I); 25 μm for inset.
RNA Extraction and Reverse Transcription
Whole OBs, OB layers, and pools of 25 cells collected in Trizol were stored at −80°C until use for RNA extraction. Modifications of the manufacturer’s protocols were to include 250 μg/ml of glycogen as a carrier, and small reagent volumes were used to extract total RNA from the 25 cell pools. For MCL and EPL samples, the organic phase was saved for subsequent protein extraction. RNA extracted from OB layers was also DNAse digested with 1 U RQ1 RNase-free DNase I/μg RNA (Promega, Madison, WI) at 37°C for 30 min and reextracted with Trizol to eliminate any genomic DNA contamination. The yield of total RNA was determined by measuring the absorbance at 260/280 nm (Eppendorf Biophotometer). Aliquots containing 300 ng of layer RNA were denatured for 5 min at 65°C and incubated in reaction mixture for 10 min at room temperature, followed by 42°C for 50 min and 70°C for 15 min. The reaction contained 200 U Superscript II reverse transcriptase (Invitrogen Life Technologies), 2.5 μM random hexamers (Amersham Bioscences, Arlington Heights, IL), 50 mM Tris-HCl (pH 8.3), 75 mM KCl, 3 mM MgCl2, 2 U/μl RNasin (Promega), 10 mM dithiothreitol (DTT), 1 mM deoxynucleotide triphosphates (dNTPs, Invitrogen Life Technologies). Pools of 25 dissociated cells were extracted as described above, with the following differences: the whole content of RNA extraction was transcribed into cDNA in one reaction. After reverse transcription, RNA was eliminated by incubation in RNase H for 20 min at 37°C and the cDNA product stored at −80°C.
Design of PCR Primers
The primers used in this study were derived from unique mouse sequences obtained from the Genbank database and specifically designed to anneal with segments located in different exons where possible. All sequences were verified by the NCBI Blast program for negligible cross-recognition of unwanted molecules. To minimize variability in amplification reactions, they were designed to have equal or very similar GC content (50%) and melting temperature. The mGluR1 primers (forward: 5′-cgctccaacaccttcctcaacatt-3′; reverse: 5′-ggggtattgtcctcttcctccacg-3′) detect simultaneously the 1a, 1b, 1d, and 1f isoforms of this receptor; however, 1d and 1f were rarely detectable in any of the samples tested. All splice variants were visible on agarose gel at different molecular weights (Fig. 1A). Similarly, the mGluR5 primers (forward: 5′-gcacgtaggagatggcaagtcatc-3′; reverse: 5′-gggttctccttcttgttgatatgg-3′) simultaneously amplify the splicing variants mGluR5a and 5b. Other primers employed in this study recognize the olfactory marker protein (OMP; forward primer: 5′-gggagaagaagcaggatggtgaga-3′; reverse primer: atacatgaccttgcggatcttggc), glyceraldehyde-3-phosphate dehydrogenase (G3PDH; forward primer: 5′-cgtcccgtagacaaaatggtgaag; reverse primer: 3′-tttgccgtgagtggagtcatactg), and glial acidic fibrillary protein (GFAP; forward: aagccaaacacgaagctaacg; reverse: 5′-agcaagtgcctcctggtaact-3′). PCR product sizes for each of the primer pairs are shown in Figure 1A. Cycling parameters were separately tested for each primer pair in order to obtain specific products with the expected molecular weights in the exponential range of the amplification reaction. Additionally, the specificity of each RT-PCR products has been assessed by sequencing of the amplification products. We found that the sequences of mGluR1a and mGluR1b were identical to previously published mouse sequences (Zhu et al., 1999). Initially mGluR5 primers were based on known rat sequences and extrapolated to mouse with a preferred codon usage table. When sequenced, the PCR product with the expected size for mGluR5a was found to be fully included in Gene Bank mouse cDNA sequence AK032422, and the PCR product with expected size for mGluR5b contained a 96-bp insert identical to the one previously described by Minakami et al. (1995). The mGluR5 primers used in this study (see sequence above) were modified from the previously made rat primers in order to match the mouse sequence fully.
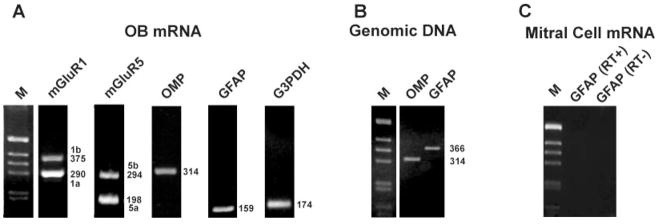
Fig. 1.
Amplification of various cDNA and genomic sequences by PCR from adult olfactory bulb and mitral cells mRNA. A: RT-PCR amplification products derived from whole olfactory bulb mRNA. The product sizes for mGluR1, mGluR5, olfactory marker protein (OMP), glial acidic fibrillary protein (GFAP), and glyceraldehyde-3-phosphate dehydrogenase primers (G3PDH) match with predicted sizes. Sequencing of all products confirmed that each primer pair amplified the correct sequences. The mGluR1 splice variants and mGluR5 splice variants are detected with single primer pairs. Markers (M) are 1-kb DNA size ladders. B: PCR amplification of genomic DNA templates. Amplification of the intronless OMP gene yielded a reaction product with the same molecular size of RNA-derived products. GFAP primers were designed to span intron V, thus yielding a larger 366-bp product if genomic contamination is present in samples. C: Representative control reactions in isolated 25-cell mitral cell pools. cDNA was amplified for 50 PCR cycles with GFAP primers in the presence (RT+) or absence (RT−) of the enzyme reverse transcriptase. No product is detectable, indicating that the mitral cell pools are free of both contaminating astrocyte cell debris and genomic DNA.
Qualitative and Semiquantitative PCR
Specific mRNAs were amplified from reverse-transcribed cDNA with Taq polymerase (Promega) in a MJ Programmable Thermal Cycler (PT200). The amplification mixture contained cDNA, 0.2–1 μM specific primers, 200 μM dNTPs, 1.5 mM MgCl2, 10 mM Tris-HCl, pH 9.0, 50 mM KCl, 0.1% Triton X-100, and 0.03 U/μl Taq polymerase in 30–50 μl volume. These mixtures were amplified for 31–45 cycles, depending on the sample and the primer pair. Amplification products were separated by agarose gel electrophoreses and the DNA bands were visualized by ethidium bromide staining of the gels (Fig. 1). Negative controls for PCR were performed using templates derived from reverse transcription reactions lacking either reverse transcriptase or total RNA and from positive controls from diluted whole bulb RNA. Signals from genomic DNA in RT-PCRs of mitral cell pools were prevented by using mGluR1 and G3PDH primers spanning at least one long intron in the corresponding genomic sequence. The GFAP primers employed in this study span a short intron, giving a 366-bp genomic fragment and a 159-bp mRNA fragment. Pools of mitral cells yielded no detectable signal for either genomic or mRNA-derived GFAP products after up to 50 PCR cycles, indicating that the mitral cell pools are free of both contaminating astrocyte cell debris and genomic DNA (Fig. 1).
Semiquantitative PCR was performed as described elsewhere (Giustetto et al., 1997; Bovetti et al., 2006). Briefly, cDNA was diluted 1:4 in water, and, for each sample, series of cDNA aliquots (2, 4, 6 μl) were amplified with Taq polymerase using specific primer pairs. After electrophoretic separation and digital image acquisition of ethidium bromide-stained gels, integrated optical intensity of individual bands was measured using the Phoretix gel analysis software (Nonlinear Inc., Durham, NC). The optical density from each band was plotted against RT volume and results from linear regression analysis calculated in the exponential phase of amplification. Results from at least three separate cDNA samples were averaged and expressed as mean ± SEM.
Western Blotting Analysis
After removal of the aqueous phase for RNA precipitation (see above), Trizol-extracted MCL and EPL samples were further processed for protein extraction according to the manufacturer’s instructions. In brief, the DNA was precipitated from the interphase with ethanol, whereas an additional precipitation with isopropyl alcohol allowed the recovery of proteins from the organic phase. The protein pellets were washed three times in a solution containing 0.3 M guanidine hydrochloride in 95% ethanol, vacuum dried, resuspended in 1% SDS, and stored at −20°C until use. Because of the small amounts of protein present in a single OB layer, a protein spot density method (Bannur et al., 1999; Mautino et al., 2004, Bovetti et al., 2006) was used to quantify proteins in extracts from single layers of the developing mouse OB. Briefly, a constant volume (1 μl) of the protein solutions was spotted on nitrocellulose paper, stained with acid Ponceau S (0.1% Ponceau S, 5% acetic acid), destained, and air dried. The intensity of the color in the spot was measured with the Phoretix analysis software (Nonlinear Inc.) and compared with dot blots of a protein standard. Samples (5 μg) were boiled in Laemmli buffer (2% SDS, 50 mM Tris-HCl, pH 7.4, 20% β-mercaptoethanol, 20% glycerol) and analyzed by 8% SDS-polyacrylamide gel electrophoresis (PAGE). Proteins were blotted onto Hybond membrane (Amersham Pharmacia, Piscataway, NJ) according to the manufacturer’s instructions. After blocking with 5% nonfat powdered milk in TBST buffer (20 mM Tris, 150 mM NaCl, 0.1% Tween 20, pH 7.4), filters were incubated for 1 hr at room temperature with primary polyclonal antibody (1 μg/ml) diluted in TBST against mGluR1a (Upstate Biotechnology, Lake Placid, NY) and primary monoclonal antibody diluted in TBST against β-actin (1:4,000; Sigma). Proteins were visualized with the appropriate peroxidase-coupled secondary antibodies (1:5,000 diluted in TBST) by using the enhanced chemiluminescence (ECL) detection system (Amersham Pharmacia).
RESULTS
mGluR Gene Expression Profiles in the OB
The present study exploited the highly laminar organization of the bulb to probe for the relative abundances of the mGluR1 and mGluR5 splice variants in the different OB layers. The ONL, GL, EPL, MCL, and GCL were microdissected from E18, P0, P4, P8, and adult mouse OBs (Fig. 2A, B). The EPL was isolated only from P8 and adult, because it could not be accurately microdissected at younger ages. Equivalent amounts of total RNA were used in each RT-PCR, and the expression of G3PDH, a housekeeping gene and key glycolytic enzyme, was similar across most samples (Fig. 3), consistent with approximately equivalent amounts of RNA loaded in each reaction.
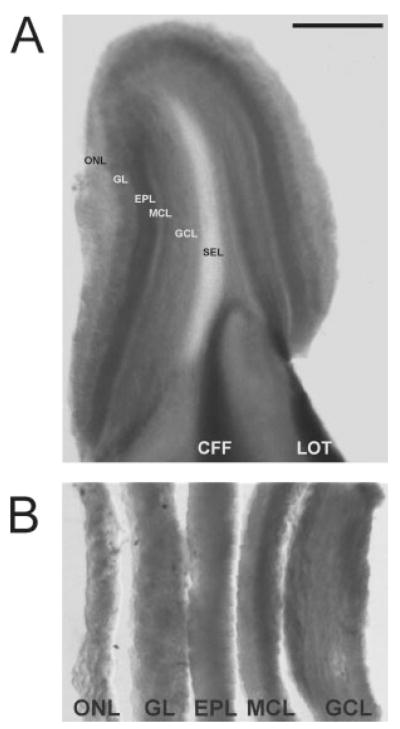
Fig. 2.
Adult olfactory bulb slice before (A) and after (B) layer microdissection. A: Each layer in the 300-μm-thick vibratome slices through the olfactory bulb is clearly visible. B: After microdissection, the different olfactory bulb layers are collected and processed for RNA extraction. ONL, olfactory nerve fiber layer; GL, glomerular layer; EPL, external plexiform layer; MCL, mitral cell layer; GCL, granule cell layer; SEL, subependymal layer. Scale bar = 1 mm in A; 200 μm in B.
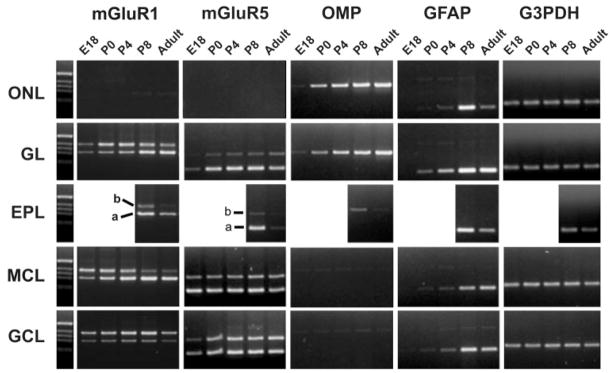
Fig. 3.
Developmental and layer mRNA expression profile for metabotropic glutamate receptor 1 (mGluR1a/b), metabotropic glutamate receptor 5 (mGluR5a/b), olfactory marker protein (OMP), glial acidic fibrillary protein (GFAP), and glyceraldehyde-3-phosphate dehydrogenase (G3PDH). RNA was extracted from OB layers of 5–10 mice at each age. Equal amounts of reverse transcription products were used in PCRs with gene-specific primers. The precision of the layer microdissection is exemplified by the presence of OMP in the ONL and GL at all ages and at only low levels in the EPL postnatally, consistent with the locations of olfactory axons. GFAP is detected first in the GL at P0 and later in the other layers, consistent with the ontology of GFAP immunohistochemistry (Bailey et al., 1999). Markers (M) are 1-kb DNA size ladders.
To assess the anatomical accuracy of the microdissections, several cell-specific markers were probed (Fig. 3). OMP protein and mRNA were present almost exclusively in ORN axons (Margolis, 1980; Buiakova et al., 1994; Vassar et al., 1994; Ressler et al., 1994; Wensley et al., 1995), and both OMP protein and mRNA are accepted markers for the presence of mature ORN axons. High levels of OMP mRNA were observed exclusively in ONL and GL layers in adult. During development, some ORN axons penetrate into the EPL (Santacana et al., 1992; Bailey et al., 1999), and, in agreement with that observation, we identified low levels of OMP mRNA in the P8 EPL. In adult, GFAP-positive astrocytes are distributed throughout most bulb layers, with the highest abundance in the GL. During development, astrocytes first differentiate in the GL at approximately P0, with only a small number present in the deeper cell layers (Bailey and Shipley, 1993; Puche and Shipley, 2001). Consistently with these observations, GFAP mRNA was first detected in GL at P0, with low levels in MCL and GCL. In adult, there was a slightly higher level of GFAP expression in the GL (Fig. 3).
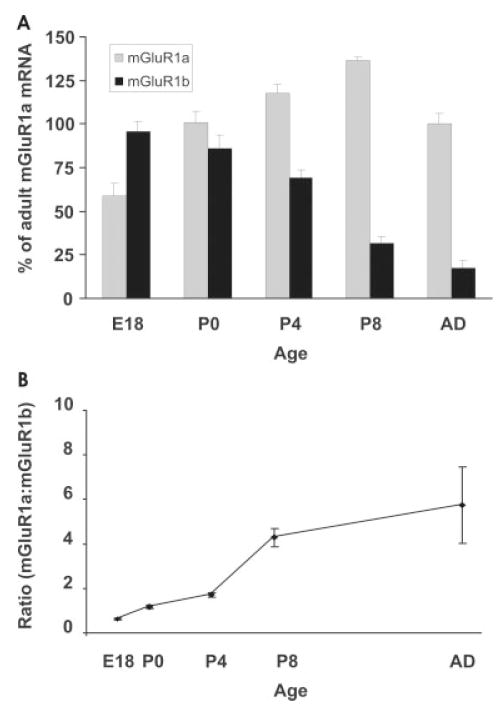
Expression of mGluR1 splice variants is highly regulated among OB layers as well as during development. In the adult bulb, mGluR1a mRNA was most abundant in the GL, EPL, and MCL, with weaker expression in the GCL, whereas mGluR1b was prominent in the GL and GCL, with very weak expression in the EPL and MCL. The developing GL showed low levels of both mGluR1a/b transcripts before birth and a marked increase in mGluR1b expression after birth, followed by a later increase in mGluR1a. In the MCL, the mGluR1 isoforms showed a prominent shift in expression pattern during development, with the 1b form almost completely replaced by 1a in the adult. We measured this temporal shift with semiquantitative PCR (Fig. 4A). Before birth, at E18, mGluR1b was the most abundant receptor isoform, constituting 62% of the total mGluR1 transcripts, with mGluR1a constituting the remaining 38% (a ratio of 0.6 mGluR1a:mGluR1b; Fig. 4B). Postnatally, the mGluR1b receptor isoform exhibited a progressive decrease that became steeper between P4 and P8, whereas mGluR1a doubled postnatally, reaching a peak at P8 and then showing a 35% decrease to adult levels. In the adult, mGluR1b constituted only 14% of the total mGluR1 mRNA (a ratio of 6.0 mGluR1a:mGluR1b; Fig. 4A, B). The temporal pattern of mGluR1a/b expression was similar to the time course of mitral cell dendritic maturation. In the GCL, mGluR1b was the predominant form of the receptor throughout development (Fig. 3).
Fig. 4.
Quantification of mGluR1a and mGluR1b transcripts in the developing mitral cell layer. A: The relative abundance of mGluR1 transcripts for each developmental stage was calculated as described in Materials and Methods and expressed as percentage vs. the adult mGluR1a value (100%). B: The ratio of mGluR1a:mGluR1b changes dramatically from 0.6 at P0 to 6.0 in adult. Each data point represents the mean value from four experiments ± SEM in both graphs.
Metabotropic glutamate receptor-5 splice variants were also differentially distributed in the bulb but did not undergo prominent shifts in splice variant expression during development (Fig. 3). Except for the ONL, mGluR5a and -b were present in all bulb layers. Interestingly, in the GL and GCL, the expression of mGluR5 increased at birth, whereas, in the MCL, it was already strongly expressed at E18. The increase in expression in the GL and GCL correlates with the postnatal increase in bulb interneurons, and we hypothesize that these cells, together with astrocytes, are the main source of mGluR5 transcripts.
mGluR1a Protein Is Differentially Compartmentalized in Adult and Developing Mitral Cells
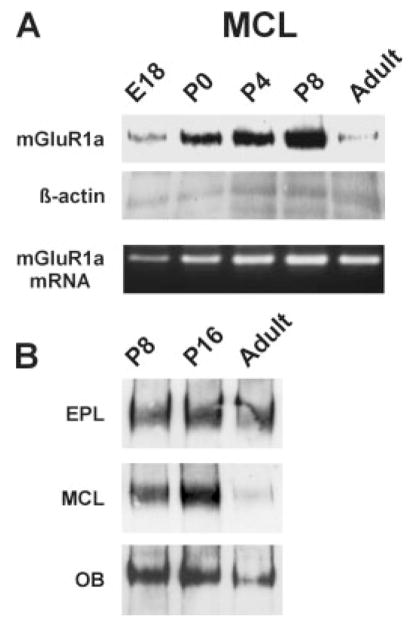
To compare expression of mGluR1a protein and mRNA, the same MCL samples were used both for mRNA and protein extraction. As shown in Figure 5A, the adult MCL exhibited a much sharper reduction in mGluR1a protein content than mRNA. This discrepancy between mRNA and protein could be interpreted as a translational down-regulation of protein synthesis. Alternatively, local dendritic synthesis or posttranslational targeting of the mGluR1a protein to dendrites in adult mitral cells could also account for this result. In this case, we would expect mGluR1a protein to be higher in the EPL, where mitral cell dendrites are located, than in the MCL. To investigate this, we isolated protein from the MCL and EPL at P8, P16, and adult. In the adult EPL, there was abundant mGluR1a protein, similar to the levels present at P8 and P16 (Fig. 5B). These findings suggest that extensive trafficking of mGluR1a mRNA and/or mGluR1a protein to dendrites may occur in the adult, whereas, in developing mitral cells, mGluR1a mRNA and protein could mostly be retained in the cell soma.
Fig. 5.
Expression of mGluR1a protein in the developing and adult olfactory bulb. A: Comparison of protein and mRNA expression in the developing mitral cell layer. The level of mGluR1a protein in the MCL is dramatically less in the adult than during development. Western blotting and RT-PCR were performed on the same samples (see Materials and Methods). B: Postnatal expression of mGluR1a protein in different layers and in the whole olfactory bulb. Expression levels in the EPL remain relatively constant, whereas the MCL decreases in the adult.
Splice Variants of mGluR1 Are Developmentally Regulated In Mitral Cells
Layer microdissections of the OB contain mixed populations of cells. For example, the MCL contains mitral cells, granule cells, astrocytes, oligodendrocytes, microglia, and some vascular elements. Indeed, cell counts suggest that the MCL contains 90% GABAergic interneurons and only about 10% mitral cells in mouse (Parrish-Aungst et al., 2007) and rat (Frazier and Brunjes, 1988). It is the large size and prominence of mitral cells, not numerical superiority, that historically yielded the layer name. Therefore, measures of gene expression in the microdissected MCL cannot be uniquely attributed to mitral cells. To overcome this problem, we visually identified and selectively harvested individual mitral cells from the OB of mice at different ages based on their expression of β-galactosidase in the GAD-LacZ-9B transgenic mouse line (Fig. 6A–C).
To confirm that β-galactosidase expression in this line of mice was restricted to mitral cells, we pressure injected the retrograde tracer cholera toxin subunit B (CTb) into the piriform cortex (Fig. 6D–F). This tracer injection labeled numerous mitral cells in the bulb but is unlikely to label all mitral cells. Double labeling for CTb and β-galactosidase showed that all CTb-labeled cell also expressed β-galactosidase (Fig. 6D–F). This indicates that most, if not all, mitral cells express β-galactosidase in this line of mice. β-Galactosidase was not observed in small soma cells, e.g., granule cells, but we cannot exclude expression in some nonmitral cells.
Immunohistochemical identification of mitral cells is incompatible with the collection of live cells; however, the fluorescent β-galactosidase substrate FDG can be used to label living cells (Fig. 6G). With this approach, acutely dissociated β-galactosidase cells from microdissected MCLs were collected for analysis of mGluR1 mRNA splice variants (Fig. 6G–I).
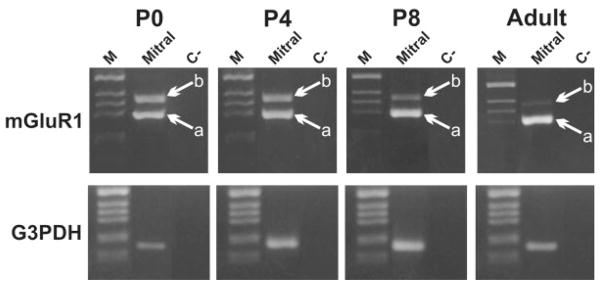
PCR amplification from single cells is often highly variable because of the small amount of mRNA in one cell. As in previous studies (Bovetti et al., 2007) we circumvented this problem by pooling 25 isolated cells. In pools of mitral cells isolated at P0 and at P4, mGluR1a and -1b were expressed at approximately the same level (Fig. 7), similar to our measurements of isolated MCLs. However, mGluR1b transcripts were down-regulated at P8, and expression was nearly undetectable in adults. This developmental regulation of mGluR1 splice variants in identified mitral cells is similar to that seen in mRNA derived from the whole MCL. Thus, the granule cells present in the MCL appear to have contributed little to the measured mGluR1 transcript expression. Alternatively, a recent report suggests that superficial granule cells may express mGluR1 (Heinbockel et al., 2007), so granule cells in the MCL might have an mGluR expression profile similar to that of mitral cells.
Fig. 7.
Representative experiment showing the developmental expression profile of mGluR1 mRNA splice variants in pools of acutely isolated mitral cells. Mitral cell pools (25 cells per pool) were labeled with FDG from the MCL of mice at different ages and were harvested under a fluorescent microscope. Pools of 25 cells were processed for RT-PCR analysis of mGluR1 splicing variants and G3PDH. Experiments were repeated in at least three or four isolated pools at each age. a, mGluR1a; b, mGluR1b.
DISCUSSION
At least two alternately spliced mGluR1 isoforms are expressed in the adult and developing OB. By using a combination of microdissected bulb layers and isolated identified cells, we show a dramatic shift in the isoform expression of mGluR1a/b in mitral cells during postnatal development. This shift temporally coincides with dramatic change in the organization of mitral cell dendrites.
Development of the OB proceeds in a sequence of cellular events involving both neurons and glia (Bailey et al., 1999; Treloar et al., 1999). At birth, all of the cellular and neuropil layers are present, but substantial changes to the neuronal circuitry occur during the first postnatal week, when mGluR1 splice variant expression changes the most. Two major cellular events occur during this early postnatal development: 1) interneuron development; the majority of juxtaglomerular and granule cell interneurons are born and integrate into the bulb during the first 14 days of postnatal development (Altman, 1969; Rosselli-Austin and Altman, 1979; Bayer, 1983); the shift in mGluR1 splice variants from mGluR1a/b to predominantly mGluR1a occurs during the first week of postnatal development; however, significant numbers of granule and juxtaglomerular cells are still migrating into the bulb after expression of mGluR1b is down-regulated; thus, changes in mGluR splice variant regulation and interneuron integration are not well correlated; and 2) dendrite development; at P0, all mitral cells have multiple apical dendrites that enter multiple glomeruli; by P8, they have a single apical dendrite that ramifies in a single glomerulus (rat, Malun and Brunjes, 1996; mouse, Lin et al., 2000); the density of synaptic contacts (Westrum, 1975; Hinds and Hinds, 1976a, b) and synaptic proteins (Bergmann et al., 1993; Berton et al., 1997) increases rapidly over the first week of postnatal development; this dendritic remodeling and synaptic protein expression correlate well with the switch in the expression of mGluR1 splice variants. Early postnatal mitral cells express equivalent levels of mGluR1a and mGluR1b at the time when mitral cells have multiple apical dendrites, but, by the time when the apical dendrites acquire an adult-like morphology, the expression of mGluR1b is down-regulated. We hypothesize that the shift to use of mGluR1a is a consequence of mitral cell dendritic maturation and that mature mitral cell synapses utilize mGluR1a rather than -1b.
The temporally restricted expression of mGluR1b in prenatal and early postnatal mitral cells suggests a developmental role for this receptor isoform. One possibility is that mGluR1b in developing neurons is involved in mediating glutamate signaling without inducing neuronal death. Excess glutamate leading to excitotoxic injury is prevalent in neurological disorders typically ascribed to ionotropic glutamate receptor activation; however, recently, mGluRs have become a target for neuroprotection (for review see Tsai and Tator, 2005). In amyotrophic lateral sclerosis (ALS), some motoneurons are thought to undergo excitotoxic injury and degenerate, whereas others are resistant. Those most vulnerable express high levels of mGluR1a, whereas the resistant motorneurons have negligible levels of mGluR1a (Laslo et al., 2001). During the first postnatal week in the OB, mitral cells may be vulnerable to excessive glutamate buildup and excitotoxic injury, because there are relatively few astrocytes, and most of the GABAergic interneurons have not yet integrated into bulb circuitry. Thus, the presence of high levels of mGluR1b in postnatal mitral cells may confer protective properties against excitotoxicity during this stage of development.
Oligonucleotide in situ hybridization for mGluR1a and mGluR1b in the adult rat olfactory bulb shows that adult mitral cells express high levels of mGluR1a, with only low levels of mGluR1b (Berthele et al., 1998). The PCR data on isolated adult mitral cells are consistent with this in situ hybridization study. Our protein measurements further suggest that, in the adult, mGluR1a protein is preferentially present in mitral cell dendrites compared with the soma. The C-terminal tail of mGluR1a contains a dominant dendritic targeting sequence (Stowell and Craig, 1999; Francesconi and Duvoisin, 2002) that could direct the protein into mitral cell dendrites. Immunohistochemical studies on adult OBs show intense mGluR1a staining in the GL (Martin et al., 1992), where mitral cell/ORN synapses occur. mGluR1a is localized to the postsynaptic side of ON to mitral cell synapses in the adult rat OB (van den Pol, 1995). Because we found high levels of mGluR1a protein in the developing MCL, we hypothesize that dendritic targeting in early postnatal mitral cells is less efficient than in the adult, consistent with other data showing differential subcellular compartmentalization of receptor proteins in mature and immature neurons (Elias et al., 2006).
The mGluR1a receptor has been implicated in adult mitral cell excitability and synaptic function. mGluR1 agonists tonically modulate adult mitral cell excitability and responsiveness to ON input (Heinbockel et al., 2004). Oscillations and synchronization of mitral cells associated with one glomerulus are triggered by mGluR1 activation (Yuan and Knöpfel, 2006). Furthermore, mGluR1 has been shown to be involved in ON-mitral cell synaptic plasticity (Ferraris et al., 1997; Casabona et al., 1998; Mutoh et al., 2005; Ennis et al., 2006). These data suggest that mGluR1 is a key modulator of mitral cell activity and that developmental shifts in mGluR1a/b isoform expression could regulate the receptor function during mitral cell maturation.
Acknowledgments
Contract grant sponsor: Maryland Stem Cell Initiative; Contract grant number: MSCRFE-0239 (to A.C.P.); Contract grant sponsor: NIDCD; Contract grant number: DC005676 (to M.T.S.); Contract grant sponsor: NIH; Contract grant number: DC03112 (to F.L.M.); Contract grant sponsor: Fondazione Cassa di Risparmio di Cuneo (to S.B.); Contract grant sponsor: University of Turin (to P.B., A.F.).
We thank Faith Scipio for genotyping and colony management of the GAD-LacZ-9B line of mice used in this study.
References
- Altman J. Autoradiographic and histological studies of postnatal neurogenesis. IV. Cell proliferation and migration in the anterior forebrain, with special reference to persisting neurogenesis in the olfactory bulb. J Comp Neurol. 1969;137:433–457. doi: 10.1002/cne.901370404. [DOI] [PubMed] [Google Scholar]
- Aroniadou-Anderjaska V, Ennis M, Shipley MT. Glomerular synaptic responses to olfactory nerve input in rat olfactory bulb slices. Neuroscience. 1997;79:425–434. doi: 10.1016/s0306-4522(96)00706-3. [DOI] [PubMed] [Google Scholar]
- Bailey MS, Shipley MT. Astrocyte subtypes in the rat olfactory bulb: morphological heterogeneity and differential laminar distribution. J Comp Neurol. 1993;328:501–526. doi: 10.1002/cne.903280405. [DOI] [PubMed] [Google Scholar]
- Bailey MS, Puche AC, Shipley MT. Development of the olfactory bulb: evidence for glia–neuron interactions in glomerular formation. J Comp Neurol. 1999;415:423–448. [PubMed] [Google Scholar]
- Bannur SV, Kulgod SV, Metkar SS, Mahajan SK, Sainis JK. Protein determination by ponceau S using digital color image analysis of protein spots on nitrocellulose membranes. Anal Biochem. 1999;267:382–389. doi: 10.1006/abio.1998.3020. [DOI] [PubMed] [Google Scholar]
- Baude A, Nusser Z, Roberts JD, Mulvihill E, McIlhinney RA, Somogyi P. The metabotropic glutamate receptor (mGluR1 alpha) is concentrated at perisynaptic membrane of neuronal subpopulations as detected by immunogold reaction. Neuron. 1993;11:771–787. doi: 10.1016/0896-6273(93)90086-7. [DOI] [PubMed] [Google Scholar]
- Bayer SA. 3H-thymidine-radiographic studies of neurogenesis in the rat olfactory bulb. Exp Brain Res. 1983;50:329–340. doi: 10.1007/BF00239197. [DOI] [PubMed] [Google Scholar]
- Bear MF, Rittenhouse CD. Molecular basis for induction of ocular dominance plasticity. J Neurobiol. 1999;41:83–91. doi: 10.1002/(sici)1097-4695(199910)41:1<83::aid-neu11>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- Bergmann M, Schuster T, Grabs D, Marqueze-Pouey B, Betz H, Traurig H, Mayerhofer A, Gratzl M. Synaptophysin and synaptoporin expression in the developing rat olfactory system. Brain Res Dev Brain Res. 1993;74:235–244. doi: 10.1016/0165-3806(93)90009-y. [DOI] [PubMed] [Google Scholar]
- Berkowicz DA, Trombley PQ, Shepherd GM. Evidence for glutamate as the olfactory receptor cell neurotransmitter. J Neurophysiol. 1994;71:2557–2561. doi: 10.1152/jn.1994.71.6.2557. [DOI] [PubMed] [Google Scholar]
- Berthele A, Laurie DJ, Platzer S, Zieglgansberger W, Tolle TR, Sommer B. Differential expression of rat and human type I metabotropic glutamate receptor splice variant messenger RNAs. Neuroscience. 1998;85:733–749. doi: 10.1016/s0306-4522(97)00670-2. [DOI] [PubMed] [Google Scholar]
- Berton F, Iborra C, Boudier JA, Seagar MJ, Marqueze B. Developmental regulation of synaptotagmin I, II, III, and IV mRNAs in the rat CNS. J Neurosci. 1997;17:1206–1216. doi: 10.1523/JNEUROSCI.17-04-01206.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bovetti S, De Marchis S, Gambarotta G, Fasolo A, Perroteau I, Puche AC, Bovolin P. Differential expression of neuregulins and their receptors in the olfactory bulb layers of the developing mouse. Brain Res. 2006;1077:37–47. doi: 10.1016/j.brainres.2006.01.034. [DOI] [PubMed] [Google Scholar]
- Bovetti S, Bovolin P, Perroteau I, Puche AC. SVZ-derived neuroblast migration to the olfactory bulb is modulated by matrix remodeling. Eur J Neurosci. 2007;25:2021–2033. doi: 10.1111/j.1460-9568.2007.05441.x. [DOI] [PubMed] [Google Scholar]
- Buiakova OI, Rama Krishna NS, Getchell TV, Margolis FL. Human and rodent OMP genes: conservation of structural and regulatory motifs and cellular localization. Genomics. 1994;20:452–462. doi: 10.1006/geno.1994.1200. [DOI] [PubMed] [Google Scholar]
- Casabona G, Knopfel T, Kuhn R, Gasparini F, Baumann P, Sortino MA, Copani A, Nicoletti F. Expression and coupling to polyphosphoinositide hydrolysis of group I metabotropic glutamate receptors in early postnatal and adult rat brain. Eur J Neurosci. 1997;9:12–17. doi: 10.1111/j.1460-9568.1997.tb01348.x. [DOI] [PubMed] [Google Scholar]
- Casabona G, Catania MV, Storto M, Ferraris N, Perroteau I, Fasolo A, Nicoletti F, Bovolin P. Deafferentation up-regulates the expression of the mGlu1a metabotropic glutamate receptor protein in the olfactory bulb. Eur J Neurosci. 1998;10:771–776. doi: 10.1046/j.1460-9568.1998.00076.x. [DOI] [PubMed] [Google Scholar]
- Catania MV, Bellomo M, Di Giorgi-Gerevini V, Seminara G, Giuffrida R, Romeo R, De Blasi A, Nicoletti F. Endogenous activation of group-I metabotropic glutamate receptors is required for differentiation and survival of cerebellar Purkinje cells. J Neurosci. 2001;21:7664–7673. doi: 10.1523/JNEUROSCI.21-19-07664.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conn PJ, Pin JP. Pharmacology and functions of metabotropic glutamate receptors. Annu Rev Pharmacol Toxicol. 1997;37:205–237. doi: 10.1146/annurev.pharmtox.37.1.205. [DOI] [PubMed] [Google Scholar]
- Elias GM, Funke L, Stein V, Grant SG, Bredt DS, Nicoll RA. Synapse-specific and developmentally regulated targeting of AMPA receptors by a family of MAGUK scaffolding proteins. Neuron. 2006;52:307–320. doi: 10.1016/j.neuron.2006.09.012. [DOI] [PubMed] [Google Scholar]
- Ennis M, Zimmer LA, Shipley MT. Olfactory nerve stimulation activates rat mitral cells via NMDA and non-NMDA receptors in vitro. Neuroreport. 1996;7:989–992. doi: 10.1097/00001756-199604100-00007. [DOI] [PubMed] [Google Scholar]
- Ennis M, Zhu M, Heinbockel T, Hayar A. Olfactory nerve-evoked, metabotropic glutamate receptor-mediated synaptic responses in rat olfactory bulb mitral cells. J Neurophysiol. 2006;95:2233–2241. doi: 10.1152/jn.01150.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferraguti F, Conquet F, Corti C, Grandes P, Kuhn R, Knopfel T. Immunohistochemical localization of the mGluR1beta metabotropic glutamate receptor in the adult rodent forebrain: evidence for a differential distribution of mGluR1 splice variants. J Comp Neurol. 1998;400:391–407. [PubMed] [Google Scholar]
- Ferraris N, Perroteau I, De Marchis S, Fasolo A, Bovolin P. Glutamatergic deafferentation of olfactory bulb modulates the expression of mGluR1a mRNA. Neuroreport. 1997;8:1949–1953. doi: 10.1097/00001756-199705260-00031. [DOI] [PubMed] [Google Scholar]
- Fotuhi M, Sharp AH, Glatt CE, Hwang PM, von Krosigk M, Snyder SH, Dawson TM. Differential localization of phosphoinositide-linked metabotropic glutamate receptor (mGluR1) and the inositol 1,4,5-trisphosphate receptor in rat brain. J Neurosci. 1993;13:2001–2012. doi: 10.1523/JNEUROSCI.13-05-02001.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francesconi A, Duvoisin RM. Alternative splicing unmasks dendritic and axonal targeting signals in metabotropic glutamate receptor 1. J Neurosci. 2002;22:2196–2205. doi: 10.1523/JNEUROSCI.22-06-02196.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazier LL, Brunjes PC. Unilateral odor deprivation: early postnatal changes in olfactory bulb cell density and number. J Comp Neurol. 1988;269:355–370. doi: 10.1002/cne.902690304. [DOI] [PubMed] [Google Scholar]
- Giustetto M, Bovolin P, Fasolo A, Bonino M, Cantino D, Sassoe-Pognetto M. Glutamate receptors in the olfactory bulb synaptic circuitry: heterogeneity and synaptic localization of N-methyl-D-aspartate receptor subunit 1 and AMPA receptor subunit 1. Neuroscience. 1997;76:787–798. doi: 10.1016/s0306-4522(96)00285-0. [DOI] [PubMed] [Google Scholar]
- Hashimoto K, Ichikawa R, Takechi H, Inoue Y, Aiba A, Mishina K, Mishina M, Hashikawa T, Konnert A, Watanabe M, Kano M. Roles of glutamate receptor delta 2 subunit (GluRdelta2) and metanotropic glutamate receptor subtype 1 (mGluR1) in climbing fiber synapse elimination during postnatal cerebellar development. J Neurosci. 2001;21:9701–9712. doi: 10.1523/JNEUROSCI.21-24-09701.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinbockel T, Heyward P, Conquet F, Ennis M. Regulation of main olfactory bulb mitral cell excitability by metabotropic glutamate receptor mGluR1. J Neurophysiol. 2004;92:3085–3096. doi: 10.1152/jn.00349.2004. [DOI] [PubMed] [Google Scholar]
- Heinbockel T, Laaris N, Ennis M. Metabotropic glutamate receptors in the main olfactory bulb drive granule cell-mediated inhibition. J Neurophysiol. 2007;97:858–870. doi: 10.1152/jn.00884.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinds JW, Hinds PL. Synapse formation in the mouse olfactory bulb. I. Quantitative studies. J Comp Neurol. 1976a;169:15–40. doi: 10.1002/cne.901690103. [DOI] [PubMed] [Google Scholar]
- Hinds JW, Hinds PL. Synapse formation in the mouse olfactory bulb. II. Morphogenesis. J Comp Neurol. 1976b;169:41–61. doi: 10.1002/cne.901690104. [DOI] [PubMed] [Google Scholar]
- Katarova Z, Mugnaini E, Sekerkova G, Mann JR, Aszodi A, Bosze Z, Greenspan R, Szabo G. Regulation of cell-type specific expression of lacZ by the 5′-flanking region of mouse GAD67 gene in the central nervous system of transgenic mice. Eur J Neurosci. 1998;10:989–999. doi: 10.1046/j.1460-9568.1998.00109.x. [DOI] [PubMed] [Google Scholar]
- Laslo P, Lipski J, Funk GD. Differential expression of group I metabotropic glutamate receptors in motoneurons at low and high risk for degeneration in ALS. Neuroreport. 2001;12:1903–1908. doi: 10.1097/00001756-200107030-00027. [DOI] [PubMed] [Google Scholar]
- Lin DM, Wang F, Lowe G, Gold GH, Axel R, Ngai J, Brunet L. Formation of precise connections in the olfactory bulb occurs in the absence of odorant-evoked neuronal activity. Neuron. 2000;26:69–80. doi: 10.1016/s0896-6273(00)81139-3. [DOI] [PubMed] [Google Scholar]
- Liu XB, Munoz A, Jones EG. Changes in subcellular localization of metabotropic glutamate receptor subtypes during postnatal development of mouse thalamus. J Comp Neurol. 1998;395:450–465. doi: 10.1002/(sici)1096-9861(19980615)395:4<450::aid-cne3>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- Malun D, Brunjes PC. Development of olfactory glomeruli: temporal and spatial interactions between olfactory receptor axons and mitral cells in opossums and rats. J Comp Neurol. 1996;368:1–16. doi: 10.1002/(SICI)1096-9861(19960422)368:1<1::AID-CNE1>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Margolis FL. A marker protein for the olfactory chemoreceptor neuron. In: Bradshaw RA, Schneider DM, editors. Proteins of the nervous system. New York: Raven; 1980. pp. 59–84. [Google Scholar]
- Martin LJ, Blackstone CD, Huganir RL, Price DL. Cellular localization of a metabotropic glutamate receptor in rat brain. Neuron. 1992;9:259–270. doi: 10.1016/0896-6273(92)90165-a. [DOI] [PubMed] [Google Scholar]
- Masu M, Tanabe Y, Tsuchida K, Shigemoto R, Nakanishi S. Sequence and expression of a metabotropic glutamate receptor. Nature. 1991;349:760–765. doi: 10.1038/349760a0. [DOI] [PubMed] [Google Scholar]
- Mautino B, Dalla Costa L, Gambarotta G, Perroteau I, Fasolo A, Dati C. Bioactive recombinant neuregulin-1, -2, and -3 expressed in Escherichia coli. Protein Expr Purif. 2004;35:25–31. doi: 10.1016/j.pep.2003.12.012. [DOI] [PubMed] [Google Scholar]
- Minakami R, Iida K, Hirakawa N, Sugiyama H. The expression of two splice variants of metabotropic glutamate receptor subtype 5 in the rat brain and neuronal cells during development. J Neurochem. 1995;65:1536–1542. doi: 10.1046/j.1471-4159.1995.65041536.x. [DOI] [PubMed] [Google Scholar]
- Munoz A, Liu XB, Jones EG. Development of metabotropic glutamate receptors from trigeminal nuclei to barrel cortex in postnatal mouse. J Comp Neurol. 1999;409:549–566. [PubMed] [Google Scholar]
- Mutoh H, Yuan Q, Knöpfel T. Long-term depression at olfactory nerve synapses. Neuroscience. 2005;25:4252–4259. doi: 10.1523/JNEUROSCI.4721-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicoletti F, Iadarola MJ, Wroblewski JT, Costa E. Excitatory amino acid recognition sites coupled with inositol phospholipid metabolism: developmental changes and interaction with alpha 1-adrenoceptors. Proc Natl Acad Sci U S A. 1986;83:1931–1935. doi: 10.1073/pnas.83.6.1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish-Aungst S, Shipley MT, Erdelyi F, Szabo G, Puche AC. Quantitative analysis of neuronal diversity in the mouse olfactory bulb. J Comp Neurol. 2007;501:825–836. doi: 10.1002/cne.21205. [DOI] [PubMed] [Google Scholar]
- Petralia RS, Wang YX, Singh S, Wu C, Shi L, Wei J, Wenthold RJ. A monoclonal antibody shows discrete cellular and subcellular localizations of mGluR1 alpha metabotropic glutamate receptors. J Chem Neuroanat. 1997;13:77–93. doi: 10.1016/s0891-0618(97)00023-9. [DOI] [PubMed] [Google Scholar]
- Pin JP, Duvoisin R. The metabotropic glutamate receptors: structure and functions. Neuropharmacology. 1995;34:1–26. doi: 10.1016/0028-3908(94)00129-g. [DOI] [PubMed] [Google Scholar]
- Puche AC, Shipley MT. Odor-induced, activity-dependent transneuronal gene induction in vitro: mediation by NMDA receptors. J Neurosci. 1999;19:1359–1370. doi: 10.1523/JNEUROSCI.19-04-01359.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puche AC, Shipley MT. Radial glia development in the mouse olfactory bulb. J Comp Neurol. 2001;434:1–12. doi: 10.1002/cne.1160. [DOI] [PubMed] [Google Scholar]
- Ressler KJ, Sullivan SL, Buck LB. Information coding in the olfactory system: evidence for a stereotyped and highly organized epitope map in the olfactory bulb. Cell. 1994;79:1245–1255. doi: 10.1016/0092-8674(94)90015-9. [DOI] [PubMed] [Google Scholar]
- Rosselli-Austin L, Altman J. The postnatal development of the main olfactory bulb of the rat. J Dev Physiol. 1979;1:295–313. [PubMed] [Google Scholar]
- Sahara Y, Kubota T, Ichikawa M. Cellular localization of metabotropic glutamate receptors mGluR1, 2/3, 5 and 7 in the main and accessory olfactory bulb of the rat. Neurosci Lett. 2001;312:59–62. doi: 10.1016/s0304-3940(01)02184-x. [DOI] [PubMed] [Google Scholar]
- Santacana M, Heredia M, Valverde F. Transient pattern of exuberant projections of olfactory axons during development in the rat. Brain Res Dev Brain Res. 1992;70:213–222. doi: 10.1016/0165-3806(92)90200-g. [DOI] [PubMed] [Google Scholar]
- Sassoe-Pognetto M, Cantino D, Panzanelli P, Verdun di Cantogno L, Giustetto M, Margolis FL, De Biasi S, Fasolo A. Presynaptic colocalization of carnosine and glutamate in olfactory neurones. Neuroreport. 1993;5:7–10. doi: 10.1097/00001756-199310000-00001. [DOI] [PubMed] [Google Scholar]
- Sekerkova G, Katarova Z, Joo F, Wolff JR, Prodan S, Szabo G. Visualization of beta-galactosidase by enzyme and immunohistochemistry in the olfactory bulb of transgenic mice carrying the LacZ transgene. J Histochem Cytochem. 1997a;45:1147–1155. doi: 10.1177/002215549704500812. [DOI] [PubMed] [Google Scholar]
- Sekerkova G, Katarova Z, Mugnaini E, Joo F, Wolff JR, Prodan S, Szabo G. Beta-galactosidase-labelled relay neurons of homotopic olfactory bulb transplants establish proper afferent and efferent synaptic connections with host neurons. Neuroscience. 1997b;80:973–979. doi: 10.1016/s0306-4522(97)00250-9. [DOI] [PubMed] [Google Scholar]
- Shigemoto R, Nakanishi S, Mizuno N. Distribution of the mRNA for a metabotropic glutamate receptor (mGluR1) in the central nervous system: an in situ hybridization study in adult and developing rat. J Comp Neurol. 1992;322:121–135. doi: 10.1002/cne.903220110. [DOI] [PubMed] [Google Scholar]
- Stowell JN, Craig AM. Axon/dendrite targeting of metabotropic glutamate receptors by their cytoplasmic carboxy-terminal domains. Neuron. 1999;22:525–536. doi: 10.1016/s0896-6273(00)80707-2. [DOI] [PubMed] [Google Scholar]
- Treloar HB, Purcell AL, Greer CA. Glomerular formation in the developing rat olfactory bulb. J Comp Neurol. 1999;413:289–304. [PubMed] [Google Scholar]
- Tsai EC, Tator CH. Neuroprotection and regeneration strategies for spinal cord repair. Curr Pharm Des. 2005;11:1211–1222. doi: 10.2174/1381612053507404. [DOI] [PubMed] [Google Scholar]
- van den Pol AN. Presynaptic metabotropic glutamate receptors in adult and developing neurons: autoexcitation in the olfactory bulb. J Comp Neurol. 1995;359:253–271. doi: 10.1002/cne.903590206. [DOI] [PubMed] [Google Scholar]
- Vassar R, Chao SK, Sitcheran R, Nunez JM, Vosshall LB, Axel R. Topographic organization of sensory projections to the olfactory bulb. Cell. 1994;79:981–991. doi: 10.1016/0092-8674(94)90029-9. [DOI] [PubMed] [Google Scholar]
- Wensley CH, Stone DM, Baker H, Kauer JS, Margolis FL, Chikaraishi DM. Olfactory marker protein mRNA is found in axons of olfactory receptor neurons. J Neurosci. 1995;15:4827–4837. doi: 10.1523/JNEUROSCI.15-07-04827.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westrum LE. Electron microscopy of synaptic structures in olfactory cortex of early postnatal rats. J Neurocytol. 1975;4:713–732. doi: 10.1007/BF01181632. [DOI] [PubMed] [Google Scholar]
- Yuan Q, Knöpfel T. Olfactory nerve stimulation-evoked mGluR1 slow potentials, oscillations, and calcium signaling in mouse olfactory bulb mitral cells. J Neurophysiol. 2006;95:3097–3104. doi: 10.1152/jn.00001.2006. [DOI] [PubMed] [Google Scholar]
- Zhu H, Ryan K, Chen S. Cloning of novel splice variants of mouse mGluR1. Brain Res Mol Brain Res. 1999;73:93–103. doi: 10.1016/s0169-328x(99)00239-9. [DOI] [PubMed] [Google Scholar]