Abstract
SUMMARY
K-Ras mutations occur frequently in epithelial cancers. Using shRNAs to deplete K-Ras in lung and pancreatic cancer cell lines harboring K-Ras mutations, two classes were identified—lines that do or do not require K-Ras to maintain viability. Comparing these two classes of cancer cells revealed a gene expression signature in K-Ras-dependent cells, associated with a well-differentiated epithelial phenotype, which was also seen in primary tumors. Several of these genes encode pharmacologically tractable proteins, such as Syk and Ron kinases and integrin beta6, depletion of which induces epithelial-mesenchymal transformation (EMT) and apoptosis specifically in K-Ras-dependent cells. These findings indicate that epithelial differentiation and tumor cell viability are associated, and that EMT regulators in “K-Ras-addicted” cancers represent candidate therapeutic targets.
SIGNIFICANCE
K-Ras is the most frequently mutated oncogene in solid tumors and when aberrantly activated, is a potent tumor initiator. However, the identification of the critical effectors of K-Ras-mediated tumorigenesis and the development of clinically effective therapeutic strategies in this setting remain challenging. We have found that cancer cell lines harboring K-Ras mutations can be broadly classified into K-Ras-dependent and K-Ras-independent groups. By establishing a gene expression signature that can distinguish these two groups, we identified genes that are specifically up-regulated in K-Ras-dependent cells and are required for their viability. Therefore, the K-Ras dependency signature has revealed several potential therapeutic targets in a subset of otherwise pharmacologically intractable human cancers.
INTRODUCTION
K-Ras is mutationally activated in approximately 20 percent of all solid tumors. However, the development of clinically effective K-Ras-directed cancer therapies has been largely unsuccessful and K-Ras mutant cancers remain among the most refractory to available treatments (Beer et al., 2002; Cox and Der, 2002; Grutzmann et al., 2004; Iacobuzio-Donahue et al., 2003; Logsdon et al., 2003; Olejniczak et al., 2007; Stearman et al., 2005; Wagner et al., 2007). K-Ras mutations occur most frequently in adenocarcinomas of the lung, pancreas and colon and mutational activation of K-Ras in these tissues is sufficient to initiate neoplasia in mice (Aguirre et al., 2003; Haigis et al., 2008; Johnson et al., 2001).
The role of oncogenic K-Ras in later stages of neoplastic progression following initiation is still poorly understood. Oncogene “addiction” is a phenomenon whereby tumors require the sustained expression and activity of a single aberrantly activated gene, despite the accumulation of multiple oncogenic lesions (Weinstein, 2002). Clinically, this is seen in BCR-ABL-expressing chronic myelogenous leukemias (CML) and EGFR mutant non-small cell lung cancer (NSCLC) (Sharma et al., 2007). Such patients, when treated with inhibitors of these activated kinases can experience impressive clinical responses, suggesting that these cancers are addicted to or dependent on single oncogenically-activated proteins. Such findings have prompted widespread efforts to develop additional “rationally-targeted” therapeutics for a variety of malignancies, potentially exploiting other settings in which oncogene addiction is involved. However, efforts to develop Ras-directed molecular therapeutics are challenged by the difficulty in selectively targeting the Ras GTPase with a small molecule. Moreover, numerous identified downstream K-Ras effectors may contribute to its role in oncogenesis (Repasky et al., 2004). Consequently, there remains a pressing need to identify pharmacologically tractable components of K-Ras driven tumorigenesis.
The goal of this study was to stratify a large panel of human cancer cell lines harboring mutant K-Ras on the basis of their requirement for sustained K-Ras function in maintaining viability and to define features of these cells that relate to their K-Ras dependency. This analysis was expected to establish phenotypic characteristics of “K-Ras addiction”, potentially revealing therapeutic targets for these largely treatment-refractory cancers.
RESULTS
An RNAi-based assay to quantify K-Ras dependency in human tumor cells
We used RNAi to determine the effects of K-Ras depletion in a panel of human tumor-derived cell lines. We first identified three different K-Ras-directed shRNA sequences that produce varying degrees of knockdown of K-Ras protein expression. The effects of K-Ras ablation on cell proliferation and viability were initially assessed in two lung cancer-derived cell lines, A549 and H358, which harbor homozygous G12S and heterozygous G12C activating K-Ras mutations, respectively. Upon K-Ras ablation, the growth of A549 cells was not significantly diminished, whereas H358 cell growth was markedly decreased (Fig. 1A). K-Ras expression was substantially reduced following shRNA expression in both cell lines and growth suppression was correlated with the level of K-Ras protein knockdown in H358 cells (Fig. 1B). Reduced K-Ras expression in A549 cells did not detectably suppress activation of the downstream Ras effectors, Akt and Erk, whereas in H358 cells, K-Ras levels were well correlated with Erk and Akt activation (Fig. 1B).
Figure 1. Differential K-Ras dependency in human cancer cell lines harboring oncogenic K-Ras.
(A) Cell growth assays following lentiviral shRNA-mediated K-Ras ablation using three different K-Ras-directed RNAi sequences (A,B,C) in A549 and H358 lung cancer cell lines. Relative cell densities were determined six days post-infection with the indicated lentiviral shRNAs and pruomycin selection. Error bars represent the standard error of the mean of three independent experiments. Lower panel corresponds to representative 96-well plates in which cell growth was measured in triplicate lentivirus-infected wells.
(B) Western blot analysis of lysates from A549 and H358 cells following RNAi-mediated K-Ras knockdown showing effects on K-Ras expression, activation status of ERK and AKT, and cleaved Caspase-3 (Casp-3). p=phospho; t=total.
(C) Rescue of K-Ras ablation-induced cell death in H358 cells. Stable lentivirally-transduced cells expressing GFP or HA-K-Ras(12V) were infected with either control, K-RasB or K-RasC shRNA-expressing lentiviruses. The K-RasB shRNA is able to knock down expression of endogenous and exogenous K-Ras, whereas K-RasC only affects endogenous K-Ras. Data are presented as the mean of two independent experiments plus standard error.
(D) Rescue of endogenous K-Ras ablation-induced cell death as assessed by both PARP and Caspase-3 cleavage by western blotting. GAPDH provides a protein loading control. Data are representative of two independent experiments.
To exclude RNAi-associated off-target effects, a “rescue” analysis was performed in which exogenous HA-tagged K-Ras(12V) was expressed in the K-Ras-dependent H358 cell line. K-Ras protein expression was ablated using two shRNAs (K-RasB and K-RasC) (Fig. 1C). The K-RasB shRNA ablates expression of both endogenous and exogenously introduced K-Ras, whereas K-RasC specifically ablates endogenous K-Ras. Exogenous K-Ras(12V) expressing cells are significantly growth inhibited by the K-RasB shRNA but are much less affected by the K-RasC shRNA (Fig. 1C). Moreover, exogenous expression of K-Ras(12V) is sufficient to prevent Caspase-3 and PARP cleavage upon ablation of endogenous K-Ras, indicating a block in apoptosis (Fig. 1D). In summary, the growth inhibitory and apoptotic effects seen upon RNAi-mediated K-Ras ablation reflect a specific requirement for K-Ras.
K-Ras dependency varies widely in K-Ras mutant cancer cell lines
To assess K-Ras dependency across a larger panel of K-Ras mutant human cancer cell lines, we established a “Ras Dependency Index” (RDI) to quantify K-Ras-dependency for individual cell lines (see Experimental Procedures). By this analysis, the greater the RDI for a given cell line, the more K-Ras-dependent the line is. RDI values for a panel of lung and pancreatic adenocarcinoma cell lines vary significantly, with two broad groups emerging, those with relatively high RDI values and those with relatively low values (Fig. 2A).
Figure 2. K-Ras dependency and Caspase-3 induction in K-Ras mutant lung and pancreas adenocarcinoma cell lines.
(A) Ras Dependency Indices (RDIs) were calculated as the inverse of the average Relative Cell Densities following K-Ras ablation with the B and C K-Ras shRNAs. RDI values are shown for a panel of lung and pancreatic cancer cell lines and plotted in order of increasing values. Data are presented as the mean and are representative of two or three independent experiments for each cell line. Error bars represent standard error. Assays were performed as described in Figure 1A.
(B) Western blot analysis showing Caspase-3 cleavage following K-Ras ablation in cell lines shown in Figure 2A. Total ERK is a loading control. Data are representative of two independent experiments for each cell line. Asterisks denote cell lines classified as being K-Ras-dependent.
(C) Quantitation of relative Caspase-3 cleavage following K-Ras ablation with the K-RasC shRNA as assessed by densitometric analysis of cleaved Caspase-3 from blots in Figure 2B. Data are shown as values normalized to cleaved Caspase-3 band density from control shRNA treated cells and presented as the mean of two densitometric readings. Error bars represent standard error.
For further analysis, we chose K-Ras ablation-induced apoptosis as a strict operational definition of “K-Ras addiction” in order to categorize cell lines more rigorously. Notably, previous studies have shown that ablation of mutant K-Ras by RNAi can affect proliferation of some pancreatic cancer cells, but does not necessarily result in apoptotic cell death (Baines et al., 2006; Fleming et al., 2005). Apoptotic responses to K-Ras ablation were assessed by measuring cleaved Caspase-3, which has been well correlated with a commitment to programmed cell death (Figs. 2B,C). Generally, induction of Caspase-3 cleavage upon K-Ras ablation was specifically seen in cell lines demonstrating RDIs greater than 2.0. We therefore defined this value of 2.0 as the “Dependency Threshold” for subsequent analyses. Thus, overall, the RDI value appears to reflect K-Ras dependency as assessed by Caspase-3 cleavage following K-Ras depletion, and RDI and relative Caspase-3 cleavage values were well correlated for each respective cell line (Fig. S1 – two tailed T-Test; p=0.0008). Based on the experimentally derived RDI values, cell lines were classified as being either K-Ras-dependent (denoted by asterisks in Fig. 2B and bold type in Fig. 2C) or K-Ras independent. PARP cleavage provided a second indicator of apoptosis and was similarly induced upon K-Ras ablation in K-Ras-dependent cell lines but not in K-Ras-independent cell lines (Fig. S2).
In SK-LU-1 and PANC-1 cells, we observed modest Caspase-3 cleavage following K-Ras knockdown using the K-RasA shRNA, but not with B or C, despite K-RasA yielding the weakest efficiency in depleting K-Ras among the three we used. We therefore classified these two cell lines as K-Ras-independent as the effect was probably due to knockdown of unintended targets by this shRNA. Knockdown of unintended targets can be a drawback of the RNAi approach and can complicate the interpretation of results from a small number of samples or a single shRNA. However, our analysis, which involves a large panel of cell lines and multiple shRNAs, permitted a straightforward categorization of cells with respect to the requirement for K-Ras expression for tumor cell viability.
Molecular and cellular differences between K-Ras-dependent and K-Ras-independent cancer cells
To identify molecular features that distinguish K-Ras-dependent and K-Ras-independent cancer cell lines, we initially analyzed whole-genome SNP (single nucleotide polymorphism) array data for common genomic alterations. The vast majority of K-Ras-dependent cell lines exhibited focal K-Ras genomic amplification. Although two of the K-Ras-independent cell lines, SK-LU-1 and H23, also demonstrated higher than diploid K-Ras copy numbers, there was a highly statistically significant correlation (r2 = 0.41; p = 1.68x10−4) between K-Ras gene copy number and K-Ras-dependency, as measured by the RDI (Fig. 3A). Moreover, K-Ras protein levels in the K-Ras-dependent cells were well correlated with K-Ras gene amplification (Fig. 3B). Significantly, the two K-Ras-independent cell lines with apparent K-Ras genomic amplification did not demonstrate elevated levels of K-Ras protein. Thus, elevated K-Ras protein expression is strongly correlated with K-Ras dependency in K-Ras mutant cancer cell lines.
Figure 3. Associations between K-Ras gene copy number, K-Ras protein overexpression, K-Ras dependency, and epithelial differentiation state.
(A) Correlation between RDI values and K-Ras copy number. K-Ras gene copy numbers in a subset of the cell lines shown in Figure 2 were determined by genomic SNP array analysis using Affymetrix Chips. RDI is shown as a function of K-Ras copy number with corresponding p-value and correlation coefficient (r2).
(B) Steady-state levels of E-cadherin and K-Ras protein were analyzed by western blotting of lysates from K-Ras-dependent and K-Ras-independent lung and pancreatic cell lines. GAPDH is a gel loading control. Data are representative of two independent experiments.
(C) Expression and subcellular localization of E-cadherin and vimentin in K-Ras-independent (red text) and K-Ras-dependent (green text) cell lines, as demonstrated by fluorescence microscopy. Green=E-cadherin; red=vimentin; blue=nuclei (Hoechst). Scale bar = 15 μM.
(D) Loss of E-cadherin in H358 cells chronically treated with TGFβ, as demonstrated by fluorescence microscopy. The resulting stable mesenchymal cell line is designated H358M. Green=E-cadherin; red=vimentin. Scale bar = 15 μM.
(E) Apoptotic response to shRNA-mediated K-Ras ablation as assessed by western blotting of Caspase-3 cleavage in H358 versus H358M cells. Data are representative of two independent experiments.
(F) RDIs for H358 versus H358M cells were derived as described previously. The graph depicts mean RDIs from two independent experiments, with error bars corresponding to standard error.
Notably, analysis of the K-Ras effectors Erk and Akt failed to reveal a correlation between K-Ras dependency and the engagement of these signaling pathways (Fig. S3). Similarly, the sensitivity of two NSCLC cell lines, A549 and H358, to pharmacologic inhibition of Mek and PI-3 kinases revealed no correlation between sensitivity to these inhibitors and K-Ras dependency (Fig. S4). However, among K-Ras-independent pancreatic cancer cell lines, there was a striking hyperactivation of Akt, which was inversely related to expression of the PTEN tumor suppressor, a negative regulator of PI-3 kinase/Akt signaling (Fig. S3). Thus, PI-3 kinase activation may contribute to loss of K-Ras dependency in a context specific manner.
Epithelial differentiation state is linked to K-Ras dependency
We noted that many of the K-Ras-dependent cells exhibit a classic epithelial morphology, whereas most K-Ras-independent cells appeared less uniformly epithelial (data not shown). We therefore examined the expression of E-cadherin, a marker of differentiated epithelia in these cell lines. Strikingly, the majority of K-Ras-independent cell lines expressed little or no E-cadherin, indicative of EMT (Fig. 3B). Furthermore, although E-cadherin is detectable in some K-Ras-independent cell lines, it is mislocalized to punctate intracellular vesicles (Fig. 3C). K-Ras-dependent cell lines, in contrast, exhibit prominent cortical E-cadherin expression. Moreover, the expression of vimentin, a mesenchymal marker, is readily detected in K-Ras-independent cell lines but is largely absent in K-Ras-dependent cell lines (Fig. 3C).
To demonstrate a causal relationship between K-Ras dependency and an epithelial phenotype, we tested the possibility that induction of EMT could affect K-Ras dependency. K-Ras-dependent H358 NSCLC cells were treated with TGFβ1, a promoter of EMT, for ten days. Consistent with EMT, the majority of TGFβ1-treated cells lost E-cadherin expression and gained vimentin expression (Fig. 3D). This resultant mesenchymal cell line was designated H358M. Whereas parental H358 cells undergo Caspase-3 cleavage following K-Ras ablation, indicative of apoptosis, the mesenchymal H358M cells failed to undergo this response (Fig. 3E). Moreover, the RDI for H358M cells is significantly lower than that of H358 cells and is below the “Dependency Threshold” of 2.0 (Fig. 3F). Thus, H358M cells bear the hallmarks of K-Ras independency. H358M cells also demonstrate reduced coupling of K-Ras to downstream signaling pathways, reminiscent of many de novo K-Ras-independent cell lines (Fig. 3E). Together, these findings support a strong link between epithelial differentiation and K-Ras-dependency for cancer cell survival.
We next sought to determine whether the reverse process of MET in K-Ras-independent cells could lead to the acquisition of K-Ras dependency. We noted that Zeb1, a transcription factor that represses E-cadherin expression (Peinado et al., 2007), is expressed specifically in K-Ras-independent cell lines but not in K-Ras-dependent cells (Fig. 4A). As expected, this is inversely correlated with E-cadherin expression. Using an shRNA to deplete expression of Zeb1 in two K-Ras-independent cell lines (A549 and PANC-1), we observed that loss of Zeb1 expression results in strong upregulation of E-cadherin (Fig. 4B). Moreover, E-cadherin in these cells prominently localized at cell-cell junctions (Fig. 4C), as observed in K-Ras-dependent cell lines. Notably, the stable cell lines in which Zeb1 had been ablated appeared to be heterogeneous, with subpopulations of cells appearing to retain strong Zeb1 expression. These cells did not undergo altered E-cadherin expression or localization and were morphologically similar to the control shRNA-treated cells.
Figure 4. Mesenchymal-to-epithelial transition and K-Ras dependency.
(A) Relationships between Zeb1 and E-cadherin protein expression in a panel of K-Ras-independent and K-Ras-dependent NSCLC cell lines as analyzed by western blotting. GAPDH is a loading control.
(B) Expression of Zeb1 and E-cadherin following stable Zeb1 ablation in a polyclonal population of A549 NSCLC and PANC-1 PDAC cells.
(C) Expression and subcellular localization of E-cadherin (green) and Zeb1 (red) in control and Zeb1 stable knockdown cells (shZeb1), as assessed by immunofluoresence. Scale bar = 15 μM.
(D) Ablation of K-Ras in control versus shZeb1 expressing A549 cells, and effects on apoptosis as assessed by Caspase-3 cleavage.
(E) Fold changes in RDI values for A549 and PANC-1 shZeb1-expressing cells. Data are normalized to control shRNA-expressing cells. Data are representative of the mean of two or three independent experiments. Error bars represent standard error.
We next tested the possibility that Zeb1 ablation, which results in MET, could reverse K-Ras dependency. We initially noted that K-Ras protein expression was elevated in the Zeb1 knockdown cells (Fig. 4D), reminiscent of cells we had previously established as K-Ras-dependent. Following ablation of K-Ras in control shRNA expressing A549 cells, we observed no Caspase-3 cleavage response. However, in Zeb1-depleted A549 cells, ablation of K-Ras resulted in a cell death response (Fig. 4D). We also noted that in Zeb1-depleted A549 and PANC-1 cells, there was a marked increase in K-Ras dependency as assessed by the RDI analysis (Fig. 4E). Together, these observations strongly suggest that the epithelial differentiation state of K-Ras mutant cancer cells is associated with dependency on K-Ras to maintain cell viability.
Establishment of an oncogenic “K-Ras dependency signature”
We next attempted to identify a gene expression “signature” associated with K-Ras dependency with a focus on genes consistently up-regulated in K-Ras-dependent cells, which could potentially correspond to therapeutic targets. Using expression data derived from a subset of cell lines previously tested for K-Ras dependency (the “training set” –Fig. S5), we performed a supervised analysis of differential gene expression between K-Ras-dependent and K-Ras-independent cell lines using the Prediction Analysis of Microarrays (PAM) algorithm (Tibshirani et al., 2002). PAM employs the “nearest shrunken centroid” method to identify genes that can accurately segregate two known training classes of samples based on cross-validation. In this case, the two classes are K-Ras-dependent and K-Ras-independent cell lines.
The PAM analysis yielded a list of differentially expressed genes ranked according to average expression within each class across the panel of tested cell lines (Fig. 5A). Thus, higher-ranking genes are, on average, more highly expressed in one class versus the other. As expected, genes highly expressed in K-Ras-dependent cell lines were relatively poorly expressed in K-Ras-independent cell lines, and vice versa. We also performed a gene functional annotation analysis using the EASE algorithm (Dennis et al., 2003; Hosack et al., 2003) for term enrichment using the K-Ras dependency signature genes (Fig. S6). Notably, this analysis demonstrated statistically significant term enrichment for facets of epithelial cell biology, again reinforcing an association between K-Ras dependency and epithelial differentiation. We also noted that both CDH1, which encodes E-cadherin, and TCF8, which encodes Zeb1, were represented in the signature established by the PAM algorithm (Table S1).
Figure 5. Derivation of a K-Ras dependency signature.
(A) Listing of genes differentially expressed in K-Ras-dependent versus K-Ras-independent cancer cell lines as generated by the PAM algorithm. Bars represent “shrunken centroids”, whose lengths are proportional to average expression of each gene across all cell lines. Red bars correspond to genes expressed at higher levels in K-Ras-independent cells (IND) and green bars correspond to genes expressed at higher levels in K-Ras-dependent cells (DEP). Each instance of a gene in the signature represents a distinct probe set on the microarray platform. Thus, three different SYK probes and two RBM35A probes are differentially represented in the K-Ras-dependency signature.
(B) The K-Ras dependency signature is associated with a signature of activated Ras expression in human mammary epithelial cells. The 20 Ras dependency genes most over-expressed upon activated Ras transfection are depicted here.
(C) The Ras dependency signature is significantly associated with genes transcriptionally upregulated by activated Ras but not by other activated oncogenes, namely β-catenin, E2F3, Src and Myc.
(D) Predictions of K-Ras dependency for a representative “test set” of K-Ras mutant cell lines from various tissue types were made using the PAM algorithm. Nominal RDIs were calculated by plotting growth versus average values for relative K-Ras expression following RNAi, extrapolated from the training set of cell lines. Data are shown as the mean of two independent experiments with error bars representing standard error. Red=K-Ras-independent; green=K-Ras-dependent. Prediction assignments are shown below cell line names. The ‘Dependency Threshold’ of 2.0 established previously is shown as a dashed line. A two-tailed student T-test yielded a p-value of 0.009, demonstrating statistical significance of the predictive value of the K-Ras dependency signature.
(E) Heat map showing hierarchical clustering of K-Ras dependency signature gene expression in a cross-tissue panel of K-Ras mutant cell lines. Red and blue indicate relative over or underexpression of genes, respectively. Note the bifurcation of cell lines into two broad groups, with a third subgroup clustering to the leftmost extreme of the heatmap. For cell line names, red=K-Ras-independent lines; green=K-Ras-dependent lines; black= not tested. Tissue of origin is indicated in parentheses: U=Uterus; I=Intestine; P=Pancreas; L=Lung; Bl=Blood; Th=Thymus; S=Stomach; E=Esophagus; Li=Liver.
To further explore the biology of K-Ras dependency and to verify the relevance of the cell culture-derived K-Ras dependency signature in human tissues, we employed the Oncomine Concepts Map (Table 1). We identified a significant association with genes that are up-regulated by expression of oncogenic Ras in immortalized human mammary epithelial cells (HMEC) (Bild et al., 2006), suggesting that a large component of the K-Ras dependency signature is comprised of Ras-regulated transcriptional targets (Fig. 5B). Significantly, the K-Ras dependency signature genes are specifically associated with signatures linked to oncogenic Ras, but not to other oncogenes such as β-catenin, E2F3, Src, or Myc (Fig. 5C). Notably, the K-Ras dependency signature was also strongly associated with sensitivity to apoptosis-inducing agents APO2L/TRAIL and the BCL-2 inhibitor, ABT-737, as well as to EGFR tyrosine kinase inhibitors (Table 1).
Table 1.
K-Ras dependency signature associations. The top 148 genes from the K-Ras dependency signature were compared with other cancer-related gene signatures using the Oncomine Concepts Map (www.oncomine.com). Significant selected associations are listed here. Citations are shown in parentheses.
| Type | Concept Name | Overlap | Odds Ratio | p-Value |
|---|---|---|---|---|
| Cell Line | Non-Small Cell Lung Cancer Cell Line Gefitinib Sensitivity - Top 5% over- expressed in Sensitive (Coldren et al. 2006) | 76 | 46.55 | 4.2E-73 |
| Cell Line | Cell Line APO2L/TRAIL Sensitivity - Top 10% over-expressed in Highly Sensitive (Wagner et al., 2007) | 86 | 13.01 | 1.30E-47 |
| Cell Line | Small Cell Lung Carcinoma Cell Line ABT-737 Sensitivity - Top 10% over- expressed in Sensitive (Olejniczak et al., 2007) | 60 | 11.07 | 2.40E-31 |
| Cell Line | Human Mammary Epithelial Cells Oncogene Transfection - Top 10% over-expressed in Activated H-Ras vs. control (Bild et al., 2006) | 45 | 4 | 4.40E-12 |
| Cancer vs. Normal | Pancreas Type - Top 10% over- expressed in Pancreatic Adenocarcinoma (Iacobuzio-Donahue et al., 2003) | 43 | 5.52 | 2.60E-15 |
| Cancer vs. Normal | Pancreas Type - Top 1% over- expressed in Pancreatic Adenocarcinoma (Logsdon et al., 2003) | 7 | 24.59 | 9.20E-08 |
| Cancer vs. Normal | Pancreas Type - Top 10% over- expressed in Microdissected Pancreatic Tumor Epithelia (Grutzmann et al., 2004) | 32 | 2.66 | 8.20E-06 |
| Cancer vs. Normal | Lung Type - Top 10% over-expressed in Lung Adenocarcinoma (Beer et al., 2002) | 14 | 4.86 | 2.00E-05 |
Predictive value of the K-Ras dependency signature
To test the predictive value of the K-Ras dependency signature, we utilized the facet of the PAM algorithm that classifies unknown samples into either class based on gene expression data. Initially, to generate a signature that minimized false discovery rate (FDR) and misclassification errors, we chose a value for the so-called threshold parameter of 6.0 (Fig. S5). This yielded a list of 46 genes that could be used to segregate the two classes. Using this threshold value, the misclassification error rate for the training set was zero (Figure S5).
We then used gene expression datasets for a representative cross-tissue “test set” of 18 K-Ras mutant cancer cell lines to predict the K-Ras dependency of these cell lines (Fig. S7). To empirically validate these predictions, we performed the K-Ras growth dependency assay, using the K-RasB and C shRNAs, and derived RDI values for these cell lines as described previously (Fig. 5D). Of 18 cell lines tested, 15 were classified correctly based on our previously assigned “Dependency Threshold” of 2.0, yielding a misclassification error rate of 0.17. A two-tailed Student T-test demonstrated statistical significance of the predictive value of the K-Ras dependency signature (p=0.009). Additionally, ANOVA calculations demonstrated that classifications of K-Ras-independent and K-Ras-dependent cell lines based on RDI values are non-random (Fig. S1; p = 4.35x10−8).
To visualize the expression of the K-Ras dependency signature genes across a large panel of K-Ras mutant cancer cell lines, we broadened the signature by identifying 250 probe sets (Table S1) for differentially expressed genes. We then generated a “heat map” by performing a hierarchical clustering analysis of these probe sets across a large panel of K-Ras mutant cell lines (Fig. 5E). The resultant heat map revealed a clear bifurcation of cell lines into two broad clusters, which generally segregate into K-Ras-dependent and K-Ras-independent cell lines, with the three previously misclassified cell lines falling outside their respective cluster. A third cluster is apparent upon close inspection to the leftmost extreme of the heat map, with cell lines that seem to express genes that are associated with both K-Ras-dependency and K-Ras-independency. Together, these results indicate that a gene expression signature associated with dependency on oncogenic K-Ras can predict a state of K-Ras addiction.
Validation of K-Ras dependency genes
To validate the differential gene expression data derived from comparative microarrays, we analyzed protein or mRNA levels corresponding to a subset of genes that were ranked highly within the K-Ras dependency signature. We first analyzed the expression of differentially-expressed genes, SYK, ITGB6 and MST1R, whose protein products, the Syk tyrosine kinase, integrin beta6 subunit, and the RON receptor tyrosine kinase, respectively, correspond to pharmacologically tractable targets. Syk, integrin beta6 and RON protein levels were relatively high in both lung and pancreatic cancer K-Ras-dependent cell lines (Fig. 6A). Some K-Ras-independent cell lines expressed Syk, integrin beta6, or RON, but not all three. Notably, several of the lines used to assess Syk and integrin beta6 expression were not in the training set used to generate the K-Ras dependency signature. We also examined whether expression of Syk, integrin beta6, and Ron-β are regulated by K-Ras. Ablation of K-Ras in H358 cells using the K-RasB and C shRNAs resulted in reduced Syk and RON-β expression but did not affect integrin beta6 expression (Fig. 6B), indicating that expression of a subset of K-Ras dependency signature genes is under the transcriptional or post-transcriptional control of oncogenic K-Ras.
Figure 6. K-Ras dependency genes are required for viability and epithelial differentiation of K-Ras-dependent cancer cells.
(A) Differential expression of the Syk tyrosine kinase, integrin beta6 and the RON receptor tyrosine kinase, β subunit in lung and pancreatic cancer cell lines. Black text=K-Ras-independent; gray text=K-Ras-dependent. Syk expression in BJAB B-lymphocyte cells is a positive control. GAPDH is a gel loading control. Data are representative of two independent experiments.
(B) Expression of Syk, Integrin β6 and RON-β proteins following knockdown of K-Ras expression in H358 cells (t-ERK is a gel loading control).
(C) Cell growth assays following shRNA-mediated knockdown of three K-Ras dependency genes: SYK, ITGB6 (integrin β6), and MST1R (RON). Data are presented as means and are representative of three independent experiments. Error bars represent standard error.
(D) Knockdown of SYK, ITGB6 and MST1R in K-Ras-independent and K-Ras-dependent cells and effects on E-cadherin expression and apoptosis. Caspase-3 induction and E-cadherin expression were analyzed by western blotting, and total ERK (t-ERK) is a loading control. Data are representative of two independent experiments.
(E) Correlation between RDI values and IC50 values for R406, a Syk kinase inhibitor, in a panel of K-Ras mutant cancer cell lines. Cells were treated with 0.016 μM to 10 μM R406 for 3 days, and relative cell densities were quantified. Dark grey symbols represent K-Ras-independent lines and light grey represent K-Ras-dependent lines. Data are represented as the mean of two independent experiments. (p-value = 0.0095)
(F) Effects R406 on Syk autophosphorylation at Y525/526 (p-SYK) and effects on cell death as assessed by Caspase-3 cleavage. Total Syk (t-Syk) and total Erk (t-ERK) serve as loading controls.
We then investigated the expression of two additional genes in the signature. ANKRD22 is a gene with unknown function predicted to encode a 22 kilodalton protein with three tandem ankyrin repeat motifs. Since ANKRD22 antibodies are not available, we analyzed its mRNA level. As predicted, K-Ras-independent A549 lung and PATU8988T pancreas cells expressed little or no ANKRD22 mRNA whereas K-Ras-dependent H358 lung and YAPC pancreas cells expressed readily detectable ANKRD22 mRNA (Fig. S8). We also confirmed the elevated expression of PROM2, which encodes prominin 2, in K-RAS-dependent cell lines compared to K-RAS-independent cell lines (Fig. S8).
Together, these findings confirm that the gene expression differences identified by microarray analysis are associated with K-Ras dependency.
K-Ras dependency genes are required for epithelial differentiation and cell survival
Since the PAM algorithm ranks genes by average expression across the entire class, we hypothesized that highly ranked genes may play functional roles in the context of K-Ras dependency. Using shRNAs to ablate the expression of MST1R, ITGB6, and SYK genes, we observed clear differential growth inhibitory effects on K-Ras-dependent cell lines versus K-Ras-independent cell lines (Fig. 6C). Moreover, depletion of ITGB6 or SYK resulted in loss of E-cadherin expression, indicative of EMT, and Caspase-3-associated cell death in K-Ras-dependent H358 NSCLC cells and YAPC PDAC cells, but not in K-Ras-independent A459 and SW1990 cells (Fig. 6D). Knockdown of MST1R resulted in EMT and cell death in H358 cells but not in the YAPC pancreatic cancer cell line, suggesting that RON may play context specific roles in the setting of K-Ras dependency. Ablation of ANKRD22 similarly resulted in reduced E-cadherin levels with concomitant apoptosis in K-Ras-dependent cells (Fig. S8). In contrast, knock down of SMAD4 expression, a gene not represented in the K-Ras dependency signature, in H358 cells did not affect E-cadherin expression or induce apoptosis (Fig. S8).
The function of Syk as a protein kinase makes it an attractive potential therapeutic target. We therefore tested the efficacy of a pharmacologic Syk kinase inhibitor, R406, which is undergoing clinical testing in rheumatoid arthritis and B-cell lymphoma (Braselmann et al., 2006). IC50 values for growth inhibition by R406 were measured in a panel of K-Ras-dependent and K-Ras-independent cell lines. We found a statistically significant difference (P-value = 0.0095) between the IC50 values for growth inhibition by R406 and the RDI values for each respective cell line tested (Fig. 6E). Thus, overall, K-Ras-dependent cell lines demonstrated substantially greater sensitivity to pharmacologic Syk inhibition than K-Ras-independent cell lines.
To elucidate potential mechanisms underlying this differential drug sensitivity, we analyzed the signaling consequences of Syk inhibition in several cell lines. PA-TU-8988T cells did not display any basal Syk autophosphorylation. In A549 K-Ras-independent cells as well as H358 and YAPC K-Ras-dependent cells, there was an R406 dose-dependent inhibition of Syk autophosphorylation on Y525/526 (Fig. 6F). However, inhibition of Syk was accompanied by a dose-dependent induction of Caspase-3 cleavage only in H358 and YAPC cells but not in A549 and PA-TU-8988T cells. Therefore, Syk plays an anti-apoptotic role specifically in the setting of K-Ras dependency. These findings implicate several of the genes within the K-Ras dependency signature in determining the epithelial character of cancer cells, and suggest that some of these may constitute therapeutic targets in a subset of K-Ras mutant tumors.
Expression of K-Ras dependency signature genes in lung and pancreatic tumors
We next attempted to analyze the expression profile of K-Ras dependency signature genes in primary lung tumors of sqaumous carcinoma and adenocarcinoma subtypes, using publicly available gene expression data (Bild et al., 2006). We assigned a ‘Ras Dependency Score’ based on the average expression of the signature genes on a per sample basis. We first categorized the samples based on K-Ras mutational status and histological grading (Fig. 7A). The Ras dependency score was highest in tumor samples that harbored K-Ras mutations. Of these K-Ras mutant cancers, all had been classified as well-to-moderately differentiated. Conversely, K-Ras mutant tumors classified as poorly-differentiated exhibited relatively low Ras dependency scores. Thus, expression of the Ras dependency signature genes is found predominantly in K-Ras mutant tumors classified as well-to-moderately differentiated, consistent with the cell culture findings, demonstrating that the signature is associated with epithelial differentiation state.
Figure 7. Expression of K-Ras dependency genes is associated with a well differentiated tumor phenotype.
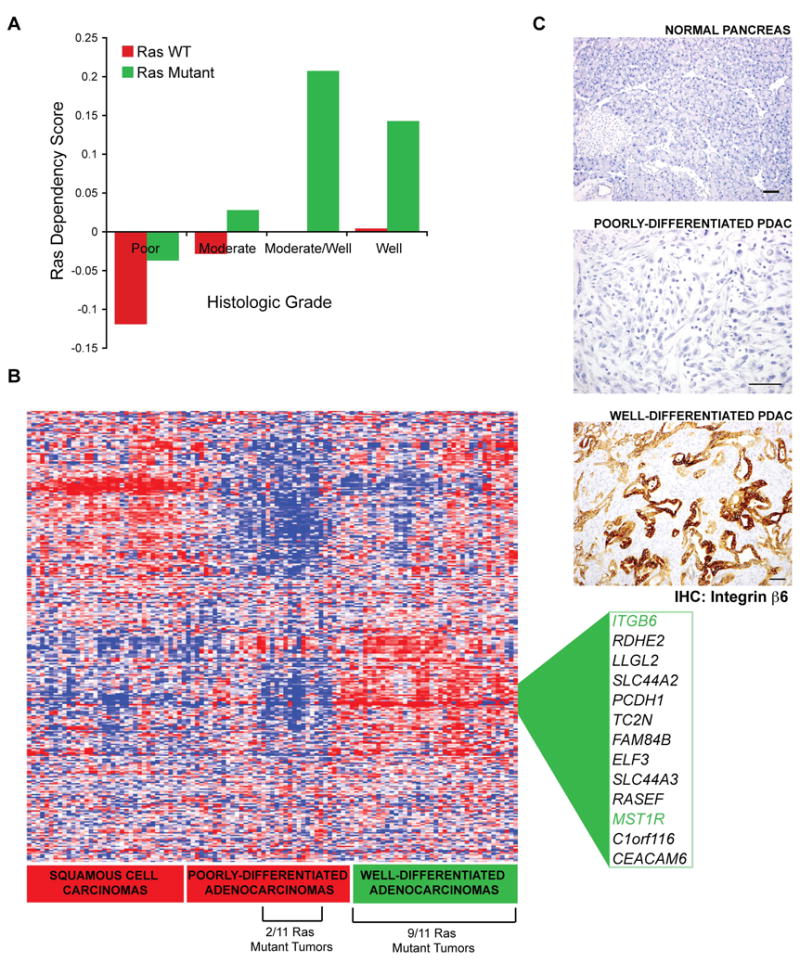
(A) The Ras Dependency Score, a measure of average median centered gene expression values of the top ranking K-Ras dependency signature genes, is shown as a function of histologic grading and K-Ras mutation status in a panel of human lung cancer specimens. Green bars=K-Ras mutant tumors, and red bars=tumors with wild-type K-Ras (WT).
(B) Hierarchical clustering analysis of gene expression profiles of K-Ras dependency signature genes from the same panel of human lung cancer specimens as in Figure 7A. Two major clusters of overexpressed genes show enrichment for either well-differentiated adenocarcinomas or squamous cell carcinomas, with 9 of 11 K-Ras mutant tumors falling within the well-differentiated adenocarcinoma cluster. Genes differentially expressed in well-differentiated adenocarcinomas are listed and ITGB6 and MST1R, two characterized genes, are highlighted in green.
(C) Expression of integrin β6, as assessed by immunohistochemistry in normal mouse pancreatic tissue versus mutant K-Ras driven pancreatic cancers (PDACs), that were classified as either poorly or well-differentiated, based on glandular ductal morphology. Sections were co-stained with hematoxylin. The field shown is representative of 5 high-power fields from independent mouse tumors. The upper and lower panels are shown at 125X magnification, and the middle panel is shown at 250X. Scale bars = 50 μM.
We then performed hierarchical clustering analyses of gene expression datasets from human primary lung adenocarcinomas and squamous cell carcinomas represented in Figure 7A, using the top 325 genes from the PAM analysis that were differentially expressed in K-Ras-dependent cell lines (Fig. 7B). Strikingly, we observed two distinct clusters in the resulting heat map. The first cluster, which comprises a distinct subset of dependency signature genes, was comprised almost entirely of squamous cell lung cancers. Conversely, the second cluster was comprised of tumor samples that were predominantly classified as well differentiated or well-to-moderately differentiated adenocarcinomas (Fig. 7B). This cluster contained two of the genes we had characterized previously as potential therapeutic targets, ITGB6 and MST1R. The absence of SYK from this cluster may be explained by the fact that it is highly expressed in endothelial and hematopoietic cells, which are typically present in “contaminating” stromal tissue within tumor specimens. Integrin beta6 and RON are not expressed in B-lymphocyte cells, whereas Syk is expressed strongly (Fig. 6A). Thus, differential SYK expression may not be apparent in tumor samples. Significantly, 9 of 11 K-Ras mutant samples fell into the well-differentiated tumor cluster, whereas 2 of 11 fell outside the cluster, suggesting that a subset of K-Ras mutant cancers do not express the K-Ras dependency signature, consistent with the cell line findings.
We also analyzed the expression of integrin beta6 in a K-Ras-driven mouse pancreatic cancer model (Aguirre et al., 2003) that exhibits tumors with varying degrees of differentiated ductal morphology. Staining was scored blindly on a 0–3 scale and the proportion of positive cells in at least 5 high power fields was determined. Normal pancreatic acinar cells did not stain (score 0) and normal ductal cells showed weak staining (score 1). Analysis of a series of 31 tumors revealed that strongly positive staining (score 2 or 3) was present in ductal (differentiated) elements (75.3 % +/− 20.6% of cells showed positive staining) whereas only weak or absent staining (score 0 or 1) was observed in poorly-differentiated anaplastic or sarcomatoid elements (Fig. 7C). These findings further support a relationship between expression of K-Ras dependency genes and epithelial differentiation in tumors.
DISCUSSION
By examining addiction to oncogenic K-Ras in a quantitative manner, we were able to classify K-Ras mutant cancer cells into two groups based on K-Ras dependency for cell viability. This classification yielded a gene expression signature that allows for the accurate prediction of K-Ras dependency across tissue types. As a predictive tool, the K-Ras dependency signature did yield a low misclassification error rate. This could reflect, in part, a bias in the training set of cell lines, which consisted predominantly of lung adenocarcinoma-derived cell lines. Therefore, a K-Ras dependency signature that includes a larger number of lines representing additional tumor types could serve to further refine a more broadly applicable K-Ras dependency signature for predictive purposes.
This expression signature is significantly associated with gene expression profiles from K-Ras human mutant tumor samples classified as well differentiated. Thus, the in vitro derived signature, which is associated with epithelial differentiation, is also associated with the differentiation state of tumors in vivo. Upon further refinement of the signature, expression of subsets of K-Ras dependency signature genes may prove useful as biomarkers for the treatment of cancers with specific molecular profiles. We have shown that ITGB6, in particular, is strongly associated with a well-differentiated K-Ras driven cancer phenotype, and efforts to target the activity of integrin beta6 are currently underway. Syk and RON, two kinases that our findings have also implicated as potential therapeutic targets, have previously established roles in cancer (Lu et al., 2007; Sada et al., 2001). Thus, comparing gene expression profiles between cancer cell lines based on oncogene dependency provides a strategy for context-specific drug target discovery.
We also document a correlation between K-Ras addiction and K-Ras genomic amplification. K-Ras amplification is observed in lung and pancreatic tumor specimens (Aguirre et al., 2004; Weir et al., 2007), and may provide a useful biomarker of response to therapeutics that target K-Ras-addicted human cancers. Interestingly, oncogene addiction in other settings is also associated with genomic amplification, most notably in the cases of MYC in many cancer types, EGFR in gliomas and lung cancers, MET in gastric and lung cancers, and HER2 in breast cancers (Collins and Groudine, 1982; Houldsworth et al., 1990; Tal et al., 1988; Wong et al., 1987).
We have established that K-Ras dependency is strongly linked to epithelial differentiation status. Upon EMT, K-Ras dependency is reduced, and conversely, by MET, K-Ras dependency is gained. We have assessed the mutational status of well-established tumor suppressor genes and oncogenes, other than K-Ras, and found no clear associations with K-Ras dependency (Figure S9). Therefore, the mechanism underlying loss or gain of K-Ras dependency is potentially epigenetic in nature, which may be affirmed by recent analysis of EMT and epithelial plasticity as epigenetic phenomena (Dumont et al., 2008).
The observed relationship between K-Ras addiction, TGFβ signaling, and epithelial differentiation is particularly interesting in the context of pancreatic adenocarcinomas, which undergo frequent homozygous deletion of the TGFβ signaling component Smad4 (~50 % of cases) (Hezel et al., 2006). Deletion of Smad4 in cooperation with K-Ras mutational activation accelerates tumor progression in a mouse model of pancreatic cancer (Bardeesy et al., 2006). TGFβ induces both EMT and growth arrest or apoptosis in a subset of cancers, and Smad4 appears to be required. Loss of Smad4 results in a well-differentiated tumor histopathology, suggesting disruption of TGFβ-driven EMT. Smad4 loss or loss of TGFβ response in general and K-Ras addiction in pancreatic adenocarcinomas may be associated, as we have observed that differentiated epithelial-like cancer cells remain K-Ras-dependent. Thus, K-Ras genomic amplification and Smad4 deletion may correspond to important biomarkers of responsiveness to K-Ras-directed therapeutics.
The described findings raise the possibility that associations between oncogene dependency and epithelial differentiation may extend beyond K-Ras addiction. Indeed, it was recently reported that FGFR addiction in a mouse model of prostate cancer is irreversible when tumors have undergone EMT (Acevedo et al., 2007). This is reminiscent of the observed K-Ras independency in mesenchymal K-Ras mutant cell lines. We also found a significant association between the K-Ras dependency signature and a gene expression profile of sensitivity to EGFR inhibitors in NSCLC (Table 1) (Coldren et al., 2006). Like K-Ras, EGFR is mutated and amplified in NSCLC, contributing to sensitivity to EGFR kinase inhibitors. Moreover, insensitivity to EGFR inhibitors in lung and liver cancers has been associated with EMT, further supporting a link between EMT and loss of oncogene addiction (Fuchs et al., 2008; Thomson et al., 2005). The notion that poorly-differentiated tumors are generally more drug resistant and are associated with poorer prognosis has been widely recognized in clinical oncology (Shah and Gallick, 2007), and our findings may provide some mechanistic insight into this observation.
EXPERIMENTAL PROCEDURES
Lentiviral shRNA experiments
293T cells were seeded (2 x 105 cells per ml) in 6 well plates. shRNA constructs were from the Broad RNAi Consortium and Clone IDs are shown in Table S2. Lentiviral particles were generated using a three plasmid system, as described previously (Moffat et al., 2006; Naldini et al., 1996). To standardize lentiviral transduction assays, viral titers were measured in a benchmark cell line, A549. For growth assays, titers corresponding to multiplicities of infection (MOIs) of 5 and 1 in A549 cells were employed. For K-Ras knockdown, cells were plated on day zero at 3x104 cells/ml in 96 or 12 well plates. Cells were spin infected, as described previously (Moffat et al., 2006). 24 hours post-infection, cells were treated with 1 μg/ml puromycin for 3 days to eliminate uninfected cells. Media was replaced and after 2 more days, cells were fixed with 4% formaldehyde and stained with 1 μM Syto60 dye for 1 hour. Syto60 fluorescence was quantified with a LiCor fluorescence scanner in the IR700 channel. Alternatively, cells were harvested for western blot analysis.
Derivation of the Ras Dependency Index (RDI)
Weighted averages for relative cell densities for MOIs of 5 and 1 with the K-Ras B and C shRNAs were calculated. The inverse of these averages were then calculated. This number was multiplied by the transduction efficiency for each respective cell line (the proportion of cells expressing the control shRNA following puromycin selection), yielding the RDI value.
Rescue of K-Ras ablation-induced cell death
pWPI-HA-K-Ras(12V) was generated by Gateway Cloning (Didier Trono, Ecole Polytechnique de Lausanne), which encodes a tandem IRES-GFP cassette. pLenti6-V5-GFP was used as a control. H358 NSCLC cells were infected with recombinant lentiviruses as described above. Cells expressing GFP or K-Ras(12V) were sorted for high GFP fluorescence intensity by FACS. The top 5% GFP-expressing cells were selected and expanded. These stable polyclonal cell lines were then subjected to K-Ras shRNA infection as described above.
Antibodies
The following antibodies were used– K-Ras (Calbiochem, OP-24), PARP (BD Pharmigen, 4C10-5), H-Ras (Abcam, Y132), phospho-ERK (Cell Signaling, 9101), total ERK (Cell Signaling, 9102), phospho AKT (Biosource), total AKT (Cell Signaling, 9272), GAPDH (Chemicon), cleaved Caspase-3 (Cell Signaling, 9661), E-cadherin (BD Pharmigen), Vimentin (Santa Cruz, H-84), Zeb1 (Santa Cruz, H-102), PTEN (Cell Signaling, 9552), phospho-Tyrosine (Cell Signaling, 9411), phospho-Syk (Y525/Y526) (Cell Signaling, 2710), total Syk (Cell Signaling, 2712), integrin beta-6 (Santa Cruz, H110), RON-β (Santa Cruz, C-20), Integrin beta-6 for IHC (Stromedix, Cambridge, MA), Prominin2 (Neuromics Inc.).
Immunofluorescence microscopy
Cells were fixed in EM grade 4% formaldehyde and permeablized with 0.1% Triton X-100. Staining with primary antibodies was carried out overnight at 4°C. For mouse monoclonal antibodies, a Cy3-conjugated goat anti-mouse secondary antibody was used (Jackson Laboratories). For rabbit polyclonal antibodies, FiTC-conjugated goat anti-rabbit secondary antibody was used (Chemicon Inc.). Nuclei were visualized using Hoechst 33342 dye (Molecular Probes). Micrographs were captured on an IX81 Spinning Disk Deconvolution Microscope equipped with a 40X Plan-Apo Oil objective lens. Digital images were processed with Slidebook and Adobe Photoshop CS4.
SNP and Gene Expression Microarray Analyses
K-Ras gene copy number analysis was performed with SNP array data using the Affymetrix GeneChip Human Mapping 500K Array Set. Raw data were converted to copy numbers using PLASQ (Probe-Level Allele-Specific Quantitation) algorithm (LaFramboise et al., 2005). Comparative whole-genome expression profiling was performed on Affymetrix U133 X3P Microarrays. Expression data were normalized using GCRMA (Bolstad et al., 2003). SNP and gene expression data as well as raw cel files are publicly available via NCBI (Accession # GSE15126). The PAM algorithm (Tibshirani et al., 2002) was used to generate a gene expression signature to differentiate K-Ras-dependent from K-Ras-independent cell lines (parameters for the algorithm are shown in Supplemental Experimental Procedures). To identify associations of the K-Ras dependency signature with published gene expression datasets, the Oncomine Concepts Map was used (Rhodes et al., 2004). The integrated software was used to generate heat-maps showing associations with Ras transcriptional targets. To generate heat-maps for hierarchical clustering of the signature genes in K-Ras mutant cell lines, the “R” gene expression analysis software was used. Normalized gene expression data for Ras_dependency genes were obtained from Oncomine. Average linkage hierarchical clustering was performed using Cluster/ Treeview (Eisen et al., 1998). To compute a Ras Dependency score, normalized_expression data for each Ras dependency gene was median-centered and_then median-centered expression values were averaged.
Immunohistochemistry of mouse pancreatic cancers
For immunohistochemical staining we employed tumor sections from the Pdx1-Cre LSL-KrasG12D Ink4a/Arf Lox/Lox mouse model of pancreatic cancer (Aguirre et al., 2003) and from control wild type mouse. All mice were housed in a pathogen-free environment at the Massachusetts General Hospital and were handled in strict accordance with Good Animal Practice as defined by the Office of Laboratory Animal Welfare, and all animal experiments were done with approval from Massachusetts General Hospital Subcommittee on Research Animal Care. Mice with signs of tumors (palpable abdominal mass, lethargy, weight loss) were euthanized, and the tumors were fixed overnight with neutral buffered 10% formalin. Tissues were incubated with pepsin (00-3008; Zymed, San Francisco, CA) 10 min at 37°C and blocked with 15ul/ml goat serum in TBS/Tween 20 + 0.1% BSA. Primary antibody was added to TBS/Tween 20 + 0.1% BSA and tissues were incubated for 60 min at room temperature. For immunostaining on mouse tissue, sections were incubated with a human/mouse chimeric form of the anti-integrin αVβ6 mAb, 2A1 (2.1 ul/ml) and an anti-human biotinylated secondary antibody (BA-3000; Vector Laboratories). Avidin-biotin complex-horseradish peroxidase (Vector kit PK-4000) was applied to sections and incubated 30 min at room temperature, and 3,3'-diaminobenzidine substrate was prepared as directed (SK-4100; Vector Laboratories) and applied to sections for 5 min at room temperature. Tissue sections were stained with Mayer’s hematoxylin for 1 min and rinsed in water and PBS.
Supplementary Material
Acknowledgments
A.S. was supported by a NRSA T32 post-doctoral fellowship. The studies were supported by NIH RO1 CA109447 to J.S. We thank Marie Classon, Jeff Engelman and Rushika Perara for comments on the manuscript. We thank Michael Rothenberg for construction of the pWPI-K-Ras(12V) expression vector and for advice regarding lentiviral use. We thank Maria Varadi for technical assistance. We thank Nathanael Gray for providing the R406 Syk inhibitor. We thank Sridhar Ramaswamy and Andrew Yee for advice with the application of gene expression signatures as predictive tools. We thank Vikram Deshpande for assistance with IHC analysis.
Footnotes
Disclosure Statement S.V. and L.K. have been or are in the employ of Biogen Idec. S.V. is currently employed by Stromedix Inc. These companies currently have an anti-integrin αVβ6 monoclonal antibody in clinical development.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Acevedo VD, Gangula RD, Freeman KW, Li R, Zhang Y, Wang F, Ayala GE, Peterson LE, Ittmann M, Spencer DM. Inducible FGFR-1 activation leads to irreversible prostate adenocarcinoma and an epithelial-to-mesenchymal transition. Cancer Cell. 2007;12:559–571. doi: 10.1016/j.ccr.2007.11.004. [DOI] [PubMed] [Google Scholar]
- Aguirre AJ, Bardeesy N, Sinha M, Lopez L, Tuveson DA, Horner J, Redston MS, DePinho RA. Activated Kras and Ink4a/Arf deficiency cooperate to produce metastatic pancreatic ductal adenocarcinoma. Genes Dev. 2003;17:3112–3126. doi: 10.1101/gad.1158703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguirre AJ, Brennan C, Bailey G, Sinha R, Feng B, Leo C, Zhang Y, Zhang J, Gans JD, Bardeesy N, et al. High-resolution characterization of the pancreatic adenocarcinoma genome. Proc Natl Acad Sci U S A. 2004;101:9067–9072. doi: 10.1073/pnas.0402932101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baines AT, Lim KH, Shields JM, Lambert JM, Counter CM, Der CJ, Cox AD. Use of retrovirus expression of interfering RNA to determine the contribution of activated K-Ras and ras effector expression to human tumor cell growth. Methods Enzymol. 2006;407:556–574. doi: 10.1016/S0076-6879(05)07045-X. [DOI] [PubMed] [Google Scholar]
- Bardeesy N, Cheng KH, Berger JH, Chu GC, Pahler J, Olson P, Hezel AF, Horner J, Lauwers GY, Hanahan D, DePinho RA. Smad4 is dispensable for normal pancreas development yet critical in progression and tumor biology of pancreas cancer. Genes Dev. 2006;20:3130–3146. doi: 10.1101/gad.1478706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beer DG, Kardia SL, Huang CC, Giordano TJ, Levin AM, Misek DE, Lin L, Chen G, Gharib TG, Thomas DG, et al. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat Med. 2002;8:816–824. doi: 10.1038/nm733. [DOI] [PubMed] [Google Scholar]
- Bild AH, Yao G, Chang JT, Wang Q, Potti A, Chasse D, Joshi MB, Harpole D, Lancaster JM, Berchuck A, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439:353–357. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- Braselmann S, Taylor V, Zhao H, Wang S, Sylvain C, Baluom M, Qu K, Herlaar E, Lau A, Young C, et al. R406, an orally available spleen tyrosine kinase inhibitor blocks fc receptor signaling and reduces immune complex-mediated inflammation. J Pharmacol Exp Ther. 2006;319:998–1008. doi: 10.1124/jpet.106.109058. [DOI] [PubMed] [Google Scholar]
- Coldren CD, Helfrich BA, Witta SE, Sugita M, Lapadat R, Zeng C, Baron A, Franklin WA, Hirsch FR, Geraci MW, Bunn PA., Jr Baseline gene expression predicts sensitivity to gefitinib in non-small cell lung cancer cell lines. Mol Cancer Res. 2006;4:521–528. doi: 10.1158/1541-7786.MCR-06-0095. [DOI] [PubMed] [Google Scholar]
- Collins S, Groudine M. Amplification of endogenous myc-related DNA sequences in a human myeloid leukaemia cell line. Nature. 1982;298:679–681. doi: 10.1038/298679a0. [DOI] [PubMed] [Google Scholar]
- Cox AD, Der CJ. Ras family signaling: therapeutic targeting. Cancer Biol Ther. 2002;1:599–606. doi: 10.4161/cbt.306. [DOI] [PubMed] [Google Scholar]
- Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- Dumont N, Wilson MB, Crawford YG, Reynolds PA, Sigaroudinia M, Tlsty TD. Sustained induction of epithelial to mesenchymal transition activates DNA methylation of genes silenced in basal-like breast cancers. Proc Natl Acad Sci U S A. 2008;105:14867–14872. doi: 10.1073/pnas.0807146105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming JB, Shen GL, Holloway SE, Davis M, Brekken RA. Molecular consequences of silencing mutant K-ras in pancreatic cancer cells: justification for K-ras-directed therapy. Mol Cancer Res. 2005;3:413–423. doi: 10.1158/1541-7786.MCR-04-0206. [DOI] [PubMed] [Google Scholar]
- Fuchs BC, Fujii T, Dorfman JD, Goodwin JM, Zhu AX, Lanuti M, Tanabe KK. Epithelial-to-mesenchymal transition and integrin-linked kinase mediate sensitivity to epidermal growth factor receptor inhibition in human hepatoma cells. Cancer Res. 2008;68:2391–2399. doi: 10.1158/0008-5472.CAN-07-2460. [DOI] [PubMed] [Google Scholar]
- Grutzmann R, Pilarsky C, Ammerpohl O, Luttges J, Bohme A, Sipos B, Foerder M, Alldinger I, Jahnke B, Schackert HK, et al. Gene expression profiling of microdissected pancreatic ductal carcinomas using high- density DNA microarrays. Neoplasia. 2004;6:611–622. doi: 10.1593/neo.04295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigis KM, Kendall KR, Wang Y, Cheung A, Haigis MC, Glickman JN, Niwa-Kawakita M, Sweet-Cordero A, Sebolt-Leopold J, Shannon KM, et al. Differential effects of oncogenic K-Ras and N-Ras on proliferation, differentiation and tumor progression in the colon. Nat Genet. 2008;40:600–608. doi: 10.1038/ngXXXX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hezel AF, Kimmelman AC, Stanger BZ, Bardeesy N, Depinho RA. Genetics and biology of pancreatic ductal adenocarcinoma. Genes Dev. 2006;20:1218–1249. doi: 10.1101/gad.1415606. [DOI] [PubMed] [Google Scholar]
- Hosack DA, Dennis G, Jr, Sherman BT, Lane HC, Lempicki RA. Identifying biological themes within lists of genes with EASE. Genome Biol. 2003;4:R70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houldsworth J, Cordon-Cardo C, Ladanyi M, Kelsen DP, Chaganti RS. Gene amplification in gastric and esophageal adenocarcinomas. Cancer Res. 1990;50:6417–6422. [PubMed] [Google Scholar]
- Iacobuzio-Donahue CA, Maitra A, Olsen M, Lowe AW, van Heek NT, Rosty C, Walter K, Sato N, Parker A, Ashfaq R, et al. Exploration of global gene expression patterns in pancreatic adenocarcinoma using cDNA microarrays. Am J Pathol. 2003;162:1151–1162. doi: 10.1016/S0002-9440(10)63911-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson L, Mercer K, Greenbaum D, Bronson RT, Crowley D, Tuveson DA, Jacks T. Somatic activation of the K-ras oncogene causes early onset lung cancer in mice. Nature. 2001;410:1111–1116. doi: 10.1038/35074129. [DOI] [PubMed] [Google Scholar]
- LaFramboise T, Weir BA, Zhao X, Beroukhim R, Li C, Harrington D, Sellers WR, Meyerson M. Allele-specific amplification in cancer revealed by SNP array analysis. PLoS Comput Biol. 2005;1:e65. doi: 10.1371/journal.pcbi.0010065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logsdon CD, Simeone DM, Binkley C, Arumugam T, Greenson JK, Giordano TJ, Misek DE, Kuick R, Hanash S. Molecular profiling of pancreatic adenocarcinoma and chronic pancreatitis identifies multiple genes differentially regulated in pancreatic cancer. Cancer Res. 2003;63:2649–2657. [PubMed] [Google Scholar]
- Lu Y, Yao HP, Wang MH. Multiple variants of the RON receptor tyrosine kinase: biochemical properties, tumorigenic activities, and potential drug targets. Cancer Lett. 2007;257:157–164. doi: 10.1016/j.canlet.2007.08.007. [DOI] [PubMed] [Google Scholar]
- Moffat J, Grueneberg DA, Yang X, Kim SY, Kloepfer AM, Hinkle G, Piqani B, Eisenhaure TM, Luo B, Grenier JK, et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124:1283–1298. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- Naldini L, Blomer U, Gallay P, Ory D, Mulligan R, Gage FH, Verma IM, Trono D. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272:263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- Olejniczak ET, Van Sant C, Anderson MG, Wang G, Tahir SK, Sauter G, Lesniewski R, Semizarov D. Integrative genomic analysis of small-cell lung carcinoma reveals correlates of sensitivity to bcl-2 antagonists and uncovers novel chromosomal gains. Mol Cancer Res. 2007;5:331–339. doi: 10.1158/1541-7786.MCR-06-0367. [DOI] [PubMed] [Google Scholar]
- Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer. 2007;7:415–428. doi: 10.1038/nrc2131. [DOI] [PubMed] [Google Scholar]
- Repasky GA, Chenette EJ, Der CJ. Renewing the conspiracy theory debate: does Raf function alone to mediate Ras oncogenesis? Trends Cell Biol. 2004;14:639–647. doi: 10.1016/j.tcb.2004.09.014. [DOI] [PubMed] [Google Scholar]
- Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sada K, Takano T, Yanagi S, Yamamura H. Structure and function of Syk protein-tyrosine kinase. J Biochem. 2001;130:177–186. doi: 10.1093/oxfordjournals.jbchem.a002970. [DOI] [PubMed] [Google Scholar]
- Shah AN, Gallick GE. Src, chemoresistance and epithelial to mesenchymal transition: are they related? Anticancer Drugs. 2007;18:371–375. doi: 10.1097/CAD.0b013e32801265d7. [DOI] [PubMed] [Google Scholar]
- Sharma SV, Bell DW, Settleman J, Haber DA. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer. 2007;7:169–181. doi: 10.1038/nrc2088. [DOI] [PubMed] [Google Scholar]
- Stearman RS, Dwyer-Nield L, Zerbe L, Blaine SA, Chan Z, Bunn PA, Jr, Johnson GL, Hirsch FR, Merrick DT, Franklin WA, et al. Analysis of orthologous gene expression between human pulmonary adenocarcinoma and a carcinogen-induced murine model. Am J Pathol. 2005;167:1763–1775. doi: 10.1016/S0002-9440(10)61257-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tal M, Wetzler M, Josefberg Z, Deutch A, Gutman M, Assaf D, Kris R, Shiloh Y, Givol D, Schlessinger J. Sporadic amplification of the HER2/neu protooncogene in adenocarcinomas of various tissues. Cancer Res. 1988;48:1517–1520. [PubMed] [Google Scholar]
- Thomson S, Buck E, Petti F, Griffin G, Brown E, Ramnarine N, Iwata KK, Gibson N, Haley JD. Epithelial to mesenchymal transition is a determinant of sensitivity of non-small-cell lung carcinoma cell lines and xenografts to epidermal growth factor receptor inhibition. Cancer Res. 2005;65:9455–9462. doi: 10.1158/0008-5472.CAN-05-1058. [DOI] [PubMed] [Google Scholar]
- Tibshirani R, Hastie T, Narasimhan B, Chu G. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc Natl Acad Sci U S A. 2002;99:6567–6572. doi: 10.1073/pnas.082099299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner KW, Punnoose EA, Januario T, Lawrence DA, Pitti RM, Lancaster K, Lee D, von Goetz M, Yee SF, Totpal K, et al. Death-receptor O-glycosylation controls tumor-cell sensitivity to the proapoptotic ligand Apo2L/TRAIL. Nat Med. 2007;13:1070–1077. doi: 10.1038/nm1627. [DOI] [PubMed] [Google Scholar]
- Weinstein IB. Cancer. Addiction to oncogenes--the Achilles heal of cancer. Science. 2002;297:63–64. doi: 10.1126/science.1073096. [DOI] [PubMed] [Google Scholar]
- Weir BA, Woo MS, Getz G, Perner S, Ding L, Beroukhim R, Lin WM, Province MA, Kraja A, Johnson LA, et al. Characterizing the cancer genome in lung adenocarcinoma. Nature. 2007;450:893–898. doi: 10.1038/nature06358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong AJ, Bigner SH, Bigner DD, Kinzler KW, Hamilton SR, Vogelstein B. Increased expression of the epidermal growth factor receptor gene in malignant gliomas is invariably associated with gene amplification. Proc Natl Acad Sci U S A. 1987;84:6899–6903. doi: 10.1073/pnas.84.19.6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.








