Abstract
Single molecule localization (SML) is a powerful tool to measure the position and trajectory of molecules in numerous systems, with nanometer accuracy. This technique has been recently utilized to overcome the diffraction limit in optical imaging. So far, super-resolution imaging by SML was demonstrated using photoactivable or photoswitchable fluorophores, as well as diffusive fluorophore probes in solution. All these methods, however, rely on special fluorophore or object properties. In this Letter, we propose and demonstrate a new super-resolution technique attainable for a bio/dielectric structure on a metal substrate. A sub-diffraction-limited image is obtained by randomly adsorbed fluorescent probe molecules on a liquid–solid interface, while the metal substrate, quenching the unwanted fluorescent signal, provides a significantly enhanced imaging contrast. As this approach does not use specific stain techniques, it can be readily applied to general dielectric objects, such as nanopatterned photoresist, inorganic nanowires, subcellular structures, etc.
The resolution of conventional optical microscopy is always restricted by the diffraction limit, due to the decay of high spatial frequency evanescent waves associated with sub-wavelength objects. Exciting developments that have been proposed to overcome this limit include near-field scanning optical microscopy (NSOM),1,2 stimulated emission depletion (STED) microscopy,3,4 silver slab superlens,5 saturated structured illumination microscopy (SSIM),6 etc. In particular, the recent achievements in single molecule localization provided a new alternative to realize ultrahigh resolution by numerically fitting the center position of the fluorescent molecules.7,8 However, a continuous tone super-resolution image requires a large number of molecules within the area of interest, whereas single molecule imaging demands a low concentration (no more than one molecule within a diffraction limited area). To overcome this problem, photoactivable/switchable fluorophore9–12 and diffusive molecules in solution13 methods were proposed. Indeed, these methods achieved relatively high concentration of fluorescent molecules, while still able to realize single molecule imaging by exciting only few molecules at each frame of measurement. However, all these methods are limited by the stain technique9–12 or to lipid bilayer objects.13
Here, we report a new way for single-molecule continuous tone super-resolution imaging that does not depend on or limited by specific fluorophore or stain technique. Our method utilizes dynamic adsorption of fluorescent molecules on a liquid–solid interface (as shown in Figure 1). The fluorescent signals from molecules sparsely adsorbed on the surface are recorded by the CCD camera, while the rest are blurred out by fast diffusion to a uniform background. Furthermore, we apply a metal substrate to quench the fluorescent signals that are not from the surface of the dielectric objects to be imaged (Figure 1a). Combining these two methods, we can detect only the local reporters adsorbed on the object surfaces, providing the necessary contrast to form an image. Since this is a physical process (not relying on specific chemical bonding), this method requires no special fluorophore or stain techniques and, thus, in principle can be applied to most dielectric objects on a metal substrate.
Figure 1.
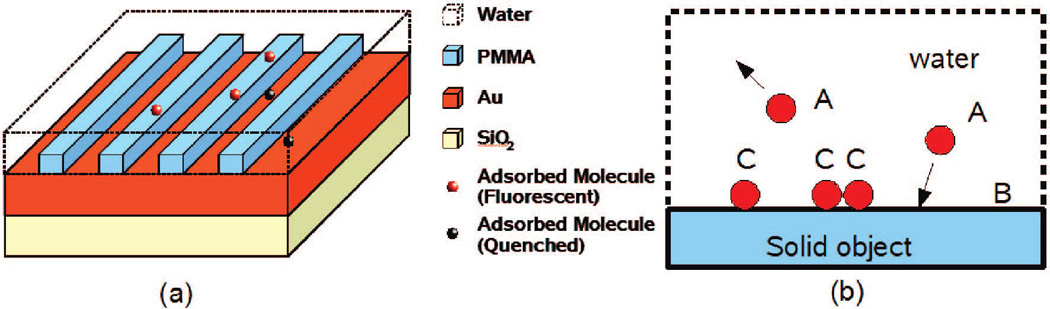
Principle of super-resolution imaging by randomly adsorbed molecular probes. (a) Schematic drawing shows nano-objects (gratings in this case) on a Au metal surface. Only the molecules (red) adsorbed on the dielectric surface emit a fluorescent signal to the far-field. The molecules on the metal surface (black) are quenched and cannot be detected. (b) Physical adsorption of molecules at a solid–liquid interface. A denotes the molecules diffusing freely in liquid, B denotes the surface vacancy, and C denotes the adsorbed molecules.
Physical adsorption of molecules on liquid/solid (or gas/solid) interface is known to be random and reversible. The fluorescent molecules act as a probe only when adsorbed and become immobilized on the interface. Consider the following chemical system (Figure 1b): a fluorescent molecule, A, is in solution and can bind to an available site on the surface, B, to become a bound species C.
| (1) |
At equilibrium, the adsorption rate equals the desorption rate, giving rise to the equilibrium constant (determined by the binding energy and temperature) K = kads/kdes = [C]/[B][A], where kads and kdes are the rates of adsorption and desorption, respectively, and the brackets indicate molecule concentration. The average lifetime of a molecule on the interface, τ, is hence 1/kdes. According to the Langmuir isotherm,14 the surface coverage
| (2) |
is proportional to the bulk molecule concentration at low concentration, [A] ≪ 1/K. This provides a simple and efficient method to tune the surface density of the fluorescent molecules.
In order to obtain sub-diffraction-limited resolution, we utilize dynamical physical adsorption so as to “stretch in time” the large molecule concentration required for the super-resolution. Namely, a large effective concentration is achieved by integrating over the total sampling time, while at each measured frame the average molecule concentration is no more than one molecule per diffraction limited area. This can be realized by diluting the molecule concentration in the solution according to eq 2. The signal, recorded by the CCD camera can be maximized by optimizing the recording time to match the lifetime, τ of the single adsorbed molecule.
As the result, images taken at different times will record different molecules randomly distributed on the surface, where subsequent adsorption events are separated into different image frames. After a long enough time, the probes will eventually cover all the surface locations and retrieve a complete image.
While the above method can provide ultrahigh resolution, a sufficient image contrast is required to generate a meaningful image. We therefore introduce a unique mechanism to generate and enhance the image contrast, relying on the quenching effect in metals. Consider a dielectric nano-object on a metal (e.g., Au) surface, as shown in Figure.1a. The immersion of the sample into liquid solution forms two different interfaces; the molecules adsorbed on the metal/solution interface will be effectively quenched,15 and only those adsorbed on the dielectric/solution interface will emit photons to the far-field to be collected by the microscope objective lens. This is to say, the Au thin film acts as substrate and any dielectric nanostructures on the substrate are imaged by the molecules.
This concept is experimentally demonstrated by imaging a PMMA nanograting with 110 nm/220 nm line width and 400 nm period in Figure 3a. The 60 nm thick sample was fabricated by E-beam lithography, and the measurement was done using an inverted fluorescent microscope (Nikon TE2000). A continuous wave laser beam (532 nm) was delivered through a 100× oil immersion objective (NA = 1.49) to the sample surface with an average intensity of 4 kW/cm2. The fluorescent signal was collected by the same objective and filtered by a dichroic mirror and a long pass (620 nm) filter and imaged onto a CCD camera by additional 1.5× amplification. The physical pixel size of the 512 × 512 CCD is 16 µm and, hence, represents 107 nm at the image plane. The sample was placed in a liquid container which was firmly attached on a motorized stage. The concentration of probe molecule (Rhodamine Phalloidin) was 0.1 nM. Silica beads (20 µm diameter) were dispersed on the optical window (bottom surface) of the container to generate a small gap between the window and sample surface, which will allow the free diffusion of dye molecules and also keep the background signal low. Figure 2a depicts a typical far-field single molecule image captured with 500 ms exposure time. On average, there are only several diffraction-limited fluorescent dots within one image. Those dots are separated far enough, and we assume each of them is from a single molecule based on the bulk concentration. The diffraction limited single molecule image was then fitted with a two-dimensional (2D) Gaussian function to find the center location and intensity (Figure 2b). By use of statistical theory,16,17 the localization accuracy of single molecule detection is 〈(Δx)2〉 = 4π1/2s3b2/aN2, where s is the standard deviation of point spread function, N is the total number of photons collected from a single molecule, b is the background signal, and a is the normalized pixel size of the CCD. The average total photon number collected from a single rhodamine phalloidin molecule is about 104, and the background signal is about 60 photons per pixel. A typical position distribution of a fixed reference particle (with similar intensity as probe signals) is shown in Figure 2c. The full width at half-maximum (FWHM) of these 2000 measurements is 18 nm, which is slightly larger than the theoretical localization accuracy of 13 nm calculated from the signal-to-noise (S/N) ratio.
Figure 3.
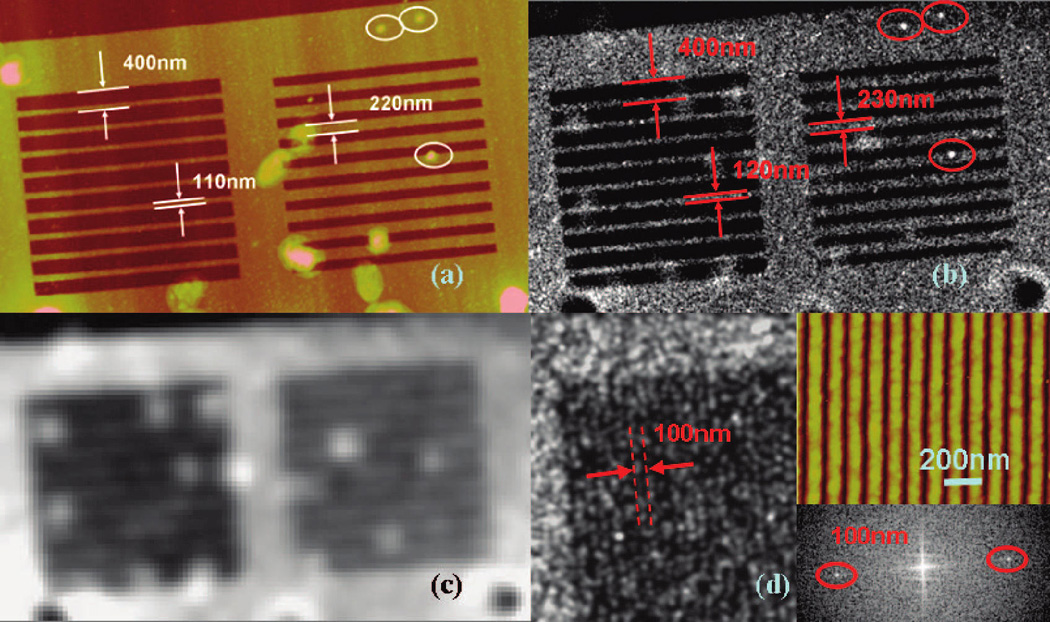
Experimental demonstration of super-resolution imaging. (a) AFM image of the PMMA nanograting structure as the object. The thickness of the PMMA is 60 nm. (b) Computer-rendered super-resolution image from sample in panel a. Each single molecule was rendered using a 2D Gaussian shape PSF with 18 nm fwhm. (c) Fluorescent image from unprocessed data in (b). (d) Super-resolution image of 100 nm period PMMA nanograting. Left image is the experimental result, the top-right image is the AFM measurement, and the bottom-right image is the 2D Fourier transform of the left image (two distinct peaks corresponding to the special frequency of the grating are encircled by red lines).
Figure 2.
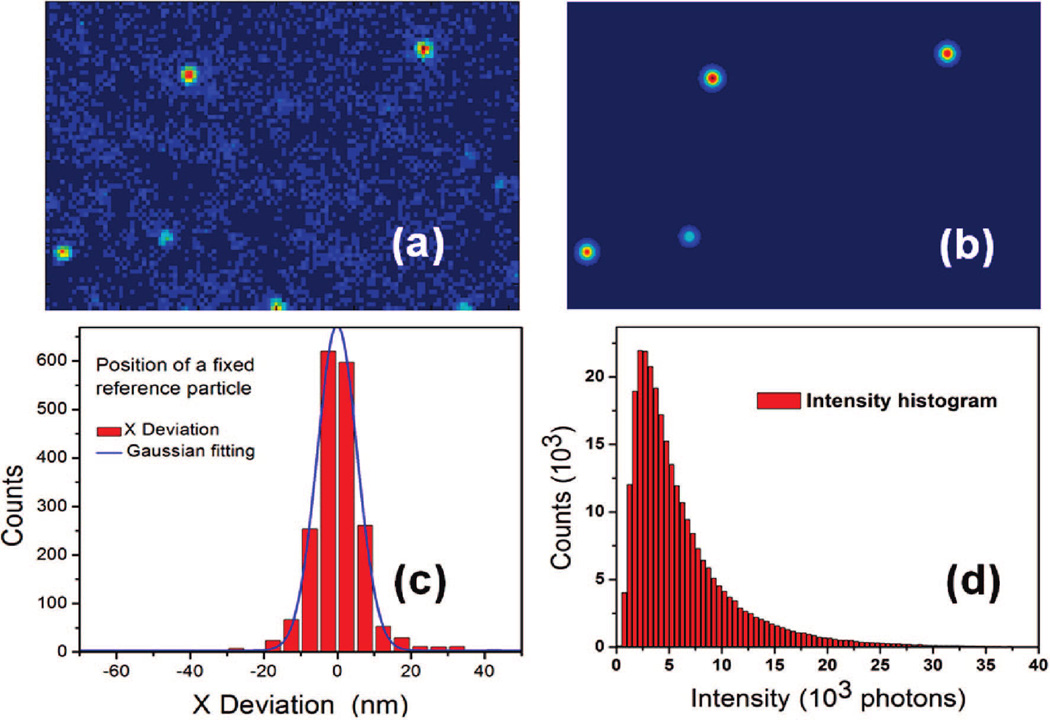
(a) Single molecule fluorescent image captured by the CCD. (b) 2D Gaussian function fitting of the experimental data in (a). The center position and intensity of each single molecule was derived from the fitting. (c) Position distribution of subsequence measurements of a fixed reference particle on the substrate. The standard deviation indicates the resolution of our method, which is about 18 nm. (d) Histogram of single molecule signal from the sample in Figure 3. A threshold was set to cut those measurements with too low S/N ratios.
The sample shown in Figure 3a was tested with our super-resolution procedure by recording about 3 × 105 useful single molecule signals within 10 h of the experiment. We have analyzed the data using a least-squares fitting algorithm and improved the localization precision by setting an intensity threshold to select only those molecules with sufficient signal-to-noise ratios. The final result was rendered using the fitted center position and Gaussian shape PSF with 18 nm fwhm, as shown in Figure 3b (see also the movie in Supporting Information). Without using such image processing, the result is equivalent to traditional fluorescent imaging, as shown in Figure 3c.
The measured line widths of the PMMA gratings from the super-resolution image are about 120 nm (left grating) and 230 nm (right grating), only 10 nm broader than the line width measured by atomic force microscopy (AFM). It is worth mentioning that our method can even retrieve the defects on PMMA structures as shown in the circled region in Figure 3a. We also tested a PMMA sample of 100 nm period grating on a Au surface, as shown in Figure 3d. The Fourier transform of the 2D image has two distinctive peaks at 100 nm spatial frequency. This clearly demonstrates the strength of this method in imaging subdiffraction limited dielectric objects.
In conclusion, we demonstrated a super-resolution technique utilizing the single molecule localization of random adsorbed probe molecules on a liquid–solid interface. The adsorption of a molecule on the surface effectively turns on the fluorescent signal at a random location, and the subsequent desorption/depletion turns such a signal off. This on–off process makes it possible to detect the position of molecules beyond the diffraction limit.18 Similar to other imaging techniques utilizing single molecule localization,9–12 the resolution of this method is not limited by the diffraction but by the signal-to-noise ratio of single molecule detection. We have demonstrated 18 nm localization resolution and 100 nm imaging resolution at 650 nm wavelength, which can be further improved by selecting brighter molecules with longer lifetime. The dynamic nature of physical adsorption makes this method very versatile, and in principle it could be applied to most dielectric nano-objects on a metal surface.
Acknowledgment
We thank Dr. Guy Bartal for useful discussion and help with the manuscript. This work is supported by NSF Nanoscale Science and Engineering Center (DMI-0327077), National Institutes of Health through the NIH Roadmap for Medical Research (PN2 EY108228).
Footnotes
Supporting Information Available: A Quicktime video showing super-resolution imaging. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Betzig E, Trautman JK. Science. 1992;257:189. doi: 10.1126/science.257.5067.189. [DOI] [PubMed] [Google Scholar]
- 2.Pohl DW, Courjon D. Near Field Optics. Kluwer: Dordrecht; 1993. [Google Scholar]
- 3.Hell SW, Wichmann J. Opt. Lett. 1994;19:780. doi: 10.1364/ol.19.000780. [DOI] [PubMed] [Google Scholar]
- 4.Westphal V, Seeger J, Salditt T, Hell SW. J. Phys. B: At., Mol. Opt. Phys. 2005;38:S695. [Google Scholar]
- 5.Fang N, Lee H, Sun C, Zhang X. Science. 2005;308:534. doi: 10.1126/science.1108759. [DOI] [PubMed] [Google Scholar]
- 6.Gustafsson MGL. Proc. Natl. Acad. Sci. U.S.A. 2005;102:13081. doi: 10.1073/pnas.0406877102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qu XH, Wu D, Mets L, Scherer NF. Proc. Natl. Acad. Sci. U.S.A. 2004;101:11298. doi: 10.1073/pnas.0402155101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yildiz A, Forkey JN, McKinney SA, Ha T, Goldman YE, Selvin PR. Science. 2003;300:2061. doi: 10.1126/science.1084398. [DOI] [PubMed] [Google Scholar]
- 9.Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, Davidson MW, Lippincott-Schwartz J, Hess HF. Science. 2006;313:1642. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 10.Hess ST, Girirajan TPK, Mason MD. Biophys. J. 2006;91:4258. doi: 10.1529/biophysj.106.091116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rust MJ, Bates M, Zhuang XW. Nat. Methods. 2006;3:793. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bates M, Huang B, Dempsey GT, Zhuang X. Science. 2007;317:1749. doi: 10.1126/science.1146598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sharonov A, Hochstrasser RM. Proc. Natl. Acad. Sci. U.S.A. 2006;103:18911. doi: 10.1073/pnas.0609643104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Langmuir I. J. Am. Chem. Soc. 1916;38:2221. [Google Scholar]
- 15.Vasilev K, Knoll W, Kreiter M. J. Chem. Phys. 2004;120:3439. doi: 10.1063/1.1640341. [DOI] [PubMed] [Google Scholar]
- 16.Cheezum MK, Walker WF, Guilford WH. Biophys. J. 2001;81:2378. doi: 10.1016/S0006-3495(01)75884-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thompson RE, Larson DR, Webb WW. Biophys. J. 2002;82:2775. doi: 10.1016/S0006-3495(02)75618-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hell SW. Science. 2007;316:1153. doi: 10.1126/science.1137395. [DOI] [PubMed] [Google Scholar]


