Abstract
Polymorphisms in genes critical to cell cycle control are outstanding candidates for association with ovarian cancer risk; numerous genes have been interrogated by multiple research groups using differing tagging SNP sets. In order to maximize information gleaned from existing genotype data, we conducted a combined analysis of five independent studies of invasive epithelial ovarian cancer. Up to 2,120 cases and 3,382 controls were genotyped in the course of two collaborations at a variety of SNPs in 11 cell cycle genes (CDKN2C, CDKN1A, CCND3, CCND1, CCND2, CDKN1B, CDK2, CDK4, RB1, CDKN2D, CCNE1) and one gene region (CDKN2A-CDKN2B). Because of the semi-overlapping nature of the 123 assayed tagging SNPs, we performed multiple imputation based on fastPHASE using data from White non-Hispanic study participants and participants in the international HapMap Consortium and NIEHS SNPs Program. Logistic regression assuming a log-additive model was performed on combined and imputed data. We observed strengthened signals in imputation-based analyses at several SNPs, particularly CDKN2A-CDKN2B rs3731239, CCND1 rs602652, rs3212879, rs649392, and rs3212891, CDK2 rs2069391, rs2069414, and rs17528736, and CCNE1 rs3218036. These results lend evidence to a role of cell cycle genes in ovarian cancer etiology, suggest a reduced set of SNPs to target in additional cases and controls, and exemplify the utility of imputation in candidate gene studies.
Introduction
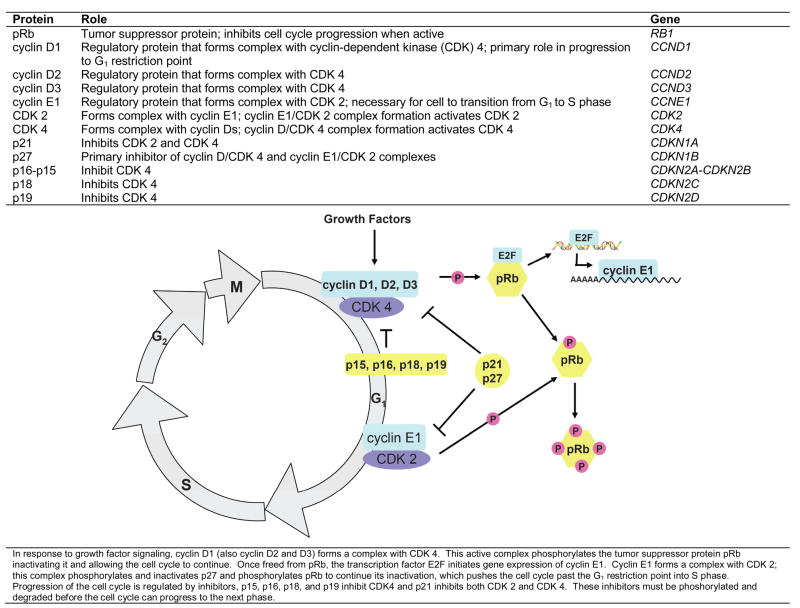
Because genes regulating cell cycle control are excellent candidates for cancer risk, multiple groups have targeted these genes for etiologic investigation. Progression of cells from G1 phase to S phase to G2 phase is closely regulated by the retinoblastoma protein (pRb), cyclins, cyclin-dependent kinases (CDKs), and CDK inhibitors (Figure 1). Loss of growth control is a key trait of cancerous cells which result from abnormalities in cell replication which is controlled by cell cycle genes (1, 2). Inhibitors of the cyclin/CDK complexes also regulate cell cycle progression by controlling the activation of these complexes (3, 4). Studies of inherited variation in cell cycle genes suggest that genotypes in this pathway may be associated with risk of breast cancer (5, 6), prostate cancer (7), lung cancer (8), bladder cancer (9, 10), and oral cancer (11), but not necessarily colorectal cancer (12).
Figure 1.
Cell Cycle Control
Evidence for a role of cell cycle variants in ovarian cancer comes from several lines of evidence. Overexpression of cyclins D1, D2, and E1 and deletion of cyclin-dependent kinase inhibitors 2A (p16) and 2B (p15) have been observed in ovarian cancers (13–15). In addition, ovarian cancers frequently have altered retinoblastoma protein (pRb) which regulates the G1 to S phase transition when cells either arrest development or proliferate (16). The complex interplay of cyclins D1, D2, D3, and E1, cyclin-dependent kinases 2 and 4, cyclin-dependent kinase inhibitors 1A (p21) and 1B (p27), CDK4 inhibitors 2A (p16), 2B (p15), 2C (p18), and 2D (p19), and pRb suggests that perturbation of any of these molecules via germline variation may predispose a woman to ovarian carcinogenesis. Finally, previous reports of inherited variation and ovarian cancer survival have shown suggestive results (17, 18).
Improved precision of disease-risk estimates associated with single-nucleotide polymorphisms (SNPs) in cell cycle control genes can be obtained with pooled analyses of several study populations. The use of tagSNPs and htSNPs has facilitated cost-savings in individual studies; however, tagging SNP sets often vary across studies due to the use of different algorithms, parameter values, data sources, and genotyping platforms (19). In the context of failed genotyping or multiple genome-wide platforms, several tools for imputation and analysis of missing genotypes have been developed (20–26). Though potentially informative, these tools have not been routinely applied to the candidate gene setting where multiple studies targeted differing, but correlated SNPs.
Here, we analyze data from five ovarian cancer study populations which, as part of two collaborations, tagged common variation in 11 cell cycle genes and in one gene region (CDKN2C, CDKN1A, CCND3, the CDKN2A-CDKN2B region, CCND1, CCND2, CDKN1B, CDK2, CDK4, RB1, CDKN2D, and CCNE1). A total of 123 SNPs were genotyped but only 24 SNPs were genotyped in both collaborations; because SNPs sets were correlated but differed, we combined data using multiple imputation (17, 27). We present results of observed and imputed analyses of ovarian cancer risk, suggest SNPs worthy of additional genotyping, and provide guidance for the application of this and other imputation methods.
Methods
Study Populations
The first genotyping effort (28, 29) utilized subjects recruited into two case-control studies at Mayo Clinic in Rochester, MN (MAY) and at Duke University in Durham, NC (NCO). At Mayo Clinic, cases were women over age 20 years with histologically-confirmed epithelial ovarian cancer living in the Upper Midwest and enrolled within one year of diagnosis. Controls without ovarian cancer and who had at least one intact ovary were recruited from among those seen for general medical examinations and frequency-matched to cases on age and region of residence. At Duke University, cases were women between age 20 and 74 years with histologically-confirmed primary epithelial ovarian cancer identified using the North Carolina Central Cancer Registry’s rapid case ascertainment system. Controls without ovarian cancer and who had at least one intact ovary were identified from the same 48-county region as the cases using list-assisted randomdigit dialing and frequency matched to cases on race and age. DNA was extracted from blood using the Gentra AutoPure LS Purgene salting out methodology (Gentra, Minneapolis, MN), and for Duke University participants, DNA was whole-genome amplified with the REPLI-G protocol (Qiagen Inc, Valencia CA) (29). Non-White and Hispanic participants and cases with borderline tumors were excluded from analysis (one NCO case with unknown race was assumed to be White non-Hispanic, and one NCO case with unknown tumor behavior was assumed to be invasive); additional details are provided elsewhere (30).
The second genotyping collaboration (17, 27) used cases and controls from three different studies: the SEARCH ovarian cancer study from East Anglia, United Kingdom (SEA), the MALOVA cancer study from Denmark (MAL), and the GEOCS study from Stanford University in Palo Alto, CA (STA). The SEARCH ovarian cancer study included invasive epithelial ovarian cancer cases collected from the East Anglian and West Midlands cancer registries, and controls randomly selected from European Prospective Investigation into Cancer and Nutrition (EPIC)-Norfolk cohort study. The MALOVA study contained invasive ovarian cancer cases and population controls randomly drawn from a defined study area in Denmark. The GEOCS study ascertained participants from six counties in northern California including invasive ovarian cancer cases and age-matched controls obtaining using random-digit dialing. Non-White and Hispanic participants and cases with borderline tumors were excluded from analysis (33 SEA cases and one SEA control with unknown race were assumed to be White non-Hispanic, and 75 SEA cases with unknown tumor behavior were assumed to be invasive); additional study participant details are provided elsewhere (31, 32).
SNP Selection
The first collaboration (MAY+NCO) identified tagSNPs within five kb of each gene using the algorithm of ldSelect (33) to bin pairwise-correlated SNPs at r2 ≥ 0.80 with minor allele frequency (MAF) ≥ 0.05 among 60 unrelated Utah Residents with Northern and Western European Ancestry (CEU) genotyped as part of the international HapMap Consortium release 20 (HapMap, mapped to NCBI build 35) (34). Within LD bins, tagSNPs with the maximum Illumina-provided SNP_Score (San Diego, CA) were selected. In addition to tagSNPs, putative-functional SNPs were included (within 1 kb upstream, 5′ UTR, 3′ UTR, or non-synonymous) with MAF ≥ 0.05 identified in Ensembl version 34 and Illumina-provided SNP_Score > 0.6. Sixty SNPs were selected.
The second collaboration (SEA+MAL+STA) used the multimarker tagging algorithm of Tagger (35) to bin SNPs pairwise-correlated or correlated with combinations of SNPs with MAF ≥ 0.05 and Rs2 ≥ 0.80 (36). CEU data from HapMap (October 2005) were used as well as resequencing data when available (October 2005) from the National Institute of Environmental Health Sciences (NIEHS) SNPs Program (37). Analysis of NIEHS SNPs used 62 individuals thought to have the least amount of African ancestry from a panel of 90 individuals (PDR90); additional information is provided elsewhere (17). Eighty-seven SNPs were selected.
Genotyping
For MAY+NCO, genotyping of 1,086 genomic and 1,282 WGA DNA samples (total = 2,368 including duplicates and laboratory controls) on 2,051 unique study participants was performed at Mayo Clinic using the Illumina GoldenGate™ BeadArray assay and BeadStudio software for automated genotype clustering and calling according to a standard protocol (38). Samples with call rates below 90% and SNPs with call rates below 95% were excluded. Of 2,051 participants genotyped, 10 were later found to be ineligible and were excluded, and 74 samples failed. Among SNPs with an overall call rate ≥ 95%, concordance was 99.99% between duplicates of genomic DNA, 99.97% between duplicates of WGA DNA and 99.16% between genomic and WGA DNA, indicating adequate genotyping of WGA DNA (29).
SEA+MAL+STA samples were genotyped using the Taqman 7900HT Sequence Detection System according to the manufacturer’s instructions. Each assay was carried out using 10 ng DNA in a 5 μl reaction with Taqman Universal PCR Master Mix (Applied Biosystems, Warrington United Kingdom), forward and reverse primers, and FAM- and VIC-labeled probes designed by Applied Biosystems (ABI Assay-by-Design). Primer and probe sequences and assay conditions used for each polymorphism analyzed are available upon request. All assays were carried out in 384-well arrays with 12 duplicate samples in each plate for quality control. Genotypes were determined using Allelic Discrimination Sequence Detection software (Applied Biosystems). Call rates ranged from 94.5% to 99.5% for all the studies and SNPs and overall concordance between duplicate samples was > 99% (17, 27).
Other Data Sources and Harmonization of Alleles
To impute missing study participant genotypes, we utilized data from study participants as well as updated data from the sources originally used to identify tagging SNPs: 60 unrelated CEU individuals in HapMap version 21a (SNPs within 10 kb of each gene using genome build 36.3, http://hapmap.org/downloads/genotypes/2007-01/) and 62 individuals with minimal evidence of African ancestry from NIEHS SNPs (resequenced regions, http://egp.gs.washington.edu/finished_genes.html, September 2007). A total of 911 SNPs were identified from MAY+NCO, SEA+MAL+STA, HapMap CEU, and NIEHS SNPs including 395 SNPs with genotype data from two or more sources.
To verify allele consistency for 395 SNPs with genotype data from two or more sources, we reviewed study-designated allele names and MAFs across sources. We found that, for 270 SNPs (68%), genotypes were easily combined across studies (similar MAF, identical nomenclature); for 112 SNPs (28%), genotypes were combined following an obvious strand reversal for at least one data source (similar MAF, reverse strand nomenclature); and for four SNPs (1%), genotypes were clearly inconsistent or of a non-obvious nature (e.g., C>G, A>T and MAF > 0.40) and excluded for at least one source (and data remained for two or more sources). For five SNPs (1%), genotypes that were clearly inconsistent or of a non-obvious nature were excluded for at least one source and only one source remained (thus not requiring allele harmonizing), and, for four SNPs (1%), genotypes that were clearly inconsistent or of a non-obvious nature were excluded for all sources and not used in analyses. Thus, a resulting 391 harmonized SNPs were merged with 516 SNPs available from only one source. One SNP (CCND3 rs1051130) was then excluded due to HWE p-value < 0.001 in the SEA+MAL+STA controls, leaving 901 SNPs included in the final analytical dataset (122 SNPs genotyped by MAY+NCO or SEA+MAL+STA). SNPs are tallied per gene and per population in Table 1; a complete listing of analyzed SNPs, MAFs, and call rates is provided in Supplemental Table 1.
Table 1.
Cell Cycle Genes
| Number of SNPs | |||||||
|---|---|---|---|---|---|---|---|
| Gene | Chr. | Start (bp) | Size (kb) | MAY+NCO† | SEA+MAL+STA‡ | CEU§ | NIEHS SNPs|| |
| CDKN2C | 1 | 51,206,196 | 6.7 | 1 | 2 | 7 | 19 |
| CDKN1A | 6 | 36,754,465 | 8.6 | 4 | 11 | 40 | 42 |
| CCND3 | 6 | 42,010,649 | 113.8 | 7 | 6 | 87 | 32 |
| CDKN2A-CDKN2B | 9 | 21,957,751 | 41.6 | 9 | 17 | 76 | 96 |
| CCND1 | 11 | 69,165,054 | 13.4 | 5 | 7 | 16 | 53 |
| CCND2 | 12 | 4,253,199 | 31.6 | 4 | 14 | 83 | 102 |
| CDKN1B | 12 | 12,761,576 | 5.0 | 8 | 8 | 17 | 15 |
| CDK2 | 12 | 54,646,826 | 6.0 | 7 | 2 | 23 | 24 |
| CDK4 | 12 | 56,428,270 | 4.1 | 3 | 2 | 11 | 26 |
| RB1 | 13 | 47,775,884 | 178.1 | 8 | 11 | 171 | 197 |
| CDKN2D | 19 | 10,538,138 | 2.5 | 2 | 2 | 15 | 5 |
| CCNE1 | 19 | 34,994,741 | 12.3 | 2 | 4 | 19 | 48 |
|
|
|||||||
| 60 | 86 | 565 | 659 | ||||
Source for tagging SNPs was HapMap release 20.
Source for tagging SNPs was NIEHS SNPs, except CCND2 and CDKN1B which used HapMap and CDKN2C which used both HapMap and NIEHS SNPs (October 2005); CCND3 rs1051130 excluded due to HWE p < 0.001.
HapMap release 21a, within 10 kb of gene start or stop.
September 2007.
Statistical Analysis
We ran a series of association analyses using observed data from each collaboration (MAY+NCO and SEA+MAL+STA), observed data combined across both collaborations, and imputed data. To impute missing genotypes, we used a hidden Markov model as implemented in fastPHASE (39), with 25 iterations and 20 random starts of the EM algorithm. Five runs of fastPHASE were conducted using different random seeds. A logistic regression model was then fit to each of the five imputed datasets for each SNP of interest and the resulting parameter and variance estimates were extracted. Results were combined across imputation runs using standard multiple imputation techniques computing both the within and between imputation variation (40). The use of multiple imputation methods allowed us to estimate the variance due to imputation and incorporate this into our overall SNP variance estimates. In general, this imputation-based variance component was small (mean=1.7 × 10−5). The variance component was largest for SNPs genotyped among MAY+NCO participants only (mean=3.7 × 10−5), slightly smaller for SNPs genotyped as SEA+MAL+STA only (mean=1.3 × 10−5), and practically equal to zero for SNPs genotyped in both collaborations (mean=1.1 × 10−7). Because of an observed slightly greater MAF discrepancy between study participants and NIEHS SNPs participants than between study participants and HapMap participants, imputations were also carried out excluding NIEHS SNPs data.
Associations between genotypes and ovarian cancer risk were assessed using logistic regression to estimate odds ratios (ORs) and 95% confidence intervals (CIs) assuming an ordinal (log-additive) genotypic effect. For imputation-based analyses, we modeled the observed number of copies of the minor allele for participants with non-missing genotypes for a given SNP and the estimated most-likely number of copies of the minor allele for subjects with imputed genotypes. Association tests were two-sided, adjusted for the potential confounding effects of age and study population (MAY, NCO, SEA, MAL, and STA), and carried out using the SAS software system (SAS Institute, Inc., Cary, NC). Adjustment of p-values due to multiple testing was not conducted as interpretation was based on relative changes in results due to imputation.
Results
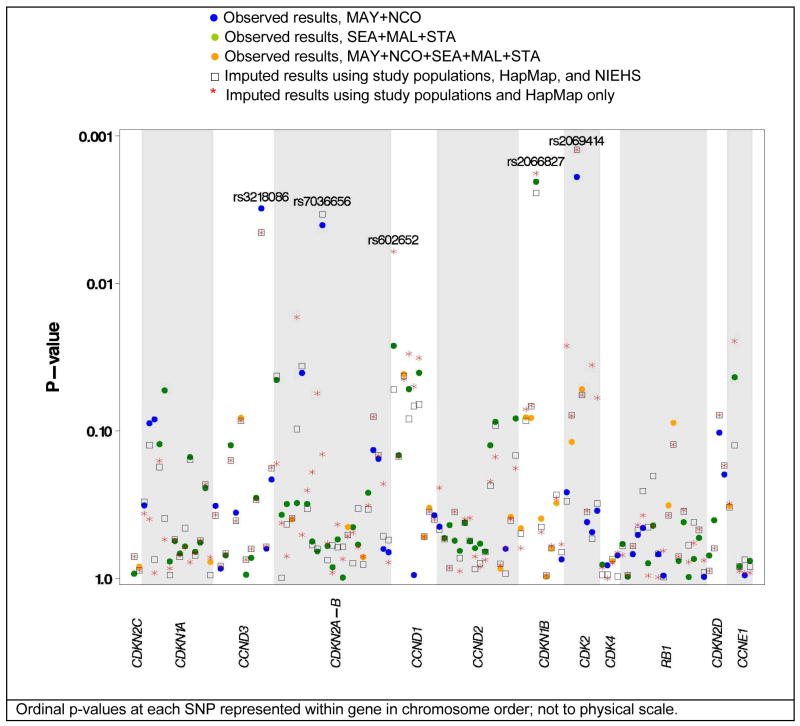
Characteristics of 5,502 study participants (2,210 cases, 3,382 controls) are shown in Table 2. The study populations were generally similar, although MAY participants were older (no upper age limit had been used), STA participants included more oral contraceptive users, and SEA included fewer known serous cases. Using observed data only, four SNPs were associated with risk of invasive ovarian cancer at p < 0.01 (Figure 2): CCND3 rs3218086 (MAY+NCO; increased risk), CDKN2A-CDKN2B rs7036656 (MAY+NCO; decreased risk), CDKN1B rs2066827 (SEA+MAL+STA; decreased risk), and CDK2 rs2069414 (MAY+NCO; increased risk) (Table 3). Risk was associated at p < 0.05 with two additional SNPs in the MAY+NCO population (CDKN1A rs7767246 and CDKN2A-CDKN2B rs2811709, both decreased risk) and seven additional SNPs in SEA+MAL+STA (CDKN2A-CDKN2B rs3731257, decreased risk; CCND1 rs602652, rs603695, and rs7178 increased risk; CCND1 rs321891, decreased risk; CCNE1 rs3218036 increased risk; RB1 rs2854344, decreased risk; Table 3) (17, 27). No SNPs typed in both populations were significant in combined analysis without reaching significance in one of the study populations. Of four SNPs typed in both populations and significant in only one population, two yielded ORs in opposite directions for null combined ORs (CDKN1A rs7767246, CCND1 rs7178), two were significant (p < 0.05) only in the larger SEA+MAL+STA study (when combined, CCND1 rs603965 remained at p < 0.05, RB1 rs2854344 lost significance).
Table 2.
Characteristics of 5,502 White non-Hispanic Invasive Cases and Controls
| MAY | NCO | MAL | SEA | STA | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Case (N=287) |
Control (N=462) |
Case (N=382) |
Control (N=479) |
Case (N=447) |
Control (N=1,221) |
Case (N=717) |
Control (N=852) |
Case (N=287) |
Control (N=368) |
|
| Age | ||||||||||
| Mean (SD) | 61.6 (12.5) | 60.0 (13.0) | 56.8 (10.7) | 54.8 (12.0) | 59.9 (10.6) | 56.8 (11.5) | 55.7 (10.0) | 52.7 (8.3) | 51.2 (8.7) | 48.3 (10.3) |
| Age Quartile, years | ||||||||||
| <=46 | 30 (11%) | 69 (15%) | 68 (18%) | 123 (26%) | 53 (12%) | 243 (20%) | 112 (16%) | 202 (24%) | 82 (29%) | 137 (37%) |
| 47–53 | 52 (18%) | 78 (17%) | 82 (22%) | 118 (25%) | 83 (19%) | 293 (24%) | 178 (25%) | 323 (38%) | 76 (27%) | 105 (29%) |
| 54–62 | 70 (24%) | 113 (25%) | 108 (28%) | 80 (17%) | 114 (26%) | 247 (20%) | 236 (33%) | 205 (24%) | 110 (38%) | 104 (28%) |
| 63+ | 135 (47%) | 202 (44%) | 124 (33%) | 158 (33%) | 197 (44%) | 438 (36%) | 191 (27%) | 122 (14%) | 19 (7%) | 22 (6%) |
| Age Menarche | ||||||||||
| Mean (SD) | 12.8 (1.53) | 13.2 (4.47) | 12.5 (1.45) | 12.7 (1.41) | 13.6 (1.69) | 13.6 (1.63) | 12.9 (1.95) | 12.9 (1.52) | 12.5 (1.44) | 12.8 (1.54) |
| Menopause Status | ||||||||||
| Pre/Peri | 70 (24%) | 108 (23%) | 73 (19%) | 154 (32%) | 74 (17%) | 373 (31%) | 240 (34%) | 557 (65%) | 14 (5%) | 182 (50%) |
| Post | 209 (73%) | 327 (71%) | 273 (72%) | 316 (66%) | 280 (63%) | 848 (70%) | 414 (58%) | 294 (35%) | 147 (51%) | 129 (35%) |
| Unknown | 8 (3%) | 27 (5.9%) | 36 (9%) | 9 (2%) | 93 (21%) | 0 (0%) | 63 (9%) | 1 (<1%) | 126 (44%) | 57 (16%) |
| Ever used OC | ||||||||||
| Yes | 133 (46%) | 261 (57%) | 240 (63%) | 328 (69%) | 146 (33%) | 686 (56%) | 297 (41%) | 580 (68%) | 209 (73%) | 311 (85%) |
| No | 139 (48%) | 164 (36%) | 131 (34%) | 147 (31%) | 207 (46%) | 535 (44%) | 399 (56%) | 271 (32%) | 76 (27%) | 55 (15%) |
| Unknown | 15 (5%) | 37 (8%) | 11 (3%) | 4 (<1%) | 94 (21%) | 0 (0%) | 21 (3%) | 1 (<1%) | 2 (<1%) | 2 (<1%) |
| Number of Live Births | ||||||||||
| 0 | 47 (16%) | 64 (14%) | 77 (20%) | 62 (13%) | 54 (12%) | 72 (6%) | 117 (16%) | 111 (13%) | 70 (24%) | 59 (16%) |
| 1–2 | 97 (34%) | 153 (33%) | 185 (48%) | 268 (56%) | 157 (35%) | 510 (42%) | 343 (48%) | 387 (45%) | 111 (39%) | 156 (42%) |
| 3+ | 137 (48%) | 218 (47%) | 116 (31%) | 149 (31%) | 143 (32%) | 639 (52%) | 223 (31%) | 353 (4%) | 106 (37%) | 151 (41%) |
| Unknown | 6 (2%) | 27 (6%) | 4 (1%) | 0 (0%) | 93 (21%) | 0 (0%) | 34 (5%) | 1 (0.1%) | 0 (0%) | 2 (<1%) |
| Histology | ||||||||||
| Serous | 168 (59%) | n.a. | 234 (61%) | n.a. | 275 (62%) | n.a. | 254 (35%) | n.a. | 159 (55%) | n.a. |
| Endometrioid | 58 (20%) | n.a. | 58 (15%) | n.a. | 56 (13%) | n.a. | 129 (18%) | n.a. | 44 (15%) | n.a. |
| Mucinous | 9 (3%) | n.a. | 22 (6%) | n.a. | 43 (10%) | n.a. | 97 (14%) | n.a. | 24 (8%) | n.a. |
| Clear Cell | 18 (6%) | n.a. | 32 (8%) | n.a. | 33 (7%) | n.a. | 62 (9%) | n.a. | 22 (8%) | n.a. |
| Other/Unknown Epithelial* | 43 (15%) | n.a. | 67 (18%) | n.a. | 40 (9%) | n.a. | 175 (24%) | n.a. | 38 (13%) | n.a. |
Values are presented as number (percent) unless otherwise indicated.
Includes papillary, undifferentiated, mixed histologies, and other unknown epithelial adenocarcinomas.
Figure 2.
Significance of Ordinal Odds Ratios using Observed and Imputed Data
Table 3.
Cell Cycle SNPs and Risk of Ovarian Cancer
| OR (95% CI) |
|||||||
|---|---|---|---|---|---|---|---|
| Gene | SNP | Distance (bp) | MAY+NCO | SEA+MAL+STA | Combined | Combined Imputed: HapMap+NIEHS | Combined Imputed: HapMap Only |
| CDKN2C | rs3176459 | N.A. | -- | 1.00 (0.90 – 1.10) | 1.00 (0.90 – 1.10) | 0.98 (0.89 – 1.08) | 0.98 (0.89 – 1.08) |
| rs12855 | 2,846 | 1.02 (0.80 – 1.30) | 1.01 (0.86 – 1.19) | 1.01 (0.89 – 1.16) | 1.01 (0.88 – 1.15) | 1.01 (0.88 – 1.15) | |
|
| |||||||
| CDKN1A | rs1977172 | N.A. | 1.11 (0.90 – 1.38) | -- | 1.11 (0.90 – 1.38) | 1.12 (0.90 – 1.39) | 1.09 (0.91 – 1.31) |
| rs3829963 | 2,632 | 1.19 (0.97 – 1.46) | -- | 1.19 (0.97 – 1.46) | 1.17 (0.96 – 1.42) | 1.06 (0.92 – 1.22) | |
| rs733590 | 817 | 0.87 (0.75 – 1.02) | -- | 0.87 (0.75 – 1.02) | 0.98 (0.87 – 1.10) | 1.00 (0.93 – 1.08) | |
| rs762624 | 385 | -- | 1.09 (0.98 – 1.21) | 1.09 (0.98 – 1.21) | 1.08 (0.97 – 1.20) | 1.08 (0.97 – 1.20) | |
| rs2395655 | 108 | -- | 1.10 (1.00 – 1.21) | 1.10 (1.00 – 1.21) | 1.04 (0.95 – 1.13) | 1.03 (0.95 – 1.11) | |
| rs3176331 | 1,829 | -- | 1.02 (0.89 – 1.18) | 1.02 (0.89 – 1.18) | 1.00 (0.87 – 1.16) | 1.01 (0.88 – 1.17) | |
| rs3176336 | 1,291 | -- | 0.97 (0.88 – 1.07) | 0.97 (0.88 – 1.07) | 0.97 (0.88 – 1.07) | 0.97 (0.88 – 1.07) | |
| rs3176343 | 1,451 | -- | 1.05 (0.84 – 1.30) | 1.05 (0.84 – 1.30) | 1.04 (0.84 – 1.29) | 1.04 (0.84 – 1.29) | |
| rs1801270 | 1,704 | -- | 0.95 (0.78 – 1.15) | 0.95 (0.78 – 1.15) | 0.93 (0.77 – 1.13) | 0.95 (0.79 – 1.15) | |
| rs3176352 | 368 | -- | 1.08 (0.97 – 1.20) | 1.08 (0.97 – 1.20) | 1.08 (0.97 – 1.20) | 1.01 (0.92 – 1.11) | |
| rs1059234 | 1,258 | -- | 0.96 (0.79 – 1.16) | 0.96 (0.79 – 1.16) | 0.96 (0.79 – 1.17) | 0.96 (0.79 – 1.16) | |
| rs6457937 | 754 | -- | 0.92 (0.68 – 1.24) | 0.92 (0.68 – 1.24) | 0.91 (0.68 – 1.23) | 0.91 (0.68 – 1.23) | |
| rs3176359 | 391 | -- | 1.61 (0.72 – 3.56) | 1.61 (0.72 – 3.56) | 1.63 (0.73 – 3.61) | 1.63 (0.73 – 3.61) | |
| rs7767246 | 4,473 | 0.82 (0.69 – 0.99) | 1.11 (0.98 – 1.25) | 1.01 (0.92 – 1.12) | 1.00 (0.91 – 1.11) | 1.02 (0.92 – 1.12) | |
|
| |||||||
| CCND3 | rs2479726 | N.A. | 0.92 (0.79 – 1.08) | -- | 0.92 (0.79 – 1.08) | 0.93 (0.80 – 1.09) | 0.93 (0.80 – 1.09) |
| rs3828855 | 2,310 | 0.98 (0.76 – 1.25) | -- | 0.98 (0.76 – 1.25) | 0.97 (0.76 – 1.25) | 0.97 (0.76 – 1.25) | |
| rs3218114 | 1,946 | -- | 0.98 (0.86 – 1.10) | 0.98 (0.86 – 1.10) | 0.97 (0.86 – 1.10) | 0.97 (0.86 – 1.10) | |
| rs3218110 | 157 | -- | 1.09 (0.98 – 1.22) | 1.09 (0.98 – 1.22) | 1.08 (0.97 – 1.21) | 1.08 (0.97 – 1.21) | |
| rs3218108 | 146 | 0.93 (0.79 – 1.09) | -- | 0.93 (0.79 – 1.09) | 0.93 (0.79 – 1.10) | 0.93 (0.79 – 1.10) | |
| rs9529 | 352 | 0.91 (0.78 – 1.07) | 0.93 (0.84 – 1.03) | 0.93 (0.85 – 1.01) | 0.93 (0.85 – 1.01) | 0.93 (0.85 – 1.01) | |
| rs2479717 | 2,167 | -- | 1.00 (0.90 – 1.12) | 1.00 (0.90 – 1.12) | 1.02 (0.92 – 1.13) | 1.02 (0.92 – 1.13) | |
| rs3218092 | 1,487 | -- | 0.98 (0.87 – 1.11) | 0.98 (0.87 – 1.11) | 0.97 (0.86 – 1.10) | 0.97 (0.86 – 1.10) | |
| rs1410492 | 1,194 | -- | 1.06 (0.95 – 1.18) | 1.06 (0.95 – 1.18) | 1.06 (0.95 – 1.18) | 1.06 (0.95 – 1.18) | |
| rs3218086 | 2,209 | 1.32 (1.10 – 1.58) | -- | 1.32 (1.10 – 1.58) | 1.31 (1.09 – 1.57) | 1.31 (1.09 – 1.57) | |
| rs3218085 | 115 | 1.17 (0.63 – 2.17) | -- | 1.17 (0.63 – 2.17) | 1.18 (0.63 – 2.20) | 1.18 (0.63 – 2.20) | |
| rs9381100 | 1,006 | 0.90 (0.76 – 1.06) | -- | 0.90 (0.76 – 1.06) | 0.89 (0.75 – 1.05) | 0.89 (0.75 – 1.05) | |
|
| |||||||
| CDKN2A-B | rs3731257 | N.A. | -- | 0.89 (0.80 – 1.00) | 0.89 (0.80 – 1.00) | 0.89 (0.80 – 1.00) | 0.93 (0.85 – 1.03) |
| rs3088440 | 1,938 | -- | 1.08 (0.91 – 1.28) | 1.08 (0.91 – 1.28) | 1.00 (0.86 – 1.16) | 1.07 (0.91 – 1.26) | |
| rs11515 | 40 | -- | 1.07 (0.94 – 1.23) | 1.07 (0.94 – 1.23) | 1.06 (0.92 – 1.21) | 0.98 (0.87 – 1.10) | |
| rs3731249 | 2,717 | 0.92 (0.62 – 1.35) | 0.89 (0.67 – 1.19) | 0.91 (0.72 – 1.14) | 0.90 (0.72 – 1.14) | 0.90 (0.72 – 1.14) | |
| rs3731239 | 3,302 | -- | 1.05 (0.95 – 1.16) | 1.05 (0.95 – 1.16) | 1.08 (0.99 – 1.18) | 1.11 (1.02 – 1.20) | |
| rs2811709 | 5,933 | 0.80 (0.64 – 0.99) | -- | 0.80 (0.64 – 0.99) | 0.79 (0.64 – 0.99) | 0.96 (0.85 – 1.08) | |
| rs4074785 | 1,432 | -- | 1.09 (0.92 – 1.28) | 1.09 (0.92 – 1.28) | 1.08 (0.92 – 1.28) | 1.10 (0.93 – 1.29) | |
| rs3731222 | 2,331 | -- | 0.96 (0.83 – 1.10) | 0.96 (0.83 – 1.10) | 0.96 (0.84 – 1.11) | 0.92 (0.82 – 1.04) | |
| rs3731211 | 2,933 | -- | 0.98 (0.88 – 1.09) | 0.98 (0.88 – 1.09) | 0.97 (0.88 – 1.08) | 0.92 (0.84 – 1.00) | |
| rs7036656 | 3,610 | 0.79 (0.67 – 0.93) | -- | 0.79 (0.67 – 0.93) | 0.79 (0.67 – 0.92) | 0.94 (0.86 – 1.02) | |
| rs3731197 | 914 | -- | 1.03 (0.93 – 1.13) | 1.03 (0.93 – 1.13) | 1.02 (0.93 – 1.11) | 1.03 (0.93 – 1.13) | |
| rs3218020 | 6,501 | -- | 0.99 (0.89 – 1.10) | 0.99 (0.89 – 1.10) | 0.98 (0.89 – 1.07) | 1.00 (0.91 – 1.09) | |
| rs2811712 | 163 | -- | 1.05 (0.90 – 1.22) | 1.05 (0.90 – 1.22) | 1.04 (0.89 – 1.21) | 0.95 (0.84 – 1.08) | |
| rs3218012 | 625 | -- | 1.00 (0.91 – 1.10) | 1.00 (0.91 – 1.10) | 0.98 (0.90 – 1.06) | 0.98 (0.89 – 1.08) | |
| rs3218009 | 97 | 1.19 (0.96 – 1.46) | 0.99 (0.86 – 1.14) | 1.05 (0.93 – 1.17) | 1.04 (0.93 – 1.17) | 1.04 (0.93 – 1.17) | |
| rs3218005 | 1,490 | -- | 1.06 (0.91 – 1.25) | 1.06 (0.91 – 1.25) | 1.02 (0.88 – 1.18) | 0.95 (0.84 – 1.09) | |
| rs3217992 | 2,976 | -- | 0.97 (0.88 – 1.08) | 0.97 (0.88 – 1.08) | 0.96 (0.87 – 1.05) | 0.98 (0.90 – 1.06) | |
| rs1063192 | 144 | 1.09 (0.94 – 1.25) | 0.98 (0.89 – 1.08) | 1.01 (0.94 – 1.10) | 1.01 (0.93 – 1.09) | 1.02 (0.94 – 1.10) | |
| rs3217986 | 1,963 | -- | 1.10 (0.93 – 1.29) | 1.10 (0.93 – 1.29) | 1.08 (0.92 – 1.27) | 1.09 (0.92 – 1.28) | |
| rs2069418 | 4,368 | 1.11 (0.97 – 1.28) | -- | 1.11 (0.97 – 1.28) | 1.13 (0.98 – 1.31) | 1.13 (0.98 – 1.31) | |
| rs575427 | 1,779 | 0.84 (0.66 – 1.07) | -- | 0.84 (0.66 – 1.07) | 0.83 (0.65 – 1.07) | 0.91 (0.79 – 1.04) | |
| rs13298881 | 574 | 0.94 (0.75 – 1.19) | -- | 0.94 (0.75 – 1.19) | 0.92 (0.73 – 1.17) | 1.09 (0.95 – 1.25) | |
| rs10811640 | 1,360 | 0.97 (0.84 – 1.12) | -- | 0.97 (0.84 – 1.12) | 0.96 (0.83 – 1.10) | 0.99 (0.91 – 1.07) | |
|
| |||||||
| CCND1 | rs602652 | N.A. | -- | 1.11 (1.01 – 1.23) | 1.11 (1.01 – 1.23) | 1.08 (1.00 – 1.17) | 1.14 (1.04 – 1.25) |
| rs3862792 | 214 | -- | 0.79 (0.58 – 1.09) | 0.79 (0.58 – 1.09) | 0.79 (0.58 – 1.09) | 0.79 (0.58 – 1.09) | |
| rs603965 | 54 | 1.01 (0.88 – 1.17) | 1.12 (1.02 – 1.23) | 1.09 (1.00 – 1.17) | 1.08 (1.00 – 1.17) | 1.08 (1.00 – 1.17) | |
| rs3212879 | 571 | -- | 0.91 (0.83 – 1.00) | 0.91 (0.83 – 1.00) | 0.93 (0.86 – 1.01) | 0.90 (0.82 – 0.99) | |
| rs649392 | 1,312 | 1.00 (0.87 – 1.14) | -- | 1.00 (0.87 – 1.14) | 0.93 (0.86 – 1.01) | 0.92 (0.85 – 1.00) | |
| rs3212891 | 714 | -- | 0.90 (0.82 – 1.00) | 0.90 (0.82 – 1.00) | 0.93 (0.86 – 1.00) | 0.90 (0.82 – 0.99) | |
| rs678653 | 1,230 | 1.01 (0.87 – 1.17) | 0.96 (0.87 – 1.06) | 0.97 (0.90 – 1.06) | 0.97 (0.90 – 1.06) | 0.97 (0.90 – 1.06) | |
| rs7178 | 2,293 | 0.83 (0.64 – 1.09) | 1.21 (1.02 – 1.45) | 1.08 (0.93 – 1.24) | 1.07 (0.93 – 1.24) | 1.07 (0.93 – 1.24) | |
| rs11603541 | 3,343 | 0.90 (0.72 – 1.13) | -- | 0.90 (0.72 – 1.13) | 0.90 (0.72 – 1.14) | 0.91 (0.72 – 1.14) | |
|
| |||||||
| CCND2 | rs1049606 | N.A. | 1.06 (0.92 – 1.22) | -- | 1.06 (0.92 – 1.22) | 1.04 (0.94 – 1.15) | 1.08 (0.95 – 1.22) |
| rs3217795 | 3,028 | -- | 0.95 (0.80 – 1.12) | 0.95 (0.80 – 1.12) | 0.95 (0.80 – 1.12) | 0.95 (0.80 – 1.12) | |
| rs3217805 | 2,020 | -- | 0.96 (0.87 – 1.06) | 0.96 (0.87 – 1.06) | 1.01 (0.92 – 1.11) | 1.01 (0.92 – 1.11) | |
| rs3217820 | 2,288 | -- | 1.03 (0.93 – 1.14) | 1.03 (0.93 – 1.14) | 0.95 (0.87 – 1.05) | 0.95 (0.87 – 1.05) | |
| rs3217852 | 7,394 | -- | 0.97 (0.87 – 1.09) | 0.97 (0.87 – 1.09) | 0.98 (0.88 – 1.10) | 0.99 (0.89 – 1.10) | |
| rs3217862 | 1,321 | -- | 0.95 (0.83 – 1.08) | 0.95 (0.83 – 1.08) | 0.95 (0.84 – 1.08) | 0.95 (0.83 – 1.07) | |
| rs3217863 | 409 | -- | 1.05 (0.88 – 1.26) | 1.05 (0.88 – 1.26) | 1.05 (0.89 – 1.25) | 1.08 (0.91 – 1.28) | |
| rs3217869 | 474 | -- | 1.02 (0.93 – 1.13) | 1.02 (0.93 – 1.13) | 1.01 (0.92 – 1.10) | 1.02 (0.93 – 1.10) | |
| rs3217901 | 5,419 | -- | 1.03 (0.93 – 1.13) | 1.03 (0.93 – 1.13) | 1.01 (0.93 – 1.10) | 1.01 (0.93 – 1.10) | |
| rs3217906 | 415 | -- | 1.03 (0.92 – 1.14) | 1.03 (0.92 – 1.14) | 1.02 (0.92 – 1.14) | 1.02 (0.92 – 1.13) | |
| rs3217916 | 2,869 | -- | 0.92 (0.83 – 1.02) | 0.92 (0.83 – 1.02) | 0.94 (0.85 – 1.04) | 0.95 (0.87 – 1.03) | |
| rs3217925 | 2,966 | -- | 0.91 (0.81 – 1.01) | 0.91 (0.81 – 1.01) | 0.92 (0.83 – 1.01) | 0.94 (0.85 – 1.02) | |
| rs3217926 | 44 | 1.03 (0.89 – 1.20) | 0.98 (0.89 – 1.08) | 0.99 (0.92 – 1.08) | 0.99 (0.91 – 1.07) | 0.99 (0.91 – 1.07) | |
| rs1049612 | 1,079 | 1.04 (0.90 – 1.20) | -- | 1.04 (0.90 – 1.20) | 0.99 (0.88 – 1.12) | 1.03 (0.93 – 1.13) | |
| rs3217933 | 238 | 0.95 (0.80 – 1.12) | 1.09 (0.97 – 1.21) | 1.04 (0.95 – 1.14) | 1.04 (0.95 – 1.14) | 1.04 (0.95 – 1.14) | |
| rs3217936 | 1,952 | -- | 0.91 (0.82 – 1.01) | 0.91 (0.82 – 1.01) | 0.94 (0.85 – 1.02) | 0.94 (0.87 – 1.03) | |
|
| |||||||
| CDKN1B | rs3759216 | N.A. | 1.05 (0.91 – 1.21) | 1.02 (0.93 – 1.13) | 1.03 (0.95 – 1.12) | 1.03 (0.95 – 1.11) | 1.02 (0.94 – 1.10) |
| rs3759217 | 366 | 1.06 (0.86 – 1.31) | 1.14 (0.99 – 1.32) | 1.11 (0.99 – 1.26) | 1.11 (0.99 – 1.25) | 1.12 (0.99 – 1.26) | |
| rs34330 | 2,243 | 0.94 (0.80 – 1.11) | 0.91 (0.81 – 1.02) | 0.92 (0.84 – 1.01) | 0.92 (0.84 – 1.01) | 0.92 (0.84 – 1.01) | |
| rs2066827 | 404 | -- | 0.84 (0.75 – 0.94) | 0.84 (0.75 – 0.94) | 0.84 (0.76 – 0.94) | 0.85 (0.77 – 0.94) | |
| rs34329 | 2,134 | 1.12 (0.96 – 1.30) | 1.00 (0.90 – 1.11) | 1.04 (0.95 – 1.13) | 1.03 (0.95 – 1.13) | 1.03 (0.95 – 1.12) | |
| rs3093736 | 68 | 1.07 (0.72 – 1.59) | 0.97 (0.75 – 1.26) | 1.00 (0.80 – 1.24) | 0.99 (0.80 – 1.23) | 0.99 (0.80 – 1.23) | |
| rs7330 | 1,616 | 1.05 (0.91 – 1.21) | 1.01 (0.92 – 1.11) | 1.02 (0.94 – 1.10) | 1.02 (0.94 – 1.10) | 1.02 (0.94 – 1.11) | |
| rs1420023 | 1,194 | 0.84 (0.68 – 1.05) | 0.98 (0.84 – 1.14) | 0.94 (0.83 – 1.06) | 0.93 (0.82 – 1.06) | 0.93 (0.82 – 1.06) | |
| rs34322 | 3,459 | 0.98 (0.85 – 1.13) | -- | 0.98 (0.85 – 1.13) | 0.97 (0.84 – 1.12) | 1.02 (0.94 – 1.11) | |
|
| |||||||
| CDK2 | rs2069391 | N.A. | 1.16 (0.89 – 1.52) | -- | 1.16 (0.89 – 1.52) | 1.15 (0.88 – 1.50) | 1.21 (1.02 – 1.43) |
| rs2069408 | 4,443 | 0.94 (0.80 – 1.09) | 0.94 (0.84 – 1.04) | 0.94 (0.86 – 1.02) | 0.93 (0.85 – 1.01) | 0.93 (0.85 – 1.01) | |
| rs2069414 | 1,378 | 1.55 (1.18 – 2.04) | -- | 1.55 (1.18 – 2.04) | 1.58 (1.20 – 2.09) | 1.58 (1.20 – 2.09) | |
| rs1045435 | 461 | 1.17 (0.90 – 1.53) | 1.14 (0.97 – 1.34) | 1.15 (1.00 – 1.32) | 1.14 (1.00 – 1.31) | 1.14 (1.00 – 1.31) | |
| rs11171710 | 1,918 | 1.06 (0.92 – 1.22) | -- | 1.06 (0.92 – 1.22) | 1.07 (0.93 – 1.23) | 1.05 (0.95 – 1.17) | |
| rs17528736 | 440 | 1.15 (0.78 – 1.70) | -- | 1.15 (0.78 – 1.70) | 1.13 (0.77 – 1.67) | 1.23 (1.01 – 1.48) | |
| rs773108 | 1,393 | 0.93 (0.80 – 1.08) | -- | 0.93 (0.80 – 1.08) | 0.92 (0.79 – 1.08) | 0.92 (0.85 – 1.00) | |
|
| |||||||
| CDK4 | rs2069506 | N.A. | -- | 0.99 (0.89 – 1.09) | 0.99 (0.89 – 1.09) | 1.00 (0.92 – 1.08) | 0.99 (0.90 – 1.09) |
| rs2069502 | 1,811 | 1.02 (0.88 – 1.18) | -- | 1.02 (0.88 – 1.18) | 1.00 (0.92 – 1.08) | 1.00 (0.89 – 1.12) | |
| rs2270777 | 491 | 0.90 (0.78 – 1.03) | 1.03 (0.93 – 1.13) | 0.99 (0.91 – 1.07) | 0.99 (0.91 – 1.07) | 0.99 (0.91 – 1.07) | |
| rs2072052 | 1,563 | 1.03 (0.89 – 1.19) | -- | 1.03 (0.89 – 1.19) | 1.00 (0.92 – 1.08) | 1.03 (0.89 – 1.20) | |
|
| |||||||
| RB1 | rs1981434 | N.A. | -- | 0.97 (0.87 – 1.08) | 0.97 (0.87 – 1.08) | 0.97 (0.88 – 1.08) | 0.98 (0.90 – 1.07) |
| rs2854345 | 10,082 | -- | 1.00 (0.89 – 1.12) | 1.00 (0.89 – 1.12) | 1.00 (0.88 – 1.12) | 1.00 (0.88 – 1.12) | |
| rs4151467 | 28,687 | 1.07 (0.78 – 1.45) | -- | 1.07 (0.78 – 1.45) | 1.09 (0.80 – 1.48) | 1.09 (0.80 – 1.48) | |
| rs7329938 | 12,056 | 1.08 (0.87 – 1.33) | -- | 1.08 (0.87 – 1.33) | 1.08 (0.88 – 1.33) | 1.09 (0.88 – 1.35) | |
| rs4151510 | 13,196 | 1.08 (0.88 – 1.34) | -- | 1.08 (0.88 – 1.34) | 1.08 (0.95 – 1.24) | 1.10 (0.89 – 1.36) | |
| rs399413 | 3,394 | -- | 1.02 (0.91 – 1.14) | 1.02 (0.91 – 1.14) | 0.96 (0.88 – 1.06) | 1.00 (0.91 – 1.09) | |
| rs4151540 | 7,091 | -- | 0.96 (0.86 – 1.07) | 0.96 (0.86 – 1.07) | 0.94 (0.85 – 1.03) | 0.96 (0.86 – 1.07) | |
| rs9568036 | 16,276 | 1.03 (0.89 – 1.19) | -- | 1.03 (0.89 – 1.19) | 1.03 (0.89 – 1.19) | 1.00 (0.91 – 1.10) | |
| rs198604 | 12,127 | 1.00 (0.84 – 1.17) | -- | 1.00 (0.84 – 1.17) | 1.00 (0.85 – 1.18) | 0.98 (0.89 – 1.07) | |
| rs4151551 | 1,376 | 1.10 (0.85 – 1.41) | 1.06 (0.90 – 1.25) | 1.07 (0.93 – 1.23) | 1.06 (0.93 – 1.22) | 1.06 (0.93 – 1.22) | |
| rs2854344 | 12,254 | 0.97 (0.73 – 1.28) | 0.81 (0.66 – 0.99) | 0.87 (0.74 – 1.02) | 0.88 (0.75 – 1.04) | 0.88 (0.75 – 1.04) | |
| rs425834 | 14,800 | -- | 1.04 (0.80 – 1.36) | 1.04 (0.80 – 1.36) | 1.05 (0.81 – 1.37) | 1.05 (0.81 – 1.37) | |
| rs4151611 | 35,438 | -- | 0.91 (0.72 – 1.14) | 0.91 (0.72 – 1.14) | 0.90 (0.71 – 1.13) | 0.89 (0.71 – 1.13) | |
| rs4151620 | 1,128 | -- | 1.00 (0.85 – 1.17) | 1.00 (0.85 – 1.17) | 1.04 (0.90 – 1.20) | 1.02 (0.87 – 1.20) | |
| rs3092904 | 2,422 | -- | 0.98 (0.87 – 1.10) | 0.98 (0.87 – 1.10) | 0.96 (0.86 – 1.07) | 0.97 (0.89 – 1.07) | |
| rs4151636 | 5,253 | -- | 0.93 (0.74 – 1.17) | 0.93 (0.74 – 1.17) | 0.92 (0.73 – 1.16) | 0.92 (0.73 – 1.15) | |
| rs990814 | 2,843 | 1.00 (0.85 – 1.17) | -- | 1.00 (0.85 – 1.17) | 1.01 (0.86 – 1.18) | 0.98 (0.90 – 1.08) | |
|
| |||||||
| CDKN2D | rs3218222 | N.A. | -- | 1.02 (0.91 – 1.14) | 1.02 (0.91 – 1.14) | 1.01 (0.91 – 1.12) | 1.01 (0.91 – 1.12) |
| rs1465702 | 1,951 | -- | 1.10 (0.88 – 1.39) | 1.10 (0.88 – 1.39) | 1.05 (0.86 – 1.29) | 1.05 (0.86 – 1.29) | |
| rs1465701 | 210 | 1.15 (0.97 – 1.35) | -- | 1.15 (0.97 – 1.35) | 1.16 (0.98 – 1.36) | 1.16 (0.98 – 1.36) | |
| rs17677316 | 1,399 | 0.90 (0.76 – 1.06) | -- | 0.90 (0.76 – 1.06) | 0.89 (0.75 – 1.05) | 0.89 (0.75 – 1.05) | |
|
| |||||||
| CCNE1 | rs997669 | N.A. | 0.97 (0.84 – 1.13) | 1.07 (0.97 – 1.18) | 1.04 (0.96 – 1.13) | 1.04 (0.96 – 1.13) | 1.04 (0.96 – 1.13) |
| rs3218036 | 1,201 | -- | 1.11 (1.00 – 1.23) | 1.11 (1.00 – 1.23) | 1.07 (0.98 – 1.17) | 1.12 (1.01 – 1.24) | |
| rs3218038 | 211 | -- | 1.03 (0.80 – 1.33) | 1.03 (0.80 – 1.33) | 1.02 (0.84 – 1.24) | 1.02 (0.79 – 1.32) | |
| rs1406 | 9,217 | 0.99 (0.84 – 1.18) | -- | 0.99 (0.84 – 1.18) | 0.98 (0.89 – 1.08) | 1.00 (0.92 – 1.10) | |
| rs3218076 | 158 | -- | 1.02 (0.91 – 1.13) | 1.02 (0.91 – 1.13) | 1.01 (0.92 – 1.11) | 1.00 (0.92 – 1.10) | |
Bold indicates p < 0.05
On the whole, imputed results did not differ from results of combined analysis of observed data; p-values increased by a mean of 0.001 using HapMap+NIEHS and decreased by a mean of 0.001 using HapMap only. For SNPs genotyped in both collaborations, the impact of imputation on results was minimal as only those participants that failed genotyping were impacted. For SNPs genotyped in only one collaboration, use of only HapMap led to slightly greater discrepancy between observed and imputed p-values, more often leading to greater significance, than use of HapMap+NIEHS which varied p-values to a lesser degree (Supplemental Figure 1). For SNPs selected using NIEHS and HapMap, genotyped in SEA+MAL+STA, and imputed for MAY+NCO samples, p-values from imputation-based analysis increased by a mean of 0.004 using NIEHS+HapMap and a mean of 0.01 using HapMap only. For SNPs selected using HapMap, genotyped in MAY+NCO, and imputed for SEA+MAL+STA samples, p-values decreased by a mean of 0.01 using NIEHS+HapMap and a mean of 0.03 using HapMap only. Thus, the largest overall difference in results occurred when imputation took place on the largest number of samples (SEA+MAL+STA), and generally results became more significant. For example, the OR for CDK2 rs2069414 increased from 1.55 (95% CI 1.18–2.04 p = 0.002) in MAY+NCO to an imputed OR of 1.58 (95% CI 1.20–2.09 p = 0.001), and the OR for CCND1 rs649392 decreased from 1.00 (95% CI 0.87–1.14 p = 0.95) in MAY+NCO to an imputed OR of 0.92 (95% CI 0.85–1.00 p = 0.05) (Table 3). In addition, two correlated CDK2 SNPs, rs2069391 and rs17528736 (MAY+NCO controls, r2=0.38; CEU r2=0.56), became significant at p < 0.05 when imputed with HapMap data; the OR for rs17528736 increased from 1.15 (95% CI 0.78–1.70 p = 0.48) in MAY+NCO to an imputed OR of 1.23 (95% CI 1.01–1.48 p = 0.04). Both of these SNPs are uncorrelated with CDK2 rs2069414 (r2’s < 0.01 in MAY+NCO and HapMap) indicative of independent associations. These results suggest that additional risk alleles in the SEA+MAL+STA population were correlated with genotyped SNPs in MAY+NCO, and imputation increased power to detect associations.
Novel SNPs of interest also came to light with imputation of MAY+NCO data, notably CDKN2A-CDKN2B rs3731239 which increased from an OR of 1.05 (95% CI 0.95–1.16; p = 0.31) to an OR of 1.11 (95% CI 1.02–1.20 p = 0.02) using HapMap and CCND1 rs602652 which increased from an OR of 1.11 (95% CI 1.01–1.23 p = 0.03) to an OR of 1.14 (95% CI 1.04–1.25 p = 0.001) using HapMap (Figure 2). Additional SNPs in CCND1, CDKN2A-CDKN2B, CDK2, and CCNE1 became more significant with HapMap-based imputation in MAY+NCO although point estimates remained similar (Table 3, Figure 2). As above, these results suggest that additional risk alleles in the MAY+NCO population were correlated with genotyped SNPs in SEA+MAL+STA and imputation increased power to detect associations.
CCND1 results warrant particular attention. LD patterns are similar across populations (Supplemental Figure 2). Using NIEHS SNPs data (the study with maximal coverage of the nine SNPs genotyped by either MAY+NCO or SEA+MAL+STA) there was strong correlation between rs3212879, rs649392, and rs3212891 (r2 values > 0.86), rs602652 (r2 values > 0.75), and rs603965 (r2 values > 0.64). Combined analysis of observed study participant data yielded three p-values < 0.05, two resulted from SEA+MAL+STA data alone and one used data from MAY+NCO as well. Use of imputation with study participant and HapMap data increased the number of significant results from three to five p-values < 0.05, even though HapMap did not genotype two of these SNPs (rs602652 and insertion/deletion polymorphism rs321879, Supplemental Table 1). These findings remind us that the underlying haplotype structure used to impute genotypes relies on all available data, here, a total of 20 SNPS with data from MAY+NCO, SEA+MAL+STA, or HapMap. An additional 39 SNPs were covered by NIEHS SNPs; inclusion of these data attenuated ORs and resulted in only one p-value < 0.05 (rs603965). Whether these results are closer to the truth, given that NIEHS SNPs participants were only presumed to be White non-Hispanic, remains to be verified by additional genotyping and fine-mapping.
In summary, our imputation-based analysis of SNPs in key cell cycle genes did not reveal novel SNPs worthy of follow-up in CDKN2C, CDKN1A, CCND3, CCND2, CDKN1B, CDK4, RB1, or CDKN2D, but suggested a handful of SNPs in CDKN2A-CDKN2B (rs3731239), CCND1 (the correlated SNPs rs602652, rs3212879, rs649392, and rs3212891), CDK2 (rs2069414 and the correlated SNPs rs17528736 and rs2069391), and CCNE1 (rs3218036) which merit genotyping in the unassayed sample population.
Discussion
Here, we report on analysis of 2,120 invasive ovarian cancer cases and 3,382 controls of White non-Hispanic ethnicity successfully genotyped on up to 122 SNPs but only 24 SNPs with maximally-genotyped participants. Using data on 901 regional SNPs genotyped in study, HapMap or NIEHS SNPs participants, we applied a Hidden Markov Model to estimate underlying haplotypes and impute missing genotypes among study participants. Analysis of imputation-based data revealed additional evidence of association with risk of ovarian cancer for SNPs in several genes. In particular, we find that additional genotyping is warranted in the genes encoding p16 and p15 (CDKN2A-CDKN2B), shown to be overexpressed and methylated in ovarian cancer, respectively (1, 41); cyclin D1 (CCND1), shown to be abnormally expressed in ovarian cancer (42); CDK 2 (CDK2), shown to inhibit G1 arrest in ovarian cancer cells (43); and cyclin E1(CCNE1), which is overexpressed in ovarian cancer (15). Several of the SNPs associated with ovarian cancer risk here have been studied in relation to risk of breast, prostate, lung, bladder, and oral cancers (5–11). Of particular interest is a SNP in the region of CDKN2A-CDKN1B rs3731239 that was found to be associated with decreased breast cancer risk (6) and had showed a protective association in the current analysis.
Our combined, imputation-based analysis strengthens existing interrogations in which tagging SNPs are typed in one collaboration and the most suggestive single SNPs are brought to a consortium for replication. For example, based on results from the SEA+MAL+STA collaboration alone, four of the currently-assessed SNPs were genotyped in over 3,500 cases and 5,700 controls by the Ovarian Cancer Association Consortium (CCND1 rs7178 and rs603965, CDKN1B rs2066827, and CDKN2A-CDKN2B rs3731257), and CDKN1B rs2066827 and CDKN2A-CDKN2B rs3731257 remained associated (17). More recently, one of the RB1 SNPs genotyped in SEA+MAL+STA (rs2854344) was assessed by the Ovarian Cancer Association Consortium using over 4,600 cases and 8,100 controls and found to replicate, despite null results in MAY+NCO; another SNP in CDKN2A (rs2811712) did not replicate (18). Combining multiple tagging SNP studies using imputation when necessary will assist preliminary candidate gene studies by (a) improving power of “phase I” analyses and (b) highlighting specific SNPs to do “fill-in” genotyping prior to consortium genotyping. Here, data suggest additional SNPs to interrogate in MAY+NCO or SEA+MAL+STA study populations for maximal discriminatory power prior to selection of SNPs in future large-scale genotyping efforts.
These analyses have the potential strengths of theoretically improved sample size at no additional genotyping cost. However, several caveats are warranted. As with non-imputation-based analysis, the benefit of larger sample size may increase the potential for study heterogeneity. In addition, analyses make similar assumptions as in tagging SNP selection including that the populations used to estimate underlying haplotypes are similar to study populations of interest, an assumption which is not always testable. In the current analysis, ethnicity of NIEHS SNPs participants was genetically inferred, which may be particularly problematic. Here, we also assumed that linkage disequilibrium is similar among cases and controls and across all studies. Analyses also assume that the densely-typed population is of sufficient sample size. Violation of these assumptions can impair inference of results. Finally, it is worth noting that merging genotype data across multiple studies and publicly-available data requires great effort to harmonize alleles; a conservative approach excluding genotypes which are not easily combined is recommended.
We make modest suggestions for future imputation-based analysis. Imputation of genotypes has typically relied on single imputation; however, this approach ignores the variation in estimation due to the imputation. An accepted alternative is the use of multiple imputation, in which a number of “imputed” datasets are created and then analyzed using standard statistical methods and models (40, 44–47) allowing one to estimate the amount of variation attributable to the imputation procedure. In general, the imputation variance components for our study were small. The tool that we used (fastPHASE) only provides the “most likely genotype” as the imputed value. Multiple imputation based on this most likely genotype may not capture the total amount of variation due to imputation (i.e., if the posterior probability is large for a particular genotype, one wouldn’t see as much variation in most likely genotype due to imputation). Allowing for imputation of a quantitative value, such as an allele “dosage” variable with possible values ranging from 0 to 2, may better capture the variation due to imputation. In the current analyses, use of HapMap data only (without NIEHS SNPs data) strengthened many associations, suggesting that either (a) the NIEHS SNPs samples were not appropriate to use as reference (if associations are true) or (b) the NIEHS SNPs samples provided increased power to discriminate true from false associations (if associations are false). Additional genotyping is underway to examine the accuracy of imputed genotypes and the consistency of ovarian cancer association signals in CDKN2A-CDKN2B, CCND1, CDK2, and CCNE1 seen with imputed data. Although developed primarily for genome-wide association studies, we conclude that pooling genotypes and using imputation techniques may also strengthen our understanding of key candidate ovarian cancer pathways.
Acknowledgments
Support was provided by the Mayo Foundation, the Fraternal Order of Eagles, the Minnesota Ovarian Cancer Alliance, R01-CA88868, R01-CA122443, R01-CA76016, and Department of Defense DAMD17-02-1-0666. SJR is funded by the Mermaid component of the Eve Appeal, HS is funded by a grant from WellBeing of Women, PDPP is a Senior Clinical Research Fellow of Cancer Research UK. Part of this research was funded by a Cancer Research UK project grant (no. C8804/A7058). Part of this work was undertaken at UCLH/UCL who received a proportion of funding from the Department of Health’s NIHR Biomedical Research Centres funding scheme.
References
- 1.Nam EJ, Kim YT. Alteration of cell-cycle regulation in epithelial ovarian cancer. Int J Gynecol Cancer. 2008 doi: 10.1111/j.1525-1438.2008.01191.x. epub. [DOI] [PubMed] [Google Scholar]
- 2.Rakoff-Nahoum S. Why cancer and inflammation? The Yale journal of biology and medicine. 2006;79:123–30. [PMC free article] [PubMed] [Google Scholar]
- 3.Cooper GM, Hausman RE. The Cell: A Molecular Approach. Sunderlander: Sinauer Associates, Inc.; 2007. [Google Scholar]
- 4.Sherr CJ, Roberts JM. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 1999;13:1501–12. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- 5.Pharoah PD, Tyrer J, Dunning AM, Easton DF, Ponder BA. Association between common variation in 120 candidate genes and breast cancer risk. PLoS Genet. 2007;3:e42. doi: 10.1371/journal.pgen.0030042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Driver KE, Song H, Lesueur F, et al. Association of single-nucleotide polymorphisms in the cell cycle genes with breast cancer in the British population. Carcinogenesis. 2008;29:333–41. doi: 10.1093/carcin/bgm284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang BL, Zheng SL, Isaacs SD, et al. A polymorphism in the CDKN1B gene is associated with increased risk of hereditary prostate cancer. Cancer Res. 2004;64:1997–9. doi: 10.1158/0008-5472.can-03-2340. [DOI] [PubMed] [Google Scholar]
- 8.Hosgood HD, 3rd, Menashe I, Shen M, et al. Pathway-based evaluation of 380 candidate genes and lung cancer susceptibility suggests the importance of the cell cycle pathway. Carcinogenesis. 2008;29:1938–43. doi: 10.1093/carcin/bgn178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ye Y, Yang H, Grossman HB, Dinney C, Wu X, Gu J. Genetic variants in cell cycle control pathway confer susceptibility to bladder cancer. Cancer. 2008;112:2467–74. doi: 10.1002/cncr.23472. [DOI] [PubMed] [Google Scholar]
- 10.Wu X, Gu J, Grossman HB, et al. Bladder cancer predisposition: a multigenic approach to DNA- repair and cell-cycle-control genes. Am J Hum Genet. 2006;78:464–79. doi: 10.1086/500848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang M, Spitz MR, Gu J, et al. Cyclin D1 gene polymorphism as a risk factor for oral premalignant lesions. Carcinogenesis. 2006;27:2034–7. doi: 10.1093/carcin/bgl048. [DOI] [PubMed] [Google Scholar]
- 12.Hazra A, Chanock S, Giovannucci E, et al. Large-scale evaluation of genetic variants in candidate genes for colorectal cancer risk in the Nurses’ Health Study and the Health Professionals’ Follow-up Study. Cancer Epidemiol Biomarkers Prev. 2008;17:311–9. doi: 10.1158/1055-9965.EPI-07-0195. [DOI] [PubMed] [Google Scholar]
- 13.Milde-Langosch K, Ocon E, Becker G, Loning T. p16/MTS1 inactivation in ovarian carcinomas: high frequency of reduced protein expression associated with hyper-methylation or mutation in endometrioid and mucinous tumors. Int J Cancer. 1998;79:61–5. doi: 10.1002/(sici)1097-0215(19980220)79:1<61::aid-ijc12>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 14.Kudoh K, Ichikawa Y, Yoshida S, et al. Inactivation of p16/CDKN2 and p15/MTS2 is associated with prognosis and response to chemotherapy in ovarian cancer. Int J Cancer. 2002;99:579–82. doi: 10.1002/ijc.10331. [DOI] [PubMed] [Google Scholar]
- 15.Schildkraut JM, Moorman PG, Bland AE, et al. Cyclin E overexpression in epithelial ovarian cancer characterizes an etiologic subgroup. Cancer Epidemiol Biomarkers Prev. 2008;17:585–93. doi: 10.1158/1055-9965.EPI-07-0596. [DOI] [PubMed] [Google Scholar]
- 16.Li SB, Schwartz PE, Lee WH, Yang-Feng TL. Allele loss at the retinoblastoma locus in human ovarian cancer. J Natl Cancer Inst. 1991;83:637–40. doi: 10.1093/jnci/83.9.637. [DOI] [PubMed] [Google Scholar]
- 17.Gayther SA, Song H, Ramus SJ, et al. Tagging single nucleotide polymorphisms in cell cycle control genes and susceptibility to invasive epithelial ovarian cancer. Cancer Res. 2007;67:3027–35. doi: 10.1158/0008-5472.CAN-06-3261. [DOI] [PubMed] [Google Scholar]
- 18.Ramus SJ, Vierkant RA, Johnatty SE, et al. Consortium analysis of 7 candidate SNPs for ovarian cancer. Intl J Cancer. 2008;123:380–8. doi: 10.1002/ijc.23448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goode EL, Fridley BL, Sun Z, et al. Comparison of tagging single-nucleotide polymorphism methods in association analyses. BMC Proc. 2007;1 (Suppl 1):S6. doi: 10.1186/1753-6561-1-s1-s6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun YV, Kardia SL. Imputing missing genotypic data of single-nucleotide polymorphisms using neural networks. Eur J Hum Genet. 2008;16:487–95. doi: 10.1038/sj.ejhg.5201988. [DOI] [PubMed] [Google Scholar]
- 21.Foulkes AS, Yucel R, Reilly MP. Mixed modeling and multiple imputation for unobservable genotype clusters. Stat Med. 2008;27:2784–801. doi: 10.1002/sim.3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Servin B, Stephens M. Imputation-based analysis of association studies: candidate regions and quantitative traits. PLoS Genet. 2007;3:e114. doi: 10.1371/journal.pgen.0030114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roberts A, McMillan L, Wang W, Parker J, Rusyn I, Threadgill D. Inferring missing genotypes in large SNP panels using fast nearest-neighbor searches over sliding windows. Bioinformatics. 2007;23:i401–7. doi: 10.1093/bioinformatics/btm220. [DOI] [PubMed] [Google Scholar]
- 24.Dai JY, Ruczinski I, LeBlanc M, Kooperberg C. Imputation methods to improve inference in SNP association studies. Genet Epidemiol. 2006;30:690–702. doi: 10.1002/gepi.20180. [DOI] [PubMed] [Google Scholar]
- 25.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–13. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 26.Yu Z, Schaid DJ. Methods to impute missing genotypes for population data. Hum Genet. 2007;122:495–504. doi: 10.1007/s00439-007-0427-y. [DOI] [PubMed] [Google Scholar]
- 27.Song H, Ramus SJ, Shadforth D, et al. Common variants in RB1 gene and risk of invasive ovarian cancer. Cancer Res. 2006;66:10220–6. doi: 10.1158/0008-5472.CAN-06-2222. [DOI] [PubMed] [Google Scholar]
- 28.Sellers TA, Huang Y, Cunningham J, et al. Association of single nucleotide polymorphisms in glycosylation genes with risk of epithelial ovarian cancer. Cancer Epidemiol Biomarkers Prev. 2008;17:397–404. doi: 10.1158/1055-9965.EPI-07-0565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cunningham JM, Sellers TA, Schildkraut JM, et al. Performance of amplified DNA in an Illumina GoldenGate BeadArray assay. Cancer Epidemiol Biomarkers Prev. 2008;17:1781–9. doi: 10.1158/1055-9965.EPI-07-2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sellers TA, Schildkraut JM, Pankratz VS, et al. Estrogen bioactivation, genetic polymorphisms, and ovarian cancer. Cancer Epidemiol Biomarkers Prev. 2005;14:2536–43. doi: 10.1158/1055-9965.EPI-05-0142. [DOI] [PubMed] [Google Scholar]
- 31.Song H, Ramus SJ, Quaye L, et al. Common variants in mismatch repair genes and risk of invasive ovarian cancer. Carcinogenesis. 2006;27:2235–42. doi: 10.1093/carcin/bgl089. [DOI] [PubMed] [Google Scholar]
- 32.Auranen A, Song H, Waterfall C, et al. Polymorphisms in DNA repair genes and epithelial ovarian cancer risk. Int J Cancer. 2005;117:611–8. doi: 10.1002/ijc.21047. [DOI] [PubMed] [Google Scholar]
- 33.Carlson CS, Eberle MA, Rieder MJ, Yi Q, Kruglyak L, Nickerson DA. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am J Hum Genet. 2004;74:106–20. doi: 10.1086/381000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Frazer KA, Ballinger DG, Cox DR, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–61. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.de Bakker PI, Yelensky R, Pe’er I, Gabriel SB, Daly MJ, Altshuler D. Efficiency and power in genetic association studies. Nat Genet. 2005;37:1217–23. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- 36.Stram DO. Tag SNP selection for association studies. Genet Epidemiol. 2004;27:365–74. doi: 10.1002/gepi.20028. [DOI] [PubMed] [Google Scholar]
- 37.National Institute of Environmental Health Sciences Environmental Genome Project. [Accessed October, 2005]; http://egp.gs.washington.
- 38.Oliphant A, Barker DL, Stuelpnagel JR, Chee MS. BeadArray technology: enabling an accurate, cost-effective approach to high-throughput genotyping. Biotechniques. 2002;(Suppl):56–8. 60–1. [PubMed] [Google Scholar]
- 39.Scheet P, Stephens M. A fast and flexible statistical model for large-scale population genotype data: applications to inferring missing genotypes and haplotypic phase. Am J Hum Genet. 2006;78:629–44. doi: 10.1086/502802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Little R, Rubin D. Statistical Analysis with Missing Data. New York: Wiley; 2002. [Google Scholar]
- 41.Yang HJ, Liu VW, Wang Y, Tsang PC, Ngan HY. Differential DNA methylation profiles in gynecological cancers and correlation with clinico-pathological data. BMC Cancer. 2006;6:212. doi: 10.1186/1471-2407-6-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Suh DS, Yoon MS, Choi KU, Kim JY. Significance of E2F-1 overexpression in epithelial ovarian cancer. Int J Gynecol Cancer. 2008;18:492–8. doi: 10.1111/j.1525-1438.2007.01044.x. [DOI] [PubMed] [Google Scholar]
- 43.Shin JS, Hong SW, Lee SL, et al. Serum starvation induces G1 arrest through suppression of Skp2- CDK2 and CDK4 in SK-OV-3 cells. Int J Oncol. 2008;32:435–9. [PubMed] [Google Scholar]
- 44.Tanner MA, Wong WH. The calculation of posterior distributions by data augmentation. Journal of the American Statistical Association. 1987;82:526–640. [Google Scholar]
- 45.Schafer JL. Monographs on Statistics and Applied Probability. Boca Raton: Chapman and Hall/CRC; 1997. Incomplete Multivariate Data. [Google Scholar]
- 46.Hopke PK, Liu C, Rubin DP. Multiple imputation for multivariate data with missing and below- threshold measurements: time-series concentrations of pollutants in the Arctic. Biometrics. 2001:22–33. doi: 10.1111/j.0006-341x.2001.00022.x. [DOI] [PubMed] [Google Scholar]
- 47.Fridley BL, de Andrade M. Missing phenotype data imputation in pedigree data analysis. Genet Epidemiol. 2008;32:52–60. doi: 10.1002/gepi.20261. [DOI] [PubMed] [Google Scholar]