Abstract
The complex molecular communications that occur between neoplastic and stromal cells within the tumor microenvironment play an integral role in breast cancer pathogenesis. Carcinoma-associated fibroblasts (CAFs) produce tumor-enhancing factors and have been strongly implicated in breast cancer development. Similar to the way in which tumors have been compared to “wounds that never heal”, CAFs have been equated to activated fibroblasts, which are present in inflammatory environments, where they aid in wound healing through tissue remodeling and repair. Matrix metalloproteinase-1 (MMP-1) and G-protein coupled receptor, CXCR4, are elevated in these activated fibroblasts, where they facilitate angiogenesis and matrix degradation, processes that are also vital to breast cancer metastasis. In this study, we investigated MMP-1 and CXCR4 expression in normal human mammary fibroblasts (HMFs) exposed to soluble breast cancer factors. Historically, elevated CXCR4 expression is associated with breast cancer cells. However, we show that soluble factors secreted by SUM102 breast cancer cells stimulated the expression of MMP-1 and CXCR4 in HMFs. As a result these stromal cells acquired an invasive and migratory phenotype. To confirm the clinical relevancy of our findings, we analyzed CAFs obtained from primary breast cancers. These cells also displayed elevated MMP-1 and CXCR4 levels compared to counterpart fibroblasts, and were more invasive and migratory. Together, our data suggest that soluble breast cancer factors initiate the transdifferentiation of normal HMFs to tumor-promoting CAFs, and that through the induction of MMP-1 and CXCR4 levels, these cells exhibit an invasive and migratory phenotype.
Keywords: tumor microenvironment, SDF-1, MMPs, invasion, migration
INTRODUCTION
For the past three decades, cancer research has focused predominantly on the characteristics of malignant epithelial cells. However, recently, clinical and experimental evidence have revealed that tumor development is intimately related to the complex interactions that transpire within the tumor microenvironment (1, 2). Neoplastic epithelial cells engage in reciprocal molecular dialogues with surrounding stromal cells, including inflammatory cells, vascular cells and fibroblasts, resulting in the production of stromal-derived tumor-aiding factors, such as growth factors, chemokines, cytokines, proteases, and vascular-stimulating factors (3, 4). These molecular communications at the site of the primary tumor enable its growth and dissemination.
Breast cancer represents one example of a carcinoma in which the collaborative efforts from malignant epithelial cells and stromal cells within the primary tumor coordinate carcinogenesis, and ultimately, metastasis to distant organs. Women have a 12% risk of developing breast cancer during their lifetimes, and a 5% risk of breast carcinoma-related death (5). Mortality results from frequent metastasis, which reduces long-term survival (+5 years) from approximately 90% to 5% (6). A xenograft model of breast cancer has shown that breast carcinoma cells depend on stromal cells to metastasize (7). This may be explained by the finding that the stroma plays a paramount role in supporting breast tumor growth and vascularization (8), both of which are prerequisite to metastasis (9). Because of the importance of the tumor/stromal interactions in promoting breast cancer metastasis, it is imperative to elucidate the molecular events that occur between cancer cells and adjacent stroma at the site of the primary tumor.
Of particular interest are the stromal fibroblasts found within mammary carcinomas, which by volume constitute the major cellular component of the tumor microenvironment (1, 10). Termed carcinoma-associated fibroblasts (CAFs), these cells have been connected to several tumor-promoting events, including: angiogenesis (8), anti-tumor immunosuppression (11), genetic instability (12), epithelial-to-mesenchymal transition (EMT) (13), and metastasis (7). Although the importance of these cells in tumor progression has been acknowledged, CAFs have not been clearly and universally defined, and are only roughly characterized by their enhanced production of tumor-promoting factors.
Similar to the way in which a tumor is equated to a “wound that never heals” (14), CAFs have been compared to “activated fibroblasts” or myofibroblasts, which are present in inflammatory environments, where they play a critical role in angiogenesis and the production of components of the extracellular matrix (ECM) (3, 4). Several studies suggest that myofibroblasts constitute the majority of CAFs, as determined by alpha smooth muscle actin (αSMA), a marker for differentiated myofibroblasts (15, 16). In contrast, other data suggest that not all CAFs within the tumor microenvironment are myofibroblasts. For example, Holliday and colleagues established that only 35% of CAFs isolated from primary breast carcinomas were αSMA-positive, and that this mirrored the expression of αSMA in normal breast fibroblasts (17). Yet other studies characterize CAFs by elevated stromal derived factor-1 (SDF-1) expression (8, 18). Sugimoto and colleagues demonstrate that the expression of commonly used “CAF-markers”, including αSMA, vimentin, and fibroblast activation protein (FAP) do not overlap with each other. Thus, they conclude that CAFs are a heterogeneous population and that the use of these markers alone will not suffice to identify all CAFs (19). We speculate, therefore, that multiple CAF subpopulations exist and suggest that they evolve from a variety of origins.
Indeed, Orimo and Weinberg have proposed three models by which CAFs are derived in human carcinomas: (1) clonal selection of a group of fibroblasts or progenitors that have acquired genetic alterations; (2) transdifferentiation of normal stromal fibroblasts, without acquiring genetic alterations; and (3) differentiation of stromal fibroblasts recruited from specialized progenitor cells in circulation (20). Studies support the notion that CAFs are either derived from bone marrow progenitor cells (7, 21, 22) and/or that they evolve from the conversion of normal mammary fibroblasts (8) or other resident cells, such as endothelial cells or epithelial cancer cells (23).
We are interested in the mechanism(s) by which breast cancer cells solicit normal, human resident mammary fibroblasts (HMFs) to produce inflammatory and tumor-promoting factors, and thus become activated and CAF-like, and/or mature CAFs. Little is known about the molecular determinants of the transformation of an HMF to a CAF (8, 24). In the current study, we examined the transdifferentiation of primary normal resident HMFs to activated fibroblasts, or to acquire CAF-like phenotype, in response to breast cancer secreted factors. We measured factors that have previously been affiliated with CAFs and/or the activated fibroblasts found in sites of inflammation (8, 25, 26). Our data demonstrate that SUM102 breast cancer cells, which were derived from an intraductal breast carcinoma, secrete factors that stimulate normal HMFs to express tumor-enhancing, inflammatory-related factors, including matrix metalloproteinase-1 (MMP-1) and the G-protein coupled receptor, CXCR4.
The neutral protease, MMP-1, is recognized as a critical factor in breast cancer metastasis (27–29), and its roles as a mediator of matrix degradation and as a modulator of cell behavior have been established (30–33). However, no study has examined MMP-1 expression and function during the transition from an HMF to a CAF, nor has the function of this protease been identified in CAF isolates from breast cancer patients, although it has been associated with matrix remodeling by activated fibroblasts in inflammation (34).
The overexpression of chemokines and their receptors, such as CXCR4, is also well documented as a key stimulus in promoting breast cancer tumorigenesis (35). Classically, CXCR4 is markedly upregulated on breast cancer cells, allowing them to migrate to targeted distant organs containing cells that secrete its ligand, stromal derived factor-1 (SDF-1) (36). However, it has also been documented that CXCR4 expression is elevated in fibroblasts in inflammatory environments other than cancer. For example, synovial fibroblasts from patients with rheumatoid arthritis display constitutively expressed CXCR4 mRNA and cell surface protein (37). Similarly, studies show that fibroblasts from mice with pulmonary fibrosis express CXCR4 and respond chemotactically to SDF-1 (38, 39). Because the cancer microenvironment has been equated to a chronic site of inflammation (14), we propose that like activated fibroblasts in inflammatory conditions, CAFs also increase CXCR4 levels and that this contributes to breast cancer progression.
Our study illustrates that MMP-1 and CXCR4 mRNA and protein levels are increased in primary HMFs treated with factors secreted by breast cancer cells, and that MMP-1 and CXCR4 are functionally important for downstream signaling, cell migration, and matrix degradation. Notably, we use normal, primary HMFs in these studies to avoid potential artifacts, such as a spontaneous increase in CXCR4 following immortalization with telomerase (40). Importantly, we also demonstrate increased MMP-1 and CXCR4 expression and function in CAF isolates from primary breast tumors, compared to counterpart, non-tumor-associated fibroblasts from the same patient, signifying that our findings are clinically relevant. Together, our data suggest that breast cancer cells secrete growth factors and chemokines that stimulate normal resident HMFs to become CAF-like.
RESULTS
Human mammary fibroblasts promote tumor engraftment and growth of 102 breast cancer cells
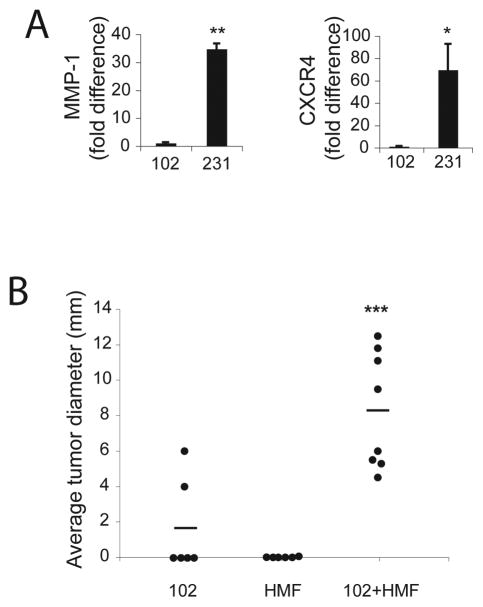
SUM102 (102) breast cancer cells were derived from an intraductal breast carcinoma and are ER-/PR-, suggesting that they are phenotypically aggressive (41, 42). Since MMP-1 and CXCR4 mediate breast cancer invasion and metastasis by MDA-MB 231 (231) breast cancer cells in vivo (43), we compared MMP-1 and CXCR4 gene expression in 102 breast cancer cells to that of the well-established, aggressive 231 breast cancer cells. Compared to 102 cells, MMP-1 and CXCR4 mRNA was greater in 231 cells by approximately 35-fold and 70-fold, respectively (Figure 1A), indicating that the 102 cells do not express the same parameters as the 231 cells. Unlike 231 cells, which readily form tumors in vivo, when 102 cells were injected orthotopically into the 4th mammary fat pad of nude mice, they were weakly tumorigenic (Figure 1B), suggesting that tumorigenesis in these cells may depend on stromal/tumor cell interactions.
Figure 1.
Human mammary fibroblasts (HMFs) enhance tumor engraftment and growth of SUM102 breast cancer cells in vivo. A. SUM102 and MDA-231 cell lines were serum-starved for 24h, and total RNA was collected for real-time RT-PCR quantification of MMP-1 and CXCR4 mRNA. MDA-231 cells display relatively higher MMP-1 (**, P<0.01) and CXCR4 (*, P<0.05) mRNA levels than SUM102 breast cancer cells. Values were normalized to HPRT and calculated using the 2−(ΔΔC(t)) method (see Materials and Methods). Columns, mean fold difference; bars, SD. B. Nude mice were injected orthotopically with 2×106 SUM102 breast cancer cells, 2×106 HMFs, or 1×106 of each cell line (102+HMF). Data represent average tumor diameter (mm) six months following injection. Bars, mean. ***, P=0.0006 compared to tumors formed by injection of SUM102 cells alone.
Reports have documented that either non-cancer associated, normal human mammary fibroblasts (HMFs) or carcinoma associated fibroblasts (CAFs) enhance breast tumor growth when injected simultaneously with mammary carcinoma cells into nude mice using an orthotopic model (8, 44). We used this model to examine the functional ramifications of the molecular interactions between weakly tumorigenic 102 breast cancer cells and HMFs isolated from cancer free mammary tissues: 102 breast cancer cells were injected orthotopically into nude mice in the presence or absence of HMFs (Figure 1B). After 6 months, 100% of mice with concomitant injections of 102 breast cancer cells and HMFs developed tumors, while only ~33% of mice injected with 102 cells alone displayed tumors of modest size (~5mm) (P=0.0027) (Table 1). Moreover, when 102 breast cancer cells were co-injected with HMFs, larger tumors (~10mm) formed than when 102 cells were injected alone (***P=0.0006) (Figure 1B and Table 1). Cell number could not explain this finding, since twice as many 102 cells were injected into mice receiving only the tumor cells. Further, mice injected with HMFs alone yielded no tumors (Figure 1B and Table 1). Thus, we conclude that tumor formation and growth are augmented when mice were co-injected with 102 cells and HMFs, suggesting that 102 breast cancer progression is enhanced by the presence of human mammary fibroblasts. Soluble factors secreted by breast cancer cells up-regulate specific tumor-enhancing genes in nearby fibroblasts (3). We hypothesized therefore, that the weakly tumorigenic 102 breast cancer cells secrete soluble factors that aid in the transdifferentiation of resident mammary fibroblasts to tumor-promoting, activated fibroblasts, that are CAFs or CAF-like, and that subsequently, these activated stromal cells up-regulate inflammatory mediators that facilitate tumor progression.
Table 1.
Tumor formation and diameter in mice injected with HMFs, 102 cells, or both simltaneously.
| % tumor formation | > 10mm diameter | average diameter (mm) | |
|---|---|---|---|
| 102 | 33.3% | 0% | 1.67 |
| HMF | 0% | 0% | 0 |
| 102+HMF | 100% | 50% | 8.28 |
102 breast cancer cells stimulate the expression of tumor-promoting genes that are associated with cancer and inflammation in normal mammary fibroblasts
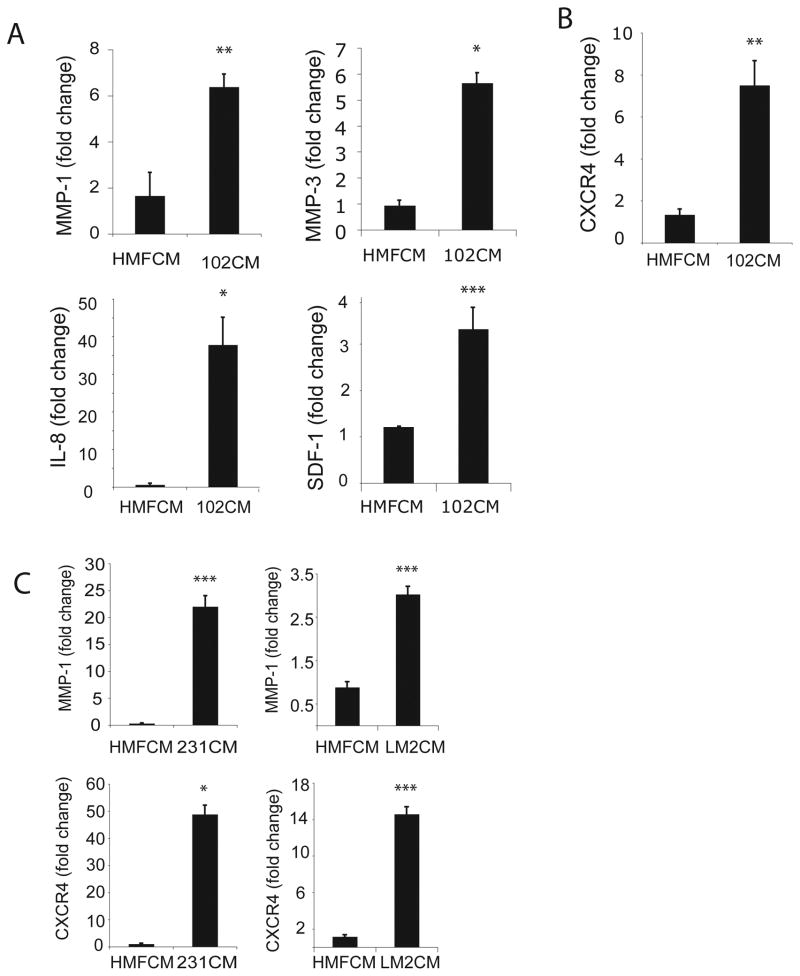
To determine whether factors produced by 102 carcinoma cells affect HMF gene expression, causing these stromal cells to gain an activated, CAF-like phenotype, we used real-time RT-PCR to measure the expression of a panel of genes known to be associated with CAFs and/or activated fibroblasts. Compared to control HMFs, treated with HMF cell conditioned media (HMFCM), HMFs treated with 102 cell conditioned media (102CM) exhibited significantly increased levels of several genes that have been previously associated with activated fibroblasts present in inflammatory situations and CAFs: MMP-1, MMP-3, IL-8, and SDF-1 (3, 8, 18, 45, 46) (Figure 2A). Notably, these genes also promote breast cancer invasion and metastasis (13, 36, 43, 47). As expected when studying primary cells from an out-bred population, we observed inherent differences in the responsiveness of HMFs treated with 102CM. Nonetheless, of the 12 primary HMF samples tested, ~70% were responsive.
Figure 2.
Soluble factors from SUM102 breast cancer cells stimulate the expression of inflammatory and tumor-promoting genes in normal human mammary fibroblasts (HMFs). HMF cultures were treated for 24h with HMF conditioned media (CM) (control) or 102CM. A. Real-time RT-PCR quantification of HMF mRNA for genes associated with inflammation and CAFs, including MMP-1, MMP-3, IL-8, SDF-1. B. Real-time RT-PCR quantification of HMF mRNA for CXCR4, which is commonly linked to inflammatory, activated fibroblasts, but has not previously associated with CAFs. C. Real-time RT-PCR quantification of MMP-1 and CXCR4 mRNA in HMFs exposed to factors from MDA-MB 231 breast cancer cells (231CM) or LM2 cells (LM2CM). Values were normalized to HPRT and calculated using the 2−(ΔΔC(t)) method, and are presented as fold change from control conditions; bars, SD. *, P<0.05; **, P<0.01; ***P<0.005. Results are representative of five independent experiments.
These data suggest that HMFs become activated or CAF-like in response to 102CM, and thus acquire an enhanced tumor-promoting phenotype following exposure to soluble breast cancer factors. However, 102CM did not induce other genes, including several classic CAF-related genes: FAP, αSMA, vimentin, and VEGF within 24h (data not shown) (19, 48). This finding suggests that resident HMFs, which become activated and CAF-like, may represent a distinct sub-population of CAFs in the tumor microenvironment. It is also feasible that HMFs may require additional environmental cues, which are absent in vitro, in order to transform into mature CAFs. Alternatively, other studies have shown that fibroblast precursor cells only express CAF-related genes, such SDF-1, αSMA and FAP, after 30 days of exposure to breast cancer CM (18).
Perhaps most interestingly, we found that HMFs exhibited elevated CXCR4 mRNA levels in response to 102CM (Figure 2B), since overexpression of this receptor is usually associated with invasive breast cancer cells (36) (see Figure 1A), while CAFs are recognized for secreting increased amounts of its ligand, SDF-1 (8). As noted earlier, we also detected SDF-1 gene induction in HMFs exposed to 102CM (see Figure 2A). Elevated CXCR4 expression on breast cancer cells has been correlated with augmented angiogenesis and directional migration (8, 36). Thus, CXCR4 induction in resident fibroblasts implies that these cells may also contribute to tumorigenesis, invasion and migration. We conclude that 102CM induces the expression of genes associated with activated fibroblasts and/or CAFs, and we will focus on MMP-1 and CXCR4 because of their documented importance in breast cancer progression (8, 36, 49, 50). To emphasize the broad significance of this finding, media conditioned by 231 breast cancer cells, or their lung metastatic derivative, LM2 cells, also induced MMP-1 (top panels) and CXCR4 (bottom panels) mRNA in HMFs within 24h (Figure 2C). Due to the low levels of MMP-1 and CXCR4 in the 102 cells, compared to those in the aggressive 231 cells (see Figure 1A) and LM2 cells (data not shown), the 102 cells represent a stronger model for examining the functional contribution of stromal-derived MMP-1 and CXCR4 in breast cancer progression. Thus, we focus on the effects made by 102 breast cancer cells on surrounding fibroblasts.
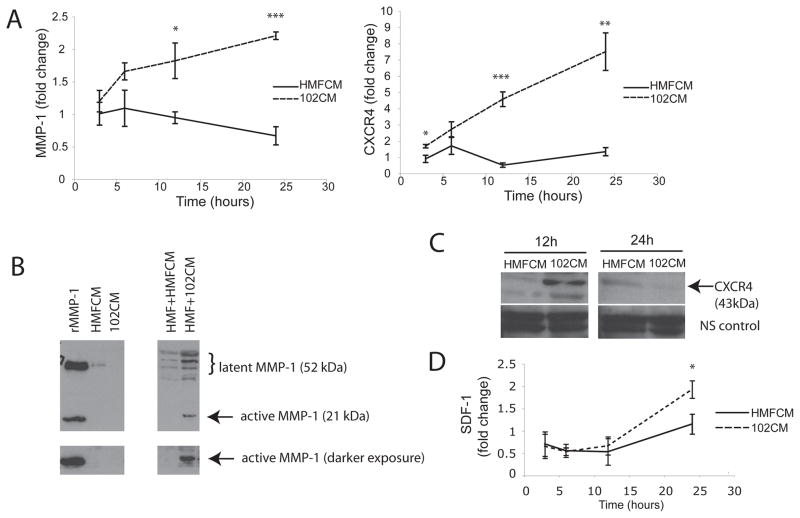
Time course of MMP-1 and CXCR4 mRNA and protein levels in HMFs exposed to soluble 102 breast cancer factors
To define the time course of MMP-1 and CXCR4 induction in HMFs, mRNA was measured following treatment of these cells with 102CM or control HMFCM. Both MMP-1 and CXCR4 mRNA in HMFs increased steadily from 3h to 24h of exposure to 102CM (Figure 3A), while levels of these mRNAs were unchanged in HMFs treated with control media, HMFCM. Both MMP-1 and CXCR4 gene expression returned to original levels within 3h upon removal of the breast cancer cell CM (data not shown). Depending on the HMF sample, the relative fold change in CXCR4 between 102CM-treated HMFs and control HMFs ranged from 10-fold to over 50-fold, while fold change in MMP-1 induction ranged from 2-fold to 10-fold (data not shown).
Figure 3.
Time course of MMP-1 and CXCR4 mRNA and protein levels in HMFs exposed to factors from SUM102 breast cancer cells. A. HMFs were treated with control HMFCM or 102CM for the indicated times, and then MMP-1 and CXCR4 mRNA was measured using real-time RT-PCR. Values were normalized to HPRT and calculated using the 2−(ΔΔC(t)) method. *, P=0.05; **, P<0.01; ***, P<0.005. B. HMFs were treated with HMFCM or 102CM for 24h, then proteins in media were precipitated and immunoblotted with antibodies against MMP-1. Recombinant MMP-1 (rMMP-1) and each CM alone serve as controls (left). Multiple banding patterns around latent MMP-1 represent glycosylated forms of MMP-1. C. Lysates were harvested from HMFs treated with HMFCM or 102CM for 12h or 24h and immunoblotted with anti-CXCR4 antibodies. Multiple banding patterns represent glycosylated forms of CXCR4. Nonspecific (NS) bands serve as a loading control. D. HMFs were treated with control HMFCM or 102CM for the indicated times and then SDF-1 mRNA was measured using real-time RT-PCR. Values were normalized to HPRT and calculated using the 2−(ΔΔC(t)) method. Columns, mean; bars, SD. *, P<0.05. Experiments were conducted three times.
Western blot analysis confirmed that protein levels correlated with gene induction. Neither HMFCM, nor 102CM alone generated measurable levels of latent or active MMP-1 (Figure 3B, left panel), confirming that basal MMP-1 levels in HMFs and 102 cells are low. However, 102CM triggered the secretion of both latent and active MMP-1 in HMFs, compared to HMFs treated with control media, HMFCM (Figure 3B, right panel). Bands larger than 52kDa in the CM represent glycosylated MMP-1 (51). Similarly, high levels of CXCR4 protein were detected in HMFs treated with 102CM after 12h, while little CXCR4 was detected in HMFs treated with HMFCM (Figure 3C). HMF-derived CXCR4 protein diminished by 24h of treatment with 102CM (Figure 3C), possibly due to the increase in SDF-1 mRNA (Figure 3D), which typically ranged from 2- to 10-fold (data not shown). Indeed, the kinetic profile of SDF-1 is such that if SDF-1 levels begin to increase at 16h, it is possible that this chemokine is generated and binds CXCR4, causing internalization and degradation of this receptor by 24h ((52) and see Discussion). Media conditioned by 231 and LM2 breast cancer cells also induced MMP-1 and CXCR4 protein levels in HMFs within 24h (data not shown).
Functional significance of increased stromal MMP-1 in the absence of breast cancer cell-derived MMP-1
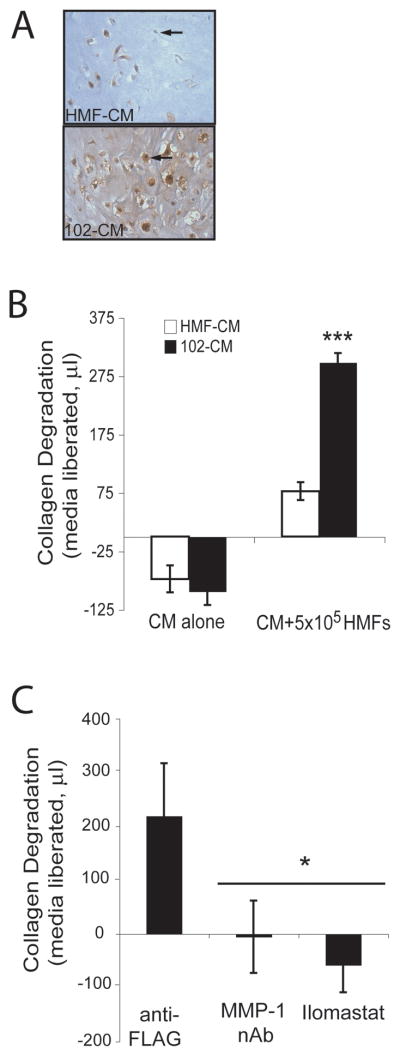
To demonstrate the functional importance of MMP-1 in HMFs, we used an in vitro collagen degradation assay (32, 50), in which HMFs were embedded in a collagen gel and incubated with control HMFCM or 102CM. Immunohistochemistry detected MMP-1 protein produced by HMFs embedded in a collagen matrix when subjected to either HMFCM or 102CM for 6h (Figure 4A). While HMFs treated with 102CM stained positively for MMP-1, only a few cells treated with HMFCM were partially stained (see arrows), despite the equal number of cells per gel. Furthermore, collagen surrounding HMFs treated with 102CM displayed punctate MMP-1 staining throughout, indicating that these cells actively secreted MMP-1. In contrast, MMP-1 was not present in HMF-embedded gels treated with HMFCM.
Figure 4.
HMF-derived MMP-1 degrades type I collagen. A. HMFs were embedded into a type I collagen matrix, treated with HMFCM or 102CM for 6h and then were stained using immunohistochemical and anti-MMP-1 antibodies. B. Collagen-embedded HMFs were treated with HMFCM or 102CM for 48h. Collagen degradation was calculated by weighing the media released from the collagen gels (see Materials and Methods). C. Collagen-embedded HMFs were treated with 102CM in the presence of control anti-FLAG antibody (1μg/mL), an MMP-1 neutralizing antibody (1μg/mL), or illomastat (25μM) for 48h. Collagen degradation was calculated by weighing the media released from the collagen gels (see Materials and Methods). The negative values on the y-axis result from evaporation of media. Columns, mean; bars, SD. A representative experiment out of three is presented. ***, P<0.0001 compared to controls (B) and *, P<0.05 versus anti-FLAG control condition (C).
While very little degradation resulted when collagen-embedded HMFs were treated with HMFCM for 48h, collagen degradation by HMFs treated with 102CM was extensive, as measured by the amount of media liberated from the collagen as it was degraded (***P<0.0001) (Figure 4B). The negative values on the y-axis result from evaporation of media. To confirm MMP-1 as the agent responsible for the collagen degradation, an MMP-1 neutralizing antibody was added to the collagen, and HMF-driven degradation of collagen was prevented (Figure 4C). Similarly, addition of a general MMP inhibitor, Ilomastat, abrogated collagen degradation (*P<0.05) (Figure 4C). Again, the negative values on the y-axis result from evaporation of media. These data imply that weakly invasive 102 breast cancer cells, which do not produce MMP-1 (Figure 3B), can exploit nearby fibroblasts to produce MMP-1, resulting in the degradation of surrounding ECM.
CXCR4 on HMFs stimulates downstream signaling and allows cells to migrate in response to exogenous SDF-1
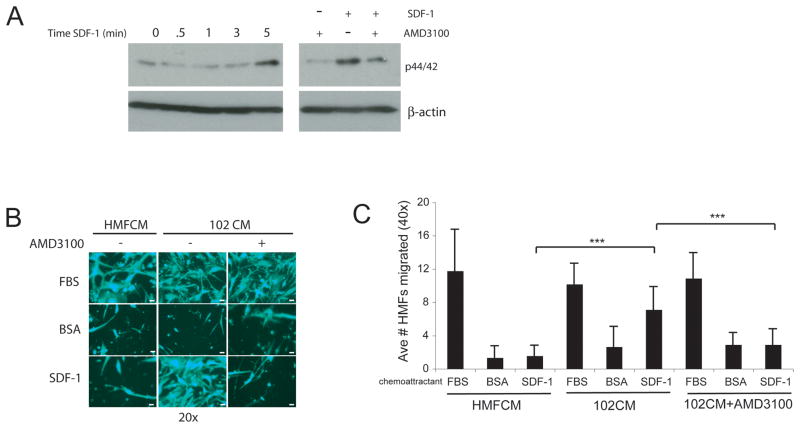
It has been demonstrated that cell-surface CXCR4 may be present on cells, but unresponsive to SDF-1 stimulation, as measured by downstream signaling (53). Since SDF-1 is the only known ligand for CXCR4 (54, 55), we determined whether SDF-1 successfully signaled through HMF CXCR4 by measuring the activation of p44/42 (Erk1/2) (56), a component of the mitogen activated protein kinase (MAPK) pathway. Addition of SDF-1 to HMFs for 5 minutes stimulated the phosphorylation of p44/42, indicating the activation of this pathway (Figure 5A, left panel). To confirm SDF-1 signaling through activation of CXCR4, cells were pretreated with a CXCR4 inhibitor, AMD3100, and subsequently treated with SDF-1 for 0 or 5 minutes. Inhibition of CXCR4 blunted p44/42 phosphorylation by SDF-1 (Figure 5A, right panel), allowing us to conclude that upon binding to CXCR4, SDF-1 stimulates downstream signal transduction in HMFs. The partial inhibition observed with AMD3100 most likely reflects the fact that this MAPK pathway is activated by other mechanisms in these cells ((57) and data not shown).
Figure 5.
HMF CXCR4 stimulates downstream signaling in response to SDF-1 and allows cells to migrate toward ligand, SDF-1. A. HMFs were exposed to 102CM for 24h and then treated with 100ng/mL SDF-1 for the indicated amount of time, or pretreated with AMD3100 (5ug/mL) for 30 minutes and then treated with SDF-1 for 0 or 5 minutes. Whole cellular lysates were analyzed by immunoblotting for phosphorylated p44/42. B. HMFs were plated in FluoroBlok transwells and subjected to 102CM for 12h, after which BSA (negative vehicle control), FBS (positive control), or SDF-1 was added to the bottom chamber. 16h later, cells were stained with Calcein AM and migrated cells were visualized using a mercury bulb. Scale bars, 25μm. C. HMFs were plated in FluoroBlok transwells and subjected to 102CM for 12h, after which time, cells were pretreated with or without 5μg/mL AMD3100 for 30 minutes and then 0.1% BSA (negative control for SDF-1), FBS (positive control), or SDF-1 (see Materials and Methods) was added to the bottom chamber. 18h later, cells were stained with Calcein AM and and cells were enumerated in 6 fields of view (40x) per condition to quantify migration. Columns, mean; bars, SD. ***, P<0.005. Data are representative of three individual experiments.
Standard chemotaxis assays were conducted to investigate the downstream function of the SDF-1/CXCR4 signaling axis in HMFs stimulated with HMFCM or 102CM. These functional experiments also confirm that the increased total CXCR4 protein observed by western blot analysis results in increased surface CXCR4. Figure 5B represents a qualitative analysis of these results, which are quantified in Figure 5C. As expected, HMFs migrated toward FBS (positive control), but not toward BSA (negative vehicle control). Although HMFs treated with control HMFCM failed to migrate toward SDF-1, HMFs exposed to 102CM, and which therefore have increased CXCR4 levels (see Figure 3), migrated readily toward SDF-1 (***P=0.0001) (Figure 5B and C). To corroborate CXCR4 as the key mediator in HMF migration toward SDF-1, the CXCR4 antagonist, AMD3100, was used to block CXCR4 signaling. Blocking CXCR4 reduced HMF migration toward SDF-1 by ~2–3 fold, as is evident in Figure 5B and C (***P=0.0016). Taken together, these data confirm that breast cancer cells secrete factors that not only induce CXCR4 protein levels in HMFs, but also that this receptor responds to its ligand by transmitting downstream signals and enhancing migration.
MMP-1 and CXCR4 mRNA and protein are increased in primary CAFs compared to their counterpart fibroblasts and HMFs
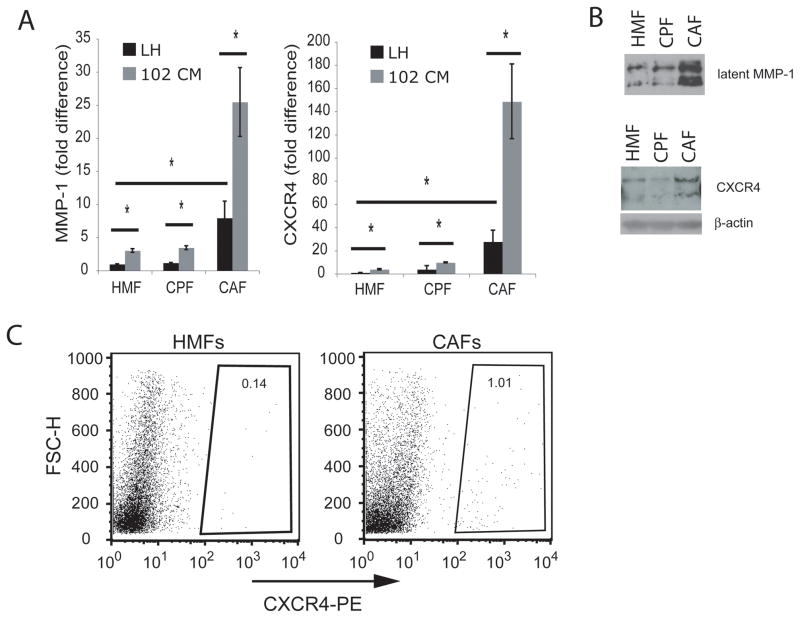
To ascertain whether our observations were clinically relevant, we investigated MMP-1 and CXCR4 expression and function in CAFs isolated from primary human breast tumors, their respective (non-cancer-associated) counterpart fibroblasts (CPFs), and age-matched, HMFs from patients without cancer (Figure 6A). Basal MMP-1 and CXCR4 mRNA levels in CPFs were similar to those in normal, unstimulated HMFs, suggesting that non-tumor associated fibroblasts collected from patients with breast cancer have comparable MMP-1 and CXCR4 gene expression to those observed in HMFs taken from patients not afflicted with cancer. In contrast, basal expression of these genes was significantly higher in CAFs, demonstrating that clinically derived CAFs have constitutively increased levels of MMP-1 and CXCR4 mRNA (*P<0.05) (Figure 6A). 102CM further stimulated MMP-1 and CXCR4 mRNA levels in HMFs, CPFs, and CAFs, suggesting that regardless of cell type, soluble factors from breast cancer cells stimulate the expression of these tumor-promoting genes (Figure 6A). To confirm their status as CAFs, we examined these cells for increased expression of a CAF-recognized gene, FAP, and found it to be elevated compared to FAP mRNA in CPFs (data not shown).
Figure 6.
CAF isolates from clinical specimens have increased MMP-1 and CXCR4 levels compared to their counterpart fibroblasts (CPFs) and HMFs. A. Primary cell lines (HMFs, CPFs, or CAFs) were treated with serum-free media (LH, see Materials and Methods) or 102CM for 24h, after which time RNA was harvested. MMP-1 and CXCR4 mRNAs were measured using real-time RT-PCR. Values were normalized to HPRT and calculated using the 2−(ΔΔC(t)) method. Columns, fold change; bars, SD. *, P<0.05. B. Following 24h of incubation with serum-free media, protein within the media were precipitated, and primary cells were lysed for immunoblotting analyses of MMP-1 and CXCR4, respectively. C. CXCR4 surface expression on HMFs and CAFs were analyzed using a PE-conjugated anti-CXCR4 antibody and flow cytometry.
Constitutive MMP-1 (top panel) and CXCR4 (bottom panel) protein levels were also increased in CAFs compared to HMFs and CPFs (Figure 6B). The multiple banding pattern for both MMP-1 and CXCR4 protein results from glycosylation (51, 58). Flow cytometry detected increased CXCR4 surface expression on ~1.0% of CAFs, compared to ~0.1% of HMFs (Figure 6C). The percentage of cells expressing CXCR4 over time is likely higher than the values obtained, because CXCR4 is rapidly internalized (52), making it difficult to detect. Importantly, flow cytometry revealed that 10-fold more CAFs express CXCR4 than HMFs, and of these cells, the mean fluorescence intensity (MFI) of CAFs was consistently greater than the MFI of HMFs (data not shown). Because CXCR4 is rapidly internalized, this method of determining CXCR4 surface expression may be less definitive than a functional assay (see below).
Migratory and invasive capabilities are increased in primary CAFs compared to their counterpart fibroblasts and HMFs
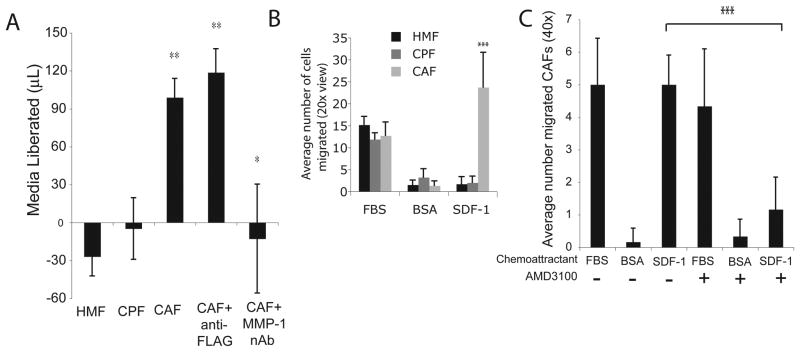
To measure the functionality of CAF-derived MMP-1, we assessed the ability of these cells to degrade collagen in the collagen degradation assay. HMFs and CPFs treated with naive, serum-free medium did not degrade the type I collagen. In contrast, CAFs readily degraded the collagen matrix, except when MMP-1 was neutralized with an antibody (Figure 7A). The negative values on the y-axis result from evaporation of media. These findings confirm and expand on previous studies, demonstrating that CAFs derived from clinical specimens not only maintain increased MMP-1 levels, but also that this fibroblast-derived enzyme degrades surrounding matrix, suggesting a role for these cells in vivo.
Figure 7.
CAF isolates from clinical specimens have increased MMP-1 and CXCR4 and as a result are more invasive and migratory than counterpart fibroblasts (CPFs) and HMFs. A. Primary cells were embedded in a collagen matrix for 48h in the presence or absence of anti-FLAG antibody control (1μg/mL) or an MMP-1 neutralizing antibody (1 μg/mL). Media were then weighed to quantify matrix degradation (see Materials and Methods). The negative values on the y-axis result from evaporation of media. Columns, mean; bars, SD. *, P<0.05; **, P<0.01. B. Primary cells were serum-starved 24h. FBS, 0.1% BSA (negative control for SDF-1), or SDF-1 (100ng/mL) was then added to bottom chamber and cells were allowed to migrate for 18h. Migrated cells were stained with Calcein AM and counted in 6 fields of view (20x) per condition to quantify migration. Columns, mean; bars, SD. ***, P<0.001. C. CAFs were plated in transwells and serum-starved 24h. Cells that received AMD3100 (5μg/mL) were treated 30 minutes prior to the addition of chemoattractants FBS (positive control), BSA (negative vehicle control), or SDF-1 to bottom chambers. Migrated cells were stained with Calcein AM and cells were counted in 6 fields of view (40x) per condition to quantify migration. Columns, mean; bars, SD. ***, P<0.001. Results are representative of individual experiments, using CAFs from different patients.
To investigate whether CAF-expressed CXCR4 promoted chemotaxis toward SDF-1, these cells were examined in a standard migration assay. CAFs migrated readily toward SDF-1, while CPFs and HMFs did not (**P<0.001) (Figure 7B), and that the degree of migration corresponded with the relative levels of CXCR4 in these cells. Each of the primary cell lines migrated toward 10% FBS, the positive control, but were not attracted toward the negative vehicle control, 0.1% BSA (Figure 7B). Importantly, blocking CXCR4 function with AMD3100 abrogated CAF migration (***P=0.001) (Figure 7C), indicating that CAF-derived CXCR4 is necessary for the migration toward SDF-1 and demonstrating that CXCR4 expressed on these cells is functional.
Thus, we conclude that CAF specimens derived from clinical material express increased MMP-1 and CXCR4, which potentiate the invasive and migratory phenotype of these cells. These results validate the clinical relevancy of our findings with HMFs treated with 102CM, since HMFs treated with 102CM share characteristics with CAFs obtained from patient specimens. Further, the data support the concept that inflammatory factors provided by breast cancer cells may initiate the HMF-to-CAF transdifferentiation.
DISCUSSION
Some CAFs have been compared to the activated fibroblasts, or myofibroblasts, present in sites of wound healing and chronic inflammation. Myofibroblasts at sites of inflammation due to rheumatoid arthritis and idiopathic pulmonary fibrosis have increased MMP-1 (59, 60) and CXCR4 expression (37, 61), which facilitate cellular invasion and migration. Taken together, these observations support our hypothesis that MMP-1 and CXCR4 may also be elevated in fibroblasts that are found within the breast tumor microenvironment and suggest that these factors are induced as an HMF becomes more activated and CAF-like.
In this report, we provide evidence to support this hypothesis. First, our data show that SUM102 breast cancer cells, which express low levels of MMP-1 and CXCR4, require mammary fibroblasts to grow in vivo (Figure 1). Second, the SUM102 cells secrete factors that stimulate the expression of several inflammatory factors, including MMP-1 and CXCR4, in adjacent stromal cells (Figures 1 and 2). This suggests that 102 breast cancer cells may solicit normal, resident HMFs to facilitate tumor growth in vivo. Further, increased MMP-1 and CXCR4 enhanced the invasive and migratory behavior of HMFs, verifying functional contributions of these cells to tumor progression (Figures 4 and 5). Finally, we validated these findings by demonstrating that mammary CAF isolates from patients with breast cancer have amplified basal MMP-1 and CXCR4 levels, which augment their invasive and migratory potential, respectively (Figures 6 and 7). In vivo, this phenotype may permit CAFs to degrade matrix as they migrate within the tumor microenvironment. Our findings emphasize the possible clinical relevance of MMP-1 and CXCR4 induction in CAFs, and demonstrate that normal HMFs exposed to soluble breast cancer factors acquire phenotypic characteristics shared with CAF specimens (Figures 6 and 7), indicating that breast cancer cells may initiate the conversion of resident HMFs to CAFs.
The process of matrix invasion by malignant cells requires both degradation of the matrix and cellular migration (62). Studies conducted previously in our laboratory demonstrated that MMP-1 derived from breast cancer cells is necessary for tumor growth and invasion at the primary site (32, 50). Since MMP-1 is secreted, it is reasonable to propose that stromal-derived MMP-1 functions similarly to that produced by the breast cancer cells. Furthermore, not all breast cancer cells produce MMP-1 (63), increasing the importance of CAF-derived MMP-1 in tumor progression.
CXCR4 up-regulation is most commonly linked to breast cancer epithelial cells (36); on the other hand, the ligand for CXCR4, SDF-1, has been classified as CAF-related (8). In this report, we have identified CXCR4 as a novel CAF-associated gene. This finding, coupled with the discovery that CAFs express increased levels of SDF-1 (8), imply that an autocrine feedback loop may exist (64–67). It is possible that SDF-1 produced by CAFs binds to and signals through, CAF CXCR4, potentially causing important downstream effects. Specifically, the SDF-1/CXCR4 signaling axis results in a multitude of cellular functions: migration, homing, adhesion, proliferation, differentiation, and the induction of MMPs (68–71). For example, SDF-1 signaling through CXCR4 on the activated fibroblasts in patients with rheumatoid arthritis results in their activation, migration and proliferation (37, 72). Several breast cancer cell lines also secrete SDF-1 (65, 73); thus, it is feasible that the SDF-1/CXCR4 axis functions not only in an autocrine manner, but also in paracrine fashion within the tumor microenvironment, where SDF-1 produced by tumor cells binds to CXCR4 on surrounding CAFs and vice-versa.
Breast cancer cells over-expressing CXCR4 home to organs constitutively expressing SDF-1, such as the lungs, liver and bone (36), yet the in vivo function of CXCR4 on CAFs is currently unknown. Nonetheless, the in vivo function of CXCR4 on fibroblasts in other inflammatory diseases has been well documented (39, 61) and cancers have been likened to a wound that does not heal (14). For example, in patients with idiopathic pulmonary fibrosis, fibroblast precursor cells expressing CXCR4 have been shown to home to the lungs, which express elevated levels of SDF-1 (39, 61), thereby indicating that activated fibroblasts have the ability to migrate in vivo. Since we show that CXCR4 on CAFs is functional and that these cells are endowed with the ability to migrate towards SDF-1, we speculate that CAFs may migrate throughout the microenvironment of the primary tumor, degrading collagen through the secretion of MMP-1.
A comprehensive list of CAF marker designations has not been definitively established. Instead, these cells have been associated with a variety of markers. Because CAFs may arise from various sources, it is not surprising that these cells could express different markers, and as a result, contribute to carcinogenesis differently. We did not observe increased levels of FAP, vimentin, and αSMA in HMFs exposed to soluble breast cancer factors. However, we document that normal HMFs increase several other CAF-related genes (see Figure 2A), including IL-8, SDF-1, MMP-3 and MMP-1. In addition, we demonstrated increased CXCR4 in CAFs and in HMFs in response to factors secreted by 102 breast cancer cells, a factor which has not previously been associated with CAFs. Since MMP-1 and CXCR4 are elevated in CAF isolates from the clinic, we suggest that the induction of specific genes by 102CM may represent an early transition phase of an HMF into a CAF and that these gene products facilitate breast cancer cell invasion and dissemination. Additionally, perhaps the high constitutive levels of MMP-1 and CXCR4 seen in the CAFs taken from patients have resulted from genetic alterations made within these cells (74).
Despite reports documenting that normal HMFs or CAFs enhance tumor growth when injected simultaneously with mammary carcinoma cells into nude mice (8, 44), it has also been proposed that normal HMFs can suppress breast tumor formation in vivo (8). Indeed, then, it would be reasonable to propose that early in tumor formation normal, resident mammary fibroblasts suppress tumor growth, but that following the breast cancer cell-induced transition of HMFs to CAFs, these cells enhance tumor pathogenesis. Alternatively, it is possible that HMFs require prolonged treatment with breast cancer cell CM. This concept coincides with data from Mishra and colleagues, who showed that fibroblast precursor cells become fully CAF-like (as determined by SDF-1, αSMA and FAP expression) after 30 days of exposure to breast cancer CM (18).
In conclusion, we demonstrate that CAFs display amplified MMP-1 and CXCR4 and that normal HMFs acquire enhanced MMP-1 and CXCR4 levels in response to soluble breast cancer agents. Our data suggest that breast cancer cells may initiate the conversion of normal, resident HMFs to CAFs.
MATERIALS AND METHODS
Cell lines and cell culture
SUM102 breast cancer cells (Asterand, Detroit, MI) were cultured in Ham’s F-12 medium supplemented with 2% FBS, 5μg/mL insulin, 10ng/mL epidermal growth factor, 1ug/mL hydrocortisone, Hepes Buffer, 100U/mL penicillin and 100ug/mL streptomycin. MDA-MB 231 breast cancer cells (American Type Culture Collection, ATCC, Rockville, MD) and their lung metastatic derivative (LM2, a kind gift from Dr. Massague at Memorial Sloan-Kettering Cancer Center) were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM) 50/50 F-12 (Life Technologies, Inc.) supplemented with 10% fetal bovine serum (FBS) (HyClone), penicillin 100U/mL, streptomycin 100ug/mL, and L-glutamine. Primary human mammary fibroblasts (HMFs) were isolated from women who had undergone reduction mammoplasty surgery (Dartmouth Hitchcock Medical Center) and were cultured in DMEM supplemented with 10% FBS, penicillin 100U/mL, streptomycin 100ug/mL, and L-glutamine. All patients were enlisted in an IRB-approved protocol. Primary carcinoma-associated fibroblasts (CAFs) and their respective counterpart fibroblasts (CPFs) (Asterand) were cultured in DMEM supplemented with 5% FBS, penicillin 100U/mL, streptomycin 100ug/mL, and L-glutamine. All cells were cultured at 37°C and in 5% CO2. All primary cells were used between passages 2 and 6.
Breast tumorigenesis in vivo
Cells were suspended in 100μl of Matrigel® (BD Biosciences), and female mice (strain nu/nu, ~49 days old, Charles River, Wilmington, MA) were injected orthotopically (4th mammary fat pad) with 2×106 SUM102 cells, 2×106 HMFs, or 1×106 of each cell line simultaneously. Eight mice were injected concomitantly with both cell lines, while six mice were injected with each cell line alone. Tumors were measured biweekly with calipers, and mice were sacrificed when tumor diameter exceeded 12mm, or at the termination of the study (6 months). The Dartmouth College Institutional Animal Care and Use Committee approved animal studies.
Generation of conditioned media
To condition media, cells were plated at 5×106 cells (102 breast cancer cells) or 1×106 (HMFs) per 150mm plate (~85% confluence) and left to seed for 24h in serum-containing media. The cells were then rinsed three times with Hank’s Buffered Saline Solution (HBSS) and 15mL of serum-free Ham’s F12 media, containing 0.2% lactalbumin hydrolysate (LH), were added per 150mm culture plate and conditioned for 24h at 37°C. After incubation, media were transferred to a conical tube, centrifuged at 1500rpm for 5 minu tes to pellet cellular debris, and then decanted into a new conical tube. Conditioned media (CM) were stored at −80°C until needed for experiments, at which time the media were thawed and added directly to cells.
Quantitative real-time RT-PCR
Total cellular RNA was purified using Qiagen’s RNeasy kit (Qiagen, Valencia, CA) as described by the manufacturer. Total cellular RNA was reverse transcribed using the TaqMan Reverse Transcriptase reagent kit (Applied Biosystems, Foster City, CA), as directed by the manufacturer. Real-time PCR reactions were performed using 250ng of input RNA per reaction, as described (30, 32, 50, 75). Data were averaged and normalized to the housekeeping gene, hypoxanthine-phosphoribosyl transferase (HPRT). mRNA values were calculated using 2−(ΔΔC(t)) (76) and are presented as fold change compared to control conditions. Primer sequences were: CXCR4: 5′-TGGGTGGTTGTGTTCCAGTTT-3′ (forward) and 5′-ATGCAATAGCAGGACAGGATGA-3′ (reverse); HPRT: 5′-AGCTTGCTGGTGAAAAGGAC-3′ (forward) and 5′-CCAGATGTTTCCAAACTCAACTTGA-3′ (reverse); IL-8: 5′-CTCTGGCAGCCTTCCTGATT3′ (forward) and 5′-TATGCACTGACATCTAAGTTCTTTAGC-3′ (reverse); MMP-1: 5′-AGCTAGCTCAGGATGACATTGATG-3′ (forward) and 5′-GCCGATGGGCTGGACAG-3′ (reverse); MMP-3: 5′-TTCCGCCTGTCTCAAGATGATAT-3′ (forward) and 5′-AAAGGACAAAGCAGGATCACAGTT-3′ (reverse); SDF-1: 5′-CACATGTTGAACCTCTTGTTTAAAAGC-3′ (forward) and 5′-AACGCCAAGGTCGTGGTCGTGCTG-3′ (reverse). cDNAs were assayed in triplicate.
In vitro collagen degradation assay
This assay was carried out as previously described (32, 50). Briefly, neutralized collagen was mixed with equivalent volumes of cells to yield 5×105 cells/mL and 500μl of the collagen/cell mixture was added to each well of a 24-well plate. After the collagen solidified at 37°C (~15 minutes), 0.5mL of control HMF cell conditioned media (HMFCM) or 102 cell conditioned media (102CM) containing ~0.00003% trypsin to activate latent MMPs was added carefully to each well in the presence or absence of anti-FLAG antibody control (1μg/mL, Sigma), an MMP-1 neutralizing antibody (1μg/mL, Chemicon) or Illomastat (25μM, Chemicon). After 48h at 37°C, the overlying media were removed and weighed. The media released from a collagen gel were calculated by weighing the total media collected minus the original media added (0.5mL=0.5g). Samples were analyzed in triplicate and experiments were repeated at least three times. Assays stained for MMP-1 using immunohistochemistry (IHC) were terminated at 6h, so that gels embedded with HMFs and treated with 102CM were not entirely degraded. Collagen gels were removed from wells with a rubber spatula, fixed in 1090 neutral buffered formaldehyde and processed for histological staining in the Pathology Department at Dartmouth Hitchcock Medical Center (DHMC). Goat MMP-1 antibody (R&D Systems, Minneapolis, MN) was diluted to 1:100.
Immunoblotting and antibodies
Proteins from HMFs and 102 breast cancer cells were analyzed by SDS-PAGE (32, 50, 75). MMP-1 protein was precipitated with trichloroacetic acid (TCA) from 1mL media, while CXCR4 was analyzed in whole cell lysates, which were suspended in 2x Laemmli lysis buffer (Sigma). Blots were probed with an MMP-1 antibody (Chemicon, Billerica, MA) diluted 1:2000, a CXCR4 anitbody (Chemicon) diluted 1:2000, phospho-specific antibodies (Cell Signaling Technology, Inc., Danvers, MA) diluted 1:1000, or actin antibodies (Calbiochem, San Diego, CA) diluted 1:10,000. Secondary antibodies were diluted 1:2000 in the appropriate blocking buffer.
SDF-1/CXCR4 signaling
HMFs were plated at 2.5×105 per 60 mm culture plate and allowed to seed overnight. After 24h, the cells were rinsed with HBSS and 102CM was added to all plates for 8h, after which time SDF-1α (100ng/ml, R&D Systems) was added for 0, 0.5, 1, 3, or 5 minutes and then cells were lysed in 150μl Laemmli buffer and prepared for immunoblotting for p44/42. Samples treated with AMD3100 (5μg/ml, Sigma Aldrich, St. Louis, MO), a small molecule inhibitor of CXCR4, were pretreated for 30 minutes, and then either vehicle control (0.1% BSA in 1X PBS) or SDF-1α (100ng/ml) was added for 5min.
Migration assays
HMFs (5×104) were plated with media containing serum FluoroBlok™ inserts (BD Falcon, 6.5mm, 8.0μm pore) as described previously (77), and allowed to seed overnight. The following morning, cells were rinsed with HBSS and treated with either HMFCM or 102CM. After 8h, DMEM containing 10% FBS (positive control), 0.1% BSA in PBS (negative vehicle control), or SDF-1α (100ng/mL) was added to the bottom chambers. Samples treated with AMD3100 (5μg/mL) were pretreated 30 minutes prior to adding attractants. Cells were permitted to migrate for approximately 18h, after which time the transwell inserts were exposed to the fluoroscent dye, Calcein AM (4μg/mL in HBSS, BD Bioscience, Franklin Lakes, NJ), to visualize and count only cells that had migrated through the membrane. Migration assays with CAFs were conducted similarly, but rather than using FluoroBlok™ inserts, standard transwell inserts (Corning, 6.5mm, 8.0μm pore) were used. To analyze migration with this method, transwell inserts were exposed to Calcein AM for 30 minutes, and then cells on the bottom were detached by incubating inserts in pre-warmed trypsin with agitation for 15 minutes at 37° C. Medium containing 10% FBS was added to neutralize trypsin and pipetted to disperse cells evenly off the membrane. Cells were incubated for 3h and then were visualized by GFP fluorescence using excitation from a 100W Mercury lamp on an Olympus 1×50 inverted phase contrast microscope and counted to quantify cell migration.
Flow cytometry
HMFs or CAFs were grown to confluence in 150mm plates and serum-starved overnight prior to being harvested for analysis of surface CXCR4 by flow cytometry. On the day of harvest, cells were rinsed with 1XPBS and incubated in acidic glycine buffer (0.05M glycine HCl buffer, pH 3, w/0.1M NaCl) for 1minute to ensure that stable receptors were stabilized, and then rinsed twice in 1X PBS. Pre-warmed Cell Stripper (a non-enzymatic cell dissociation solution from Sigma) together with gentle scraping removed cells from the plates, and cells were immediately added to cold PBS with 10% FBS and 0.1% sodium azide and centrifuged. The supernatant was decanted and cells were either left unstained or stained with an anti-human CXCR4-PE-conjugated antibody (R&D Systems, Cat. #FAB170P) (diluted 1:50) for 30 minutes at 4°C and in the dark. After staining cells, they were fixed in 4% paraformaldahyde for 20 minutes and subsequently washed 3 times in cold PBS with 10% FBS. Cells were then resuspended in PBS supplemented with 10% FBS and 0.1% sodium azide, and were analyzed using FacScan.
Statistical analysis
Student’s t test determined statistical significance. All samples were prepared in triplicate and experiments were conducted at least three times. Statistical power at the P<0.05 was considered significant.
Acknowledgments
Grant support: NIH grants AR26599 and CA77267 (C.E. Brinckerhoff) and NIH grant T32-CA009658 (S.M. Eck)
Footnotes
There are no conflicts of interest associated with this submission.
References
- 1.Bhowmick NA, Neilson EG, Moses HL. Stromal fibroblasts in cancer initiation and progression. Nature. 2004;432:332–337. doi: 10.1038/nature03096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coussens LM, Fingleton B, Matrisian LM. Matrix metalloproteinase inhibitors and cancer: trials and tribulations. Science. 2002;295:2387–2392. doi: 10.1126/science.1067100. [DOI] [PubMed] [Google Scholar]
- 3.Mueller MM, Fusenig NE. Friends or foes - bipolar effects of the tumour stroma in cancer. Nat Rev Cancer. 2004;4:839–849. doi: 10.1038/nrc1477. [DOI] [PubMed] [Google Scholar]
- 4.Serini G, Gabbiani G. Mechanisms of myofibroblast activity and phenotypic modulation. Exp Cell Res. 1999;250:273–283. doi: 10.1006/excr.1999.4543. [DOI] [PubMed] [Google Scholar]
- 5.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 6.Greenberg PA, Hortobagyi GN, Smith TL, Ziegler LD, Frye DK, Buzdar AU. Long-term follow-up of patients with complete remission following combination chemotherapy for metastatic breast cancer. J Clin Oncol. 1996;14:2197–2205. doi: 10.1200/JCO.1996.14.8.2197. [DOI] [PubMed] [Google Scholar]
- 7.Karnoub AE, Dash AB, Vo AP, et al. Mesenchymal stem cells within tumour stroma promote breast cancer metastasis. Nature. 2007;449:557–563. doi: 10.1038/nature06188. [DOI] [PubMed] [Google Scholar]
- 8.Orimo A, Gupta PB, Sgroi DC, et al. Stromal fibroblasts present in invasive human breast carcinomas promote tumor growth and angiogenesis through elevated SDF-1/CXCL12 secretion. Cell. 2005;121:335–348. doi: 10.1016/j.cell.2005.02.034. [DOI] [PubMed] [Google Scholar]
- 9.Folkman J. The role of angiogenesis in tumor growth. Semin Cancer Biol. 1992;3:65–71. [PubMed] [Google Scholar]
- 10.Desmouliere A, Guyot C, Gabbiani G. The stroma reaction myofibroblast: a key player in the control of tumor cell behavior. Int J Dev Biol. 2004;48:509–517. doi: 10.1387/ijdb.041802ad. [DOI] [PubMed] [Google Scholar]
- 11.Djouad F, Plence P, Bony C, et al. Immunosuppressive effect of mesenchymal stem cells favors tumor growth in allogeneic animals. Blood. 2003;102:3837–3844. doi: 10.1182/blood-2003-04-1193. [DOI] [PubMed] [Google Scholar]
- 12.Kurose K, Hoshaw-Woodard S, Adeyinka A, Lemeshow S, Watson PH, Eng C. Genetic model of multi-step breast carcinogenesis involving the epithelium and stroma: clues to tumour-microenvironment interactions. Hum Mol Genet. 2001;10:1907–1913. doi: 10.1093/hmg/10.18.1907. [DOI] [PubMed] [Google Scholar]
- 13.Radisky DC, Levy DD, Littlepage LE, et al. Rac1b and reactive oxygen species mediate MMP-3-induced EMT and genomic instability. Nature. 2005;436:123–127. doi: 10.1038/nature03688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dvorak HF. Tumors: wounds that do not heal. Similarities between tumor stroma generation and wound healing. N Engl J Med. 1986;315:1650–1659. doi: 10.1056/NEJM198612253152606. [DOI] [PubMed] [Google Scholar]
- 15.Ronnov-Jessen L, Petersen OW, Bissell MJ. Cellular changes involved in conversion of normal to malignant breast: importance of the stromal reaction. Physiol Rev. 1996;76:69–125. doi: 10.1152/physrev.1996.76.1.69. [DOI] [PubMed] [Google Scholar]
- 16.Sappino AP, Skalli O, Jackson B, Schurch W, Gabbiani G. Smooth-muscle differentiation in stromal cells of malignant and non-malignant breast tissues. Int J Cancer. 1988;41:707–712. doi: 10.1002/ijc.2910410512. [DOI] [PubMed] [Google Scholar]
- 17.Holliday DL, Hughes S, Shaw JA, Walker RA, Jones JL. Intrinsic genetic characteristics determine tumor-modifying capacity of fibroblasts: matrix metalloproteinase-3 5A/5A genotype enhances breast cancer cell invasion. Breast Cancer Res. 2007;9:R67. doi: 10.1186/bcr1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mishra PJ, Mishra PJ, Humeniuk R, et al. Carcinoma-associated fibroblast-like differentiation of human mesenchymal stem cells. Cancer Res. 2008;68:4331–4339. doi: 10.1158/0008-5472.CAN-08-0943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sugimoto H, Mundel TM, Kieran MW, Kalluri R. Identification of fibroblast heterogeneity in the tumor microenvironment. Cancer Biol Ther. 2006;5:1640–1646. doi: 10.4161/cbt.5.12.3354. [DOI] [PubMed] [Google Scholar]
- 20.Orimo A, Weinberg RA. Stromal fibroblasts in cancer: a novel tumor-promoting cell type. Cell Cycle. 2006;5:1597–1601. doi: 10.4161/cc.5.15.3112. [DOI] [PubMed] [Google Scholar]
- 21.Direkze NC, Hodivala-Dilke K, Jeffery R, et al. Bone marrow contribution to tumor-associated myofibroblasts and fibroblasts. Cancer Res. 2004;64:8492–8495. doi: 10.1158/0008-5472.CAN-04-1708. [DOI] [PubMed] [Google Scholar]
- 22.Ishii G, Sangai T, Oda T, et al. Bone-marrow-derived myofibroblasts contribute to the cancer-induced stromal reaction. Biochem Biophys Res Commun. 2003;309:232–240. doi: 10.1016/s0006-291x(03)01544-4. [DOI] [PubMed] [Google Scholar]
- 23.De Wever O, Demetter P, Mareel M, Bracke M. Stromal myofibroblasts are drivers of invasive cancer growth. Int J Cancer. 2008;123:2229–2238. doi: 10.1002/ijc.23925. [DOI] [PubMed] [Google Scholar]
- 24.Kalluri R, Zeisberg M. Fibroblasts in cancer. Nat Rev Cancer. 2006;6:392–401. doi: 10.1038/nrc1877. [DOI] [PubMed] [Google Scholar]
- 25.Mueller L, Goumas FA, Affeldt M, et al. Stromal fibroblasts in colorectal liver metastases originate from resident fibroblasts and generate an inflammatory microenvironment. Am J Pathol. 2007;171:1608–1618. doi: 10.2353/ajpath.2007.060661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sato T, Sakai T, Noguchi Y, Takita M, Hirakawa S, Ito A. Tumor-stromal cell contact promotes invasion of human uterine cervical carcinoma cells by augmenting the expression and activation of stromal matrix metalloproteinases. Gynecol Oncol. 2004;92:47–56. doi: 10.1016/j.ygyno.2003.09.012. [DOI] [PubMed] [Google Scholar]
- 27.Eltarhouny SA, Elsawy WH, Radpour R, Hahn S, Holzgreve W, Zhong XY. Genes controlling spread of breast cancer to lung “gang of 4”. Exp Oncol. 2008;30:91–95. [PubMed] [Google Scholar]
- 28.Gupta GP, Minn AJ, Kang Y, et al. Identifying site-specific metastasis genes and functions. Cold Spring Harb Symp Quant Biol. 2005;70:149–158. doi: 10.1101/sqb.2005.70.018. [DOI] [PubMed] [Google Scholar]
- 29.Okuyama N, Matsumine A, Kosugi R, Wakabayashi H, Uchida A. Matrix metalloproteinase-1 is a crucial bone metastasis factor in a human breast cancer-derived highly invasive cell line. Oncol Rep. 2008;20:1497–1504. [PubMed] [Google Scholar]
- 30.Blackburn JS, Rhodes CH, Coon CI, Brinckerhoff CE. RNA Interference Inhibition of Matrix Metalloproteinase-1 Prevents Melanoma Metastasis by Reducing Tumor Collagenase Activity and Angiogenesis. Cancer Res. 2007;67:10849–10858. doi: 10.1158/0008-5472.CAN-07-1791. [DOI] [PubMed] [Google Scholar]
- 31.Brinckerhoff CE, Matrisian LM. Matrix metalloproteinases: a tail of a frog that became a prince. Nat Rev Mol Cell Biol. 2002;3:207–214. doi: 10.1038/nrm763. [DOI] [PubMed] [Google Scholar]
- 32.Eck SM, Hoopes PJ, Petrella BL, Coon CI, Brinckerhoff CE. Matrix metalloproteinase-1 promotes breast cancer angiogenesis and osteolysis in a novel in vivo model. Breast Cancer Res Treat. 2008 doi: 10.1007/s10549-008-0085-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Visse R, Nagase H. Matrix metalloproteinases and tissue inhibitors of metalloproteinases: structure, function, and biochemistry. Circ Res. 2003;92:827–839. doi: 10.1161/01.RES.0000070112.80711.3D. [DOI] [PubMed] [Google Scholar]
- 34.Sorsa T, Konttinen YT, Lindy O, et al. Collagenase in synovitis of rheumatoid arthritis. Semin Arthritis Rheum. 1992;22:44–53. doi: 10.1016/0049-0172(92)90048-i. [DOI] [PubMed] [Google Scholar]
- 35.de Jong JS, van Diest PJ, van der Valk P, Baak JP. Expression of growth factors, growth inhibiting factors, and their receptors in invasive breast cancer. I: An inventory in search of autocrine and paracrine loops. J Pathol. 1998;184:44–52. doi: 10.1002/(SICI)1096-9896(199801)184:1<44::AID-PATH984>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 36.Muller A, Homey B, Soto H, et al. Involvement of chemokine receptors in breast cancer metastasis. Nature. 2001;410:50–56. doi: 10.1038/35065016. [DOI] [PubMed] [Google Scholar]
- 37.Garcia-Vicuna R, Gomez-Gaviro MV, Dominguez-Luis MJ, et al. CC and CXC chemokine receptors mediate migration, proliferation, and matrix metalloproteinase production by fibroblast-like synoviocytes from rheumatoid arthritis patients. Arthritis Rheum. 2004;50:3866–3877. doi: 10.1002/art.20615. [DOI] [PubMed] [Google Scholar]
- 38.Hashimoto N, Jin H, Liu T, Chensue SW, Phan SH. Bone marrow-derived progenitor cells in pulmonary fibrosis. J Clin Invest. 2004;113:243–252. doi: 10.1172/JCI18847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Phillips RJ, Burdick MD, Hong K, et al. Circulating fibrocytes traffic to the lungs in response to CXCL12 and mediate fibrosis. J Clin Invest. 2004;114:438–446. doi: 10.1172/JCI20997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qu Y, Mao M, Li X, et al. Enhanced migration and CXCR4 over-expression in fibroblasts with telomerase reconstitution. Mol Cell Biochem. 2008;313:45–52. doi: 10.1007/s11010-008-9740-6. [DOI] [PubMed] [Google Scholar]
- 41.Anders C, Carey LA. Understanding and treating triple-negative breast cancer. Oncology (Williston Park) 2008;22:1233–1239. discussion 1239–1240, 1243. [PMC free article] [PubMed] [Google Scholar]
- 42.Irvin WJ, Jr, Carey LA. What is triple-negative breast cancer? Eur J Cancer. 2008;44:2799–2805. doi: 10.1016/j.ejca.2008.09.034. [DOI] [PubMed] [Google Scholar]
- 43.Minn AJ, Gupta GP, Siegel PM, et al. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Noel A, De Pauw-Gillet MC, Purnell G, Nusgens B, Lapiere CM, Foidart JM. Enhancement of tumorigenicity of human breast adenocarcinoma cells in nude mice by matrigel and fibroblasts. Br J Cancer. 1993;68:909–915. doi: 10.1038/bjc.1993.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Forbes K, Webb MA, Sehgal I. Growth factor regulation of secreted matrix metalloproteinase and plasminogen activators in prostate cancer cells, normal prostate fibroblasts and normal osteoblasts. Prostate Cancer Prostatic Dis. 2003;6:148–153. doi: 10.1038/sj.pcan.4500640. [DOI] [PubMed] [Google Scholar]
- 46.Schutz A, Schneidenbach D, Aust G, Tannapfel A, Steinert M, Wittekind C. Differential expression and activity status of MMP-1, MMP-2 and MMP-9 in tumor and stromal cells of squamous cell carcinomas of the lung. Tumour Biol. 2002;23:179–184. doi: 10.1159/000064034. [DOI] [PubMed] [Google Scholar]
- 47.Yao C, Lin Y, Chua MS, et al. Interleukin-8 modulates growth and invasiveness of estrogen receptor-negative breast cancer cells. Int J Cancer. 2007;121:1949–1957. doi: 10.1002/ijc.22930. [DOI] [PubMed] [Google Scholar]
- 48.Konstantinopoulos PA, Vandoros GP, Karamouzis MV, Gkermpesi M, Sotiropoulou-Bonikou G, Papavassiliou AG. EGF-R is expressed and AP-1 and NF-kappaB are activated in stromal myofibroblasts surrounding colon adenocarcinomas paralleling expression of COX-2 and VEGF. Cell Oncol. 2007;29:477–482. doi: 10.1155/2007/831416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Minn AJ, Kang Y, Serganova I, et al. Distinct organ-specific metastatic potential of individual breast cancer cells and primary tumors. J Clin Invest. 2005;115:44–55. doi: 10.1172/JCI22320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wyatt CA, Geoghegan JC, Brinckerhoff CE. Short hairpin RNA-mediated inhibition of matrix metalloproteinase-1 in MDA-231 cells: effects on matrix destruction and tumor growth. Cancer Res. 2005;65:11101–11108. doi: 10.1158/0008-5472.CAN-05-2446. [DOI] [PubMed] [Google Scholar]
- 51.Saarinen J, Welgus HG, Flizar CA, Kalkkinen N, Helin J. N-glycan structures of matrix metalloproteinase-1 derived from human fibroblasts and from HT-1080 fibrosarcoma cells. Eur J Biochem. 1999;259:829–840. doi: 10.1046/j.1432-1327.1999.00105.x. [DOI] [PubMed] [Google Scholar]
- 52.Signoret N, Oldridge J, Pelchen-Matthews A, et al. Phorbol esters and SDF-1 induce rapid endocytosis and down modulation of the chemokine receptor CXCR4. J Cell Biol. 1997;139:651–664. doi: 10.1083/jcb.139.3.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mitra P, De A, Ethier MF, et al. Loss of chemokine SDF-1alpha-mediated CXCR4 signalling and receptor internalization in human hepatoma cell line HepG2. Cell Signal. 2001;13:311–319. doi: 10.1016/s0898-6568(01)00156-5. [DOI] [PubMed] [Google Scholar]
- 54.Ma Q, Jones D, Borghesani PR, et al. Impaired B-lymphopoiesis, myelopoiesis, and derailed cerebellar neuron migration in CXCR4- and SDF-1-deficient mice. Proc Natl Acad Sci U S A. 1998;95:9448–9453. doi: 10.1073/pnas.95.16.9448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zou YR, Kottmann AH, Kuroda M, Taniuchi I, Littman DR. Function of the chemokine receptor CXCR4 in haematopoiesis and in cerebellar development. Nature. 1998;393:595–599. doi: 10.1038/31269. [DOI] [PubMed] [Google Scholar]
- 56.Roland J, Murphy BJ, Ahr B, et al. Role of the intracellular domains of CXCR4 in SDF-1-mediated signaling. Blood. 2003;101:399–406. doi: 10.1182/blood-2002-03-0978. [DOI] [PubMed] [Google Scholar]
- 57.Burger M, Hartmann T, Krome M, et al. Small peptide inhibitors of the CXCR4 chemokine receptor (CD184) antagonize the activation, migration, and antiapoptotic responses of CXCL12 in chronic lymphocytic leukemia B cells. Blood. 2005;106:1824–1830. doi: 10.1182/blood-2004-12-4918. [DOI] [PubMed] [Google Scholar]
- 58.Zhou H, Tai HH. Characterization of recombinant human CXCR4 in insect cells: role of extracellular domains and N-glycosylation in ligand binding. Arch Biochem Biophys. 1999;369:267–276. doi: 10.1006/abbi.1999.1368. [DOI] [PubMed] [Google Scholar]
- 59.Checa M, Ruiz V, Montano M, Velazquez-Cruz R, Selman M, Pardo A. MMP-1 polymorphisms and the risk of idiopathic pulmonary fibrosis. Hum Genet. 2008;124:465–472. doi: 10.1007/s00439-008-0571-z. [DOI] [PubMed] [Google Scholar]
- 60.Jeong JG, Kim JM, Cho H, Hahn W, Yu SS, Kim S. Effects of IL-1beta on gene expression in human rheumatoid synovial fibroblasts. Biochem Biophys Res Commun. 2004;324:3–7. doi: 10.1016/j.bbrc.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 61.Andersson-Sjoland A, de Alba CG, Nihlberg K, et al. Fibrocytes are a potential source of lung fibroblasts in idiopathic pulmonary fibrosis. Int J Biochem Cell Biol. 2008;40:2129–2140. doi: 10.1016/j.biocel.2008.02.012. [DOI] [PubMed] [Google Scholar]
- 62.Friedl P, Wolf K. Proteolytic and non-proteolytic migration of tumour cells and leucocytes. Biochem Soc Symp. 2003:277–285. doi: 10.1042/bss0700277. [DOI] [PubMed] [Google Scholar]
- 63.Giambernardi TA, Grant GM, Taylor GP, et al. Overview of matrix metalloproteinase expression in cultured human cells. Matrix Biol. 1998;16:483–496. doi: 10.1016/s0945-053x(98)90019-1. [DOI] [PubMed] [Google Scholar]
- 64.Barbero S, Bonavia R, Bajetto A, et al. Stromal cell-derived factor 1alpha stimulates human glioblastoma cell growth through the activation of both extracellular signal-regulated kinases 1/2 and Akt. Cancer Res. 2003;63:1969–1974. [PubMed] [Google Scholar]
- 65.Kang Y, He W, Tulley S, et al. Breast cancer bone metastasis mediated by the Smad tumor suppressor pathway. Proc Natl Acad Sci U S A. 2005;102:13909–13914. doi: 10.1073/pnas.0506517102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Marchesi F, Monti P, Leone BE, et al. Increased survival, proliferation, and migration in metastatic human pancreatic tumor cells expressing functional CXCR4. Cancer Res. 2004;64:8420–8427. doi: 10.1158/0008-5472.CAN-04-1343. [DOI] [PubMed] [Google Scholar]
- 67.Raman D, Baugher PJ, Thu YM, Richmond A. Role of chemokines in tumor growth. Cancer Lett. 2007;256:137–165. doi: 10.1016/j.canlet.2007.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Alsayed Y, Ngo H, Runnels J, et al. Mechanisms of regulation of CXCR4/SDF-1 (CXCL12)-dependent migration and homing in multiple myeloma. Blood. 2007;109:2708–2717. doi: 10.1182/blood-2006-07-035857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Busillo JM, Benovic JL. Regulation of CXCR4 signaling. Biochim Biophys Acta. 2007;1768:952–963. doi: 10.1016/j.bbamem.2006.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhou Y, Larsen PH, Hao C, Yong VW. CXCR4 is a major chemokine receptor on glioma cells and mediates their survival. J Biol Chem. 2002;277:49481–49487. doi: 10.1074/jbc.M206222200. [DOI] [PubMed] [Google Scholar]
- 71.Luker KE, Luker GD. Functions of CXCL12 and CXCR4 in breast cancer. Cancer Lett. 2006;238:30–41. doi: 10.1016/j.canlet.2005.06.021. [DOI] [PubMed] [Google Scholar]
- 72.Nanki T, Nagasaka K, Hayashida K, Saita Y, Miyasaka N. Chemokines regulate IL-6 and IL-8 production by fibroblast-like synoviocytes from patients with rheumatoid arthritis. J Immunol. 2001;167:5381–5385. doi: 10.4049/jimmunol.167.9.5381. [DOI] [PubMed] [Google Scholar]
- 73.Lee BC, Lee TH, Avraham S, Avraham HK. Involvement of the chemokine receptor CXCR4 and its ligand stromal cell-derived factor 1alpha in breast cancer cell migration through human brain microvascular endothelial cells. Mol Cancer Res. 2004;2:327–338. [PubMed] [Google Scholar]
- 74.Hawsawi NM, Ghebeh H, Hendrayani SF, et al. Breast carcinoma-associated fibroblasts and their counterparts display neoplastic-specific changes. Cancer Res. 2008;68:2717–2725. doi: 10.1158/0008-5472.CAN-08-0192. [DOI] [PubMed] [Google Scholar]
- 75.Petrella BL, Lohi J, Brinckerhoff CE. Identification of membrane type-1 matrix metalloproteinase as a target of hypoxia-inducible factor-2 alpha in von Hippel-Lindau renal cell carcinoma. Oncogene. 2005;24:1043–1052. doi: 10.1038/sj.onc.1208305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 77.Petrella BL, Brinckerhoff CE. Tumor cell invasion of von Hippel Lindau renal cell carcinoma cells is mediated by membrane type-1 matrix metalloproteinase. Mol Cancer. 2006;5:66. doi: 10.1186/1476-4598-5-66. [DOI] [PMC free article] [PubMed] [Google Scholar]