Abstract
Antigen presenting molecules play an important role in both innate and adoptive immune responses by priming and activating T cells. Among them, CD1 molecules have been identified to present both exogenous and endogenous lipid antigens to CD1-restricted T cells. The involvement of CD1-restricted T cells in autoimmune diseases and in defense against infectious diseases, however, remains largely unknown. Identifying novel antigenic lipids that bind to CD1 molecules and understanding the role of CD1-restricted T cells should lead to the successful development of vaccines, because the lipids can be used as antigens and also as adjuvants. In this paper, we have constructed functional recombinant human CD1 dimeric proteins and established a competitive ELISA assay to measure the lipid binding to CD1 molecules using the CD1 dimers. By using the competitive ELISA assay, we were able to show that the lipid extracts from murine malaria parasites can actually be loaded onto CD1 molecules. In addition, we have demonstrated that artificial antigen-presenting cells, which consist of magnetic beads coated with CD1d dimer and anti-CD28 antibody, stimulated and expanded human invariant NKT cells as efficiently as autologous immature DCs. A set of the tools presented in the current study should be valuable for screening various CD1 molecule-binding lipid antigens and for isolating CD1-restricted T cells.
Keywords: CD1, lipid antigen, artificial antigen presenting cells, NKT cells
1. Introduction
Antigen presentation by antigen-presenting cells (APCs) to naïve T cells is a fundamental process in developing adoptive immune response. T cells recognize a large variety of antigenic molecules, such as peptides and lipids, in the context of antigen-presenting molecules. MHC class I/II molecules present peptide antigens, whereas CD1 molecules present lipid antigens of both self and pathogens to T cells (Rosat et al., 1999; Moody et al., 2000; Shamshiev et al., 2002; Wu et al., 2003; Agea et al., 2005; Vincent et al., 2005; Tsuji 2006; Russano et al., 2007). CD1 is a family of cell surface glycoproteins that are distantly related in structure to MHC molecules. In humans, five CD1 molecules (CD1a-e) are encoded on chromosome 1 and these molecules can be divided into group I (CD1a, b and c), group II (CD1d) and CD1e.
Among CD1 molecules, the physiological role of CD1d has been studied most intensively for the following reasons: (1) potent ligands for CD1d, such as α-galactosylceramide (α-GalCer) and its analogs have been identified (Kawano et al., 1997; Miyamoto et al., 2001; Schmieg et al., 2003; Yu et al., 2005), (2) mouse model can be used to evaluate the function of CD1d in vivo, because functional CD1d molecules (CD1d1 and CD1d2) are expressed in mice (Bradbury et al., 1988), (3) humans and mice have a unique T cell subpopulation called invariant NKT cells (iNKT cells), which recognizes CD1d-ligand complex. iNKT cells have a specific TCR α-chain (Vα24 in humans; Vα14 in mice) (Lantz and Bendelac, 1994), and play a variety of roles in immune system by producing Th1 and Th2 cytokines rapidly upon activation (reviewed by Van Kaer, 2007; Cerundolo et al., 2009). Therefore, CD1d ligands have a wide range of biological activities in vivo such as anti-tumor (Kawano et al., 1998), anti-autoimmune disease (Sharif et al. 2001) and adjuvant activity (Gonzalez-Aseguinolaza et al., 2002).
On the other hand, group I CD1-restricted T cells have been shown to have a variety of TCR α-chain (Grant et al., 1999), and, in some cases, lipid antigen-specific T cells can be detected only in people who have exposed to the pathogen which contains the lipid antigens (Moody et al., 2000; Agea et al., 2005).
Self lipids and pathogen-derived lipids, such as mycolic acid (Beckman et al., 1994), phosphatidylinositol mannosides (Sieling et al., 1995) and sulphatide (Shamshiev et al., 2002), have been identified as group I CD1 ligands and shown to be able to stimulate group I CD1-restricted T cells. One of the most characterized group I CD1 ligand is mycobacterial lipid, and an immunization of guinea pigs, which have functional group I CD1s (Dascher et al., 1999), with mycobacterial lipid antigens has been shown to induce the activation of CD1-restricted T cells and improve pulmonary pathology in an aerosol tuberculosis challenge model (Dascher et al., 2003). In addition, CD1-restricted T cells seem to be involved in an allergic reaction, because pollen lipids-reactive, CD1-restriced T cells were detected in allergic subjects during pollinating season (Agea et al., 2005). These studies indicate that CD1-restricetd T cells play a crucial role in protection to pathogens, allergic and autoimmune diseases in a similar manner to that played by MHC-restricted T cells (reviewed by Brigl and Brenner, 2004; Barral and Brenner, 2007).
Although the important role of CD1 molecules in immune system has been suggested, only a limited number of studies have actually been done to identify and isolate lipid antigens from pathogens or self tissues. One of the reasons for this might be that less experimental tools are available for the analysis of CD1-binding lipids and CD1-restricted T cells. For example, several methods, such as fluorescent lipid probes (Im et al., 2004), Isoelectric-Forcusing Electrophoresis (IFE) (Cantu et al., 2003) and Surface Plasmon Resonance (SRP) (Naidenko et al., 1999), have been applied to measure the binding of the lipid to CD1 molecules, but some of these assays have required direct labeling of the lipids with a tag molecule like biotin, and, therefore, are not suitable for high throughput screening.
In this paper, we constructed recombinant human CD1 (hCD1) dimeric proteins, which are covalently linked beta-2 microglobulin (β2m) at N-terminus and mouse IgG1 at C-terminus, and showed that these hCD1 dimers are functional in staining and stimulating human CD1-restricted T cells. Using these CD1 dimers, we established a novel competitive ELISA assay to measure the binding of the lipid to CD1 molecules and a bead-based artificial antigen-presenting cell (aAPC) system to stimulate and expand CD1-restricted T cells.
2. Materials and Methods
2.1 Cell lines
Murine myeloma cell line, J558L, was kindly provided by Dr. Jonathan P. Schneck. J558L cells were maintained in RPMI-1640 (Invitrogen, Carlsbad, CA, USA) supplemented with 10%FBS, 1mM sodium pyruvate, 100μM 2-mercaptoethanol, 100μg/mL Gentamicin, 20 IU/mL penicillin, and 20mg/mL streptomycin. Human invariant NKT cell lines were generated as described previously (Kinjo et al., 2006). Briefly, Vα24+ T cells were isolated from PBMC using magnetic beads (Miltenyi Biotec, Gladbach, Germany) coupled to mouse monoclonal anti-human Vα24 antibody (Immunotech Research, Quebec, Canada) and were cultured for 10–14 days with α-GalCer-pulsed autologous immature DC in the presence of 10IU/mL human IL-2 (Roche, Basel, Switzerland). After a single cycle of stimulation, approximately 90% of NKT cells were shown to be Vα24+ cells by FACS.
2.2 Lipid antigens
18:1 Biotinyl PE (1,2-Dioleoyl-sn-Glycero-3-Phosphoethanolamine-N-(Biotinyl)) and 18:1 Caproylamine PE (1,2-Dioleoyl-sn-Glycero-3-Phosphoethanolamine-N-(hexanoylamine)) were purchased from Avanti Polar Lipids (Alabaster, AL, USA). Mycolic acid was from Sigma-Aldrich (St. Louis, MO, USA). α-galactosylceramide (α-GalCer) was a gift from the Kirin pharmaceutical research corporation (Gunma, Japan).
2.3 Generation of hCD1-mIgG dimers
The whole coding regions of human CD1b, CD1c, CD1d and beta-2 microglobulin (β2m) were PCR-amplified from human thymus cDNA (Clontech, Mountain View, CA, USA) and cloned into pcDNA3.1 (Invitrogen, Carlsbad, CA, USA). In this cloning step, XhoI site in CD1b was mutated by introducing a synonymous mutation at this site. To construct expression vectors of β2m-CD1-mouse IgG (mIgG) dimers, β2m coding region without a signal sequence was connected in frame to cDNA corresponding to α1-α3 domains of CD1s with a flexible peptide linker sequence (GGGSGGSGSGGGA) (Im et al., 2004) by two-step PCR. Briefly, β2m with a flexible linker sequence was amplified with a forward primer containing a MluI site and a reverse primer containing the linker sequence at 5′ region. α1-α3 domains of CD1s with the linker sequence were amplified with forward primers containing the linker sequence and reverse primers containing a XhoI site. The PCR products were connected by second PCR using the β2m forward primer and the CD1 reverse primers. The products were digested by MluI and XhoI, and subcloned at the same site of pT7blue vector (Novagen, Madison, WI, USA). After confirming the sequence, the MluI-XhoI fragments were cloned in frame into the upstream of the murine IgG1-VH region in pXIg plasmid (Dal Porto et al., 1993). J558L cells, which are deficient in synthesizing Ig H chains, but capable of producing Igλ chains, were transfected with the plasmids by nucleofector (Lonza Cologne AG, Cologne, Germany) and stable clones were isolated by a limiting dilution method in the presence of 0.8mg/mL G418 (Invitrogen, Carlsbad, CA, USA).
To construct expression vectors of CD1-mIgG dimers, α1-α3 domains of CD1s were amplified with forward primers containing MluI site and reverse primers containing XhoI site, and inserted into the same site of pX/Ig. J558L cells were stably transfected with β2m/pcDNA3.1/Zeo (+) and CD1/pX/Ig plasmids, and stable clones were isolated in the presence of 0.8mg/mL G418 and 1mg/mL Zeocin (Invitrogen, Carlsbad, CA, USA). Stable clones expressing high amount of CD1-IgG dimers were selected by ELISA, and used for large scale production of CD1-IgG dimers. For CD1 dimers production, the clones were cultured in Hybridoma-SFM medium (Invitrogen, Carlsbad, CA, USA), and the culture supernatant was collected and applied onto Protein G sepharose 4B column (GE Healthcare, Giles, United Kingdom). The column was washed with 0.1M glycine buffer (pH4.5) and CD1-IgG dimers were eluted by 0.1M glycine buffer (pH2.8).
2.4 Staining of human iNKT cells with hCD1-mIgG dimers
One microgram of human CD1d-mouse IgG dimer was incubated with 1.4 μg α-GalCer in 50μL of PBS at 37°C for overnight to load the lipid onto hCD1d-mIgG dimer and used to stain 2×105 Human iNKT cells on ice for 30 min. Then the cells were washed with PBS containing 5% FBS twice and incubated with phycoerythrin-labeled rat anti-mouse IgG1 antibody (A85-1) and APC (allophycocyanin)-labeled anti-human CD3ε antibody (BD Biosciences, San Diego, CA, USA) on ice for 30 min. After washing, the stained cells were analyzed with FACSCalibur System (BD Biosciences, San Diego, CA, USA). Flow cytometry data was analyzed with FlowJo v8.8 software (Tree Star, Inc, Ashland, OR).
2.5 Stimulation of human iNKT cells with hCD1-mIgG dimers
96 well U bottom plates (Becton Dickinson, Franklin Lakes, NJ, USA) were coated for overnight with 1μg CD1d dimer loaded with α-GalCer as described above. The plates were washed with PBS twice, and then 3×104 human iNKT cells were added to the plates. After 24 hour culture, the culture supernatant was collected and IFN-γ concentration was measured by ELISA (eBioscience, San Diego, CA, USA).
2.6 Competitive ELISA assay
Nunc MaxiSorp flat-bottom 96 well plates (Thermo Fisher Scientific, Waltham, MA, USA) were coated with 100μL of Goat anti-mouse IgG Fc gamma antibody (Biomedia, New York, NY, USA) (10μg/mL in 0.1 M NaHCO3, pH 9.6) for overnight at 4°C, and the plates were washed with PBST (PBS containing 0.05% Tween-20) three times and blocked with 1x assay diluent (eBioscience, San Diego, CA, USA) for 1 hour. The plates were washed three times with PBST and 100μL of lipid-CD1 dimer mixture was added to the plates immediately after washing. The mixture solution was prepared by mixing CD1dimer (5μg/mL) and lipids in the presence of Biotinyl PE (2μg/mL) in PBS. In view of the slow kinetics of Biotinyl PE binding to hCD1d (Fig. 3C), we incubated the mixture for overnight at 37°C. After that, the plates were washed three times with PBST and CD1-Biotinyl PE complex was detected with HRP-labeled Avidin (eBioscience, San Diego, CA, USA). Kd of Biotinyl PE was determined by titrating the amount of Biotinyl PE to reach the maximum binding in the absence of competitors and applying the following equation to the data; Y=(Bmax* X)/(Kd + X). Kd of Biotinyl PE and Ki of PE and α-GalCer were calculated using GraphPad Prism (Ver.4.02) (GraphPad Software, Inc., La Jolla, CA, USA).
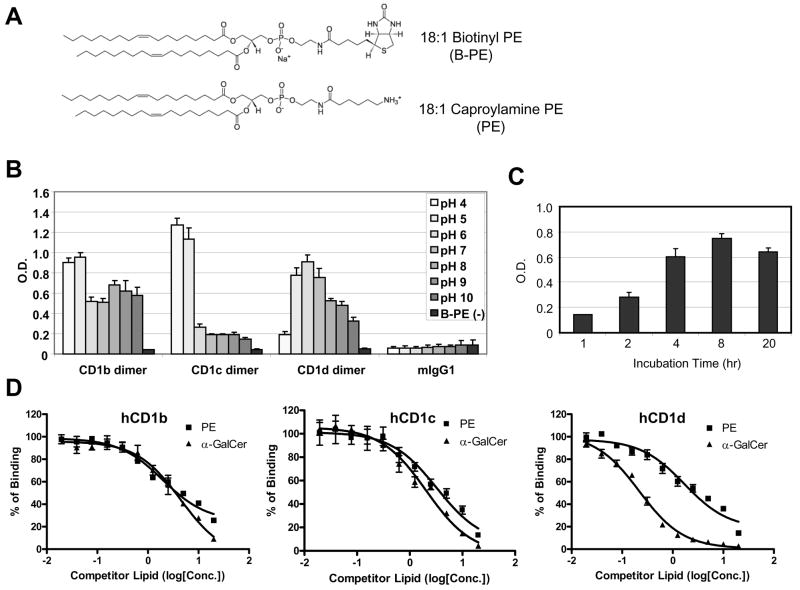
Fig. 3. Competitive ELISA assay to detect the binding of the lipid to hCD1 molecules.
A) Chemical structures of lipids used in the competitive ELISA assay. 18:1 Biotinyl PE was used as a detector lipid and 18:1 Caproylamine was used as a competitor lipid. B) Binding of Biotinyl PE to human CD1 molecules in different pH conditions. The pH of phosphate buffer saline was adjusted with HCl or NaOH to the indicated pH. Biotinyl PE (1μg/mL) was incubated with hCD1 dimers in the indicated pH conditions for overnight at 37°C in ELISA plates coated with anti-mouse IgG Fc antibody. mIgG1 is mouse isotype control IgG1, which has the same structure as hCD1 dimers except for the β2m-CD1 portion. B-PE (−) is the well without biotinylated PE at pH 7. The assay was done in triplicate and the error bars indicate Standard Deviation (SD). C) Kinetics of Biotinyl PE binding to hCD1d dimer. Biotinyl PE (1μg/mL) was incubated with hCD1 dimers for the indicated time at 37°C in ELISA plates coated with anti-mouse IgG Fc antibody. The assay was done in triplicate and the error bars indicate Standard Deviation (SD). D) Results of the competition ELISA assays with PE or α-GalCer. Biotinyl PE (2μg/mL) was incubated with hCD1b (left), hCD1c (middle) or hCD1d (right) dimer in the presence of the competitors at the indicated concentration in ELISA plates described in C). The assays were done in triplicate and the error bars indicate Standard Deviation (SD). X-axis represents concentration of a competitor lipid (log [μM]).
2.7 Lipid extraction from malaria parasites
Malaria lipids were extracted from blood stages of Plasmodium yoelii parasites. The parasites were collected from heparin-treated blood from malaria-infected C57BL/6 mice (Marchesini et al., 2000). In brief, the blood was diluted with PBS and applied onto Whatman CF11 (GE Healthcare, Giles, United Kingdom) cellulose column to remove leucocytes and platelets, and RBCs were collected from the flow through fraction. Then, infected RBCs were separated from uninfected RBCs by percoll centrifugation. The purified infected RBCs were treated with saponin to destroy RBC membrane and the parasites were pelleted and washed with PBS. The purified parasites were sonicated in PBS, lyophilized, and extracted with Chloroform-MeOH (2:1). The extract was dried and partitioned between water (Aqueous fraction) and water-saturated 1-butanol (1:2) (Organic fraction). The insoluble pellet after Chloroform-MeOH extraction was dried and extracted with water (Protein fraction). All of the extracts were dried or lyophilized and resuspended in 100% methanol or Water.
2.8 Artificial antigen-presenting cells (aAPCs) coated with hCD1d dimer
M-450 Tosylactivated Dynabeads (Dynal Biotech ASA, Oslo, Norway) were coated with human CD1d dimer with or without anti-human CD28 antibody (BD Biosciences, San Diego, CA, USA) as described in the instruction manual, and used as artificial APCs coated with hCD1d dimer, hCD1d-aAPC. In brief, the beads were washed with Buffer 1 (0.1M Sodium Borate, pH 8.5) twice and resuspended in Buffer 1 at the concentration of 4×108 beads/mL. CD1dimer (40–200μg/mL) and anti-human CD28 antibody (200–400μg/mL) was added to the beads, and the mixture was incubated at 37 °C with rotating for 30 min. After incubation, BSA was added to the beads at 0.1% w/v and the mixture was incubated at 37 °C with rotating for overnight. The Beads were washed twice for 5min with Buffer 2 (0.1% BSA, 2mM EDTA in PBS), and then incubated at 37°C for overnight again in Buffer 3 (0.2M Tris, 0.1% BSA, pH 8.5) to block the beads. After blocking, the beads were washed once for 5min with Buffer 2 and resuspended in Buffer 2. For lipid loading, the beads were washed with PBS twice and incubated with lipid for overnight with rotating.
To stimulate human iNKT cell, the CD1d-aAPCs loaded with α-GalCer were resuspended in medium and added to 2×104 NKT cells in a 96 well U-bottom plate at the ratio of 1:3 (iNKT cells : Beads). IFN-γ in the culture supernatant was measured 24 hours after stimulation, as described above.
For human iNKT cell expansion assay, 1×107 human iNKT cells were suspended in 1mL of PBS containing 0.1% BSA, and, then, CFSE was added to the cells at a final concentration of 0.5μM. The cells were incubated at 37°C for 5 min and washed three times with medium. 5×105 CFSE-labeled iNKT cells were seeded in 48 well plates, cultured with hCD1d-aAPCs loaded with α-GalCer, or autologous human DCs pulsed with α-GalCer, for one week and then analyzed by a flow cytometric assay.
2.9 Statistical Analysis
All data were expressed as the mean ± standard deviation (S.D.) of triplicate determinations for each group. Statistical analysis of experimental and control data was evaluated by Student’s t-test. A value of P < 0.05 was considered statistically significant.
3. Results
3.1 Generation of human CD1 dimeric proteins
Recombinant human CD1 (hCD1) proteins are useful tools to analyze hCD1-restricted T cells and to screen lipids which can be presented by hCD1 molecules. Therefore, we generated two different types of hCD1-IgG fusion proteins, namely β2m/hCD1-IgG and β2m-hCD1-IgG. β2m/hCD1-IgG is hCD1-IgG fusion protein that is noncovalently associated with human β2m, which is secreted as a complex from a stable cell line transfected with β2m and hCD1-IgG expression vectors (Fig. 1A, left). β2m-hCD1-IgG is hCD1-IgG fusion protein, which has human β2m covalently linked to the N-terminus of hCD1 with a flexible linker sequence (Fig. 1A, right). For the production of hCD1-IgG fusion proteins, stably transfected J558L cells were cultured in the serum free medium, and the fusion proteins were purified from the culture supernatants with Protein G sepharose column.
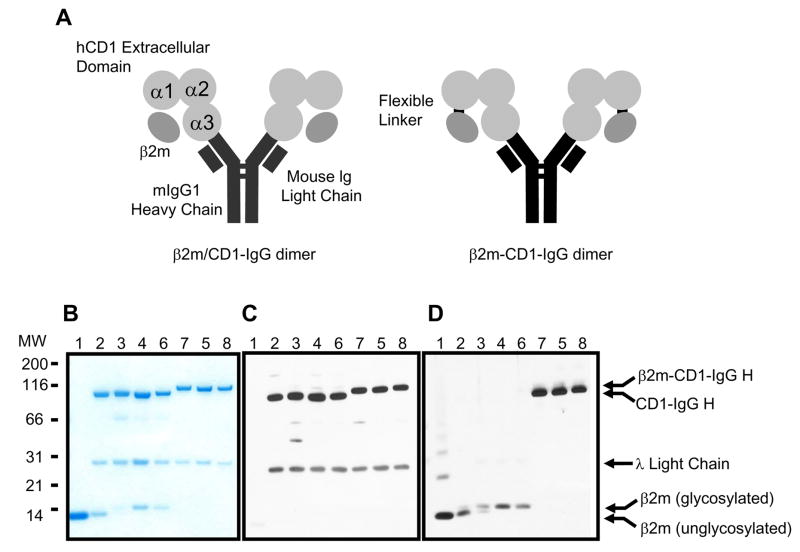
Fig. 1. Recombinant Human CD1-mouse IgG dimers associated with human β2-microglobulin.
A) Schematic structures of human CD1 dimers, β2m/CD1-IgG dimer (left) and β2m-CD1-IgG dimer (right). α1, α2, α3 represent the extracellular domains of human CD1 molecules. The Flexible linker is the sequence (GGGSGGSGSGGGA) used to link β2m to the N-terminus of human CD1 molecules. B) SDS-PAGE and CBB staining of β2m/CD1-IgG and β2m-CD1-IgG dimers. Lane 1: recombinant human β2m (BD Biosciences, San Diego, CA, USA), 2: recombinant human CD1d dimer (BD Biosciences, San Diego, CA, USA), 3: β2m/CD1b-IgG dimer, 4: β2m/CD1c-IgG dimer, 5: β2m/CD1d-IgG dimer, 6: β2m-CD1b-IgG dimer, 7: β2m-CD1c-IgG dimer, 8: β2m-CD1d-IgG dimer. C) Western blotting with HRP-labeled anti-mouse IgG antibody. The lower bands are mouse immunoglobulin λ light chains produced by J558L cells and associated with CD1-immunoglobulin heavy chains. D) Western blotting with anti-human β2m antibody.
SDS-PAGE analysis of the fusion protein showed that the purity was comparable to the commercially available hCD1d-IgG dimer (BD Biosciences, San Diego, CA, USA) (Fig. 1B). β2m could be purified with a protein G column from the supernatants of β2m/hCD1-IgG expressing cells, indicating that noncovalent binding of β2m to hCD1 is relatively strong. The structures of the proteins were confirmed by immunoblot analysis using anti-mouse IgG antibody (Fig. 1C) and anti-human β2m antibody (Fig. 1D). The difference in size of β2m came from the difference in glycosylation (Tubbs et al., 2001). The upper bands of β2m-hCD1-IgGs were recognized by both anti-β2m antibody and anti-mouse IgG antibody, indicating that β2m was covalently bound to hCD1-IgGs.
3.2 Functional analysis of recombinant hCD1d dimers
To evaluate the functionality of the recombinant hCD1 dimers, we first stained human iNKT cells with α-GalCer-loaded β2m/hCD1d-IgG or β2m-hCD1d-IgG. Human iNKT cells were stained only with α-GalCer-loaded CD1d dimers, and there was no difference in the intensity of the staining between β2m/hCD1d-IgG and β2m-hCD1d-IgG (Fig. 2A).
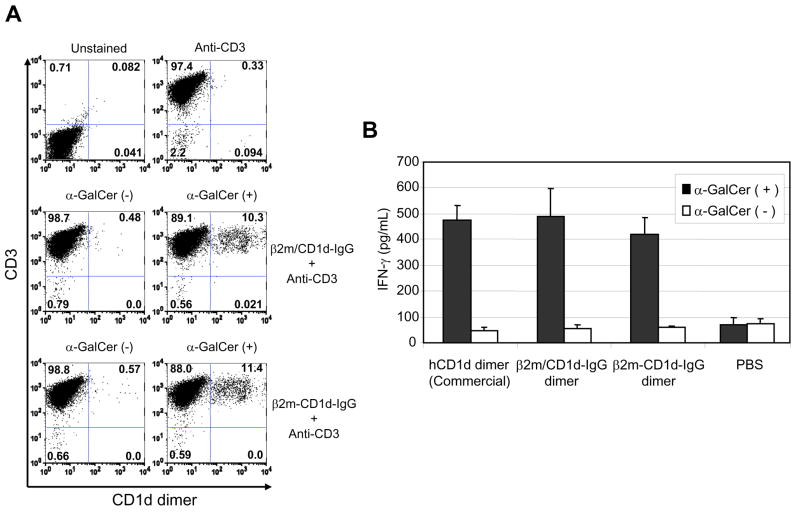
Fig. 2. Staining and stimulation of human iNKT cells with β2m/CD1d-IgG dimer or β2m-CD1d-IgG dimer.
A) The CD1d dimers were incubated with or without α-GalCer and then used to stain human iNKT cells. The dimers were detected with phycoerythrin-labeled rat anti-mouse IgG antibody. APC (allophycocyanin)-labeled anti-human CD3ε antibody was used to identify T cell populations. B) Human iNKT cells were cultured on 96 well plates coated with α-GalCer-loaded or unloaded CD1d dimers for 24 hours and IFN-γ concentration in the culture supernatants was measured by ELISA assay. Recombinant human CD1d dimer (BD Biosciences, San Diego, CA, USA) was used as a positive control. The assay was done in triplicate and the error bars indicate Standard Deviation (SD).
Next, we cultured human iNKT cells on a plate coated with β2m/hCD1d-IgG or β2m-hCD1d-IgG to test whether these dimers can activate iNKT cells in a ligand dependent manner. We could detect IFN-γ secretion only when human iNKT cells were cultured in a well coated with α-GalCer-loaded CD1d dimers, but not with unloaded CD1 dimers (Fig. 2B). Again there was no difference in the ability to induce IFN-γ secretion between β2m/hCD1d-IgG and β2m-hCD1d-IgG.
These results showed that β2m-hCD1d-IgG, which has a covalently bound β2m at N-terminus, has an equal functionality to β2m/hCD1d-IgG in terms of recognition by TCR and stimulating TCR. Based on these observations, we decided to use β2m-hCD1d-IgG for further experiments because protein expression level of these CD1 dimers was higher than that of β2m/hCD1d-IgG (data not shown), and β2m-hCD1d-IgG were expected to be more stable due to the covalent linkage between β2m and hCD1 molecules.
3.3 Competitive ELISA assay to measure the binding of lipid to hCD1 molecules
We sought to establish a novel assay system to detect the binding of the lipid to hCD1 molecules for the purpose of screening lipid antigens from pathogens or self organs. For this purpose, we utilized a competitive assay, in which a tagged lipid was used as an indicator for the competition in binding to hCD1 molecules. 18:1 Biotinyl PE (Fig. 3A) was selected as the indicator lipid, because of the ability of PE to bind hCD1 molecules (Rauch et al., 2003), as well as of its commercial availability.
Within a cell, the loading of the lipid onto CD1 molecules is known to occur in late endosomes and lysosomes (Kang and Cresswell, 2004), which are acidic compartments, and therefore lipid-loading onto CD1b and CD1c has been shown to be more efficient in an acidic condition in a cell-free loading system (Im et al., 2004). To test whether the binding of Biotinyl PE to hCD1 molecules is a physiological binging or not, we first measured the binding of Biotinyl PE to hCD1 molecules in different pH conditions (pH 4–10) without a competitor lipid. As we expected, Biotinyl PE could bind more strongly to hCD1 molecules in acidic conditions. The optimal pH for CD1b and CD1c was pH4-5, whereas that for CD1d was pH5-7. These observations were consistent with the previous reports (Im JS et al., 2004; Prigozy TI et al., 2001). The binding was considered CD1 molecule-specific, because mouse IgG1 failed to bind to Biotinyl PE (Fig. 3B). We then measured the kinetics of Biotinyl PE binding to hCD1d under the optimal condition. In agreement to a previous report (Cantu et al., 2003), we found that the kinetics of the lipid loading onto CD1 molecules in vitro is a very slow process, e.g. 8 hrs to reach a peak level (Fig. 3C).
It is imperative to see whether the binding of Biotinyl PE can be competed with unlabeled PE or α-GalCer, the most well characterized ligand for CD1d. Because PE has the same structure as Biotinyl PE except for the polar head (Fig. 3A), the equilibrium dissociation constant in a competition assay (Ki) of unlabeled PE should be around the Kd of Biotinyl PE if the binding is mediated by the acyl chains. In fact, we observed a dose dependent competition by PE and Ki of unlabeled PE was almost equal to Kd of Biotinyl PE (Fig. 3D and Table 1). These suggest that the lipid binding to hCD1 molecules in our assay occurs in a lipid structure specific fashion. Interestingly, Ki of α-GalCer for CD1d (0.097 μM) was about one log lower than Ki of unlabeled PE (1.46 μM), although there was little difference in these Ki for CD1b or CD1c (Table 1). The Ki of α-GalCer obtained in this assay was similar to Kd of α-GalCer binding to mouse CD1d measured using surface Plasmon resonance (Kd = 0.34μM) (Naidenko OV et al., 1999). This result clearly showed that the competitive ELISA assay can reflect lipid-selectivity by CD1 molecules, which are indispensable for screening of lipid-ligands.
Table 1.
Ki (equilibrium dissociation constant in a competition assay) of PE and aGalCer for CD1 molecules. Ki was calculated based on the data in Figure 3C, D and E. Kds of B-PE (CD1b: 2.69 (1.11–4.26), CD1c: 2.28 (0.76–3.79), CD1d: 1.69 (0.64–2.87)) was determined by titrating the amount B-PE in the absence of competitor lipids (data not shown) and used to calculate Kis of the competitor lipids by the equation (Ki=IC50/(1+[B-PE]/Kd)). The figures in the bracket represent 95% confidential interval (95%CI).
| CD1b Ki (μM) | CD1c Ki (μM) | CD1d Ki (μM) | |
|---|---|---|---|
| PE | 1.19 (0.77–1.84) | 2.26 (1.15–4.45) | 1.46 (0.86–2.47) |
| α-GalCer | 2.84 (1.72–4.69) | 1.37 (0.90–2.08) | 0.097 (0.07–0.14) |
3.4 Lipid extraction from malaria parasites
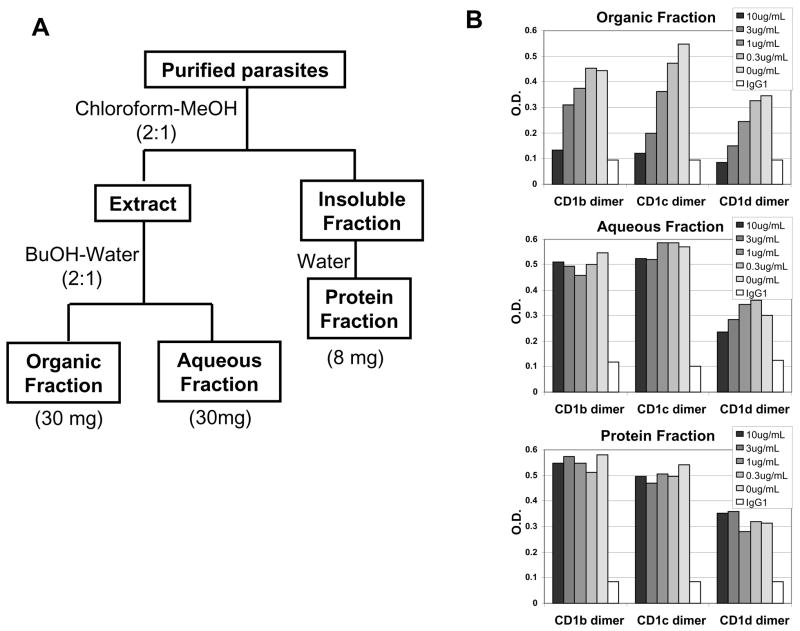
Next, we extracted lipids from the blood stages of malaria parasites and tested whether the malarial lipids can be loaded onto hCD1 molecules. Blood stages of murine malaria parasites, Plasmodium yoelii 17NXL, were prepared from infected red blood cells (RBCs) and extracted with organic solvents and/or water in order to obtain three fractions of the malarial extract, i.e. organic, aqueous and protein fractions (Fig. 4A), following a modified Folch method (Folch J et al., 1957). Starting from10mL blood, we obtained 30mg of organic fraction, 30mg of aqueous fraction and 8mg of protein fraction (data not shown). These three fractions were then subjected onto the competitive ELISA assay, and the organic fraction was shown to contain lipids that can bind CD1b, c and d molecules (Fig. 4B). The aqueous fraction, which might contain highly glycosylated lipids, could weakly inhibit Biotinyl PE binding to CD1d.
Fig. 4. Binding of malarial lipid extracts to hCD1 molecules.
A) Schematic flow of lipid extraction from blood stages of Plasmodium yoelii 17NXL stain. B) Competitive ELISA assay with the malarial extract fractions. The lyophilized each fraction was dissolved in MeOH (Organic and Aqueous fraction) or water (Protein fraction) and used as a competitor in the assay described in Fig. 3.
3.5 Stimulation and expansion of human iNKT cells with hCD1d-aAPCs
In most studies, CD1-restricted T cells were expanded and cloned ex vivo with a lipid antigen presented by autologous DCs for further analysis. This technique, however, cannot be applied in the following cases; (1) autologous DCs are not available, (2) a lipid antigen is in a crude extract that cannot exclude protein contamination, (3) CD1-restricted T cells are either autoreactive and/or self-lipid reactive.
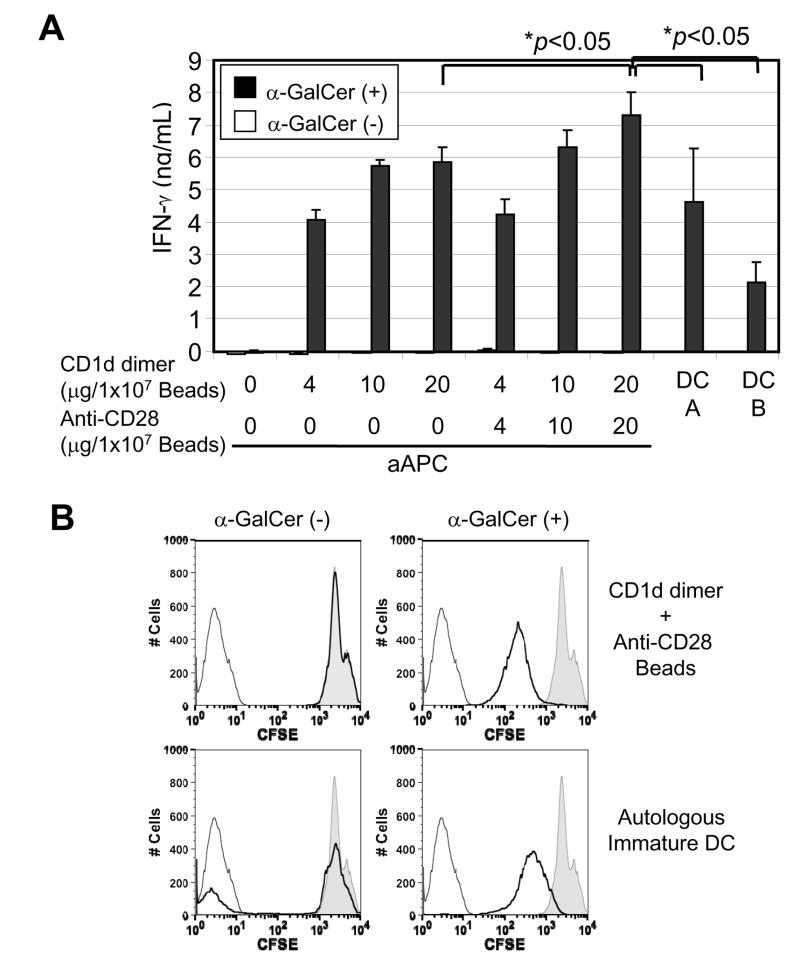
To overcome these hurdles, we prepared hCD1d-aAPCs, which are magnetic beads coated with hCD1d dimer with or without anti-CD28 antibody, and tested their functionality as APCs in stimulating and expanding human iNKT cells. We found that α-GalCer-loaded, but not unloaded hCD1d-aAPCs strongly stimulated human iNKT cells to produce IFN-γ (Fig. 5A). The presence of anti-CD28 antibody, which activates the co-stimulatory pathway, slightly but significantly (p<0.05) augmented the level of IFN-γ production at the highest dose. Also the IFN-γ secretion by iNKT cells stimulated with aAPCs was significantly superior to that stimulated with α-GalCer-pulsed, autologous human DCs.
Fig. 5. Functional analysis of hCD1d-aAPC.
A) IFN-γsecretion by human iNKT cells stimulated with α-GalCer-loaded, hCD1d-aAPC. The indicated amount of human CD1d dimer was immobilized to magnetic beads with or without anti-human CD28 antibody. After blocking, the beads were incubated with or without α-GalCer, washed with medium, and added to human iNKT cells. DC A and DC B were α-GalCer-pulsed, autologous immature DCs prepared by culturing human CD14+ monocytes in the presence of GM-CSF and IL-4. The immature DCs were incubated with 1μg/mL α-GalCer for one hour and added to human NTK cell at a ratio of 1:1 (DC:NKT). The assay was done in triplicate and the error bars indicate Standard Deviation (SD). B) Expansion of human iNKT cells with α-GalCer-loaded hCD1d-aAPC. Human iNKT cells were labeled with CFSE and then cultured with α-GalCer-loaded, hCD1d-aAPC or α-GalCer pulsed, immature DC, as described in A), for one week, and the intensity of CFSE was measured by a flow cytometric analysis. The thin black line shows unlabeled iNKT cells. The shadow shows human iNKT cells cultured without APCs. The bold black line shows human iNKT cells cultured with the indicated APCs.
Human iNKT cells stimulated with α-GalCer-loaded hCD1d-aAPCs showed remarkable proliferation, as determined by CFSE assay, and the ability of aAPCs to induce human iNKT cell proliferation was comparable or even superior to that of human DCs (Fig. 5B). Unloaded aAPCs had no effect on inducing human iNKT cell proliferation.
4. Discussion
In this paper, we generated β2m-hCD1-IgG fusion protein having human β2m covalently linked to the N-terminus of hCD1, and established a competitive ELISA assay to measure the binding of lipids to human CD1 molecules and the ability of aAPCs to stimulate and expand human CD1-restricted T cells. Furthermore, we extracted malarial lipids from the extracts of blood stages of rodent malaria parasites and demonstrated that malarial lipids can be loaded onto CD1 molecules.
Recent reports have shown that lipid antigen presentation by CD1 molecules and CD1-restricted T cells are involved in antimicrobial immunity (Nieuwenhuis et al., 2002), allergy (Agea et al., 2005) and autoimmune diseases (Forestier et al., 2005). However, the progress of the research on lipid antigens and CD1-restricted T cells, in particular group I CD1-restricted T cells, has been dawdling compared to the research on peptide antigens and MHC class I/II-restricted T cells. One of the possible reasons for this is that fewer tools have been available for research on CD1-restricted T cells up to date. For example, although MHC class I-binding peptide candidates are commonly screened using a prediction algorithm in combination with genomic, transcript and protein databases (Antes et al., 2006), such tools are not available for CD1-binding lipids.
Our current study has provided a useful platform to overcome this obstacle. If the relationship between the lipid structures, e.g. the length of acyl chain, and their binding ability to CD1 molecules is analyzed systematically and the lipid structure-binding ability information is combined with the CD1 protein structures, it would be possible to create an algorithm for predicting the binding affinity of the lipid of your interest to CD1 molecules. Some features of our competitive assay could make this approach possible. First, direct modification of the lipids to be tested, such as biotinylation, is not required in this assay. Second, this assay is based on ELISA assay, and, therefore, is easily applied to High Throughput Screening (HTS) systems. The covalent link between CD1s and β2m in CD1 dimers is ideal for this kind of analysis, because the covalent link makes the dimers more uniform, thus contributing to the reproducibility among assays.
aAPCs, which are magnetic beads coated with an antigen-presenting molecule and an anti-CD28 antibody, could be a powerful tool to investigate antigen-presenting molecule-restricted T cells (Oelke et al., 2003). We would like to note that, although application of bead-based aAPC in NKT research has previously been reported (Huang et al., 2004; Webb et al., 2008), our current paper is the first report to demonstrate that hCD1d-aAPCs, consisting of beads coated with α-GalCer-loaded, human CD1d molecules, were able to stimulate human iNKT cells to secrete IFN-γ and expanded iNKT cells at a similar degree compared to α-GalCer-pulsed DCs. The advantage of aAPCs is that hCD1d-aAPCs can activate and expand CD1-restricted T cells in solely a lipid-CD1 complex specific manner. Autologus DCs are widely used in research on CD1-restricted T cells, but they have many functions in processing/presenting antigens and stimulating T cells. For example, if crude lipid extract is used as a lipid antigen, it is difficult to exclude the possibility that a small amount of contaminated protein antigens, which might be processed and loaded onto MHC molecules, might stimulate and expand other T cell populations. In addition, other lipids/proteins may trigger TLRs and other receptors expressed by DCs and indirectly activate T cells. In this regard, CD1-aAPCs not only have an advantage over autologous DCs in establishing antigen-specific CD1-restricted T cell clones, but also can be applicable to other CD1-restricted T cell assays, including an ELISPOT assay, as well as the analysis of TCR-mediated signal transductions in CD1-restricted T cells.
Hence, the use of recombinant CD1-dimer is expected to significantly contribute to the advancement of the research in the field of CD1 molecules and CD1-restricted T cells. However, it should be noted that physical properties of lipids, such as solubility and ability to form micelle, might affect the lipid-loading efficiency in a cell free system (Brigl and Brenner, 2004). Within a cell, insoluble lipids can be loaded onto CD1 molecules with the aid of lipid-binding proteins, Saposins (Kang and Cresswell, 2004; Zhou et al., 2004; Winau et al., 2004). Saposin knock-out mice lack iNKT cells, and this clearly indicates that Saposin is indispensable for a natural CD1d ligand to be loaded onto CD1d in order to maintain iNKT cells in vivo. Also, the involvement of microsomal triglyceride transfer protein in endogenous and exogenous antigen presentation by CD1 molecules has recently been demonstrated (Kaser et al., 2008). In fact, we were unable to detect the binding of mycolic acid, known as a CD1b ligand, to CD1b molecule in our competitive ELISA assay (data not shown), probably due to its long acyl chain, which makes it difficult to be dissolved in aqueous solution.
The fractionation of malaria lipids and the binding assay to CD1 molecules provided a novel strategy to identify lipid antigens from pathogens. In most cases, lipid antigen-specific, CD1-restricted T cell clones were established first, and, then, lipid antigens were identified based on the reactivity of the cloned T cells. However, if we could identify lipids that can be loaded onto CD1 molecules, an active immunization with those lipids would possibly result in the induction of CD1-restricted T cells and ultimately lead to the determination of the role of the lipids against the infection in a CD1-transgenic mouse model or Guinea pig model (Hiromatsu et al., 2002; Dascher et al., 2003).
In summary, our current study has provided powerful and indispensable assay systems for the advancement of the research in the field of CD1-binding lipids and CD1-restricted T cells. Identification of a novel lipid antigen might widen the window of opportunity to develop an effective vaccine for some pathogens that might be unachievable if only proteins are used as antigens.
Acknowledgments
We thank Vincent Sahi for FACS analysis and Kirin Brewery Co. for providing α-GalCer. This work was partially supported by a grant from NIH R21 AI062842-01 (M.T.), the Irene Diamond Foundation, the Bill and Melinda Gates Foundation, the Japanese Health Sciences Foundation, and Otsuka Pharmaceutical Co. Ltd.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agea E, Russano A, Bistoni O, Mannucci R, Nicoletti I, Corazzi L, Postle AD, De Libero G, Porcelli SA, Spinozzi F. Human CD1-restricted T cell recognition of lipids from pollens. J Exp Med. 2005;202:295. doi: 10.1084/jem.20050773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antes I, Siu SW, Lengauer T. DynaPred: a structure and sequence based method for the prediction of MHC class I binding peptide sequences and conformations. Bioinformatics. 2006;22:e16. doi: 10.1093/bioinformatics/btl216. [DOI] [PubMed] [Google Scholar]
- Barral DC, Brenner MB. CD1 antigen presentation: how it works. Nat Rev Immunol. 2007;7:929. doi: 10.1038/nri2191. [DOI] [PubMed] [Google Scholar]
- Beckman EM, Porcelli SA, Morita CT, Behar SM, Furlong ST, Brenner MB. Recognition of a lipid antigen by CD1-restricted alpha beta+ T cells. Nature. 1994;372:691. doi: 10.1038/372691a0. [DOI] [PubMed] [Google Scholar]
- Bradbury A, Belt KT, Neri TM, Milstein C, Calabi F. Mouse CD1 is distinct from and coexists with TL in the same thymus. EMBO J. 1988;7:081. doi: 10.1002/j.1460-2075.1988.tb03173.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brigl M, Brenner MB. CD1: antigen presentation and T cell function. Annu Rev Immunol. 2004;22:817. doi: 10.1146/annurev.immunol.22.012703.104608. [DOI] [PubMed] [Google Scholar]
- Cantu C, 3rd, Benlagha K, Savage PB, Bendelac A, Teyton L. The paradox of immune molecular recognition of alpha-galactosylceramide: low affinity, low specificity for CD1d, high affinity for alpha beta TCRs. J Immunol. 2003;170:4673. doi: 10.4049/jimmunol.170.9.4673. [DOI] [PubMed] [Google Scholar]
- Cerundolo V, Silk JD, Masri SH, Salio M. Harnessing invariant NKT cells in vaccination strategies. Nat Rev Immunol. 2009;9:28. doi: 10.1038/nri2451. [DOI] [PubMed] [Google Scholar]
- Dal Porto J, Johansen TE, Catipovic B, Parfiit DJ, Tuveson D, Gether U, Kozlowski S, Fearon DT, Schneck JP. A soluble divalent class I major histocompatibility complex molecule inhibits alloreactive T cells at nanomolar concentrations. Proc Natl Acad Sci U S A. 1993;90:6671. doi: 10.1073/pnas.90.14.6671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dascher CC, Hiromatsu K, Naylor JW, Brauer PP, Brown KA, Storey JR, Behar SM, Kawasaki ES, Porcelli SA, Brenner MB, LeClair KP. Conservation of a CD1 multigene family in the guinea pig. J Immunol. 1999;163:5478. [PubMed] [Google Scholar]
- Dascher CC, Hiromatsu K, Xiong X, Morehouse C, Watts G, Liu G, McMurray DN, LeClair KP, Porcelli SA, Brenner MB. Immunization with a mycobacterial lipid vaccine improves pulmonary pathology in the guinea pig model of tuberculosis. Int Immunol. 2003;15:915. doi: 10.1093/intimm/dxg091. [DOI] [PubMed] [Google Scholar]
- Folch J, Lees M, Sloane Stanley GH. A simple method for the isolation and purification of total lipids from animal tissues. J Biol Chem. 1957;226:497. [PubMed] [Google Scholar]
- Forestier C, Molano A, Im JS, Dutronc Y, Diamond B, Davidson A, Illarionov PA, Besra GS, Porcelli SA. Expansion and hyperactivity of CD1d-restricted NKT cells during the progression of systemic lupus erythematosus in (New Zealand Black x New Zealand White)F1 mice. J Immunol. 2005;175:763. doi: 10.4049/jimmunol.175.2.763. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Aseguinolaza G, Van Kaer L, Bergmann CC, Wilson JM, Schmieg J, Kronenberg M, Nakayama T, Taniguchi M, Koezuka Y, Tsuji M. Natural killer T cell ligand alpha-galactosylceramide enhances protective immunity induced by malaria vaccines. J Exp Med. 2002;195:617. doi: 10.1084/jem.20011889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant EP, Degano M, Rosat JP, Stenger S, Modlin RL, Wilson IA, Porcelli SA, Brenner MB. Molecular recognition of lipid antigens by T cell receptors. J Exp Med. 1999;189:195. doi: 10.1084/jem.189.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang MM, Borszcz P, Sidobre S, Kronenberg M, Kane KP. CD1d1 displayed on cell size beads identifies and enriches an NK cell population negatively regulated by CD1d1. J Immunol. 2004;172:5304. doi: 10.4049/jimmunol.172.9.5304. [DOI] [PubMed] [Google Scholar]
- Hiromatsu K, Dascher CC, LeClair KP, Sugita M, Furlong ST, Brenner MB, Porcelli SA. Induction of CD1-restricted immune responses in guinea pigs by immunization with mycobacterial lipid antigens. J Immunol. 2002;169:330. doi: 10.4049/jimmunol.169.1.330. [DOI] [PubMed] [Google Scholar]
- Im JS, Yu KO, Illarionov PA, LeClair KP, Storey JR, Kennedy MW, Besra GS, Porcelli SA. Direct measurement of antigen binding properties of CD1 proteins using fluorescent lipid probes. J Biol Chem. 2004;279:299. doi: 10.1074/jbc.M308803200. [DOI] [PubMed] [Google Scholar]
- Kang SJ, Cresswell P. Saposins facilitate CD1d-restricted presentation of an exogenous lipid antigen to T cells. Nat Immunol. 2004:175. doi: 10.1038/ni1034. [DOI] [PubMed] [Google Scholar]
- Kaser A, Hava DL, Dougan SK, Chen Z, Zeissig S, Brenner MB, Blumberg RS. Microsomal triglyceride transfer protein regulates endogenous and exogenous antigen presentation by group 1 CD1 molecules. Eur J Immunol. 2008;38:2351. doi: 10.1002/eji.200738102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawano T, Cui J, Koezuka Y, Toura I, Kaneko Y, Motoki K, Ueno H, Nakagawa R, Sato H, Kondo E, Koseki H, Taniguchi M. CD1d-restricted and TCR-mediated activation of valpha14 NKT cells by glycosylceramides. Science. 1997;278:1626. doi: 10.1126/science.278.5343.1626. [DOI] [PubMed] [Google Scholar]
- Kawano T, Cui J, Koezuka Y, Toura I, Kaneko Y, Sato H, Kondo E, Harada M, Koseki H, Nakayama T, Tanaka Y, Taniguchi M. Natural killer-like nonspecific tumor cell lysis mediated by specific ligand-activated Valpha14 NKT cells. Proc Natl Acad Sci U S A. 1998;95:5690. doi: 10.1073/pnas.95.10.5690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinjo Y, Tupin E, Wu D, Fujio M, Garcia-Navarro R, Benhnia MR, Zajonc DM, Ben-Menachem G, Ainge GD, Painter GF, Khurana A, Hoebe K, Behar SM, Beutler B, Wilson IA, Tsuji M, Sellati TJ, Wong CH, Kronenberg M. Natural killer T cells recognize diacylglycerol antigens from pathogenic bacteria. Nat Immunol. 2006;7:978. doi: 10.1038/ni1380. [DOI] [PubMed] [Google Scholar]
- Lantz O, Bendelac A. An invariant T cell receptor alpha chain is used by a unique subset of major histocompatibility complex class I-specific CD4+ and CD4-8- T cells in mice and humans. J Exp Med. 1994;180:1097. doi: 10.1084/jem.180.3.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchesini N, Luo S, Rodrigues CO, Moreno SN, Docampo R. Acidocalcisomes and a vacuolar H+-pyrophosphatase in malaria parasites. Biochem J. 2000;347:243. [PMC free article] [PubMed] [Google Scholar]
- Miyamoto K, Miyake S, Yamamura T. A synthetic glycolipid prevents autoimmune encephalomyelitis by inducing TH2 bias of natural killer T cells. Nature. 2001;413:531. doi: 10.1038/35097097. [DOI] [PubMed] [Google Scholar]
- Moody DB, Ulrichs T, Mühlecker W, Young DC, Gurcha SS, Grant E, Rosat JP, Brenner MB, Costello CE, Besra GS, Porcelli SA. CD1c-mediated T-cell recognition of isoprenoid glycolipids in Mycobacterium tuberculosis infection. Nature. 2000;404:884. doi: 10.1038/35009119. [DOI] [PubMed] [Google Scholar]
- Naidenko OV, Maher JK, Ernst WA, Sakai T, Modlin RL, Kronenberg M. Binding and antigen presentation of ceramide-containing glycolipids by soluble mouse and human CD1d molecules. J Exp Med. 1999;190:1069. doi: 10.1084/jem.190.8.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwenhuis EE, Matsumoto T, Exley M, Schleipman RA, Glickman J, Bailey DT, Corazza N, Colgan SP, Onderdonk AB, Blumberg RS. CD1d-dependent macrophage-mediated clearance of Pseudomonas aeruginosa from lung. Nat Med. 2002:588. doi: 10.1038/nm0602-588. [DOI] [PubMed] [Google Scholar]
- Oelke M, Maus MV, Didiano D, June CH, Mackensen A, Schneck JP. Ex vivo induction and expansion of antigen-specific cytotoxic T cells by HLA-Ig-coated artificial antigen-presenting cells. Nat Med. 2003;9:619. doi: 10.1038/nm869. [DOI] [PubMed] [Google Scholar]
- Prigozy TI, Naidenko O, Qasba P, Elewaut D, Brossay L, Khurana A, Natori T, Koezuka Y, Kulkarni A, Kronenberg M. Glycolipid antigen processing for presentation by CD1d molecules. Science. 2001;291:664. doi: 10.1126/science.291.5504.664. [DOI] [PubMed] [Google Scholar]
- Rauch J, Gumperz J, Robinson C, Sköld M, Roy C, Young DC, Lafleur M, Moody DB, Brenner MB, Costello CE, Behar SM. Structural features of the acyl chain determine self-phospholipid antigen recognition by a CD1d-restricted invariant NKT (iNKT) cell. J Biol Chem. 2003;278:47508. doi: 10.1074/jbc.M308089200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosat JP, Grant EP, Beckman EM, Dascher CC, Sieling PA, Frederique D, Modlin RL, Porcelli SA, Furlong ST, Brenner MB. CD1-restricted microbial lipid antigen-specific recognition found in the CD8+ alpha beta T cell pool. J Immunol. 1999;162:366. [PubMed] [Google Scholar]
- Russano AM, Bassotti G, Agea E, Bistoni O, Mazzocchi A, Morelli A, Porcelli SA, Spinozzi F. CD1-restricted recognition of exogenous and self-lipid antigens by duodenal gammadelta+ T lymphocytes. J Immunol. 2007;178:3620. doi: 10.4049/jimmunol.178.6.3620. [DOI] [PubMed] [Google Scholar]
- Schmieg J, Yang G, Franck RW, Tsuji M. Superior protection against malaria and melanoma metastases by a C-glycoside analogue of the natural killer T cell ligand alpha-Galactosylceramide. J Exp Med. 2003;198:1631. doi: 10.1084/jem.20031192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamshiev A, Gober HJ, Donda A, Mazorra Z, Mori L, De Libero G. Presentation of the same glycolipid by different CD1 molecules. J Exp Med. 2002;195:1013. doi: 10.1084/jem.20011963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharif S, Arreaza GA, Zucker P, Mi QS, Sondhi J, Naidenko OV, Kronenberg M, Koezuka Y, Delovitch TL, Gombert JM, Leite-De-Moraes M, Gouarin C, Zhu R, Hameg A, Nakayama T, Taniguchi M, Lepault F, Lehuen A, Bach JF, Herbelin A. Activation of natural killer T cells by alpha-galactosylceramide treatment prevents the onset and recurrence of autoimmune Type 1 diabetes. Nat Med. 2001;7:1057. doi: 10.1038/nm0901-1057. [DOI] [PubMed] [Google Scholar]
- Sieling PA, Chatterjee D, Porcelli SA, Prigozy TI, Mazzaccaro RJ, Soriano T, Bloom BR, Brenner MB, Kronenberg M, Brennan PJ, et al. CD1-restricted T cell recognition of microbial lipoglycan antigens. Science. 1995;269:227. doi: 10.1126/science.7542404. [DOI] [PubMed] [Google Scholar]
- Tsuji M. Glycolipids and phospholipids as natural CD1d-binding NKT cell ligands. Cell Mol Life Sci. 2006;63:1889. doi: 10.1007/s00018-006-6073-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tubbs KA, Nedelkov D, Nelson RW. Detection and quantification of beta-2-microglobulin using mass spectrometric immunoassay. Anal Biochem. 2001;289:26. doi: 10.1006/abio.2000.4921. [DOI] [PubMed] [Google Scholar]
- Van Kaer L. NKT cells: T lymphocytes with innate effector functions. Curr Opin Immunol. 2007;19:354. doi: 10.1016/j.coi.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Vincent MS, Xiong X, Grant EP, Peng W, Brenner MB. CD1a-, b-, and c-restricted TCRs recognize both self and foreign antigens. J Immunol. 2005;175:6344. doi: 10.4049/jimmunol.175.10.6344. [DOI] [PubMed] [Google Scholar]
- Webb TJ, Giuntoli RL, 2nd, Rogers O, Schneck J, Oelke M. Ascites specific inhibition of CD1d-mediated activation of natural killer T cells. Clin Cancer Res. 2008;14:7652. doi: 10.1158/1078-0432.CCR-08-1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winau F, Schwierzeck V, Hurwitz R, Remmel N, Sieling PA, Modlin RL, Porcelli SA, Brinkmann V, Sugita M, Sandhoff K, Kaufmann SH, Schaible UE. Saposin C is required for lipid presentation by human CD1b. Nat Immunol. 2004;5:169. doi: 10.1038/ni1035. [DOI] [PubMed] [Google Scholar]
- Wu DY, Segal NH, Sidobre S, Kronenberg M, Chapman PB. Cross-presentation of disialoganglioside GD3 to natural killer T cells. J Exp Med. 2003;198:173. doi: 10.1084/jem.20030446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu KO, Im JS, Molano A, Dutronc Y, Illarionov PA, Forestier C, Fujiwara N, Arias I, Miyake S, Yamamura T, Chang YT, Besra GS, Porcelli SA. Modulation of CD1d-restricted NKT cell responses by using N-acyl variants of alpha-galactosylceramides. Proc Natl Acad Sci U S A. 2005;102:3383. doi: 10.1073/pnas.0407488102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D, Cantu C, 3rd, Sagiv Y, Schrantz N, Kulkarni AB, Qi X, Mahuran DJ, Morales CR, Grabowski GA, Benlagha K, Savage P, Bendelac A, Teyton L. Editing of CD1d-bound lipid antigens by endosomal lipid transfer proteins. Science. 2004;303:523. doi: 10.1126/science.1092009. [DOI] [PMC free article] [PubMed] [Google Scholar]