Abstract
While the role of the prolyl isomerase Pin1 in dividing cells has long been recognized, Pin1’s function in postmitotic neurons is poorly understood. We have identified a novel mechanism by which Pin1 mediates activation of the mitochondrial cell death machinery specifically in neurons. This perspective presents a sophisticated signaling pathway that triggers neuronal apoptosis upon JNK-mediated phosphorylation of the BH3-only protein BIMEL at serine 65. Pin1 is enriched at the mitochondria in neurons together with BIMEL and components of a neuron-specific JNK signaling complex and functions as a molecular switch that couples the phosphorylation of BIMEL by JNK to apoptosis specifically in neurons. We discuss how these findings relate to our understanding of the development of the nervous system and the pathogenesis of neurologic disorders.
Keywords: cell death, neuron, Pin1, BIM, JNK
The PROLYL ISOMERASE PIN1
In recent years, phosphorylation-dependent prolyl isomerization has emerged as a major post-translational signaling mechanism. A growing number of phosphorylated proteins participating in important cellular processes are regulated through prolyl isomerization-induced conformational changes. The cis-trans configuration of peptide bonds between (phosphorylated) serine or threonine residues and an adjacent proline residue can have significant effects on protein phosphorylation status, as well as protein structure, activity, and stability and therefore needs to be carefully controlled by the cell.
The isomerization of phosphorylated serine/threonine-proline motifs is efficiently catalyzed by the prolyl isomerase Pin1.1 Pin1 contains an amino-terminal WW domain that specifically interacts with phosphorylated serine/threonine motifs and a carboxyl-terminal enzymatic peptidyl prolyl isomerase domain. Pin1-interacting motifs in substrate proteins are generated by proline-dependent kinases, which are pivotal players in numerous cellular signaling pathways.2 Pin1 was originally identified as an interacting protein of the fungal mitotic kinase NIMA, pointing to an essential role for Pin1 in mitosis.3 Subsequently, numerous studies have established a role for Pin1 in cell cycle progression and proliferation. Pin1 regulates the progression of the cell cycle by interacting with a large number of mitotic phosphoproteins including Cdc25 and cyclin D as well as transcription factors such as beta-catenin.4,5 Many mitotic substrates of Pin1 are phosphorylated by the kinase Cdc2 (also known as cyclin-dependent kinase 1 {Cdk1}), a master regulator of mitosis. Interestingly, misregulation of both Cdc2 and Pin1 in dividing cells can lead to blocked mitosis and mitotic catastrophe,3,6,7 suggesting that both proteins function in the same mitotic pathways. Consistent with its role in cellular proliferation, Pin1 is overexpressed in many common types of cancer including prostate, breast, lung, and colon cancer.8 Furthermore, increased Pin1 expression correlates with poor prognosis of these tumors.8–10
Surprisingly, Pin1 levels are high in neurons, which are terminally differentiated and postmitotic cells,11,12 suggesting additional functions for Pin1 in neurons distinct from cell cycle regulation and proliferation. Pin1 has been proposed to play a role in age-dependent neurodegeneration associated with hyperphosphorylated tau.13 We have recently identified a novel neural-specific mechanism of apoptosis, whereby Pin1 activates the mitochondrial cell death machinery in neurons following trophic factor and activity deprivation.11
APOPTOSIS IN THE NERVOUS SYSTEM
Apoptosis of neurons is a conserved and fundamental process in the development of the nervous system. The nervous system is initially generated with an excess number of neurons, followed by extensive apoptosis that ensures the proper assembly of neural circuits.14,15 In the adult brain, neuronal apoptosis contributes to the pathogenesis of several common neurologic disorders including ischemia and hereditary motor neuron diseases.16–18 Given the importance of neuronal cell death in brain development and disease, the molecular underpinnings of neuronal cell death are the subject of intense scrutiny. It has become apparent that the mitochondrial apoptotic machinery plays an essential role in the control of neuronal cell death.19 Ultimately, almost all survival and death signaling pathways in neurons converge at the mitochondria where they are integrated mainly by the BCL-2 family of proteins.20 In particular, members of the pro-apoptotic BH3-only subfamily are critical for the activation of the mitochondrial cell death program by acting as upstream sentinels of cellular damage that transduce apoptotic signals to the mitochondrial cell death machinery.21
The mitochondrial BH3-only protein BIMEL is a key regulator of neuronal apoptosis following trophic factor deprivation as well as activation of the p75 cell death receptor (p75NTR).22–24 In addition to its contribution to developmental neuronal cell death, BIMEL has been implicated in Alzheimer’s disease and ischemia.25–27 The apoptotic function of BIMEL in neurons is controlled at several levels including transcriptional and post-translational mechanisms. One of the key regulators of BIMEL in neurons is the c-Jun N-terminal kinase (JNK) signaling pathway, which is one of the major signaling pathways activated by numerous apoptotic stimuli in neurons including growth factor withdrawal and excitotoxicity.28–30 In addition to upregulating bim gene expression through the activation of the transcription factor c-Jun,23,24,31 JNK also directly phosphorylates the BIMEL protein at the distinct site of serine 65.22,32 Phosphorylation of BIMEL at serine 65 activates the apoptotic function of BIMEL in neurons. Intriguingly, phosphorylation of BIMEL at the same site in proliferating non-neural cells suppresses cell death, possibly through the proteasomal degradation of phosphorylated BIMEL.33–35 The critical regulatory phosphorylation event of BIMEL at serine 65 provides therefore a foundation for the elucidation of neural-specific cell death activation. We have proposed a Pin1-dependent mechanism by which phosphorylation of BIMEL at serine 65 leads to the activation of the mitochondrial apoptotic machinery specifically in neurons.11 Our findings and their implications for neuronal function are discussed below.
PIN1 IS AN IMPORTANT MEDIATOR OF NEURONAL APOPTOSIS
Our studies shed light on the important question of how apoptosis is regulated in a cell-specific manner in the nervous system. We showed that Pin1 interacts specifically with serine 65-phosphorylated BIMEL and thereby stabilizes BIMEL, resulting in neuronal apoptosis. These findings are the first to implicate Pin1 in the direct regulation of the mitochondrial cell death machinery. Pin1 might be posited to promote apoptosis by simply binding to serine 65-phosphorylated BIMEL via its WW domain and thereby protecting BIMEL from degradation. However, we demonstrated that the isomerase activity of Pin1 is also required for Pin1’s function in neuronal apoptosis. These findings suggest that BIMEL undergoes isomerization following phosphorylation of serine 65, resulting in increased resistance of BIMEL to proteasomal degradation and hence increased stabilization of BIMEL. The underlying mechanism for this stabilization of BIMEL remains to be identified. Pin1-mediated isomerization of serine 65-phosphorylated BIMEL might change the cis-trans configuration of the BIMEL phosphodegron motif that is recognized by an undefined E3 ubiquitin ligase, thereby reducing its affinity for BIMEL. A link between Pin1-mediated isomerization and ubiquitination, although promoting degradation, has recently been reported for the Pin1 substrate cyclin E.36 It will be important in the future to characterize the mechanism of phosphorylation-dependent degradation of BIMEL in greater detail. While some studies reported proteasomal degradation of BIMEL following phosphorylation at serine 65,33,34 others did not observe clearance of serine 65-phosphorylated BIMEL.35,37 Importantly, to date no ubiquitin ligase has been identified that binds to serine 65-phosphorylated BIMEL in neurons.
In addition to interfering with the recognition by ubiquitin ligases, Pin1-mediated isomerization could result in a conformational change in serine 65-phosphorylated BIMEL that might lead to the activation of BIMEL. An attractive hypothesis is that the motif containing serine 65 might act as an (auto)inhibitory domain that regulates the apoptotic function of BIMEL. A similar mechanism has been reported for two other BCL-2 family proteins, namely BCL-2 and BCL-XL, which are both regulated by post-translational modifications including JNK-mediated phosphorylation.38–40 To investigate the effect of Pin1-mediated isomerization on the conformation of BIMEL, it will be imperative to perform structural studies on BIMEL alone and serine 65-phosphorylated BIMEL in complex with Pin1.
Our findings raised the intriguing question of how Pin1, a protein strongly expressed in proliferating cells as well as in neurons, could be the molecular switch determining the cell type-specific consequences of BIMEL phosphorylation at serine 65. We reasoned that Pin1’s function in the activation of the mitochondrial apoptotic machinery in neurons might be conferred by a distinct subcellular localization of Pin1 in neurons. In non-neural cells, Pin1 has been reported to primarily localize to the nucleus.7 Interestingly, we found that a significant proportion of Pin1 in neural but not in non-neural cells is tethered to the mitochondrial membrane. This allows Pin1 specifically in neurons to be in close proximity with its substrate BIMEL, which is also localized to the mitochondria. In contrast, in non-neural cells Pin1 and BIMEL are localized to different cellular compartments, which might allow for the prompt degradation of BIMEL following phosphorylation at serine 65.
What allows for the distinctive localization of Pin1 to the mitochondria in neurons? The selective enrichment of Pin1 at the mitochondrial membrane in neurons may be achieved through Pin1’s interaction with a neuron-specific JNK signaling complex.11 JNK signaling proteins are organized by scaffold proteins that coordinate activation and specificity of signal transduction.41 The JNK-interacting protein 3 (JIP3) is a neuron-enriched JNK scaffold protein,42,43 and significant amounts of JIP3 and other components of the JNK signaling pathway are localized at the mitochondrial membrane in neurons.11,32 Interestingly, we found that Pin1 is associated with JIP3 in neurons in a phosphorylation-dependent manner. Our findings suggest a central role for the JIP3 signaling complex in neuronal apoptosis by providing both the kinase (JNK) and the post-phosphorylation isomerase activity (Pin1) to BIMEL at the mitochondrial membrane. The organization of both of these enzymatic activities by the JIP3 complex may serve to efficiently activate BIMEL in neurons and offers an explanation for the neuron-specific activation of the mitochondrial apoptotic machinery by the ubiquitously expressed Pin1.
While we have focused on Pin1 as a mediator of BIMEL-induced neuronal apoptosis, Pin1 may have a more general role in neuronal cell death. Given Pin1’s intimate link with JIP3 and JNK signaling in neurons, it will be interesting to determine whether Pin1 regulates other substrates of JNK following apoptotic stimuli in neurons.
Remarkably, in addition to the stress-activated protein kinases, the mitotic kinase Cdc2 has emerged as a key mediator of neuronal apoptosis during brain development and disease.44 Since Cdc2 generates numerous substrates for Pin1 in proliferating cells, Pin1 might also contribute to Cdc2-induced neuronal cell death. One of the apoptotic targets of Cdc2 in neurons is the BH3-only protein BAD.45 Cdc2 phosphorylates BAD at serine 128, resulting in the inhibition of the interaction of BAD with sequestering 14-3-3 proteins and hence the activation of BAD’s apoptotic function in neurons. Interestingly, JNK also phosphorylates BAD at Serine 128 and thereyby activates BAD-mediated apoptosis in neurons.46 Similar to the phosphorylation of BIMEL at serine 65, phosphorylation of BAD at serine 128 on its own does not trigger apoptosis in proliferating cells.17,48 In light of our study, it is tempting to speculate that Pin1 acts as a more general molecular switch that couples the phosphorylation of components of the apoptotic machinery to cell death specifically in neurons. In the case of serine 128-phosphorylated BAD, it is conceivable that Pin1-mediated isomerization causes a conformational change in BAD that results in the disruption of BAD’s sequestration by 14-3-3 proteins.
The function of cell cycle proteins in neurons is not restricted to apoptosis, as cell cycle proteins have been recently shown to regulate other essential neuronal processes including axonal growth and patterning in the developing brain.49,50 It will be therefore very interesting to determine whether Pin1 also functions in other cell cycle protein-mediated processes in the nervous system.
Our study raises the attractive possibility that Pin1’s specific role in the activation of the mitochondrial apoptotic machinery in neurons might contribute to the pathogenesis of neurologic disorders. Both JNK signaling and cell cycle reactivation have been implicated in neuronal apoptosis under pathologic conditions including ischemia and neurodegeneration.44,51 The role of Pin1 in neuronal apoptosis in the mature brain remains to be established. Future studies of neurologic disease models in Pin1-deficient and -overexpressing mouse models will give important insights into the contribution of Pin1 to neuronal apoptosis in vivo. The information gained from such studies will be invaluable in determining whether manipulation of Pin1 and its interaction with JIP3 might be a viable strategy for the treatment of human neurologic diseases.
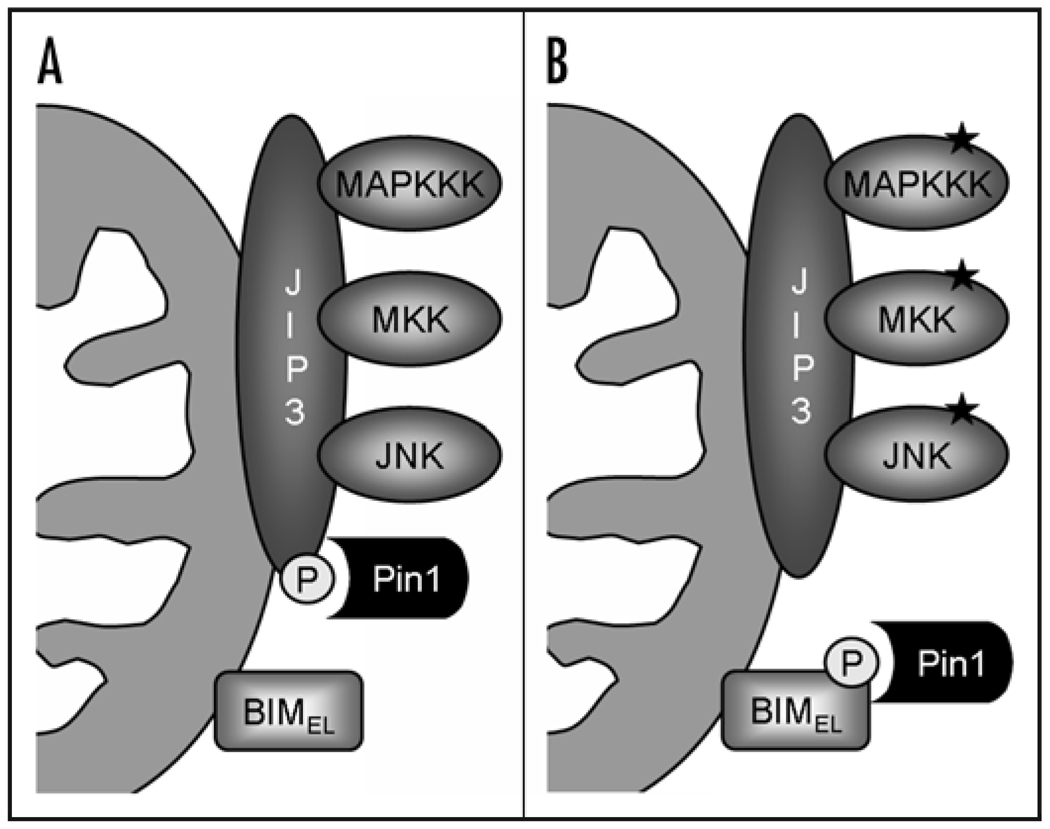
Figure 1.
Model of neural-specific Pin1-mediated activation of BIMEL following JNK-induced phosphorylation of BIMEL at serine 65. (A) In neurons, Pin1 is tethered to the mitochondria through binding to the neuron-enriched JNK-scaffolding protein JIP3, where it is in close proximity to its substrate BIMEL. (B) Upon an apoptotic stimulus, JNK signaling is activated, which results in phosphorylation of BIMEL at serine 65. Pin1 is released from JIP3 and interacts specifically with serine 65-phosphorylated BIMEL, thereby stabilizing BIMEL and activating BIMEL-mediated neuronal apoptosis.
ACKNOWLEDGEMENTS
We apologize to all colleagues whose work has not been cited due to the focus of this article. This work is supported by an NIH grant to A.B. (NS047188).
ABBREVIATIONS
- BAD
BCL-2 associated death agonist
- BCL-2
B-cell leukemia/lymphoma 2
- BIM
BCL-2-interacting mediator of cell death
- JIP3
JNK-interacting protein 3
- JNK
c-Jun N-terminal kinase
References
- 1.Lu KP, Liou YC, Zhou XZ. Pinning down proline-directed phosphorylation signaling. Trends Cell Biol. 2002;12:164–172. doi: 10.1016/s0962-8924(02)02253-5. [DOI] [PubMed] [Google Scholar]
- 2.Yaffe MB, Schutkowski M, Shen M, Zhou XZ, Stukenberg PT, Rahfeld JU, Xu J, Kuang J, Kirschner MW, Fischer G, Cantley LC, Lu KP. Sequence-specific and phosphorylation-dependent proline isomerization: A potential mitotic regulatory mechanism. Science. 1997;278:1957–1960. doi: 10.1126/science.278.5345.1957. [DOI] [PubMed] [Google Scholar]
- 3.Lu KP, Hanes SD, Hunter T. A human peptidyl-prolyl isomerase essential for regulation of mitosis. Nature. 1996;380:544–547. doi: 10.1038/380544a0. [DOI] [PubMed] [Google Scholar]
- 4.Lu KP. Pinning down cell signaling, cancer and Alzheimer’s disease. Trends Biochem Sci. 2004;29:200–209. doi: 10.1016/j.tibs.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 5.Wulf G, Finn G, Suizu F, Lu KP. Phosphorylation-specific prolyl isomerization: Is there an underlying theme? Nat Cell Biol. 2005;7:435–441. doi: 10.1038/ncb0505-435. [DOI] [PubMed] [Google Scholar]
- 6.Castedo M, Perfettini JL, Roumier T, Andreau K, Medema R, Kroemer G. Cell death by mitotic catastrophe: A molecular definition. Oncogene. 2004;23:2825–2837. doi: 10.1038/sj.onc.1207528. [DOI] [PubMed] [Google Scholar]
- 7.Lu PJ, Zhou XZ, Liou YC, Noel JP, Lu KP. Critical role of WW domain phosphorylation in regulating phosphoserine binding activity and Pin1 function. J Biol Chem. 2002;277:2381–2384. doi: 10.1074/jbc.C100228200. [DOI] [PubMed] [Google Scholar]
- 8.Bao L, Kimzey A, Sauter G, Sowadski JM, Lu KP, Wang DG. Prevalent overexpression of prolyl isomerase Pin1 in human cancers. Am J Pathol. 2004;164:1727–1737. doi: 10.1016/S0002-9440(10)63731-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fukuchi M, Fukai Y, Kimura H, Sohda M, Miyazaki T, Nakajima M, Masuda N, Tsukada K, Kato H, Kuwano H. Prolyl isomerase Pin1 expression predicts prognosis in patients with esophageal squamous cell carcinoma and correlates with cyclinD1 expression. Int J Oncol. 2006;29:329–334. [PubMed] [Google Scholar]
- 10.Kuramochi J, Arai T, Ikeda S, Kumagai J, Uetake H, Sugihara K. High Pin1 expression is associated with tumor progression in colorectal cancer. J Surg Oncol. 2006;94:155–160. doi: 10.1002/jso.20510. [DOI] [PubMed] [Google Scholar]
- 11.Becker EB, Bonni A. Pin1 mediates neural-specific activation of the mitochondrial apoptotic machinery. Neuron. 2006;49:655–662. doi: 10.1016/j.neuron.2006.01.034. [DOI] [PubMed] [Google Scholar]
- 12.Lu PJ, Wulf G, Zhou XZ, Davies P, Lu KP. The prolyl isomerase Pin1 restores the function of Alzheimer-associated phosphorylated tau protein. Nature. 1999;399:784–788. doi: 10.1038/21650. [DOI] [PubMed] [Google Scholar]
- 13.Liou YC, Sun A, Ryo A, Zhou XZ, Yu ZX, Huang HK, Uchida T, Bronson R, Bing G, Li X, Hunter T, Lu KP. Role of the prolyl isomerase Pin1 in protecting against age-dependent neurodegeneration. Nature. 2003;424:556–561. doi: 10.1038/nature01832. [DOI] [PubMed] [Google Scholar]
- 14.Levi-Montalcini R. The nerve growth factor 35 years later. Science. 1987;237:1154–1162. doi: 10.1126/science.3306916. [DOI] [PubMed] [Google Scholar]
- 15.Oppenheim RW. Cell death during development of the nervous system. Annu Rev Neurosci. 1991;14:453–501. doi: 10.1146/annurev.ne.14.030191.002321. [DOI] [PubMed] [Google Scholar]
- 16.Cleveland DW, Rothstein JD. From Charcot to Lou Gehrig: Deciphering selective motor neuron death in ALS. Nat Rev Neurosci. 2001;2:806–819. doi: 10.1038/35097565. [DOI] [PubMed] [Google Scholar]
- 17.Mattson MP. Apoptosis in neurodegenerative disorders. Nat Rev Mol Cell Biol. 2000;1:120–129. doi: 10.1038/35040009. [DOI] [PubMed] [Google Scholar]
- 18.Yuan J, Yankner BA. Apoptosis in the nervous system. Nature. 2000;407:802–809. doi: 10.1038/35037739. [DOI] [PubMed] [Google Scholar]
- 19.Putcha GV, Johnson EM., Jr Men are but worms: Neuronal cell death in C. elegans and vertebrates. Cell Death Differ. 2004;11:38–48. doi: 10.1038/sj.cdd.4401352. [DOI] [PubMed] [Google Scholar]
- 20.Gross A, McDonnell JM, Korsmeyer SJ. BCL-2 family members and the mitochondria in apoptosis. Genes Dev. 1999;13:1899–1911. doi: 10.1101/gad.13.15.1899. [DOI] [PubMed] [Google Scholar]
- 21.Huang DC, Strasser A. BH3-Only proteins-essential initiators of apoptotic cell death. Cell. 2000;103:839–842. doi: 10.1016/s0092-8674(00)00187-2. [DOI] [PubMed] [Google Scholar]
- 22.Becker EB, Howell J, Kodama Y, Barker PA, Bonni A. Characterization of the c-Jun N-terminal kinase-BimEL signaling pathway in neuronal apoptosis. J Neurosci. 2004;24:8762–8770. doi: 10.1523/JNEUROSCI.2953-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Putcha GV, Moulder KL, Golden JP, Bouillet P, Adams JA, Strasser A, Johnson EM. Induction of BIM, a proapoptotic BH3-only BCL-2 family member, is critical for neuronal apoptosis. Neuron. 2001;29:615–628. doi: 10.1016/s0896-6273(01)00238-0. [DOI] [PubMed] [Google Scholar]
- 24.Whitfield J, Neame SJ, Paquet L, Bernard O, Ham J. Dominant-negative c-Jun promotes neuronal survival by reducing BIM expression and inhibiting mitochondrial cytochrome c release. Neuron. 2001;29:629–643. doi: 10.1016/s0896-6273(01)00239-2. [DOI] [PubMed] [Google Scholar]
- 25.Biswas SC, Shi Y, Vonsattel JP, Leung CL, Troy CM, Greene LA. Bim is elevated in Alzheimer’s disease neurons and is required for beta-amyloid-induced neuronal apoptosis. J Neurosci. 2007;27:893–900. doi: 10.1523/JNEUROSCI.3524-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Engidawork E, Gulesserian T, Seidl R, Cairns N, Lubec G. Expression of apoptosis related proteins in brains of patients with Alzheimer’s disease. Neurosci Lett. 2001;303:79–82. doi: 10.1016/s0304-3940(01)01618-4. [DOI] [PubMed] [Google Scholar]
- 27.Ness JM, Harvey CA, Strasser A, Bouillet P, Klocke BJ, Roth KA. Selective involvement of BH3-only Bcl-2 family members Bim and Bad in neonatal hypoxia-ischemia. Brain Res. 2006;1099:150–159. doi: 10.1016/j.brainres.2006.04.132. [DOI] [PubMed] [Google Scholar]
- 28.Harper SJ, LoGrasso P. Signalling for survival and death in neurones: The role of stress-activated kinases, JNK and p38. Cell Signal. 2001;13:299–310. doi: 10.1016/s0898-6568(01)00148-6. [DOI] [PubMed] [Google Scholar]
- 29.Ham J, Eilers A, Whitfield J, Neame SJ, Shah B. c-Jun and the transcriptional control of neuronal apoptosis. Biochem Pharmacol. 2000;60:1015–1021. doi: 10.1016/s0006-2952(00)00372-5. [DOI] [PubMed] [Google Scholar]
- 30.Mielke K, Herdegen T. JNK and p38 stresskinases-degenerative effectors of signal-transduction-cascades in the nervous system. Prog Neurobiol. 2000;61:45–60. doi: 10.1016/s0301-0082(99)00042-8. [DOI] [PubMed] [Google Scholar]
- 31.Harris CA, Johnson EM. BH3-only Bcl-2 family members are coordinately regulated by the JNK pathway and require Bax to induce apoptosis in neurons. J Biol Chem. 2001;276:37754–37760. doi: 10.1074/jbc.M104073200. [DOI] [PubMed] [Google Scholar]
- 32.Putcha GV, Le S, Frank S, Besirli CG, Clark K, Chu B, Alix S, Youle RJ, LaMarche A, Maroney AC, Johnson EM. JNK-mediated BIM phosphorylation potentiates BAX-dependent apoptosis. Neuron. 2003;38:899–914. doi: 10.1016/s0896-6273(03)00355-6. [DOI] [PubMed] [Google Scholar]
- 33.Ley R, Ewings KE, Hadfield K, Howes E, Balmanno K, Cook SJ. Extracellular signal-regulated kinases 1/2 are serum-stimulated “Bim(EL) kinases” that bind to the BH3-only protein Bim(EL) causing its phosphorylation and turnover. J Biol Chem. 2004;279:8837–8847. doi: 10.1074/jbc.M311578200. [DOI] [PubMed] [Google Scholar]
- 34.Luciano F, Jacquel A, Colosetti P, Herrant M, Cagnol S, Pages G, Auberger P. Phosphorylation of Bim-EL by Erk1/2 on serine 69 promotes its degradation via the proteasome pathway and regulates its proapoptotic function. Oncogene. 2003;22:6785–6793. doi: 10.1038/sj.onc.1206792. [DOI] [PubMed] [Google Scholar]
- 35.Reginato MJ, Mills KR, Becker EB, Lynch DK, Bonni A, Muthuswamy SK, Brugge JS. Bim regulation of lumen formation in cultured mammary epithelial acini is targeted by oncogenes. Mol Cell Biol. 2005;25:4591–4601. doi: 10.1128/MCB.25.11.4591-4601.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Drogen F, Sangfelt O, Malyukova A, Matskova L, Yeh E, Means AR, Reed SI. Ubiquitylation of cyclin E requires the sequential function of SCF complexes containing distinct hCdc4 isoforms. Mol Cell. 2006;23:37–48. doi: 10.1016/j.molcel.2006.05.020. [DOI] [PubMed] [Google Scholar]
- 37.Harada H, Quearry B, Ruiz-Vela A, Korsmeyer SJ. Survival factor-induced extracellular signal-regulated kinase phosphorylates BIM, inhibiting its association with BAX and proapoptotic activity. Proc Natl Acad Sci USA. 2004;101:15313–15317. doi: 10.1073/pnas.0406837101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deverman BE, Cook BL, Manson SR, Niederhoff RA, Langer EM, Rosova I, Kulans LA, Fu X, Weinberg JS, Heinecke JW, Roth KA, Weintraub SJ. Bcl-xL deamidation is a critical switch in the regulation of the response to DNA damage. Cell. 2002;111:51–62. doi: 10.1016/s0092-8674(02)00972-8. [DOI] [PubMed] [Google Scholar]
- 39.Srivastava RK, Mi QS, Hardwick JM, Longo DL. Deletion of the loop region of Bcl-2 completely blocks paclitaxel-induced apoptosis. Proc Natl Acad Sci USA. 1999;96:3775–3780. doi: 10.1073/pnas.96.7.3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamamoto K, Ichijo H, Korsmeyer SJ. BCL-2 is phosphorylated and inactivated by an ASK1/Jun N-terminal protein kinase pathway normally activated at G(2)/M. Mol Cell Biol. 1999;19:8469–8478. doi: 10.1128/mcb.19.12.8469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morrison DK, Davis RJ. Regulation of MAP kinase signaling modules by scaffold proteins in mammals. Annu Rev Cell Dev Biol. 2003;19:91–118. doi: 10.1146/annurev.cellbio.19.111401.091942. [DOI] [PubMed] [Google Scholar]
- 42.Ito M, Yoshioka K, Akechi M, Yamashita S, Takamatsu N, Sugiyama K, Hibi M, Nakabeppu Y, Shiba T, Yamamoto KI. JSAP1, a novel jun N-terminal protein kinase (JNK)-binding protein that functions as a Scaffold factor in the JNK signaling pathway. Mol Cell Biol. 1999;19:7539–7548. doi: 10.1128/mcb.19.11.7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kelkar N, Delmotte MH, Weston CR, Barrett T, Sheppard BJ, Flavell RA, Davis RJ. Morphogenesis of the telencephalic commissure requires scaffold protein JNK-interacting protein 3 (JIP3) Proc Natl Acad Sci USA. 2003;100:9843–9848. doi: 10.1073/pnas.1733944100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Becker EB, Bonni A. Cell cycle regulation of neuronal apoptosis in development and disease. Prog Neurobiol. 2004;72:1–25. doi: 10.1016/j.pneurobio.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 45.Konishi Y, Lehtinen M, Donovan N, Bonni A. Cdc2 phosphorylation of BAD links the cell cycle to the cell death machinery. Mol Cell. 2002;9:1005–1016. doi: 10.1016/s1097-2765(02)00524-5. [DOI] [PubMed] [Google Scholar]
- 46.Donovan N, Becker EB, Konishi Y, Bonni A. JNK phosphorylation and activation of BAD couples the stress-activated signaling pathway to the cell death machinery. J Biol Chem. 2002;277:40944–40949. doi: 10.1074/jbc.M206113200. [DOI] [PubMed] [Google Scholar]
- 47.Berndtsson M, Konishi Y, Bonni A, Hagg M, Shoshan M, Linder S, Havelka AM. Phosphorylation of BAD at Ser-128 during mitosis and paclitaxel-induced apoptosis. FEBS Lett. 2005;579:3090–3094. doi: 10.1016/j.febslet.2005.04.067. [DOI] [PubMed] [Google Scholar]
- 48.Hashimoto A, Hirose K, Iino M. BAD detects coincidence of G2/M phase and growth factor deprivation to regulate apoptosis. J Biol Chem. 2005;280:26225–26232. doi: 10.1074/jbc.M409363200. [DOI] [PubMed] [Google Scholar]
- 49.Becker EB, Bonni A. Beyond proliferation-cell cycle control of neuronal survival and differentiation in the developing mammalian brain. Semin Cell Dev Biol. 2005;16:439–448. doi: 10.1016/j.semcdb.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 50.McClellan KA, Slack RS. Novel functions for cell cycle genes in nervous system development. Cell Cycle. 2006;5:1506–1513. doi: 10.4161/cc.5.14.2980. [DOI] [PubMed] [Google Scholar]
- 51.Johnson GL, Nakamura K. The c-jun kinase/stress-activated pathway: Regulation, function and role in human disease. Biochim Biophys Acta. 2007 doi: 10.1016/j.bbamcr.2006.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]