Abstract
To gain insight into the basic neurobiological processes regulated by lithium—an effective drug for bipolar disorder—we used Affymetrix Genome Arrays to examine lithium-induced changes in genome-wide gene expression profiles of head mRNA from the genetic model organism Drosophila melanogaster. First, to identify the individual genes whose transcript levels are most significantly altered by lithium, we analyzed the microarray data with stringent criteria (fold change > 2; p <0.001) and evaluated the results by RT-PCR. This analysis identified 12 genes that encode proteins with various biological functions, including an enzyme responsible for amino acid metabolism and a putative amino acid transporter. Second, to uncover the biological pathways involved in lithium’s action in the nervous system, we used less stringent criteria (fold change >1.2; FDR <0.05) and assigned the identified 66 lithium-responsive genes to biological pathways using DAVID (Database for Annotation, Visualization and Integrated Discovery). The gene ontology categories most significantly affected by lithium were amino acid metabolic processes. Taken together, these data suggest that amino acid metabolism is important for lithium’s actions in the nervous system, and lay a foundation for future functional studies of lithium-responsive neurobiological processes using the versatile molecular and genetic tools that are available in Drosophila.
Keywords: Drosophila, lithium, microarray, RT-PCR, gene expression, nervous system
Introduction
The alkaline metal lithium affects various developmental and physiological processes in evolutionarily diverse organisms (Phiel and Klein, 2001). In particular, lithium’s actions in the nervous system have attracted special attention because lithium is highly effective in the prophylaxis and treatment of bipolar affective disorder (Schou, 2001). In addition, recent studies suggest that chronic lithium treatment is efficacious in preventing apoptosis-dependent neuronal death (Chuang, 2004), which raises the interesting possibility that lithium might be effective in treating or preventing brain damage, either following injury or over the course of progression of neurodegenerative diseases. Despite the proven clinical usefulness and the further potential of this drug, the molecular mechanisms underlying its actions in the nervous system are still only poorly understood.
At therapeutically relevant concentrations, lithium inhibits several enzymes, including glycogen synthase kinase 3β (GSK3β) as well as inositol monophosphatase (IMPase) and related enzymes (Berridge et al., 1989; Klein and Melton, 1996). These lithium-sensitive enzymes are intimately involved in the regulation of various intracellular molecular cascades, such as the Wnt and inositol phosphate signaling pathways (Chen et al., 2000; Ding et al., 2000). Lithium-based perturbation of these signaling pathways has a significant impact on global gene expression profiles (Rowe and Chuang, 2004), and this may contribute to the therapeutic as well as toxic actions of lithium. In order to elucidate the importance of gene regulation in lithium’s actions in the nervous system, we need to identify genes whose expression is influenced by therapeutic concentrations of lithium, and to study their roles in the lithium-responsive neurobiological processes at the molecular, cellular and organismal levels.
The fruit fly Drosophila melanogaster has been a valuable genetic model system for examining fundamental problems in neurobiology. In part, this is due to the fact that Drosophila and higher vertebrates share genetic pathways for cellular signaling (Miklos and Rubin, 1996; Rubin et al., 2000). In addition, many human genes involved in brain functions and neurological disorders have fly counterparts (Reiter et al., 2001; Davis, 2005; Hamet and Tremblay, 2006). Importantly, the genetic pathways involved in lithium’s actions in the nervous system appear to be shared by Drosophila and vertebrates. For example, the administration of lithium to fruit flies and vertebrates has a similar effect on circadian clocks, and in both cases this effect involves the inhibition of glycogen synthase kinase 3β (GSK3β) (Padiath et al., 2004; Dokucu et al., 2005; Iitaka et al., 2005). Additionally, as in vertebrates, lithium has neuroprotective effects in transgenic flies that over-express either human tau proteins or a mutant form of huntingtin (Mudher et al., 2004; Berger et al., 2005). Furthermore, lithium improves the physiological, behavioral and developmental mutant phenotypes characteristic of a mouse model of Fragile X syndrome (Min et al., 2009), and likewise rescues such defects in a Drosophila model of this disease (McBride et al., 2005). These results strongly suggest that studies of the genes responsible for lithium’s actions in the Drosophila nervous system would provide important insights into the basis of lithium’s neurobiological effects in vertebrates.
In this study, we carried out a microarray-based gene expression profiling analysis of Drosophila head mRNA, to identify the genes and biological pathways of the nervous system that are significantly influenced by lithium treatment in adult animals. This study lays the foundation for future functional studies using the versatile molecular and genetic tools available in Drosophila to understand the lithium-responsive neurobiological processes.
Materials and Methods
Drosophila stock
Flies were reared at 25°C at 65% humidity, in a 12 hr:12 hr light:dark cycle, on a conventional cornmeal-based medium containing glucose, yeast and agar supplemented with the mold inhibitor methyl 4-hydroxybenzoate (0.05 %). The Canton-S (CS) strain was used as the wild-type control.
RNA extraction and microarray experiment
Newly eclosed 0–1 day old wild-type female flies were grouped into sets of 20 and placed into a vial containing regular fly food with or without 50 mM LiCl. Flies in five vials (total of 100 flies) were combined as one biological sample, and three biological replicates were prepared for each treatment condition. The fly heads were removed from bodies on a dry ice block after 24-hour treatment, and kept frozen at −80°C until used. Total RNAs were extracted from the fly heads using Trizol solution (Invitrogen, Carlsbad, CA), followed by further purification using RNasy column (Qiagen, Valencia, CA). The quality of the purified total RNA was verified using Agilent Bioanalyzer (Agilent Technologies, Stockport, Cheshire, UK). cRNA labeling and microarray experiments were carried out at the Translational Genomics Research Institute (Phoenix, AZ), using Affymetrix Drosophila Genome 2.0 Arrays (Affymetrix, Santa Clara, CA).
Microarray data analysis
Image data were quantified using the genechip-operating software Affymetrix GCOS v1.4. Gene expression data were normalized using the robust multi-array average (RMA) statistical algorithms (Irizarry et al., 2003). Besides six sets of data from wild-type flies (three biological replicates for each condition, with or without lithium treatment), additional six data sets created under the same conditions from Shudderrer mutant flies (which display neurological phenotypes that are improved by lithium treatment) (Williamson, 1982) were included in the normalization process. In this report, we have focused on the wild-type data to lay a foundation for future genetic studies on lithium-responsive processes. A heat-map was generated for the expression data corresponding to a subset of genes with fold change >1.2; FDR<0.05 (Supplementary data) using Partek GS version 6.4. (Partek Inc., St. Louis, MO). Correlation coefficients were calculated using the GeneSpring software. Cluster euclidean distance analysis (Dougherty et al., 2002) was carried out using Ward’s method, via Partek GS software (Partek Inc., St. Louis, MO). Comparisons between signals for lithium-treated and untreated groups were made using one way ANOVA. Bonferoni’s multiple comparison corrections were applied to obtain the false discovery rate (FDR). Genes were annotated and biological processes were analyzed using the Database for Annotation, Visualization and Integrated Discovery (DAVID) (http://david.abcc.ncifcrf.gov) (Dennis et al., 2003).
Reverse transcription PCR (RT-PCR)
Total RNA was extracted from 30 fly heads after 24-hour treatment with or without 50mM LiCl. Four biological replicates were analyzed for each experimental condition. cDNA was synthesized using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA) and subjected to semi-quantitative PCR amplification using gene-specific primers, most of which were designed to overlap with the locations of Affymetrix probes for the corresponding genes. The primers used for the RT-PCR were listed in Table 1. The PCR products corresponding to 1–5 ng of the original total RNA were analyzed on an agarose gel for quantification. Rp49 (a.k.a., RpL32), which encodes the ribosomal protein L32, is commonly used as the internal standard for quantitative analysis of mRNA levels in Drosophila (Gabler et al., 2005). The Rp49 transcripts were amplified from the same cDNA samples using Rp49-specific primers and used as the internal standard. PCR conditions were optimized for each gene, such that the endpoint of each PCR was in the linear range of amplifications. Transcript levels were quantified by analyzing pixel intensity of the bands using Image J software (NIH, Bethesda, MD). A fold-change for each gene was determined by calculating a ratio of the band signal intensities between lithium-treated and untreated samples. Averages of the fold-changes were presented with the standard error of the mean (SEM).
Table 1.
Primers used for RT-PCR analysis
| Gene | 5′-primer | 3′-primer |
|---|---|---|
| CG9377 | TGATCACCACCGCGCACTGC | ACATCAGCGGCACAGCGGTCA |
| GST D2 | CATCGCCGTCTATCTGGTGGA | GGCATTGTCGTACCACCTGG |
| Arc1 | CTACAGTGGGCGTGAGCCGGGCA | AGTTGATGGCGCACGGTGCAAG |
| CG1673 | TGCGCTTTTACTTCCAAGCAGCA | GGGCCTAGGTTCTACTGACGGGT |
| Nmdmc | ATCGATGGCAAGGCCATAGC | ACCAGTATCCCCGTGACCTGG |
| CG7763 | TTCGCAACAACTGCTGACAC | GATATCCACACAATTGGCCTTCG |
| CG15784 | TCCAGCTCCGATGGCGAAGTG | AGAGAACGACTGCGACGACCTG |
| Cyp309a1 | GATTCGAAACATCTGGAGCCGT | CCTGGTAATTTGAGAGGATGTGC |
| CG15088 | TTTCTTCCTCATGCTATTCGTCTTAGGCAT | GAGCGGACATATCGAGCAGCATGT |
| Lsp 1γ | ACTATCAAGCGCAGCTCCAAG | ACTAAGAGCTAGATCCAGTGTGG |
| Bin 1 | ACCGCAGAATAGGGAGACATG | ACTCCGGAAGTTGGGGTACA |
| CG5999 | TATTCTTTACTCGAAGTGGCTTCCGCA | CTCCGTCCAGTAGAGGAATGATTC |
| Rp49 | GATCGATATGCTAAGCTGTCGC | CGACCACGTTACAAGAACTCT′ |
RESULTS
In order to determine the transcriptome response to lithium treatment in Drosophila heads, wild-type female flies were treated for 24 hours with or without 50 mM LiCl. We chose this concentration of lithium because it results in an internal lithium level that is comparable to the therapeutic serum concentrations (μM ~ mM) in bipolar disorder patients treated with lithium (Padiath et al., 2004; Dokucu et al., 2005). These lithium concentrations do not cause obvious toxic effects on adult wild-type flies after 24 hr treatment, but do have significant effects on circadian rhythm in wild-type flies (Padiath et al., 2004; Dokucu et al., 2005), as well as on cellular or behavioral phenotypes in certain mutants and transformants (Williamson, 1982; Mudher et al., 2004; Berger et al., 2005; McBride et al., 2005).
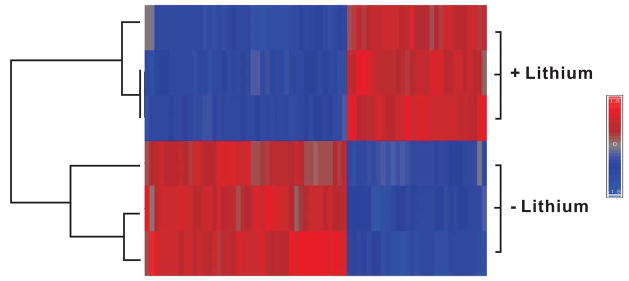
Three biological replicates were analyzed for each condition, i.e., with or without lithium treatment, using the Affymetrix Drosophila Genome 2.0 Arrays. The signal intensities of each chip were normalized using RMA algorithms, and the lithium-treated and untreated groups were compared as described in the Material and Methods section. The correlation coefficients for the three biological replicates were greater than 0.93 under both conditions, indicating that the gene expression data obtained in this study were highly reproducible. The biological replicates were well clustered when a method of hierarchical clustering based on Ward’s distance was applied to all 6 wild-type data sets (3 treated with lithium and 3 not treated with lithium; Fig. 1). Consistency among the biological replicates was visualized by generating a heat-map of expression data for the genes that are differentially regulated in response to lithium (fold change >1.2; FDR <0.05) (Figure 1).
Figure 1. Correlation of microarray data obtained from lithium-treated and untreated samples.
Euclidean distance clustering analysis was performed using Ward’s method. Three biological replicates (+Lithium or −Lithium) were well clustered. Consistency of the data is visualized by the heat-map representation of expression data for the 66 genes (represented by 71 probe sets) that are differentially regulated in response to lithium treatment (fold change >1.2; FDR <0.05).
In this study, we took two approaches to investigating lithium-induced changes in genome-wide expression profiles of Drosophila head mRNA. In the first, we identified individual genes whose transcript levels are most significantly altered in response to lithium treatment, using stringent criteria (fold change >2; p <0.001) to select differentially regulated genes. In the second, we uncovered the biological pathways that are likely involved in lithium’s actions in the nervous system, using less stringent criteria (fold change >1.2; FDR <0.05) to identify lithium-responsive genes and then assigning these genes to relevant biological pathways by DAVID.
Genes whose transcript levels are most significantly altered in response to lithium treatment
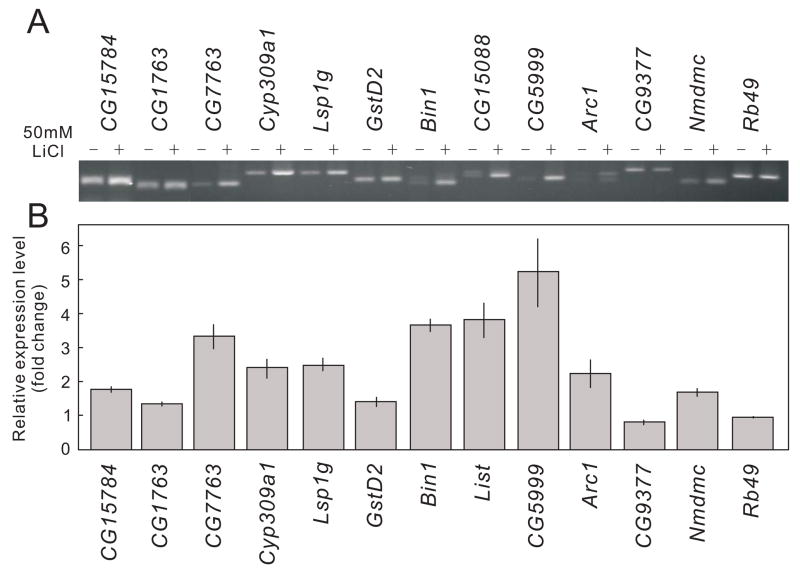
Of 18,500 Drosophila genes analyzed using the Affymetrix Drosophila Genome 2.0 Arrays, 16 genes that were represented by 18 Affymetrix probe sets responded to lithium treatment by changes of >2 (p <0.001) in their expression, in wild-type flies. Among these 16 genes, 2 and 14 were down- and up-regulated, respectively, in response to lithium treatment. The results of the microarray experiment were confirmed by RT-PCR analysis on four RNA samples independently prepared from wild-type fly heads that had or had not been treated with 50 mM lithium for 24 hr. RT-PCR analysis confirmed that the transcript levels of 12 genes differed significantly between lithium-treated and lithium-untreated samples (one down-regulated and 11 up-regulated in lithium-treated samples) (Fig. 2, Table 2). Four genes (CG31781, CG11654, CG11425 and CG18522) of the original 16 were either not amplified by RT-PCR or yielded results that were not consistent with those of the microarray analysis (data not shown). For 10 of the 12 genes identified both by microarray and RT-PCR analyses, the fold-change based on RT-PCR analysis was smaller than that estimated by microarray analysis (Table 2). This is probably because the linear range of RT-PCR analysis is narrower than that for the Affymetrix microarray method. The 12 genes identified as undergoing significant change in expression encode a variety of proteins, including ones involved in the transport (CG15088) and metabolism (CG1673) of amino acids, and others required for detoxification (GstD2) and the stress response (Arc1) (Table 2).
Figure 2. RT-PCR analysis of the genes whose expression is significantly affected by lithium treatment.
(A) A representative result of the RT-PCR analysis (carried out in quadruplicate). The PCR products were quantitated based on the pixel intensity of the corresponding bands using Image J software. (B) Ratios of signal intensity of lithium-treated and untreated samples. Average ± SEM, N=4.
Table 2.
Genes differentially expressed in response to lithiuma.
| Probe set ID | Gene | Microarray | RT-PCR | Description | |||
|---|---|---|---|---|---|---|---|
| Normalized Signal intensity (log2) b |
Fold Change |
p-value | Fold Changec |
||||
| − LiCl | + LiCl | ||||||
| Down-regulated gene | |||||||
| 1631512_at | CG9377 | 7.17 ± 0.09 | 6.05 ± 0.10 | −2.17 | 1.54E−04 | −1.30 ± 0.06 | Serine type endopeptidase |
| Up-regulated genes | |||||||
| 1630258_at | Glutathione S transferase D2 | 6.27 ± 0.21 | 7.40 ± 0.30 | 2.19 | 6.17E−03 | 1.39 ± 0.14 | Glutathione S transferase |
| 1639180_at 1639694_s_at |
Arc1 | 10.01 ± 0.08 8.83 ± 0.06 |
11.19 ± 0.35 10.00 ± 0.32 |
2.27 2.25 |
4.80E−03 3.45E−03 |
2.18 ± 0.43 | zinc ion binding; nucleic acid binding |
| 1623770_at | CG1673 | 7.10 ± 0.09 | 8.30 ± 0.04 | 2.31 | 2.60E−05 | 1.30 ± 0.10 | Branched chain amino acid transaminase |
| 1640775_a_at | NAD dependent methylenetetrahydrof olate dehydrogenase | 8.49 ± 0.11 | 9.86 ± 0.37 | 2.59 | 3.62E−03 | 1.68 ± 0.12 | methenyltetrahydrofolate cyclohydrolase/dehydrogenase/dehydrogenase |
| 1626144_at | CG7763 | 5.61 ± 0.54 | 7.20 ± 0.25 | 3.02 | 9.42E−03 | 3.28 ± 0.35 | C type lectin/sugar binding |
| 1640884_at | CG15784 | 9.16 ± 0.02 | 10.84 ± 0.57 | 3.22 | 7.06E−03 | 1.84 ± 0.02 | unknown |
| 1626324_at | Cyp309a1 | 7.69 ± 0.28 | 9.39 ± 0.37 | 3.25 | 3.22E−03 | 2.36 ± 0.28 | monooxygenase |
| 1636594_at | 7.31 ± 0.07 | 9.28 ± 0.07 | 3.91 | 4.91E−06 | Sodium:neurotransmitter | ||
| 1632986_a_at | CG15088 | 6.75 ± 0.09 | 9.26 ± 0.02 | 5.68 | 1.12E−06 | 3.74 ± 0.49 | symporter |
| 1626429_at | Larval serum protein 1 γ | 4.50 ± 0.17 | 6.69 ± 0.22 | 4.55 | 1.71E−04 | 2.41 ± 0.16 | oxygen transporter |
| 1632565_at | Bicoid interacting protein 1 | 7.64 ± 0.03 | 10.06 ± 0.73 | 5.32 | 4.73E−03 | 3.61 ± 0.18 | transcription co repressor |
| 1638182_at | CG5999 | 6.79 ± 0.32 | 10.46 ± 1.11 | 12.7 | 5.32E−03 | 5.12 ± 0.96 | UDP glucuronosyltransferase |
Genes were selected based on the following criteria: 2.0 fold or greater changes in the normalized signal intensities with p-value with 0.01 or less in Student’s t-test.
Signal intensities of each chip were normalized values using RMA algorithm and presented in log2.
Fold change values estimated from RT-PCR analyses were shown as Ave ± SEM, n=4.
Biological pathways that are significantly affected by lithium-induced alterations in gene expression
In less stringent analysis of the microarray data obtained from lithium-treated and untreated flies (fold change >1.2; FDR<0.05), we identified 71 probe sets representing 66 differentially regulated genes, including 28 that were down-regulated and 38 up-regulated (Supplementary data). These genes were assigned to biological pathways using a DAVID functional annotation tool (http://david.abcc.ncifcrf.gov). This web-accessible program annotates each gene and performs enrichment analysis that identifies the most relevant biological meanings associated with a given gene list, with output in the form of a “functional annotation chart”. Although the functional annotation chart is useful for identifying biological terms or pathways in which the analyzed genes are significantly enriched, these terms often overlap due to inherent redundancy in annotations. To improve the usefulness of the functional annotation analysis, we carried out DAVID clustering analysis. The functional annotation chart was integrated with the clustering analyses to identify biological pathways that are significantly affected by lithium-induced alterations in genome-wide gene expression.
Analysis of the identified 66 lithium-responsive genes with the DAVID functional annotation tool resulted in the identification of 13 term entries as significantly “enriched terms/pathways”, with p < 0.001 (Table 3). Among these, the gene ontology (GO) term “branched chain family amino acid metabolic process (GO: 0009081)” showed the highest fold enrichment (69.1-fold). In addition, “valine, leucine and isoleucine degradation (KEGG PATHWAY)” was identified as the term with the third highest fold enrichment (15.9-fold).
Table 3.
Functional annotation chart of the selected lithium-responsive genes analyzed by DAVID.
| Term | p-valuea | Fold Enrichmentb | |
|---|---|---|---|
| 1 | branched chain family amino acid metabolic process (GO:0009081) | 7.57E−04 | 69.1 |
| 2 | stress response (SP PIR keywords) | 9.23E−05 | 43.8 |
| 3 | valine, leucine and isoleucine degradation (KEGG pathway: dme00280) | 1.69E−05 | 15.9 |
| 4 | organic acid metabolic processc (GO:0006082) | 8.20E−06 | 5.99 |
| 5 | carboxylic acid metabolic processc (GO:0019752) | 8.20E−06 | 5.99 |
| 6 | oxidoreductase (SP PIR keywords) | 3.18E−05 | 5.83 |
| 7 | mitochondrion (GO:0005739) | 9.09E−05 | 4.29 |
| 8 | amino acid metabolic process (GO:0006520) | 8.05E−04 | 6.11 |
| 9 | oxidoreductase activity (GO:0016491) | 9.69E−04 | 3.16 |
| 10 | hydrolase (SP PIR keywords) | 4.03E−04 | 3.15 |
| 11 | cytoplasmic part (GO:0044444) | 1.66E−04 | 2.25 |
| 12 | cytoplasm (GO:0005737) | 8.09E−04 | 1.92 |
| 13 | catalytic activity (GO:0003824) | 3.48E−05 | 1.71 |
Modified Fisher Exact p-value (EASE score),
Enrichment factor for the lithium responsive genes (fold change >1.2; FDR<0.05),
The identical genes are assigned to these terms for Drosophila genome.
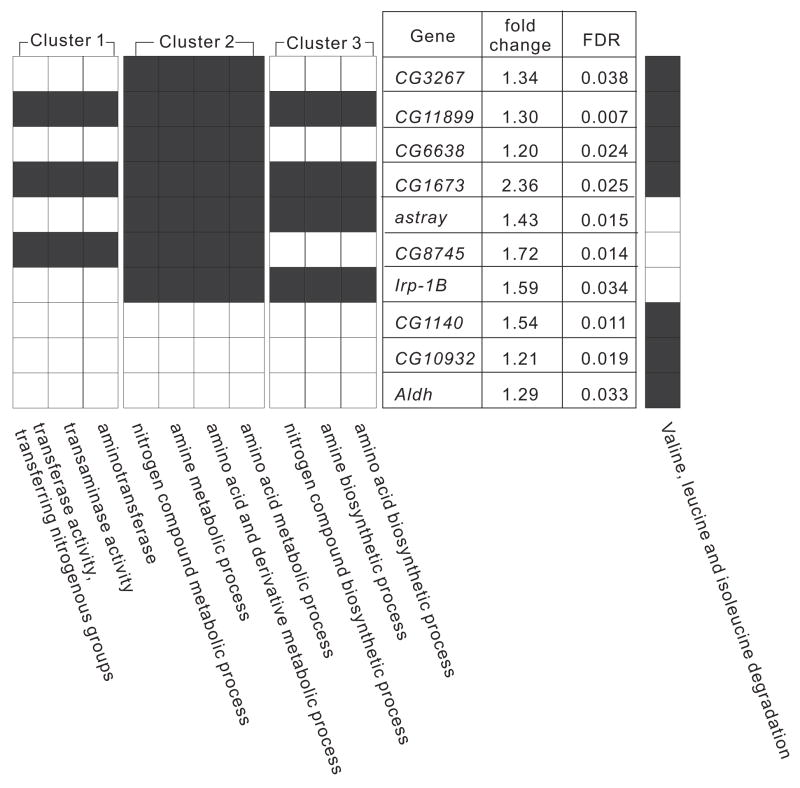
We also performed the DAVID clustering analysis under the high stringency conditions on the list of lithium-responsive genes obtained (Huang da et al., 2007). Only three clusters met the statistical criteria (p < 0.01), and all of these were related to amino acid metabolism (Table 4). Each of these clusters was composed of 3–4 GO terms, with a fold enrichment of 3.30–49.3. For example, functional cluster 2 is composed of four GO terms that apply to identical sets of seven genes. These terms were “amino acid metabolic process (GO:0006520)”, “amino acid and derivative metabolic process (GO:0006519)”, “amine metabolic process (GO:0009308)” and “nitrogen compound metabolic process (GO:0006807)”. As shown in Figure 3, these seven genes displayed increases in gene expression of 1.20–2.36-fold in response to lithium treatment. A fraction of the genes in functional cluster 2 also formed functional clusters 1 and 3. Three of the seven genes (CG3267, CG6638 and CG1673) were involved in the branched amino acid (BCAA) degradation pathway.
Table 4.
Functional annotation clustering of the selected lithium-responsive genes.
| Functional cluster 1 | |||
|---|---|---|---|
| Category | Term | p-valuea | Fold Enrichmentb |
| SP PIR keywords | aminotransferase | 0.0016 | 49.3 |
| GO:0008483 | transaminase activity | 5.79E−03 | 25.7 |
| GO:0016769 | transferase activity, transferring nitrogenous groups | 7.29E−03 | 22.8 |
|
Functional cluster 2 | |||
| GO:0006520 | amino acid metabolic process | 8.05E−04 | 6.11 |
| GO:0006519 | amino acid and derivative metabolic process | 0.0020 | 5.10 |
| GO:0009308 | amine metabolic process | 0.0145 | 3.39 |
| GO:0006807 | nitrogen compound metabolic process | 0.0165 | 3.30 |
|
Functional cluster 3 | |||
| GO:0008652 | amino acid biosynthetic process | 0.0030 | 13.4 |
| GO:0009309 | amine biosynthetic process | 0.0070 | 9.96 |
| GO:0044271 | nitrogen compound biosynthetic process | 0.0075 | 9.69 |
Modified Fisher Exact p-value (EASE score),
Enrichment factor for the lithium-responsive genes (fold change >1.2; FDR<0.05) in each category.
Figure 3. Biological pathways significantly affected by lithium treatment.
Biological pathways identified by DAVID clustering analysis, based on the genes whose expression is affected by lithium treatment. Genes that were assigned to these pathways are also shown with values for fold change in expression and false discovery rate (FDR) in response to lithium treatment. Genes that are involved in “valine, leucine, isoleucine degradation pathway (KEGG PATHWAY)” are indicated at right.
In addition to identifying the terms related to amino acid metabolic pathways, the DAVID-based functional annotation analysis identified the term “stress response” with the second highest enrichment (43.8-fold), with p= 9.23E−05 (Table 3). Two GO terms that were revealed in this analysis with highest statistical significance were “organic acid metabolism” and “carboxylic acid metabolism” (p = 8.2E−06) (Table 3). For the Drosophila melanogaster genes, these two terms share identical components. Eleven genes were assigned to these GO terms, showing a significant enrichment (6.0 times) in the corresponding biological processes.
Discussion
Characteristic features of the individual genes whose expression is significantly affected by lithium
In this study, we examined the lithium-induced alterations in genome-wide gene expression profiles in the adult head of the genetic model organism, Drosophila melanogaster. Using stringent analysis criteria, we identified 12 genes whose transcript levels are most significantly altered by lithium (Table 2). These genes can be categorized into 4 groups based on their biological function or characteristic features: (i) amino acid transport and metabolism, (ii) detoxication, stress response or self-defense reactions, (iii) psychiatric or neurological disorders, and (iv) others.
(i) Genes involved in amino acid transport and metabolism (CG15088 and CG1673)
Of the 18,500 Drosophila genes analyzed, the expression of CG15088 was most significantly affected by lithium treatment, as assessed by the p-value (p=1.12E−09 and 6.97E−13 for two Affymetrix probe sets recognizing CG15088). CG15088 encodes a putative nutrient amino acid transporter of the solute carrier 6 (SLC6) Na+/Cl−-dependent transporter family (Thimgan et al., 2006; Romero-Calderon et al., 2007; Miller et al., 2008). We have recently investigated the role of CG15088 in lithium-responsive biological processes. Using transformants expressing CG15088-specific RNAi and flies deficient in CG15088, we showed that down-regulation or complete elimination of CG15088 function results in a remarkable increase in the susceptibility of adult flies to lithium’s toxic effects, demonstrating the importance of this transporter for resistance to lithium’s toxicity. We have also obtained evidence that this is likely a consequence of its transporter activity in glia of the CNS (Kasuya et al., submitted). This finding strongly supports the expectation that our microarray analysis is effective in revealing new genes of functional significance to the lithium-responsive pathways. Further genetic studies on CG15088 are expected to lead to useful information regarding the mechanisms that underlie lithium-dependent gene regulation and resistance to lithium toxicity.
CG1673 encodes a cytosolic isoform of branched-chain-amino-acid transaminase (BCATc; EC 2.6.1.42). BCAT catalyzes transamination, which is the first reaction in the common degradation pathway for three essential branched-chain amino acids (valine, leucine, and isoleucine). BCAT is also involved in the synthesis of L-glutamate (an important excitatory neurotransmitter both in vertebrates and invertebrates) from these three branched-chain amino acids.
(ii) Genes involved in detoxication, the stress response and self-defense reactions (CG5999, GstD2, Bin1 and Arc1)
Of the 12 genes confirmed to be lithium-responsive in this analysis, CG5999 displayed the largest fold increase in expression, both by microarray and RT-PCR analyses. CG5999 encodes UDP-glucuronosyl/UDP-glucosyltransferase, which catalyzes the addition of the glycosyl group from a UTP-sugar to a small hydrophobic molecule (e.g. an apolar xenobiotic). This is an important step for detoxication, as it converts potentially toxic molecules to less harmful and more water-soluble forms.
GstD2 (CG4181) encodes an isoform of glutathione S transferase (Gst), GstD2 (EC 2.5.1.18) and is up-regulated by lithium. Gst is one of the major detoxication enzymes and plays an essential role in protecting cells against oxidative stress by conjugating the antioxidant glutathione to various oxidized molecules to convert them to nontoxic forms. There are 38 glutathione S transferase genes in the Drosophila genome (Tu and Akgul, 2005). In addition to GstD2, five other Gst genes (GstD1, D10, E6, E7 and E9) were also up-regulated in a lithium-dependent manner, with p-values of 0.005 or less, although fold changes for these genes were less than 2.
Bicoid interacting protein 1 (Bin1) (CG6046) is also highly up-regulated by lithium. Bin1 is an ortholog of the human SAP18 (Sin3 associated polypeptide p18) gene. It encodes a component of the histone acetylase complex that causes transcriptional repression by modifying histones (Zhu et al., 2001). In Arabidopsis, SAP18 loss-of-function mutants are unusually sensitive to NaCl, indicating that this protein is involved in transcriptional repression of genes under conditions of salt stress (Song and Galbraith, 2006).
A lithium-inducible gene, Activity-regulated cytoskeleton-associated protein 1 (Arc1 or CG12505) has been recognized as an immediate early gene whose expression in mammalian CNS neurons is enhanced by synaptic activity (Lyford et al., 1995). This suggests that it may play a role in neuronal plasticity. Arc1 expression is also modulated by stress. The stress-dependent modulation of Arc1 is impaired in the hippocampus-specific glucocorticoid receptor-deficient mouse (GR+/−), a genetic model of predisposition to depression (Molteni et al., 2009). In Drosophila, Arc1 expression is up-regulated by seizure (Guan et al., 2005). Drosophila Arc1 mutants have recently been shown to be more resistant to starvation than wild-type flies, and their ability to survive longer in the absence of food than wild type flies (Mattaliano et al., 2007) may be related to the fact that they lack normal starvation-induced hyperlocomotor activity. Thus, Arc1 plays a role in behavioral responses to metabolic stress in Drosophila.
(iii) Genes potentially related to psychiatric or neurological disorders (Nmdmc, CG9377, and Lsp1γ)
Nmdmc (NAD-dependent methylenetetrahydrofolate dehydrogenase) or CG18466 encodes nicotinamide adenine dinucleotide (NAD)-dependent tri-functional enzyme, and is up-regulated in response to lithium. This enzyme has distinct functional domains for each of its substrates: 5,10-methylenetetrahydrofolatedehydrogenase, 5,10-methenyltetrahydrofolatecyclohydrolase and 10-formylotetrahydrofolatesynthetase. It catalyzes three sequential reactions in the conversion of 1-carbon derivatives of tetrahydrofolate, which are substrates for biosynthesis of the purine nucleotides thymidylate and methionine (Hum et al., 1988). Recent genotype analysis of patients for bipolar disorder and schizophrenia suggests that there is a genetic association between risk of these disorders and polymorphisms in one of the human Nmdmc orthologs (Kempisty et al., 2007).
CG9377 encodes a serine proteinase and is the only down-regulated gene that has been positively scored by both microarray and RT-PCR analyses. Serine proteases and serine proteinase homologs have been suggested to participate in various defense responses (Jiang and Kanost, 2000; Kanost et al., 2001). Cytosolic serine peptidase, which hydrolyzes relatively short proline-containing peptides (prolyl oligopeptidase), has been found to be significantly down-regulated in bipolar disorder patients undergoing lithium treatment (Breen et al., 2004), although the functional significance of prolyl oligopeptidase suppression by lithium is not known.
Lithium treatment increased the expression of Larval serum protein 1 gamma (Lsp1γ) or CG6821. Lsp1γ is one of the major hemolymph proteins in larvae that serve as a nutrient reservoir for the production of adult cuticle structures. Although several different isoforms of the larval serum proteins are expressed in adult, especially in the adipose cells of the head, their function in adult flies remains unknown. Lsp1γ protein is down-regulated significantly (by ~40%) according to proteome analysis of transgenic flies expressing human wild-type α-synuclein; this implies a potential connection between Lsp1γ and a Drosophila model of Parkinson’s disease (Xun et al., 2008).
(iv) Other genes (CG15794, Cyp309a1 and CG7763)
CG15794 is significantly up-regulated in response to lithium. However, its function is not known. A 1.8kb transcript is expected to encode a polypeptide composed of 554 amino acids. It is presumably a soluble protein, and possesses 2 regions of 22 serine-residue stretches separated by ~220 amino acids. Several repetitive motifs (e.g., GXXX or HG repeats) are found throughout the protein. A blast search (http://blast.ncbi.nlm.nih.gov/Blast.cgi) indicates that similar proteins are present in other species of Drosophila, but not in vertebrate organisms.
Lithium treatment also leads to up-regulated expression of Cyp309a1 (CG9964). Cyp309a1 encodes a member of cytochrome P450 enzymes, which catalyze a diverse range of chemical reactions including the oxidization of xenobiotic compounds and the synthesis of endogenous hormones. Notably, the expression levels of several cytochrome P450 enzyme genes also increase with lithium treatment in the human lung carcinoma cell line A549 (Allagui et al., 2007). Although this enzyme may play a role in the detoxication of certain molecules, its specific functions are not yet identified. Among the 87 cytochrome P450 enzyme genes on the chip, this analysis identified cyp309a1 as the only one whose expression is significantly influenced by lithium.
The function of another up-regulated gene, CG7763 is unknown. The predicted gene product contains C-type lectin domain, suggesting that it may encode a type of sugar-binding C-type lectin. Several C-type lectins are involved in immune-system functions in vertebrates (Mukherjee et al., 2009).
Potential significance of amino acid metabolic processes in the genomic response to lithium
Our dual approaches — a direct analysis of lithium-regulated genes and an analysis of biological pathways affected by lithium treatment — have revealed the potential significance of amino acid metabolism and stress response/self-defense reaction in the genomic response to lithium. Several groups have previously performed microarray-based expression profiling in mammalian systems to examine the effects of lithium on gene regulation (Wang et al., 2004; Rojek et al., 2005; Zhang et al., 2005; Blair et al., 2006; Yazlovitskaya et al., 2006; Youngs et al., 2006; McQuillin et al., 2007; Chetcuti et al., 2008; Seelan et al., 2008). In these studies, mRNA samples were isolated from different sources (e.g., primary brain cell cultures, neuronal cell lines and brain tissues of mice, rats or humans) and various conditions were used for lithium treatment. Nevertheless, certain genes and biological pathways were identified as being lithium-responsive in multiple studies.
When mouse and human neuronal cell lines were chronically exposed to lithium, genes that positively and negatively regulate apoptosis were down- and up-regulated, respectively (Yazlovitskaya et al., 2006; Seelan et al., 2008). However, our current analysis did not show any significant change in the expression levels of pro- and anti-apoptotic genes. This discrepancy could be partially due to different durations of lithium treatment. In the mammalian studies, cells were treated with lithium for 7–33 days, whereas flies were exposed to lithium for 24 hr in our experiment.
Consistent with our results, some mammalian microarray studies revealed that genes involved in stress response/self-defense reaction and amino acid metabolism are differentially regulated by lithium. For example, antioxidant genes, such as Gst or Peroxiredoxin 2 (PRDX2), were found to be up-regulated by lithium in primary cultured rat cerebral cortical cells or human neuronal cells (Wang et al., 2004; Seelan et al., 2008). Our study also showed that the levels of several Gst transcripts are increased after lithium treatment, suggesting that up-regulation of antioxidant genes may be an evolutionarily conserved genomic response to lithium. Lithium’s influence on antioxidative systems through the activation of Gst expression may protect the brain from oxidative stress and contribute to its therapeutic action (Wang et al., 2004). In addition, the expression of mammalian genes that are related to amino acid metabolic pathways is significantly affected by lithium treatment. These include genes encoding two aminotrasferases (McQuillin et al., 2007), glutamate ammonia ligase (glutamine synthase) (Chetcuti et al., 2008) and aminobutyl aminotransferase (GABA transaminase) (Zhang et al., 2005). These findings further suggest that amino acids are important to the physiological responses to lithium in different animal species.
Our study shows that the degradation pathways for branched amino acids are specifically enhanced by lithium treatment. What are the implications of this enhanced degradation of branched amino acids for lithium’s effects on nervous system functions? One of the possibilities is its influence on the brain serotonin level. Reduced serotonergic neuronal activity is thought to contribute to the pathogenesis of bipolar affective disorder (van Praag and de Haan, 1980) and, because serotonin is synthesized from tryptophan in neurons, brain tryptophan levels play a critical role in the availability of serotonin. All large neutral amino acids, such as tryptophan and branched amino acids, are transported into the brain across the blood-brain barrier by a single transport system, which means that all of these amino acids compete for the carrier proteins (Oldendorf and Szabo, 1976). Thus, lower levels of branched amino acids result in higher tryptophan — and consequently also higher serotonin — levels in the brain. It has been reported that the tryptophan/neutral amino acid ratio is significantly lower in unipolar and bipolar patients than in control subjects (Lucca et al., 1992) and that lithium decreases plasma levels of isoleucine, leucine, and valine (Leighton et al., 1983). These observations are consistent with the hypothesis that modulation of amino acid profiles contributes to lithium’s therapeutic action. It will therefore be interesting to investigate whether in Drosophila lithium influences brain functions and behaviors by increasing levels of tryptophan and serotonin therein.
The present genome-wide gene expression study has established an important foundation for functional analyses of the mechanisms underlying lithium’s action in the nervous system, using the genetic model organism Drosophila melanogaster. As exemplified by our analysis of the lithium-inducible CG15088, Drosophila offers versatile molecular and genetic tools for functional investigation of the evolutionarily conserved genes in lithium-responsive processes. Similar studies of other genes identified in this microarray analysis are expected to reveal their functional significance in lithium’s biological actions at the molecular, cellular and organismal levels, which should provide novel insights into the basic neurobiological processes regulated by lithium in higher vertebrates including humans.
Supplementary Material
Acknowledgments
We would like to thank Dr. Tom Bair (DNA core facility, University of Iowa) for his assistance with the statistical analyses of microarray data. This study was supported by grants from the NIH (R03 MH078271), American Parkinson’s Disease Association Research and National Alliance for Research on Schizophrenia and Depression (NARSAD) to T.K.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Allagui MS, Vincent C, El Feki A, Gaubin Y, Croute F. Lithium toxicity and expression of stress-related genes or proteins in a549 cells. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 2007;1773:1107–1115. doi: 10.1016/j.bbamcr.2007.04.007. [DOI] [PubMed] [Google Scholar]
- Berger Z, Ttofi EK, Michel CH, Pasco MY, Tenant S, Rubinsztein DC, O’kane CJ. Lithium rescues toxicity of aggregate-prone proteins in drosophila by perturbing wnt pathway. Hum Mol Genet. 2005;14:3003–3011. doi: 10.1093/hmg/ddi331. [DOI] [PubMed] [Google Scholar]
- Berridge MJ, Downes CP, Hanley MR. Neural and developmental actions of lithium: A unifying hypothesis. Cell. 1989;59:411–419. doi: 10.1016/0092-8674(89)90026-3. [DOI] [PubMed] [Google Scholar]
- Blair IP, Chetcuti AF, Badenhop RF, Scimone A, Moses MJ, Adams LJ, Craddock N, Green E, Kirov G, Owen MJ, Kwok JB, Donald JA, Mitchell PB, Schofield PR. Positional cloning, association analysis and expression studies provide convergent evidence that the cadherin gene fat contains a bipolar disorder susceptibility allele. Mol Psychiatry. 2006;11:372–383. doi: 10.1038/sj.mp.4001784. [DOI] [PubMed] [Google Scholar]
- Breen G, Harwood AJ, Gregory K, Sinclair M, Collier D, St Clair D, Williams RS. Two peptidase activities decrease in treated bipolar disorder not schizophrenic patients. Bipolar Disorders. 2004;6:156–161. doi: 10.1111/j.1399-5618.2004.00100.x. [DOI] [PubMed] [Google Scholar]
- Chen RH, Ding WV, Mccormick F. Wnt signaling to beta-catenin involves two interactive components. Glycogen synthase kinase-3beta inhibition and activation of protein kinase. J Biol Chem. 2000;275:17894–17899. doi: 10.1074/jbc.M905336199. [DOI] [PubMed] [Google Scholar]
- Chetcuti A, Adams LJ, Mitchell PB, Schofield PR. Microarray gene expression profiling of mouse brain mrna in a model of lithium treatment. Psychiatr Genet. 2008;18:64–72. doi: 10.1097/YPG.0b013e3282fb0051. [DOI] [PubMed] [Google Scholar]
- Chuang DM. Neuroprotective and neurotrophic actions of the mood stabilizer lithium: Can it be used to treat neurodegenerative diseases? Crit Rev Neurobiol. 2004;16:83–90. doi: 10.1615/critrevneurobiol.v16.i12.90. [DOI] [PubMed] [Google Scholar]
- Davis RL. Olfactory memory formation in drosophila: From molecular to systems neuroscience. Annu Rev Neurosci. 2005;28:275–302. doi: 10.1146/annurev.neuro.28.061604.135651. [DOI] [PubMed] [Google Scholar]
- Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. David: Database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- Ding VW, Chen RH, Mccormick F. Differential regulation of glycogen synthase kinase 3beta by insulin and wnt signaling. J Biol Chem. 2000;275:32475–32481. doi: 10.1074/jbc.M005342200. [DOI] [PubMed] [Google Scholar]
- Dokucu ME, Yu L, Taghert PH. Lithium- and valproate-induced alterations in circadian locomotor behavior in drosophila. Neuropsychopharmacology. 2005;30:2216–2224. doi: 10.1038/sj.npp.1300764. [DOI] [PubMed] [Google Scholar]
- Dougherty ER, Barrera J, Brun M, Kim S, Cesar RM, Chen Y, Bittner M, Trent JM. Inference from clustering with application to gene-expression microarrays. J Comput Biol. 2002;9:105–126. doi: 10.1089/10665270252833217. [DOI] [PubMed] [Google Scholar]
- Gabler M, Volkmar M, Weinlich S, Herbst A, Dobberthien P, Sklarss S, Fanti L, Pimpinelli S, Kress H, Reuter G, Dorn R. Trans-splicing of the mod(mdg4) complex locus is conserved between the distantly related species drosophila melanogaster and d. Virilis. Genetics. 2005;169:723–736. doi: 10.1534/genetics.103.020842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Z, Saraswati S, Adolfsen B, Littleton JT. Genome-wide transcriptional changes associated with enhanced activity in the drosophila nervous system. Neuron. 2005;48:91–107. doi: 10.1016/j.neuron.2005.08.036. [DOI] [PubMed] [Google Scholar]
- Hamet P, Tremblay J. Genetics of the sleep-wake cycle and its disorders. Metabolism. 2006;55:S7–12. doi: 10.1016/j.metabol.2006.07.006. [DOI] [PubMed] [Google Scholar]
- Huang Da W, Sherman BT, Tan Q, Kir J, Liu D, Bryant D, Guo Y, Stephens R, Baseler MW, Lane HC, Lempicki RA. David bioinformatics resources: Expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007;35:W169–175. doi: 10.1093/nar/gkm415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hum DW, Bell AW, Rozen R, Mackenzie RE. Primary structure of a human trifunctional enzyme. Isolation of a cdna encoding methylenetetrahydrofolate dehydrogenase-methenyltetrahydrofolate cyclohydrolase-formyltetrahydrofolate synthetase. J Biol Chem. 1988;263:15946–15950. [PubMed] [Google Scholar]
- Iitaka C, Miyazaki K, Akaike T, Ishida N. A role for glycogen synthase kinase-3beta in the mammalian circadian clock. J Biol Chem. 2005;280:29397–29402. doi: 10.1074/jbc.M503526200. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Jiang H, Kanost MR. The clip-domain family of serine proteinases in arthropods. Insect Biochemistry and Molecular Biology. 2000;30:95–105. doi: 10.1016/s0965-1748(99)00113-7. [DOI] [PubMed] [Google Scholar]
- Kanost MR, Jiang H, Wang Y, Yu XQ, Ma C, Zhu Y. Hemolymph proteinases in immune responses of manduca sexta. Advances in experimental medicine and biology. 2001;484:319–328. doi: 10.1007/978-1-4615-1291-2_32. [DOI] [PubMed] [Google Scholar]
- Kempisty B, Sikora J, Lianeri M, Szczepankiewicz A, Czerski P, Hauser J, Jagodzinski PP. Mthfd 1958g>a and mtr 2756a>g polymorphisms are associated with bipolar disorder and schizophrenia. Psychiatr Genet. 2007;17:177–181. doi: 10.1097/YPG.0b013e328029826f. [DOI] [PubMed] [Google Scholar]
- Klein PS, Melton DA. A molecular mechanism for the effect of lithium on development. Proc Natl Acad Sci U S A. 1996;93:8455–8459. doi: 10.1073/pnas.93.16.8455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leighton WP, Rosenblatt S, Chanley JD. Lithium-induced changes in plasma amino acid levels during treatment of affective disorders. Psychiatry Res. 1983;8:33–40. doi: 10.1016/0165-1781(83)90136-1. [DOI] [PubMed] [Google Scholar]
- Lucca A, Lucini V, Piatti E, Ronchi P, Smeraldi E. Plasma tryptophan levels and plasma tryptophan/neutral amino acids ratio in patients with mood disorder, patients with obsessive-compulsive disorder, and normal subjects. Psychiatry Res. 1992;44:85–91. doi: 10.1016/0165-1781(92)90043-3. [DOI] [PubMed] [Google Scholar]
- Lyford GL, Yamagata K, Kaufmann WE, Barnes CA, Sanders LK, Copeland NG, Gilbert DJ, Jenkins NA, Lanahan AA, Worley PF. Arc, a growth factor and activity-regulated gene, encodes a novel cytoskeleton-associated protein that is enriched in neuronal dendrites. Neuron. 1995;14:433–445. doi: 10.1016/0896-6273(95)90299-6. [DOI] [PubMed] [Google Scholar]
- Mattaliano MD, Montana ES, Parisky KM, Littleton JT, Griffith LC. The drosophila arc homolog regulates behavioral responses to starvation. Molecular and cellular neurosciences. 2007;36:211–221. doi: 10.1016/j.mcn.2007.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcbride SM, Choi CH, Wang Y, Liebelt D, Braunstein E, Ferreiro D, Sehgal A, Siwicki KK, Dockendorff TC, Nguyen HT, Mcdonald TV, Jongens TA. Pharmacological rescue of synaptic plasticity, courtship behavior, and mushroom body defects in a drosophila model of fragile x syndrome. Neuron. 2005;45:753–764. doi: 10.1016/j.neuron.2005.01.038. [DOI] [PubMed] [Google Scholar]
- Mcquillin A, Rizig M, Gurling HM. A microarray gene expression study of the molecular pharmacology of lithium carbonate on mouse brain mrna to understand the neurobiology of mood stabilization and treatment of bipolar affective disorder. Pharmacogenet Genomics. 2007;17:605–617. doi: 10.1097/FPC.0b013e328011b5b2. [DOI] [PubMed] [Google Scholar]
- Miklos GL, Rubin GM. The role of the genome project in determining gene function: Insights from model organisms. Cell. 1996;86:521–529. doi: 10.1016/s0092-8674(00)80126-9. [DOI] [PubMed] [Google Scholar]
- Miller MM, Popova LB, Meleshkevitch EA, Tran PV, Boudko DY. The invertebrate b(0) system transporter, d. Melanogaster nat1, has unique d-amino acid affinity and mediates gut and brain functions. Insect Biochem Mol Biol. 2008;38:923–931. doi: 10.1016/j.ibmb.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min WW, Yuskaitis CJ, Yan Q, Sikorski C, Chen S, Jope RS, Bauchwitz RP. Elevated glycogen synthase kinase-3 activity in fragile x mice: Key metabolic regulator with evidence for treatment potential. Neuropharmacology. 2009;56:463–472. doi: 10.1016/j.neuropharm.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molteni R, Calabrese F, Chourbaji S, Brandwein C, Racagni G, Gass P, Riva M. Depression-prone mice with reduced glucocorticoid receptor expression display an altered stress-dependent regulation of brain-derived neurotrophic factor and activity-regulated cytoskeleton-associated protein. J Psychopharmacol. 2009 doi: 10.1177/0269881108099815. [DOI] [PubMed] [Google Scholar]
- Mudher A, Shepherd D, Newman TA, Mildren P, Jukes JP, Squire A, Mears A, Drummond JA, Berg S, Mackay D, Asuni AA, Bhat R, Lovestone S. Gsk-3beta inhibition reverses axonal transport defects and behavioural phenotypes in drosophila. Mol Psychiatry. 2004;9:522–530. doi: 10.1038/sj.mp.4001483. [DOI] [PubMed] [Google Scholar]
- Mukherjee S, Partch CL, Lehotzky RE, Whitham CV, Chu H, Bevins CL, Gardner KH, Hooper LV. Regulation of c-type lectin antimicrobial activity by a flexible n-terminal prosegment. J Biol Chem. 2009;284:4881–4888. doi: 10.1074/jbc.M808077200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldendorf WH, Szabo J. Amino acid assignment to one of three blood-brain barrier amino acid carriers. Am J Physiol. 1976;230:94–98. doi: 10.1152/ajplegacy.1976.230.1.94. [DOI] [PubMed] [Google Scholar]
- Padiath QS, Paranjpe D, Jain S, Sharma VK. Glycogen synthase kinase 3beta as a likely target for the action of lithium on circadian clocks. Chronobiol Int. 2004;21:43–55. doi: 10.1081/cbi-120027981. [DOI] [PubMed] [Google Scholar]
- Phiel CJ, Klein PS. Molecular targets of lithium action. Annu Rev Pharmacol Toxicol. 2001;41:789–813. doi: 10.1146/annurev.pharmtox.41.1.789. [DOI] [PubMed] [Google Scholar]
- Reiter LT, Potocki L, Chien S, Gribskov M, Bier E. A systematic analysis of human disease-associated gene sequences in drosophila melanogaster. Genome Res. 2001;11:1114–1125. doi: 10.1101/gr.169101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojek A, Nielsen J, Brooks HL, Gong H, Kim YH, Kwon TH, Frokiaer J, Nielsen S. Altered expression of selected genes in kidney of rats with lithium-induced ndi. American journal of physiology. 2005;288:F1276–1289. doi: 10.1152/ajprenal.00305.2004. [DOI] [PubMed] [Google Scholar]
- Romero-Calderon R, Shome RM, Simon AF, Daniels RW, Diantonio A, Krantz DE. A screen for neurotransmitter transporters expressed in the visual system of drosophila melanogaster identifies three novel genes. Dev Neurobiol. 2007;67:550–569. doi: 10.1002/dneu.20342. [DOI] [PubMed] [Google Scholar]
- Rowe MK, Chuang DM. Lithium neuroprotection: Molecular mechanisms and clinical implications. Expert Rev Mol Med. 2004;6:1–18. doi: 10.1017/S1462399404008385. [DOI] [PubMed] [Google Scholar]
- Rubin GM, Yandell MD, Wortman JR, Gabor Miklos GL, Nelson CR, Hariharan IK, Fortini ME, Li PW, Apweiler R, Fleischmann W, Cherry JM, Henikoff S, Skupski MP, Misra S, Ashburner M, Birney E, Boguski MS, Brody T, Brokstein P, Celniker SE, Chervitz SA, Coates D, Cravchik A, Gabrielian A, Galle RF, Gelbart WM, George RA, Goldstein LS, Gong F, Guan P, Harris NL, Hay BA, Hoskins RA, Li J, Li Z, Hynes RO, Jones SJ, Kuehl PM, Lemaitre B, Littleton JT, Morrison DK, Mungall C, O’farrell PH, Pickeral OK, Shue C, Vosshall LB, Zhang J, Zhao Q, Zheng XH, Lewis S. Comparative genomics of the eukaryotes. Science. 2000;287:2204–2215. doi: 10.1126/science.287.5461.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schou M. Lithium treatment at 52. J Affect Disord. 2001;67:21–32. doi: 10.1016/s0165-0327(01)00380-9. [DOI] [PubMed] [Google Scholar]
- Seelan RS, Khalyfa A, Lakshmanan J, Casanova MF, Parthasarathy RN. Deciphering the lithium transcriptome: Microarray profiling of lithium-modulated gene expression in human neuronal cells. Neuroscience. 2008;151:1184–1197. doi: 10.1016/j.neuroscience.2007.10.045. [DOI] [PubMed] [Google Scholar]
- Song CP, Galbraith DW. Atsap18, an orthologue of human sap18, is involved in the regulation of salt stress and mediates transcriptional repression in arabidopsis. Plant Mol Biol. 2006;60:241–257. doi: 10.1007/s11103-005-3880-9. [DOI] [PubMed] [Google Scholar]
- Thimgan MS, Berg JS, Stuart AE. Comparative sequence analysis and tissue localization of members of the slc6 family of transporters in adult drosophila melanogaster. J Exp Biol. 2006;209:3383–3404. doi: 10.1242/jeb.02328. [DOI] [PubMed] [Google Scholar]
- Tu CP, Akgul B. Drosophila glutathione s-transferases. Methods Enzymol. 2005;401:204–226. doi: 10.1016/S0076-6879(05)01013-X. [DOI] [PubMed] [Google Scholar]
- Van Praag HM, De Haan S. Central serotonin deficiency--a factor which increases depression vulnerability? Acta Psychiatr Scand Suppl. 1980;280:89–96. [PubMed] [Google Scholar]
- Wang JF, Shao L, Sun X, Young LT. Glutathione s-transferase is a novel target for mood stabilizing drugs in primary cultured neurons. J Neurochem. 2004;88:1477–1484. doi: 10.1046/j.1471-4159.2003.02276.x. [DOI] [PubMed] [Google Scholar]
- Williamson RL. Lithium stops hereditary shuddering in drosophila melanogaster. Psychopharmacology (Berl) 1982;76:265–268. doi: 10.1007/BF00432558. [DOI] [PubMed] [Google Scholar]
- Xun Z, Kaufman TC, Clemmer DE. Proteome response to the panneural expression of human wild-type alpha-synuclein: A drosophila model of parkinson’s disease. J Proteome Res. 2008;7:3911–3921. doi: 10.1021/pr800207h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazlovitskaya EM, Edwards E, Thotala D, Fu A, Osusky KL, Whetsell WO, Jr, Boone B, Shinohara ET, Hallahan DE. Lithium treatment prevents neurocognitive deficit resulting from cranial irradiation. Cancer research. 2006;66:11179–11186. doi: 10.1158/0008-5472.CAN-06-2740. [DOI] [PubMed] [Google Scholar]
- Youngs RM, Chu MS, Meloni EG, Naydenov A, Carlezon WA, Jr, Konradi C. Lithium administration to preadolescent rats causes long-lasting increases in anxiety-like behavior and has molecular consequences. J Neurosci. 2006;26:6031–6039. doi: 10.1523/JNEUROSCI.0580-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang WV, Jullig M, Connolly AR, Stott NS. Early gene response in lithium chloride induced apoptosis. Apoptosis. 2005;10:75–90. doi: 10.1007/s10495-005-6063-x. [DOI] [PubMed] [Google Scholar]
- Zhu W, Foehr M, Jaynes JB, Hanes SD. Drosophila sap18, a member of the sin3/rpd3 histone deacetylase complex, interacts with bicoid and inhibits its activity. Development genes and evolution. 2001;211:109–117. doi: 10.1007/s004270100135. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.