Abstract
The Ras/mitogen activated protein kinase (MAPK) pathway is essential in the regulation of the cell cycle, differentiation, growth and cell senescence, all of which are critical to normal development. It is therefore not surprising that its dysregulation has profound effects on development. A class of developmental disorders, the “RASopathies”, is caused by germline mutations in genes that encode protein components of the Ras/MAPK pathway. The vast majority of these mutations result in increased signal transduction down the Ras/MAPK pathway, but usually to a lesser extent than somatic mutations associated with oncogenesis. Each syndrome exhibits unique phenotypic features, however, since they all cause dysregulation of the Ras/MAPK pathway, there are numerous overlapping phenotypic features between the syndromes, including characteristic facial features, cardiac defects, cutaneous abnormalities, neurocognitive delay and a predisposition to malignancies. Here we review the clinical and underlying molecular basis for each of these syndromes.
Keywords: autoimmune lymphoproliferative disorder, capillary malformation-arteriovenous malformation syndrome, cardio-facio-cutaneous syndrome, Costello syndrome, BRAF, gingival fibromatosis 1 syndrome, HRAS, KRAS, Legius syndrome, LEOPARD syndrome, MAP2K1 (MEK1), MAP2K2 (MEK2), neurofibromatosis 1, neurofibromin, Noonan syndrome, NRAS, PTPN11, RASA1, RAF1/CRAF, Ras/MAPK, signal transduction pathway, SOS1, SPRED1
Introduction
A class of human genetic syndromes has emerged that are caused by germline mutations in genes which encode components of the Ras/mitogen activated protein kinase (MAPK) pathway. This pathway plays an essential role in the control of the cell cycle and differentiation, therefore its dysregulation has profound developmental consequences. These “RASopathies” each exhibit unique phenotypic features, however, many share characteristic overlapping features including craniofacial dysmorphology, cardiac malformations and cutaneous, musculoskeletal and ocular abnormalities, varying degrees of neurocognitive impairment and, in some syndromes, an increased risk of developing cancer.
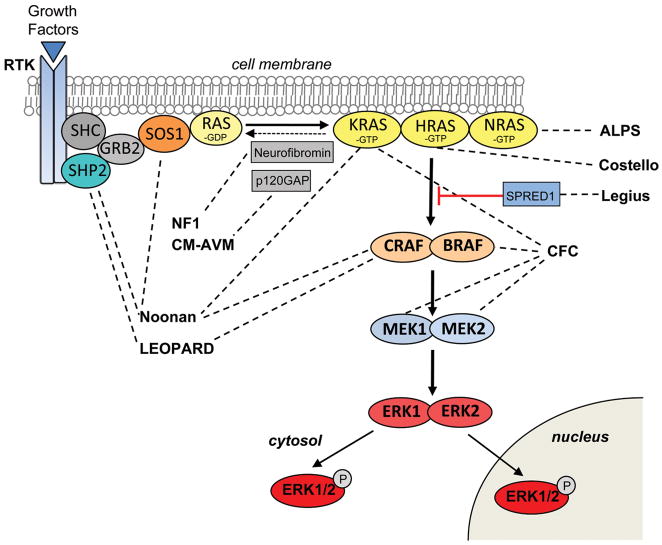
The Ras/MAPK pathway transduces extracellular input in the form of growth factors and small molecules to the intracellular environment (Figure 1). The pathway has been studied extensively in the context of oncogenesis since its dysregulation is one of the primary causes of cancer, with Ras found to be somatically mutated in approximately 20% of malignancies [1]. Ras proteins are small guanosine nucleotide-bound GTPases which comprise a critical signaling hub within the cell. Ras genes exist as a multigene family that includes HRAS, NRAS and KRAS. They are activated through growth factors binding to receptor tyrosine kinases (RTK), G-protein-coupled receptors, cytokine receptors and extracellular matrix receptors. Ras proteins cycle between an active GTP-bound form and an inactive GDP-bound form. Activation through RTK occurs with the binding of a growth factor causing RTK autophosphorylation and interaction with the adaptor protein GRB2. GRB2 is bound to SOS which is then recruited to the plasma membrane. SOS proteins are guanosine nucleotide exchange factors (GEF) that increase the Ras nucleotide exchange rate of GDP for GTP, resulting in an increase Ras in the active GTP-bound form. The Raf-mediated MAPK pathway is one of several important downstream cascades of Ras. Activated Ras leads to the activation of Raf (ARAF, BRAF, and/or CRAF) the first MAPK kinase kinase of the pathway. Raf phosphorylates and activates MEK1 and/or MEK2 (MAPK kinase), which in turn phosphorylates and activates ERK1 and/or ERK2. ERK1/2 are the ultimate effectors and exert their function on a large number of downstream molecules, both nuclear and cytosolic. ERK1/2 substrates include nuclear components, transcription factors, membrane proteins, and protein kinases that in turn control vital cellular functions including cell cycle progression, differentiation and the control of cellular growth [2].
Figure 1.
The Ras/mitogen-activated protein kinase (MAPK) signaling pathway and associated developmental syndromes (indicated by dashed lines). The MAPK signaling pathway of protein kinases is critically involved in cell proliferation, differentiation, motility, apoptosis and senescence. The Ras/MAPK pathway proteins with germline mutations in their respective genes are associated with Noonan, LEOPARD, gingival fibromatosis 1, neurofibromatosis 1, capillary malformation-arteriovenous malformation, Costello, autoimmune lymphoproliferative (ALPS), cardio-facio-cutaneous and Legius syndromes.
Noonan Syndrome
Noonan syndrome (NS) is an autosomal dominant disorder that affects approximately 1/1000 to 1/2500 newborns. NS is characterized by distinctive craniofacial features, short stature, congenital cardiac anomalies, bleeding disorders and a variable degree neurocognitive delay (for review see [3]). Individuals with NS have an increased risk of cancer. At present four genes, PTPN11 [4], KRAS [5], SOS1 [6,7] and RAF1 [8,9] harboring heterozygous germline mutations cause NS with all genes encoding various components of the Ras/MAPK pathway (Table 1). However, there are still more genes to be identified. The most common gene associated with NS is PTPN11 which accounts for approximately half of all cases. SHP2, the protein product of PTPN11, is a non-receptor protein tyrosine phosphatase composed of N-terminal and C-terminal SH2 domains and a catalytic protein tyrosine phosphatase (PTP) domain. The catalytic function of the protein is auto-inhibited through a blocking interaction between the N-SH2 and PTP domains [10]. The majority of NS-causing missense mutations in PTPN11 cluster in residues involved with the interaction between the N-SH2 and PTP domains. Mutations in this region interfere with the stability of the catalytically inactive form of SHP2 resulting in impairment of the protein’s ability to switch from the active to the inactive protein conformation [11,12] causing increased signaling down the Ras/MAPK pathway [13].
Table 1.
Genetic syndromes of the Ras/MAPK pathway
| Syndrome | Ras Pathway Gene | Chromosome Location | Protein | Protein Function | Reference |
|---|---|---|---|---|---|
| Noonan | PTPN11 | 12q24.1 | SHP2 | Phosphatase | [4] |
| SOS1 | 2p22.1 | SOS1 | RasGEF | [6] | |
| RAF1 | 3p25.1 | CRAF | Kinase | [7] | |
| KRAS | 12p12.1 | KRAS | GTPase | [8] | |
| LEOPARD | PTPN11 | 12q24.1 | SHP2 | Phosphatase | [15] |
| RAF1 | 3p25.1 | RAF1/CRAF | Kinase | [8] | |
| Gingival fibromatosis 1 | SOS1 | 2p22.1 | SOS1 | RasGEF | [20] |
| Neurofibromatosis 1 | NF1 | 17q11.2 | Neurofibromin | RasGAP | [23–25] |
| Capillary malformation- arteriovenous malformation | RASA1 | 5q14.3 | p120Gap | RasGAP | [27] |
| Costello | HRAS | 11p15.5 | HRAS | GTPase | [30] |
| Autoimmune lymphoproliferative | NRAS | 1p15.2 | NRAS | GTPase | [38] |
| Cardio-facio- cutaneous | BRAF | 7q34 | BRAF | Kinase | [41,42] |
| MAP2K1 | 15q22.31 | MEK1 | Kinase | [41] | |
| MAP2K2 | 19p13.3 | MEK2 | Kinase | [41] | |
| KRAS | 12p12.1 | KRAS | GTPase | [42] | |
| Legius | SPRED1 | 15q14 | SPRED1 | Sprouty-related, EVH1 domain- containing protein 1 | [46] |
SOS1 missense mutations are the second most common cause of NS and account for approximately 13% of cases [6,7]. SOS1 encodes the Ras-GEF protein, SOS1, which is responsible for stimulating the conversion of Ras from the inactive GDP-bound form to the active GTP-bound form. The majority of SOS1 missense mutations are located in codons encoding residues that are responsible for stabilizing the protein in an inhibited conformation. These mutations disrupt the auto-inhibition of SOS1 RasGEF activity resulting in a gain-of-function of SOS1 and a subsequent increase in the active form of Ras and increased Ras/MAPK pathway signaling [6,7].
KRAS mutations are associated with a small percentage (< 2%) of individuals with NS [5]. KRAS encodes two splice variants with KRASA expressed in a tissue specific and developmentally restricted fashion and KRASB being ubiquitously expressed. The identified KRAS mutations that cause NS produce increase signaling down the Ras/MAPK pathway through two distinct mechanisms: either by mutations that reduce the intrinsic and GAP stimulated GTPase activity [5,14] or by mutations that interfere with the binding of KRAS and guanine nucleotides. Both result in a net increase in the active GTP-bound form of KRAS. The resulting increase in signaling down the Ras/MAPK pathway is, however, less than that which occurs with oncogenic KRAS activating mutations [5].
Mutations in RAF1 also cause NS [8,9]. RAF1 encodes the protein RAF1 (aka CRAF), a serine/threonine kinase that is one of the direct downstream effectors of Ras. The majority of RAF1 mutations associated with NS cluster in two regions: in conserved region 2 flanking S259 and in conserved region 3, surrounding the activation segment. These mutations result in a CRAF gain-of-function, since the phosphorylation of residues S259 and S621 are responsible for regulation of CRAF. Rare mutations within the activation segment have lower kinase activity [8].
LEOPARD Syndrome
LEOPARD syndrome (LS) is a rare autosomal dominant disorder with a similar phenotype to NS, including a “Noonan-like” appearance as well as multiple Lentigines, EKG abnormalities, Ocular hypertelorism, Pulmonary valve stenosis, Abnormal genitalia, Retardation of growth, and Deafness (acronym LEOPARD). LS and NS are allelic disorders, caused by different heterozygous missense mutations in the same genes, PTPN11 [15,16] and RAF1 [8] (Table 1). The most common LS associated PTPN11 mutations affect amino acids in the catalytic PTP domain which result in reduced SHP2 catalytic activity [15,17]. In contrast, PTPN11 mutations associated with NS all produce SHP2 gain-of-function [12]. It has recently been proposed that the residual catalytic activity in the LS mutant SHP2 protein is sufficient to produce a gain-of function-like phenotype due to dysregulation of the protein causing continuous MAPK pathway activity during development [18].
Hereditary Gingival Fibromatosis
Hereditary gingival fibromatosis (HGF) is characterized by a slowly progressive, benign, fibrous overgrowth of the keratinized gingiva [19]. HGF is a genetically heterogenous condition with both autosomal dominant and recessive inheritance reported. One rare autosomal dominant form, HGF Type 1, is caused by an insertion mutation in the SOS1 gene [20] (Table 1). The SOS1 insertion mutation causes a frame-shift that generates 22 novel amino acids prior to a premature stop codon which abolishes four proline-rich SH3 binding domains required for GRB2 binding in the C-terminus [20]. Ectopic expression of the mutant SOS1 protein shows that the truncated protein localizes to the plasma membrane without growth factor binding causing Ras activation and sustained signaling down the Ras/MAPK pathway [21]. Interestingly, no developmental effects are observed with the SOS1 mutation associated with HGF1 as there are with the activating SOS1 mutations associated with NS.
Neurofibromatosis Type 1
Neurofibromatosis type 1 (NF1) is an autosomal dominant inherited disorder affecting approximately 1 in 3000 newborns (for review see [22]). The clinical diagnosis of NF1 is based on the presence of café-au-lait maculae, intertriginous freckling, neurofibromas and plexiform neurofibromas, iris Lisch nodules, osseous dysplasia, optic pathway glioma and/or a first-degree relative with the clinical diagnosis of NF1. In addition, individuals may have a Noonan-like facies, mild neurocognitive impairment and a predisposition to developing certain malignancies. NF1 is caused by germline mutations in the NF1 gene, with approximately half of the mutations being de novo [23–25]. NF1 encodes neurofibromin, a GTPase activating protein that is a negative regulator of Ras (Table 1). The NF1 missense mutations result in loss-of-function of neurofibromin GAP reduces RasGTPase activity and therefore an increase in active GTP-bound Ras and signaling down the Ras/MAPK pathway.
Capillary malformation – arteriovenous malformation
Capillary malformation–arteriovenous malformation syndrome (CM-AVM) is an autosomal dominant inherited disorder characterized by mutifocal capillary malformations which may be associated with arteriovenous malformations and fistulas (for review see [26]). CM-AVM syndrome is caused heterozygous inactivating mutations in the gene RASA1, which like NF1, encodes a RasGAP [27] (Table 1). The hallmark of this syndrome is the mutifocality of the malformations. AVMs can occur in many tissues including skin, muscle, bone, and in various internal organs including the heart and the brain. In addition, RASA1 mutations have been associated with individuals diagnosed with Parkes Weber syndrome and vein of Galen malformations [28]. Haploinsufficiency of p120-RasGAP, the protein product of RASA1, causes a reduction in the hydrolysis of Ras-GTP and, therefore, increases Ras/MAPK pathway signaling. Interestingly, CM-AVM patients may be at increased risk of developing tumors.
Costello Syndrome
Costello syndrome (CS) is a rare developmental disorder with multiple anomalies, including characteristic dysmorphic craniofacial features, failure to thrive, cardiac, musculoskeletal and ectodermal abnormalities and neurocognitive delay (for review see [29]). Individuals with CS are at increased risk of developing neoplasms, both benign and malignant.
Heterozygous germline mutations in HRAS cause CS [30] (Table 1). The distribution frequency of mutations reveals that more than 80% of individuals have a G12S substitution, followed by the second most common, G12A. (for review see [31]). These substitutions disrupt guanine nucleotide binding and cause a reduction in intrinsic and GAP induced GTPase activity resulting in Ras remaining in the active state [32]. In addition, less frequently observed mutations, such as K117R and A146T may result in an atypical phenotype [33,34]. Biochemical investigation of the novel HRAS mutant protein, K117R, has demonstrated normal intrinsic GTP hydrolysis and responsiveness to GTPase-activating proteins; however, the nucleotide dissociation rate is increased. The increase in the guanine nucleotide exchange rate results in a net increase in the active GTP bound form of Ras due to the higher concentration of GTP in the cell [35]. It is interesting that amino acid positions 12 and 13, the two most common positions mutated in CS, are also the most frequently mutated positions in oncogenic Ras. In fact, Ras mutations in codons 12, 13 or 61 are present in approximately 20% of all tumors [1]. However, no mutations in codon 61 have been reported in CS.
Autoimmune lymphoproliferative Syndrome
Autoimmune lymphoproliferative syndrome (ALPS) is characterized by defective lymphocyte apoptosis, an accumulation of nonmalignant lymphocytes and an increase in the risk of developing hematological malignancies [36]. Most cases of ALPS are associated with impaired extrinsic Fas-receptor mediated apoptosis caused by mutations associated with the CD95 pathway [37]. Recently, a cause of ALPS independent of the CD95 pathway has been identified as being due to a germline mutation in NRAS [38] (Table 1). The germline NRAS mutation associated with ALPS causes an activating G13D amino acid residue substitution which results in stabilization of the active GTP-bound form of NRAS resulting in increased signaling down the Ras/MAPK pathway. Increased ERK phosphorylation, due to NRAS activation, inhibits the expression of the apoptosis promoting protein BCL-2-interacting mediator of cell death (BIN) in lymphocytes. This decrease in BIN lymphocytic expression causes an inhibition of intrinsic mitochondrial apoptosis [36]. In contrast to the effects of activating mutations in HRAS and KRAS, which have profound developmental effects, the activating NRAS mutation associated with ALPS does not affect development.
Cardio-facio-cutaneous Syndrome
Cardio-facio-cutaneous syndrome (CFC), like CS, is rare, and shares many overlapping phenotypic features with NS and CS, and to some extent with NF1. CFC individuals have a Noonan-like facies, cardiac malformations, ectodermal, gastrointestinal, ocular and musculoskeletal abnormalities, with most having short stature (for review see [39]). Neurologic abnormalities are universally present to varying degrees and include hypotonia, motor delay, speech delay and/or learning disability [40]. Four genes that encode proteins in the Ras/MAPK pathway downstream of Ras, have been associated with CFC syndrome: BRAF [41,42], MAP2K1 and MAP2K2 [41], and KRAS [42] (Table 1). The role of KRAS in CFC remains unclear because KRAS mutations were also identified in individuals clinically diagnosed NS [5].
Heterozygous BRAF mutations are found in approximately 75% of mutation-positive CFC individuals (for review see [43]). BRAF is a serine/threonine protein kinase and one of the direct downstream effectors of Ras. The majority of BRAF mutations cluster in the cysteine-rich domain in exon 6 and in the protein kinase domain. In vitro functional analyses of the BRAF mutation proteins have demonstrated that most have increased kinase activity; however a few mutant proteins have kinase-impaired activities [41,42]. Increased BRAF activity results in increased signaling down the MAPK pathway. BRAF kinase impairment has also been shown to dysregulate signaling down the MAPK pathway [44], possibly through CRAF interaction [45]. Somatic mutations in BRAF have been reported in several different types of malignancies; however, the majority of CFC-associated mutations are novel.
Heterozygous missense mutations in MAP2K1 (MEK1) and MAP2K2 (MEK2) are present in approximately 25% CFC individuals in which a gene mutation has been identified (for review see [43]). MEK1 and MEK2 are threonine/tyrosine kinases, with both isoforms having the ability to activate ERK1 and ERK2. Functional studies of the mutant proteins have shown that all are activating [41]. Unlike NS, CS and NF1, it is unclear if individuals with CFC syndrome are at an increased risk for malignancies.
Legius Syndrome (Neurofibromatosis 1-Like)
Legius syndrome is an autosomal dominant disorder that shares many phenotypic features with NF1. Individuals may have café-au-lait maculae, axillary freckling, mild neurocognitive impairment and macrocephaly with some having a Noonan-like facies. However, the phenotypic features common in NF1 such as neurofibromas, iris Lisch nodules and central nervous system tumors are lacking. Legius syndrome is caused by heterozygous mutations in SPRED1 [46] (Table 1). SPRED1, encodes SPRED1 which is a member of the SPROUTY/SPRED family of proteins. SPRED1 functions as negative regulator of Ras by inhibiting phosphorylation of Raf [47]. Heterogenous SPRED1 mutations associated with NF1-like syndrome cause truncation of the protein resulting in a loss of SPRED1 function thereby increasing signaling down the Ras/MAPK pathway. It is unclear at this point whether individuals with SPRED1 germline mutations are at increased risk for developing cancer.
Conclusion
The RASopathies, caused by germline mutations in genes encoding components of the Ras/MAPK pathway, underscores the essential role the pathway plays in normal embryonic and postnatal development. In Xenopus, elevated MAPK activity initiates oocyte meiotic maturation, metaphase arrest in cleaving embryos and is essential for mesoderm induction and patterning [48,49]. Highly controlled MAPK pathway regulation is also observed during Drosophila and Zebrafish embryonic development [50,51]. In mice, transient ERK phosphorylation is found within the neural crest, peripheral nervous system, nascent blood vessels and the early forming structures of the ear, eye and heart [52].
The number of different affected genes and the diversity of mutations within each gene are reflected in the variety of affected phenotypic features present in this class of syndromes. All the mutations result in dysregulation of the Ras/MAPK pathway, with functional studies determining that the vast majority increase signaling down the pathway, and possibly result in constitutive pathway activation. Therefore, it is not surprising that many of these syndromes exhibit overlapping phenotypic features and share a predisposition to developing malignancies. Although many of the activating mutations are similar to activating somatic mutations seen in cancer, on the whole, they tend to be not as strongly activating. For example, the most common oncogenic BRAF mutation, V600E, does not occur in CFC syndrome and the specific KRAS mutations associated with NS are not the same as the known somatic mutations associated with cancer. It is likely that the strongly activating oncogenic mutations cannot be tolerated as germline mutations. Transgenic mouse models harboring Ras/MAPK gene knockouts, or oncogenic mutations in the same genes that are implicated in the Ras/MAPK developmental disorders are informative and many have demonstrated the essential role these genes play during development [53–55]. Finally, the RASopathies provide not only valuable information into the consequences of dysregulation of the Ras/MAPK pathway, but insight into its role during normal human development.
Acknowledgments
The authors thank patients and families for their ongoing support of research in genetic medicine. The authors apologize for not citing all relevant references due to space limitations. This work was supported in part by NIH grant HD048502 (K.A.R.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bos JL. ras oncogenes in human cancer: a review. Cancer Res. 1989;49:4682–4689. [PubMed] [Google Scholar]
- 2.Yoon S, Seger R. The extracellular signal-regulated kinase: multiple substrates regulate diverse cellular functions. Growth Factors. 2006;24:21–44. doi: 10.1080/02699050500284218. [DOI] [PubMed] [Google Scholar]
- 3.Shaw AC, Kalidas K, Crosby AH, Jeffery S, Patton MA. The natural history of Noonan syndrome: a long-term follow-up study. Arch Dis Child. 2007;92:128–132. doi: 10.1136/adc.2006.104547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4*.Tartaglia M, Mehler EL, Goldberg R, Zampino G, Brunner HG, Kremer H, van der Burgt I, Crosby AH, Ion A, Jeffery S, et al. Mutations in PTPN11, encoding the protein tyrosine phosphatase SHP-2, cause Noonan syndrome. Nat Genet. 2001;29:465–468. doi: 10.1038/ng772. This is the first paper to identify mutations in the PTPN11 gene being responsible for approximately 50% of Noonan syndrome cases. [DOI] [PubMed] [Google Scholar]
- 5*.Schubbert S, Zenker M, Rowe SL, Boll S, Klein C, Bollag G, van der Burgt I, Musante L, Kalscheuer V, Wehner LE, et al. Germline KRAS mutations cause Noonan syndrome. Nat Genet. 2006;38:331–336. doi: 10.1038/ng1748. This is the first report of mutations in KRAS associated with a small percentage of Noonan syndrome cases. [DOI] [PubMed] [Google Scholar]
- 6*.Roberts AE, Araki T, Swanson KD, Montgomery KT, Schiripo TA, Joshi VA, Li L, Yassin Y, Tamburino AM, Neel BG, et al. Germline gain-of-function mutations in SOS1 cause Noonan syndrome. Nat Genet. 2007;39:70–74. doi: 10.1038/ng1926. This paper, along with the following paper (#7), is the first to report mutations in SOS1 in approximately 13% of Noonan syndrome cases. [DOI] [PubMed] [Google Scholar]
- 7*.Tartaglia M, Pennacchio LA, Zhao C, Yadav KK, Fodale V, Sarkozy A, Pandit B, Oishi K, Martinelli S, Schackwitz W, et al. Gain-of-function SOS1 mutations cause a distinctive form of Noonan syndrome. Nat Genet. 2007;39:75–79. doi: 10.1038/ng1939. This paper along with the simultaneously published paper #6, are the first reports to associate SOS1 mutations with Noonan syndrome. [DOI] [PubMed] [Google Scholar]
- 8*.Pandit B, Sarkozy A, Pennacchio LA, Carta C, Oishi K, Martinelli S, Pogna EA, Schackwitz W, Ustaszewska A, Landstrom A, et al. Gain-of-function RAF1 mutations cause Noonan and LEOPARD syndromes with hypertrophic cardiomyopathy. Nat Genet. 2007;39:1007–1012. doi: 10.1038/ng2073. This is one of two papers, along with (#9), that reported RAF1 mutations causing Noonan syndrome. In addition, in this report the authors show that LEOPARD syndrome is also associated with RAF1 mutations. [DOI] [PubMed] [Google Scholar]
- 9*.Razzaque MA, Nishizawa T, Komoike Y, Yagi H, Furutani M, Amo R, Kamisago M, Momma K, Katayama H, Nakagawa M, et al. Germline gain-of-function mutations in RAF1 cause Noonan syndrome. Nat Genet. 2007;39:1013–1017. doi: 10.1038/ng2078. This is one of two papers (also #8) that first report RAF1 mutations being causative of Noonan syndrome. [DOI] [PubMed] [Google Scholar]
- 10.Hof P, Pluskey S, Dhe-Paganon S, Eck MJ, Shoelson SE. Crystal structure of the tyrosine phosphatase SHP-2. Cell. 1998;92:441–450. doi: 10.1016/s0092-8674(00)80938-1. [DOI] [PubMed] [Google Scholar]
- 11.Keilhack H, David FS, McGregor M, Cantley LC, Neel BG. Diverse biochemical properties of Shp2 mutants. Implications for disease phenotypes. J Biol Chem. 2005;280:30984–30993. doi: 10.1074/jbc.M504699200. [DOI] [PubMed] [Google Scholar]
- 12.Tartaglia M, Martinelli S, Stella L, Bocchinfuso G, Flex E, Cordeddu V, Zampino G, Burgt I, Palleschi A, Petrucci TC, et al. Diversity and functional consequences of germline and somatic PTPN11 mutations in human disease. Am J Hum Genet. 2006;78:279–290. doi: 10.1086/499925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Niihori T, Aoki Y, Ohashi H, Kurosawa K, Kondoh T, Ishikiriyama S, Kawame H, Kamasaki H, Yamanaka T, Takada F, et al. Functional analysis of PTPN11/SHP-2 mutants identified in Noonan syndrome and childhood leukemia. J Hum Genet. 2005;50:192–202. doi: 10.1007/s10038-005-0239-7. [DOI] [PubMed] [Google Scholar]
- 14.Schubbert S, Bollag G, Lyubynska N, Nguyen H, Kratz CP, Zenker M, Niemeyer CM, Molven A, Shannon K. Biochemical and functional characterization of germ line KRAS mutations. Mol Cell Biol. 2007;27:7765–7770. doi: 10.1128/MCB.00965-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15*.Digilio MC, Conti E, Sarkozy A, Mingarelli R, Dottorini T, Marino B, Pizzuti A, Dallapiccola B. Grouping of multiple-lentigines/LEOPARD and Noonan syndromes on the PTPN11 gene. Am J Hum Genet. 2002;71:389–394. doi: 10.1086/341528. This is one of two simultaneously published reports (also #16) that first identified mutations in PTPN11 causing LEOPARD syndrome. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16*.Legius E, Schrander-Stumpel C, Schollen E, Pulles-Heintzberger C, Gewillig M, Fryns JP. PTPN11 mutations in LEOPARD syndrome. J Med Genet. 2002;39:571–574. doi: 10.1136/jmg.39.8.571. This is one of two simultaneously published reports (also #15) that first identified mutations in PTPN11 causing LEOPARD syndrome. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kontaridis MI, Swanson KD, David FS, Barford D, Neel BG. PTPN11 (Shp2) mutations in LEOPARD syndrome have dominant negative, not activating, effects. J Biol Chem. 2006;281:6785–6792. doi: 10.1074/jbc.M513068200. [DOI] [PubMed] [Google Scholar]
- 18.Oishi K, Zhang H, Gault WJ, Wang CJ, Tan CC, Kim IK, Ying H, Rahman T, Pica N, Tartaglia M, et al. Phosphatase-defective LEOPARD syndrome mutations in PTPN11 gene have gain-of-function effects during Drosophila development. Hum Mol Genet. 2009;18:193–201. doi: 10.1093/hmg/ddn336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hart TC, Pallos D, Bowden DW, Bolyard J, Pettenati MJ, Cortelli JR. Genetic linkage of hereditary gingival fibromatosis to chromosome 2p21. Am J Hum Genet. 1998;62:876–883. doi: 10.1086/301797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20*.Hart TC, Zhang Y, Gorry MC, Hart PS, Cooper M, Marazita ML, Marks JM, Cortelli JR, Pallos D. A mutation in the SOS1 gene causes hereditary gingival fibromatosis type 1. Am J Hum Genet. 2002;70:943–954. doi: 10.1086/339689. This is the first report that one form of hereditary gingival fibromatosis (Type 1) is associated with a mutation in SOS1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jang SI, Lee EJ, Hart PS, Ramaswami M, Pallos D, Hart TC. Germ line gain of function with SOS1 mutation in hereditary gingival fibromatosis. J Biol Chem. 2007;282:20245–20255. doi: 10.1074/jbc.M701609200. [DOI] [PubMed] [Google Scholar]
- 22.Williams VC, Lucas J, Babcock MA, Gutmann DH, Korf B, Maria BL. Neurofibromatosis type 1 revisited. Pediatrics. 2009;123:124–133. doi: 10.1542/peds.2007-3204. [DOI] [PubMed] [Google Scholar]
- 23*.Wallace MR, Marchuk DA, Andersen LB, Letcher R, Odeh HM, Saulino AM, Fountain JW, Brereton A, Nicholson J, Mitchell AL, et al. Type 1 neurofibromatosis gene: identification of a large transcript disrupted in three NF1 patients. Science. 1990;249:181–186. doi: 10.1126/science.2134734. This paper is one of three simultaneously published reports (also # 24 and #25) that first identified mutations in NF1 as causative for neurofibromatosis type 1. [DOI] [PubMed] [Google Scholar]
- 24*.Cawthon RM, O’Connell P, Buchberg AM, Viskochil D, Weiss RB, Culver M, Stevens J, Jenkins NA, Copeland NG, White R. Identification and characterization of transcripts from the neurofibromatosis 1 region: the sequence and genomic structure of EVI2 and mapping of other transcripts. Genomics. 1990;7:555–565. doi: 10.1016/0888-7543(90)90199-5. This paper is one of three simultaneously published reports (also # 23 and #25) that first identified mutations in NF1 as causative for neurofibromatosis type 1. [DOI] [PubMed] [Google Scholar]
- 25*.Viskochil D, Buchberg AM, Xu G, Cawthon RM, Stevens J, Wolff RK, Culver M, Carey JC, Copeland NG, Jenkins NA, et al. Deletions and a translocation interrupt a cloned gene at the neurofibromatosis type 1 locus. Cell. 1990;62:187–192. doi: 10.1016/0092-8674(90)90252-a. This paper is one of three simultaneously published reports (also # 23 and #24) that first identified mutations in NF1 as causative for neurofibromatosis type 1. [DOI] [PubMed] [Google Scholar]
- 26.Boon LM, Mulliken JB, Vikkula M. RASA1: variable phenotype with capillary and arteriovenous malformations. Curr Opin Genet Dev. 2005;15:265–269. doi: 10.1016/j.gde.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 27*.Eerola I, Boon LM, Mulliken JB, Burrows PE, Dompmartin A, Watanabe S, Vanwijck R, Vikkula M. Capillary malformation-arteriovenous malformation, a new clinical and genetic disorder caused by RASA1 mutations. Am J Hum Genet. 2003;73:1240–1249. doi: 10.1086/379793. This is the first report associating capillary malformation-arteriovenous malformation syndrome with mutations in RASA1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Revencu N, Boon LM, Mulliken JB, Enjolras O, Cordisco MR, Burrows PE, Clapuyt P, Hammer F, Dubois J, Baselga E, et al. Parkes Weber syndrome, vein of Galen aneurysmal malformation, and other fast-flow vascular anomalies are caused by RASA1 mutations. Hum Mutat. 2008;29:959–965. doi: 10.1002/humu.20746. [DOI] [PubMed] [Google Scholar]
- 29.Rauen KA. HRAS and the Costello syndrome. Clin Genet. 2007;71:101–108. doi: 10.1111/j.1399-0004.2007.00743.x. [DOI] [PubMed] [Google Scholar]
- 30*.Aoki Y, Niihori T, Kawame H, Kurosawa K, Ohashi H, Tanaka Y, Filocamo M, Kato K, Suzuki Y, Kure S, et al. Germline mutations in HRAS proto-oncogene cause Costello syndrome. Nat Genet. 2005;37:1038–1040. doi: 10.1038/ng1641. This is the first published report of mutations in HRAS as causative for Costello syndrome. [DOI] [PubMed] [Google Scholar]
- 31.Tidyman WE, Rauen KA. Noonan, Costello and cardio-facio-cutaneous syndromes: dysregulation of the Ras-MAPK pathway. Expert Rev Mol Med. 2008;10:e37. doi: 10.1017/S1462399408000902. [DOI] [PubMed] [Google Scholar]
- 32.Gibbs JB, Sigal IS, Poe M, Scolnick EM. Intrinsic GTPase activity distinguishes normal and oncogenic ras p21 molecules. Proc Natl Acad Sci U S A. 1984;81:5704–5708. doi: 10.1073/pnas.81.18.5704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kerr B, Delrue MA, Sigaudy S, Perveen R, Marche M, Burgelin I, Stef M, Tang B, Eden T, O’Sullivan J, et al. Genotype-phenotype correlation in Costello syndrome; HRAS mutation analysis in 43 cases. J Med Genet. 2006 doi: 10.1136/jmg.2005.040352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zampino G, Pantaleoni F, Carta C, Cobellis G, Vasta I, Neri C, Pogna EA, De Feo E, Delogu A, Sarkozy A, et al. Diversity, parental germline origin, and phenotypic spectrum of de novo HRAS missense changes in Costello syndrome. Hum Mutat. 2007;28:265–272. doi: 10.1002/humu.20431. [DOI] [PubMed] [Google Scholar]
- 35.Denayer E, Parret A, Chmara M, Schubbert S, Vogels A, Devriendt K, Frijns JP, Rybin V, de Ravel TJ, Shannon K, et al. Mutation analysis in Costello syndrome: functional and structural characterization of the HRAS p. Lys117Arg mutation. Hum Mutat. 2008;29:232–239. doi: 10.1002/humu.20616. [DOI] [PubMed] [Google Scholar]
- 36.Oliveira JB, Fleisher T. Autoimmune lymphoproliferative syndrome. Curr Opin Allergy Clin Immunol. 2004;4:497–503. doi: 10.1097/00130832-200412000-00005. [DOI] [PubMed] [Google Scholar]
- 37.Bidere N, Su HC, Lenardo MJ. Genetic disorders of programmed cell death in the immune system. Annu Rev Immunol. 2006;24:321–352. doi: 10.1146/annurev.immunol.24.021605.090513. [DOI] [PubMed] [Google Scholar]
- 38*.Oliveira JB, Bidere N, Niemela JE, Zheng L, Sakai K, Nix CP, Danner RL, Barb J, Munson PJ, Puck JM, et al. NRAS mutation causes a human autoimmune lymphoproliferative syndrome. Proc Natl Acad Sci U S A. 2007;104:8953–8958. doi: 10.1073/pnas.0702975104. This is the first report of NRAS mutations causing one form of human autoimmune lymphoproliferative syndrome. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roberts A, Allanson J, Jadico SK, Kavamura MI, Noonan J, Opitz JM, Young T, Neri G. The Cardio-Facio-Cutaneous (CFC) syndrome: a review. J Med Genet. 2006 doi: 10.1136/jmg.2006.042796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoon G, Rosenberg J, Blaser S, Rauen KA. Neurological complications of cardio-facio-cutaneous syndrome. Dev Med Child Neurol. 2007;49:894–899. doi: 10.1111/j.1469-8749.2007.00894.x. [DOI] [PubMed] [Google Scholar]
- 41*.Rodriguez-Viciana P, Tetsu O, Tidyman WE, Estep AL, Conger BA, Cruz MS, McCormick F, Rauen KA. Germline mutations in genes within the MAPK pathway cause cardio-facio-cutaneous syndrome. Science. 2006;311:1287–1290. doi: 10.1126/science.1124642. This is one of two papers (including #41) which first report mutations in BRAF are responsible for the majority of CFC cases. In addition, this was the first report to show that mutations in MAP2K1 and MAP2K2 are responsible for approximately a quarter of CFC syndrome cases. [DOI] [PubMed] [Google Scholar]
- 42*.Niihori T, Aoki Y, Narumi Y, Neri G, Cave H, Verloes A, Okamoto N, Hennekam RC, Gillessen-Kaesbach G, Wieczorek D, et al. Germline KRAS and BRAF mutations in cardio-facio-cutaneous syndrome. Nat Genet. 2006;38:294–296. doi: 10.1038/ng1749. This is one of two reports, including #40, which first report BRAF mutations are responsible for CFC syndrome. [DOI] [PubMed] [Google Scholar]
- 43.Tidyman WE, Rauen KA. Molecular Causes of Cardio-Facio-Cutaneous Syndrome. In: Karger MZ, editor. Monographs in Human Genetics: Noonan Syndrome and Related Disorders - A matter of deregulated Ras signaling. Vol. 17 2009. pp. 73–82. [Google Scholar]
- 44.Anastasaki C, Estep AL, Marais R, Rauen KA, Patton EE. Kinase-activating and kinase-impaired cardio-facio-cutaneous syndrome alleles have activity during zebrafish development, and are sensitive to small molecule inhibitors. Human Molecular Genetics. 2009 doi: 10.1093/hmg/ddp186. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wan PT, Garnett MJ, Roe SM, Lee S, Niculescu-Duvaz D, Good VM, Jones CM, Marshall CJ, Springer CJ, Barford D, et al. Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell. 2004;116:855–867. doi: 10.1016/s0092-8674(04)00215-6. [DOI] [PubMed] [Google Scholar]
- 46*.Brems H, Chmara M, Sahbatou M, Denayer E, Taniguchi K, Kato R, Somers R, Messiaen L, De Schepper S, Fryns JP, et al. Germline loss-of-function mutations in SPRED1 cause a neurofibromatosis 1-like phenotype. Nat Genet. 2007;39:1120–1126. doi: 10.1038/ng2113. This is the first report that mutations in SPRED1 cause an NF1-like syndrome; this new syndrome was subsequently named Legius syndrome. [DOI] [PubMed] [Google Scholar]
- 47.Wakioka T, Sasaki A, Kato R, Shouda T, Matsumoto A, Miyoshi K, Tsuneoka M, Komiya S, Baron R, Yoshimura A. Spred is a Sprouty-related suppressor of Ras signalling. Nature. 2001;412:647–651. doi: 10.1038/35088082. [DOI] [PubMed] [Google Scholar]
- 48.Haccard O, Lewellyn A, Hartley RS, Erikson E, Maller JL. Induction of Xenopus oocyte meiotic maturation by MAP kinase. Dev Biol. 1995;168:677–682. doi: 10.1006/dbio.1995.1112. [DOI] [PubMed] [Google Scholar]
- 49.Curran KL, Grainger RM. Expression of activated MAP kinase in Xenopus laevis embryos: evaluating the roles of FGF and other signaling pathways in early induction and patterning. Dev Biol. 2000;228:41–56. doi: 10.1006/dbio.2000.9917. [DOI] [PubMed] [Google Scholar]
- 50.Gabay L, Seger R, Shilo BZ. MAP kinase in situ activation atlas during Drosophila embryogenesis. Development. 1997;124:3535–3541. doi: 10.1242/dev.124.18.3535. [DOI] [PubMed] [Google Scholar]
- 51.Furthauer M, Van Celst J, Thisse C, Thisse B. Fgf signalling controls the dorsoventral patterning of the zebrafish embryo. Development. 2004;131:2853–2864. doi: 10.1242/dev.01156. [DOI] [PubMed] [Google Scholar]
- 52.Corson LB, Yamanaka Y, Lai KM, Rossant J. Spatial and temporal patterns of ERK signaling during mouse embryogenesis. Development. 2003;130:4527–4537. doi: 10.1242/dev.00669. [DOI] [PubMed] [Google Scholar]
- 53.Johnson L, Greenbaum D, Cichowski K, Mercer K, Murphy E, Schmitt E, Bronson RT, Umanoff H, Edelmann W, Kucherlapati R, et al. K-ras is an essential gene in the mouse with partial functional overlap with N-ras. Genes Dev. 1997;11:2468–2481. doi: 10.1101/gad.11.19.2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mercer K, Giblett S, Green S, Lloyd D, DaRocha Dias S, Plumb M, Marais R, Pritchard C. Expression of endogenous oncogenic V600EB-raf induces proliferation and developmental defects in mice and transformation of primary fibroblasts. Cancer Res. 2005;65:11493–11500. doi: 10.1158/0008-5472.CAN-05-2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Galabova-Kovacs G, Matzen D, Piazzolla D, Meissl K, Plyushch T, Chen AP, Silva A, Baccarini M. Essential role of B-Raf in ERK activation during extraembryonic development. Proc Natl Acad Sci U S A. 2006;103:1325–1330. doi: 10.1073/pnas.0507399103. [DOI] [PMC free article] [PubMed] [Google Scholar]