Abstract
We screened exon 4 of the gene isocitrate dehydrogenase 1 (NADP+), soluble (IDH1) for mutations in 596 primary intracranial tumors of all major types. Codon 132 mutation was seen in 54% of astrocytomas and 65% of oligodendroglial tumors but in only 6% of glioblastomas (3% of primary and 50% of secondary glioblastomas). There were no mutations in any other type of tumor studied. While mutations in the tumor protein p53 gene (TP53) and total 1p/19q deletions were mutually exclusive, IDH1 mutations were strongly correlated with these genetic abnormalities. All four types of mutant IDH1 proteins showed decreased enzymatic activity. The data indicate that IDH1 mutation combined with either TP53 mutation or total 1p/19q loss is a frequent and early change in the majority of oligodendroglial tumors, diffuse astrocytomas, anaplastic astrocytomas, and secondary glioblastomas but not in primary glioblastomas.
Keywords: astrocytoma, oligodendroglioma, 1p/19q loss, oxidative stress, p53
A recent report indicated that the gene isocitrate dehydrogenase 1 (NADP+), soluble (IDH1) is somatically mutated predominantly among younger patients with glioblastomas and secondary glioblastomas (sGBs) compared with primary glioblastomas.1 sGBs arise by progression of an astrocytic tumor of lower malignancy grade (a diffuse astrocytoma WHO grade II [A] or an anaplastic astrocytoma WHO grade III [AA]), while primary (or de novo) glioblastomas (pGB) have no known precursor lesions. Thus, the question arises of whether A and AA also show this mutation or whether it is involved in progression to sGB. Glioblastomas (GB) are the most malignant as well as the most common and, consequently, the best studied of the astrocytic tumors.2 Many genetic abnormalities have been identified in GBs, including genes in the RB1 pathway (CDKN2A/B, CDK4, CDK6, RB1), the p53 pathway (p14ARF, MDM2, MDM4, TP53), the Akt pathway (PTEN, PIK3CA), and epithelial growth factor receptor gene EGFR (for review, see Collins3 and references therein). While these cellular pathways are almost always disrupted in both pGBs and sGBs, the frequency with which a particular gene is targeted differs among these GB types.4 Much less is known about other types of gliomas and other types of primary brain tumors. Rare mutations of the gene cyclin-dependent kinase inhibitor 2C (CDKN2C [p18INK4C]) or the tumor protein p53 gene (TP53) have been found in oligodendroglial tumors.5 Combined total codeletion of 1p and 19q, generally as a result of an unbalanced t(1;19) (q10;p10) translocation, is a common event among oligodendrogliomas.6 However, the molecular consequences of this translocation are unknown. We hypothesized that IDH1 mutations might be associated with A and/or AA, or even with a wider range of primary brain tumors, and carried out mutation screening of IDH1 in a series of 596 intracranial primary tumors and 15 glioma cell lines. We found that IDH1 is frequently mutated in A and AA as well as in all oligodendroglial tumor types. Among the astrocytic tumors, pilocytic astrocytomas (PAs) and pGBs are exceptions. We also demonstrate that mutated IDH1 proteins showed decreased enzymatic activity.
Materials and Methods
Tumor Materials, DNA Extraction, and Array-Comparative Genomic Hybridization
The histopathological diagnosis was made according to WHO criteria.2 The primary tumors consisted of 305 astrocytic tumors (including 38 PA, 22 A, 62 AA, 183 GB), 97 oligodendroglial tumors (including 34 oligodendrogliomas [O], 20 anaplastic oligodendrogliomas [AO], 20 oligoastrocytomas [OA], and 23 anaplastic oligoastrocytomas [AOA]), and 50 ependymal tumors (ependymoma WHO grade II, anaplastic ependymoma WHO grade III, subependymoma WHO grade I, and myxopapillary ependymoma WHO grade I), as well as 144 other intracranial tumors: 36 medulloblastomas, 8 primitive neuroectodermal tumors (PNETs), 4 dysembryoplastic neuroepithelial tumors (DNETs), 48 vestibular schwannomas, and 48 meningiomas (see Table 1). Collection, handling, and DNA/RNA extraction of tumor tissues and the patients’ blood samples were as described previously.7 The study was approved by the Ethical Committee of the Karolinska Hospital (no. 91:16), Sahlgrenska University Hospital (S339:01), and Cambridge Research Ethics Committee (Cambridge, UK; NRES Cambridgeshire 2 REC reference 03/115). In addition, 15 established glioma cell lines (U87MG, U118MG, U138MG, U178MG, U251MG, U373MG, TP265MG, TP365MG, TP483MG, TP276MG, TP336MG, T98G, H4, CCF-STTG1, A172) were included in the study.
Table 1.
Summary of genetic abnormalities
| Tumor Type | WHO Grade | No. Cases | IDH1 Mutation | TP53 Mutation | Total 1p/19q Deletion | ||||
|---|---|---|---|---|---|---|---|---|---|
| Gliomas | 452 | 119 | 142 | 54 | |||||
| Astrocytomas | 305 | ||||||||
| Pilocytic astrocytoma | I | 38 | 0 | ND | 0 | ||||
| Diffuse astrocytoma | II | 22 | 13 | 59% | 11 | 50% | 3 | 14% | |
| Anaplastic astrocytoma | III | 62 | 32 | 52% | 42 | 68% | 3 | 5% | |
| Glioblastoma | IV | 183 | 11 | 6% | 65 | 36% | 3 | 2% | |
| Primary glioblastoma | 173 | 6 | 3% | 59 | 34% | 3 | 2% | ||
| Secondary glioblastoma | 10 | 5 | 50% | 6 | 60% | 0 | |||
| Oligodendrogliomas | 97 | ||||||||
| Oligodendroglioma | II | 34 | 23 | 68% | 2 | 6% | 26 | 76% | |
| Anaplastic oligodendroglioma | III | 20 | 12 | 60% | 3 | 15% | 12 | 60% | |
| Oligoastrocytoma | II | 20 | 10 | 50% | 8 | 40% | 1 | 5% | |
| Anaplastic oligoastrocytoma | III | 23 | 18 | 78% | 11 | 48% | 6 | 26% | |
| Ependymomas | 50 | ||||||||
| Ependymoma | II | 23 | 0 | ND | 0 | ||||
| Anaplastic ependymoma | III | 7 | 0 | ND | 0 | ||||
| Subependymoma | I | 12 | 0 | ND | 0 | ||||
| Myxopapillary ependymoma | I | 8 | 0 | ND | 0 | ||||
| Other brain tumors | 144 | 0 | |||||||
| Medulloblastoma | IV | 36 | 0 | ND | 0 | ||||
| PNET | IV | 8 | 0 | ND | 0 | ||||
| DNET | I | 4 | 0 | ND | 0 | ||||
| Vestibular schwannoma | I | 48 | 0 | ND | ND | ||||
| Meningioma | I | 48 | 0 | ND | ND | ||||
| Glioma cell lines | 15 | 0 | 10 | 67% | 0 | ||||
Abbreviations: PNET, primitive neuroectodermal tumor; DNET, dysembryoplastic neuroepithelial tumor; ND, not determined.
Genetic Analysis
The genomewide copy number data were determined by comparative genomic hybridization on a 1-Mb array, as previously described.8,9 The details of the 1-Mb array data will be published elsewhere (K.I., unpublished data). Primers for IDH1 exon 4 were according to Parsons et al.1 Sequencing was performed as previously described.10 Mutation analysis of all other genes—TP53, retinoblastoma 1 (RB1), phosphatase and tensin homolog (PTEN), and cyclin-dependent kinase inhibitor 2A (CDKN2A)—was performed as previously described.7,11
IDH1 Enzymatic Assay
A wild-type IDH1 expression vector construct with a 5′ hemagglutinin (HA) tag was generated by amplifying cDNA synthesized from U251 cells (IDH1 wild type; see below) using primers PC5779 (TAGAATTCCACCATGGCTTACCCATACGATGTTCCAGATTACGCTATGTCCAAAAAAATCAGTGGC) and PC5778 (GTATGAATTCAAAGTTTGGCCTGAGCTAG) and subcloning the PCR product into pTracer-CMV2 (Invitrogen, Paisley, UK). All four IDH1 mutants identified in this study (see below) were generated by site-directed mutagenesis using a QuikChange XL site-directed mutagenesis kit followed by confirmatory sequencing (Agilent Technologies, Santa Clara, CA, USA). COS-7 kidney cells were transfected with the constructs using Lipofectamine 2000 (Invitrogen), harvested 48 h post-transfection, and lysed with the CelLytic M reagent (Sigma-Aldrich, Gillingham, UK). The supernatant was used for both the enzymatic assay and Western blotting. The IDH1 enzymatic assay was performed according to the manufacturer’s recommendation (Sigma-Aldrich). Briefly, the substrate mix (42 mM glycylglycine buffer, 0.44 mM d,l-isocitric acid, 1 mM β-nicotinamide adenine dinucleotide phosphate [NADP], 0.6 mM MnSO4) was preincubated at 37°C for 10 min. The cell lysate equivalent to 5 μg total protein was then added to the premix, and the amount of NADPH produced was monitored by spectrophotometry at 340 nm at intervals for 25 min. The enzymatic activity was determined as ΔA340nm/min. Western blotting was performed as previously described10 using a rabbit anti-HA polyclonal antibody (Abcam, Cambridge, UK), a goat anti-IDH1 monoclonal antibody (Santa Cruz Biotechnology, Santa Cruz, CA, USA), and a mouse anti–α-tubulin monoclonal antibody (Sigma-Aldrich).
Statistical Analysis
Statistical analyses were performed using Statistical Package for Social Sciences (SPSS) version 16.0 (SPSS, Inc., Chicago, IL, USA). Significance of correlations between the parameters was assessed using either the chi-square test with Yates’s correction or two-tailed Fisher’s exact test. The log-rank test was used for univariate analysis, and Cox regression was used for multivariate analysis of potential association between IDH1 mutations and overall patient survival.
Results
The entire exon 4 and adjacent intronic sequences of IDH1 were sequenced in the 596 intracranial primary tumors and 15 glioma cell lines. Mutations were confirmed as somatic by sequencing matched normal DNA from peripheral white blood cells from individual patients in selected cases. In total, 119 somatic mutations were identified. All mutations were missense mutations at codon 132 (arginine). Of these, 110 were c.395G>A (Arg132His), one c.394C>A (Arg132Ser), four c.394C>G (Arg132Gly), and four c.394C>T (Arg-132Cys) (examples of the mutations are shown in Supplementary Fig. 1S). With the exception of six tumors (five with deletion of one allele and one with gain; data not shown), 113 of 119 mutations were associated with normal copy number at IDH1 (2q33), and one retained the wild-type allele. No IDH1 mutations were identified in any of the 17 matched-blood DNA from patients with IDH1-mutated tumors. Matched normal brain tissues surrounding tumors that harbored IDH1 mutations in two cases (O24, OA9) showed no mutation, indicating that IDH1 mutations were present only in glioma cells but not in the nonneoplastic cells (Supplementary Fig. 1S).
The prevalence of mutations in each tumor type is shown in Table 1. No PA had an IDH1 mutation. A and AA showed high frequencies of mutations (59% and 52%), while GBs had a very low mutation rate overall (6%). However, when divided into primary and secondary GBs, IDH1 mutations were found in only 6 of 173 (3%) of pGB, whereas they were present in 5 of 10 (50%) of sGBs (p <0.001). Xenografts established from one pGB (GB181) retained the same mutation (c.395G>A). A majority of the four subtypes of oligodendroglial tumors (O, AO, OA, and AOA) also had IDH1 mutations: 23 of 34 O (68%), 12 of 20 AO (60%), 10 of 20 OA (50%), and 18 of 23 AOA (78%). No mutations were found among the 50 ependymal tumors, 36 medulloblastomas, 8 supratentorial PNETs, 4 DNETs, 48 vestibular schwannomas, or 48 meningiomas. None of the 15 established glioma cell lines had IDH1 mutations. The details of the IDH1 mutations in individual tumors are presented in Supplementary Table 1S.
A majority of A, AA, and sGB tumors but only about 30% of pGBs are known to have TP53 mutations, while a majority of O, AO, OA, and AOA are known to have total 1p/19q loss and very unusual TP53 mutations.7,12 When all these tumors were included in one group (A, AA, GB, O, AO, OA, and AOA; total, 364 tumors), TP53 mutations were present in 142 and combined total 1p/19q loss was present in 54 tumors (Table 1). The TP53 mutations and total 1p/19q loss were, with the exception of three AOs, mutually exclusive (Supplementary Table 1S). When we looked for correlations between these two completely different genomic abnormalities and IDH1 mutations, we found that the vast majority of IDH1 mutations (109/119, 92%) were associated with TP53 mutation, total 1p/19q loss, or both (p <0.001; including two of the three AOs that had both TP53 mutations and total 1p/19q loss). Only 10 tumors with IDH1 mutations had neither TP53 mutations nor total 1p/19q loss. However, 77 (54%) of the tumors with TP53 mutations had no abnormality of IDH1 (1 A, 14 AA, 1 sGB, 56 pGB, 1 O, 1 AO, 1 OA, and 2 AOA), and seven cases had total 1p/19q loss with no IDH1 mutation (1 A, 1 AA, 1 GB, and 4 O). One AO had both TP53 mutation and total 1p/19q loss but no IDH1 mutation. Note that there were also astrocytomas with no TP53 or IDH1 mutation and oligodendrogliomas with neither total loss of 1p/19q nor IDH1 mutation, some of which had other detectable clonal genetic abnormalities.
When any abnormality of the p53 pathway was considered (TP53 mutation, MDM2/MDM4 amplification, or p14ARF homozygous deletion),7 it was significantly associated with IDH1 mutation only in astrocytomas (A and AA, p = 0.005). Abnormalities of the RB1 pathway (RB1 mutation, CDKN2A homozygous deletion or Cyclins/CDK4/6 [cyclin-dependent kinase 4/6] amplification) were negatively associated with IDH1 mutation in adult astrocytic tumors (all A, AA, and GB; p <0.001) but not in oligodendroglial tumor groups. Among all the adult astrocytic tumors, IDH1 mutations were also negatively associated with PTEN mutations (p <0.001; no tumor had both IDH1 and PTEN mutations) and with EGFR amplifications (p <0.001; only one tumor had both an IDH1 mutation and EGFR amplification).
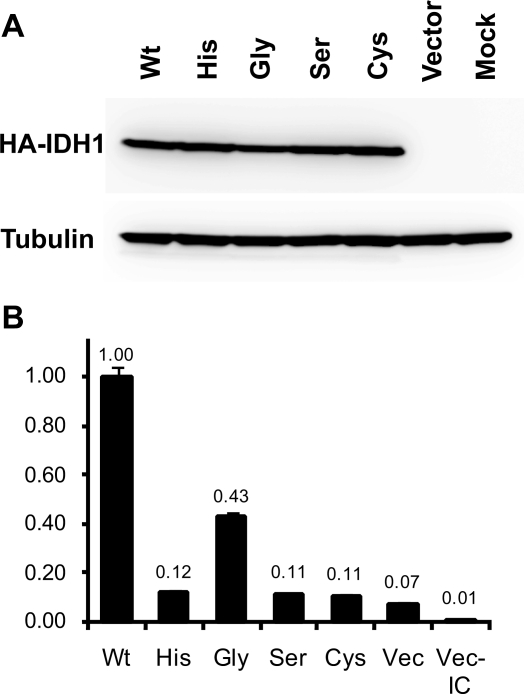
The enzymatic activity of IDH1 resulting in the production of NADPH was assayed using COS-7 cells transfected with the wild-type and all four mutant IDH1 constructs. The Arg132His, Arg132Ser, and Arg132Cys mutants showed an 8- to 9-fold decrease of enzymatic activity. Arg132Gly showed a 2.4-fold decrease compared with wild type (p <0.001, t-test; Fig. 1), thus showing a significantly higher level of activity than the other three mutants (p <0.001). All transfectants showed higher levels of activity than the negative control (vector-alone transfectants; p <0.05).
Fig. 1.
Enzymatic assay of the wild-type and mutant IDH1 (isocitrate dehydrogenase 1 [NADP+], soluble). (A) Western blotting of the COS-7 transfectants with an antihemagglutinin (anti-HA) antibody (top). The HA-tagged recombinant IDH1 (46 kDa) is strongly expressed in the wild-type (Wt) and mutant transfectants (His, Arg132His; Gly, Arg132Gly; Ser, Arg132Ser; Cys, Arg132Cys). A comparable result was obtained using a goat anti-IDH1 antibody (data not shown). The blot was reprobed with anti–α-tubulin antibody to control for lane loading (bottom). (B) Enzymatic activity of wild-type and mutant IDH1. The relative activity is shown as a ratio to wild type (Wt, 1.00). The error bar shows standard error of the quadruplicated assays. The enzymatic activities in all four mutants were significantly decreased compared with wild-type (p <0.001). The activity detected in the vector transfectants (Vec) compared with the vector transfectants without isocitrate (Vec-IC) represents the activity of endogenous IDH1 in COS-7 cells. Reproducible results were obtained from independent transfection experiments.
In addition to IDH1 mutations occurring more frequently among secondary than among primary GBs (p <0.001), such mutations were much more common in younger GB patients. The mean age of GB patients with IDH1 mutations was 41 years, compared with 56 years for GB patients with no mutations of IDH1 (t-test, p = 0.002), confirming the findings in a previous report.1
Univariate analyses using the log-rank test were used to assess the impact of IDH1 mutation on overall survival. IDH1 mutations were associated with longer overall survival among all 364 tumors (including all A, AA, GB, O, AO, OA, and AOA; p <0.001), all adult astrocytic tumors (including A, AA, and GB; p <0.001), and even among all GBs (p = 0.011; Supplementary Fig. 2S). However, Cox regression multivariate analysis including age, histological diagnosis, WHO malignancy grade, TP53, 1p/19q status, and primary/secondary GB indicated that IDH1 status was not an independent prognostic factor.
Discussion
In this study, we carried out an extensive screening for mutations of exon 4 of the IDH1 gene in a large series of human brain tumors. We identified a high frequency of IDH1 mutations in the majority of A, AA, O, AO, OA, AOA, and sGB tumors, contrasting with a mutation rate of only 3% in pGB tumors. These findings have been independently confirmed.13 No mutations were found in PAs, medulloblastomas, supratentorial PNETs, DNETs, ependymal tumors, schwannomas, or meningiomas. Thus, our results show that mutations of IDH1 and/or TP53 are the earliest common genetic abnormalities in the majority of diffuse astrocytomas (A). Single cases had one or the other, so a temporal order cannot be established. The situation is similar for the oligodendroglial tumors, where concurrent IDH1, total loss of 1p/19q, or both are the earliest common genetic abnormalities identified.
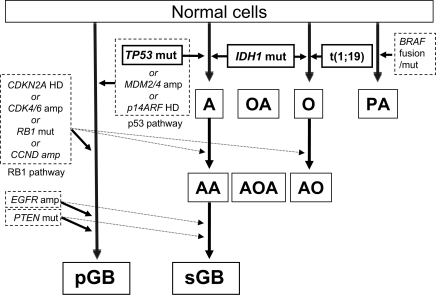
The finding that the majority of IDH1 mutations are associated with either TP53 mutation or total 1p/19q loss is striking. TP53 mutations and total 1p/19q loss are generally mutually exclusive12 and characterize WHO grade II diffuse astrocytomas and oligodendrogliomas, respectively, both of which are well known to show malignant progression. There are no known cellular consequences common to these two aberrations. However, based on these findings, the genetic models for development and progression of astrocytic and oligodendroglial tumors must be revised and include mutation of IDH1 as an early event common to both types of glioma (Fig. 2, Supplementary Table 1S). In this model, the majority of oligodendrogliomas develop through acquisition of concurrent IDH1 mutation and total 1p/19q loss (as a result of t[1;19] translocation), and grade II tumors may progress, some acquiring RB1 pathway abnormalities. The majority of diffuse astrocytomas arise through developing concomitant mutations of IDH1 and TP53. These tumors generally acquire RB1 pathway alterations when they progress to sGBs. The alternative pathway for GB development, resulting in a pGB, is via synchronous RB1 and p53 pathway disruption,7 and this does not seem to be associated with IDH1 mutation. Disruptions of the p53 pathway by mechanisms other than TP53 mutation occur predominantly among pGB. Primary GBs may also have Akt pathway disruption (e.g., by PTEN mutation/deletion) and/or EGFR amplification (with or without rearrangement), unlike other astrocytomas or oligodendrogliomas. Oligoastrocytic tumors appear to develop through pathways similar to either the grade II/III astrocytomas or the oligodendrogliomas (i.e., IDH1 mutation and either TP53 mutation or total 1p/19q loss). Pilocytic astrocytomas do not progress, and the majority have either a unique fusion gene containing the constitutively activated kinase domain of v-raf murine sarcoma viral oncogene homolog B1 (BRAF) or mutations of BRAF or neurofibromin 1 (NF1).10 They have no IDH1 mutations or any other changes described above.
Fig. 2.
A model for the development/progression of astrocytic and oligodendroglial tumors. Abbreviations: CDKN2A, cyclin-dependent kinase inhibitor 2A; HD, homozygous deletion; CDK4/6, cyclin-dependent kinase 4/6; amp, amplification; RB1, retinoblastoma 1; mut, mutation; CCND, cyclin D1/D2/D3; RB1 pathway, CDKN2A, CDK4/6, RB1, or CCND ; EGFR, epithelial growth factor receptor; PTEN, phosphatase and tensin homolog; pGB, primary glioblastoma; TP53, tumor protein p53; MDM2/4, Mdm2/4 p53 binding protein homolog (mouse); p14ARF, the p14 alternative reading frame product of cyclin-dependent kinase inhibitor 2A; p53 pathway, TP53, MDM2/4, or p14ARF; A, diffuse astrocytoma; AA, anaplastic astrocytoma; sGB, secondary glioblastoma; IDH1, isocitrate dehydrogenase 1 (NADP+), soluble; OA, oligoastrocytoma; AOA, anaplastic oligoastrocytoma; O, oligodendroglioma grade II; AO, anaplastic oligodendroglioma; t(1;19), unbalanced t(1;19)(q10;p10) translocation; PA, pilocytic astrocytoma; BRAF, v-raf murine sarcoma viral oncogene homolog B1.
IDH1 functions as a homodimer, is localized in peroxisomes and more diffusely in the cytoplasm, and catalyzes oxidative decarboxylation of isocitrate into α-ketoglutarate, generating NADPH from NADP+.14 NADPH is an important cofactor for regeneration of reduced glutathione, which is a critical component of the defense mechanism against oxidative damage.15,16 High levels of oxidative stress are reported in the brain compared with other organs. This is reflected in the common transition mutations (G:C → A:T) seen in gliomas, for example, of TP53.17 This was also the case in 81 of 142 (57%) of the TP53 mutations and 114 of 119 (95%) of the IDH1 mutations in our series, with only 5 of 119 (4%) mutations of IDH1 being of any other type.
The 119 somatic mutations, almost all of which were heterozygous, resulted in the substitution of arginine with histidine, serine, or cysteine at codon 132. We showed that the ability to generate NADPH was significantly reduced in all four mutant IDH1 proteins. These results indicate that Arg132 is critical for the enzymatic activity of IDH1. Jennings et al.18 reported that a substitution of arginine by glutamate at codon 132 in a rat ortholog of IDH1 resulted in a significant reduction in catalytic activity. Although this mutation has not been seen in humans, their observations are in line with ours. Arg132 is one of the amino acids in IDH1 that form hydrophilic interactions with the α- and β-carboxylate groups of isocitrate.19 It has also been proposed that Arg132 may contribute to a semiopen conformation of IDH1 during the transition from the inactive form to the active form of the enzyme.19 Substitution of Arg132 may therefore result in conformational changes with a consequent decrease in enzymatic activity.
Because the protein functions as a dimer, the consequences of mutation of one allele may be profound, since one mutated protein in a homodimer may result in a significant reduction in function—producing a dominant negative effect in a similar manner to p53 (functioning as a tetramer). Thus, mutation of one allele of IDH1, which results in the decreased ability to generate NADPH as shown in this study, is likely to lead to an increased susceptibility to oxidative damage and thus facilitate the accumulation of further, especially transition, mutations (G:C → A:T). Cells with reduced IDH1 expression have increased levels of oxidative stress and DNA damage.16 The finding that IDH1 mutations frequently accompany TP53 mutations in the diffuse astrocytomas is particularly notable, because loss of p53 will circumvent normal oxidative-stress–induced apoptosis, as has been demonstrated in glioma cells.20 This combination will therefore likely promote further mutations and tumor progression. It is more difficult to understand the association of IDH1 mutation with the total concurrent loss of 1p/19q.
The fact that there were no mutations in the 15 established glioma cell lines is not surprising because almost all such lines are derived from GBs, and most of these were probably pGB. Most were derived many years before the differential classification of GBs into primary and secondary, and there are no data to indicate their status.
We confirmed that IDH1 mutations occur more frequently among sGB and young patients with GB, as reported by Parsons et al.1 Univariate analyses also showed that tumors with IDH1 mutations were associated with longer overall survival of patients with any adult astrocytic or oligodendroglial tumor included in the series, with any adult astrocytic tumor alone, or with any GB. However, multivariate analysis failed to identify IDH1 status as a prognostic factor independent of tumor type, grade, or age. These results are not surprising, because IDH1 mutations are involved in both oligodendroglial and astrocytic tumors and represent only one of a diverse range of genetic abnormalities, and are therefore unlikely to be an independent prognostic factor.
IDH1 mutations appear to be rare in other common human tumors. One IDH1 mutation (Arg132Cys) in a colon cancer has been reported in a genomewide mutation analysis of a series of colon and breast cancers,21 and no mutations were found in a series of pancreatic cancers.22 Bleeker et al.23 found no mutations in IDH1 exon 4 among 559 various nonglioma human cancers, including bladder, breast, colorectal, lung, melanoma, thyroid, ovary, and pancreas. High levels of oxidative stress in the brain compared with other organs may provide strong selective pressure for tumor cells that acquired IDH1 mutations.
Our results indicate that IDH1 mutation is common in A, AA, and sGB as well as all types of oligodendroglial tumors, but infrequent in pGB, thereby helping to distinguish pGBs from the others. Consequently, the models of astrocytic and oligodendroglial tumor development/progression require some revision. We also show that the missense mutations identified at Arg132 result in decreased enzymatic activity of IDH1. Further assessments of the cellular consequences of IDH1 mutations are under way. We hope a more accurate understanding of the genetic changes and their consequences in these tumors will lead to improvements in the accuracy of diagnosis and prognosis, and the development of novel molecular targeted therapies.
It is important to note that after this paper was submitted for publication we identified somatic missense mutations at codon 172 (arginine) of IDH2 in seven tumors that had either TP53 mutation or total 1p/19q loss but no IDH1 mutations. As a result, all oligodendroglial tumors with total 1p/19q loss harbored mutations of either IDH1 or IDH2, while six oligodendroglial tumors with IDH1 mutation had neither total 1p/19q loss nor TP53 mutations. This strongly suggests that mutations of the NADP+-dependent IDH genes are the earliest changes in the development of oligodendroglial tumors.
Acknowledgments
The U251MG and U373MG glioma cell lines used in this study were obtained directly from the original depositor, and their distinct genotypes have been verified.24 We thank the Mapping Core, Map Finishing, and Microarray Facility groups of the Wellcome Trust Sanger Institute, Hinxton, UK, for clone supply and verification, and the Centre for Microarray Resources in the Department of Pathology, University of Cambridge, for printing the arrays.
This work was supported by grants from Cancer Research UK, Samantha Dickson Brain Tumour Trust, Jacqueline Seroussi Memorial Foundation for Cancer Research, and Cambridge Fund for the Prevention of Disease (CAMPOD).
References
- 1.Parsons DW, Jones S, Zhang X, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK. WHO Classification of Tumours of the Central Nervous System. 4th ed. Lyon, France: International Agency for Research on Cancer; 2007. [Google Scholar]
- 3.Collins VP. Mechanisms of disease: genetic predictors of response to treatment in brain tumors. Nat Clin Pract Oncol. 2007;4:362–374. doi: 10.1038/ncponc0820. [DOI] [PubMed] [Google Scholar]
- 4.Ohgaki H, Kleihues P. Genetic pathways to primary and secondary glioblastoma. Am J Pathol. 2007;170:1445–1453. doi: 10.2353/ajpath.2007.070011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Husemann K, Wolter M, Buschges R, Bostrom J, Sabel M, Reifenberger G. Identification of two distinct deleted regions on the short arm of chromosome 1 and rare mutation of the CDKN2C gene from 1p32 in oligodendroglial tumors. J Neuropathol Exp Neurol. 1999;58:1041–1050. doi: 10.1097/00005072-199910000-00002. [DOI] [PubMed] [Google Scholar]
- 6.Jenkins RB, Blair H, Ballman KV, et al. A t(1;19)(q10;p10) mediates the combined deletions of 1p and 19q and predicts a better prognosis of patients with oligodendroglioma. Cancer Res. 2006;66:9852–9861. doi: 10.1158/0008-5472.CAN-06-1796. [DOI] [PubMed] [Google Scholar]
- 7.Ichimura K, Bolin MB, Goike HM, Schmidt EE, Moshref A, Collins VP. Deregulation of the p14ARF/MDM2/p53 pathway is a prerequisite for human astrocytic gliomas with G1-S transition control gene abnormalities. Cancer Res. 2000;60:417–424. [PubMed] [Google Scholar]
- 8.McCabe MG, Ichimura K, Liu L, et al. High-resolution array-based comparative genomic hybridization of medulloblastomas and supratentorial primitive neuroectodermal tumors. J Neuropathol Exp Neurol. 2006;65:549–561. doi: 10.1097/00005072-200606000-00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones DT, Ichimura K, Liu L, Pearson DM, Plant K, Collins VP. Genomic analysis of pilocytic astrocytomas at 0.97 Mb resolution shows an increasing tendency toward chromosomal copy number change with age. J Neuropathol Exp Neurol. 2006;65:1049–1058. doi: 10.1097/01.jnen.0000240465.33628.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jones DT, Kocialkowski S, Liu L, et al. Tandem duplication producing a novel oncogenic BRAF fusion gene defines the majority of pilocytic astrocytomas. Cancer Res. 2008;68:8673–8677. doi: 10.1158/0008-5472.CAN-08-2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmidt EE, Ichimura K, Goike HM, Moshref A, Liu L, Collins VP. Mutational profile of the PTEN gene in primary human astrocytic tumors and cultivated xenografts. J Neuropathol Exp Neurol. 1999;58:1170–1183. doi: 10.1097/00005072-199911000-00007. [DOI] [PubMed] [Google Scholar]
- 12.Ino Y, Betensky RA, Zlatescu MC, et al. Molecular subtypes of anaplastic oligodendroglioma: implications for patient management at diagnosis. Clin Cancer Res. 2001;7:839–845. [PubMed] [Google Scholar]
- 13.Balss J, Meyer J, Mueller W, Korshunov A, Hartmann C, von Deimling A. Analysis of the IDH1 codon 132 mutation in brain tumors. Acta Neuropathol. 2008;116:597–602. doi: 10.1007/s00401-008-0455-2. [DOI] [PubMed] [Google Scholar]
- 14.Geisbrecht BV, Gould SJ. The human PICD gene encodes a cytoplasmic and peroxisomal NADP(+)-dependent isocitrate dehydrogenase. J Biol Chem. 1999;274:30527–30533. doi: 10.1074/jbc.274.43.30527. [DOI] [PubMed] [Google Scholar]
- 15.Kim SY, Park JW. Cellular defense against singlet oxygen-induced oxidative damage by cytosolic NADP+-dependent isocitrate dehydrogenase. Free Radic Res. 2003;37:309–316. doi: 10.1080/1071576021000050429. [DOI] [PubMed] [Google Scholar]
- 16.Lee SM, Koh HJ, Park DC, Song BJ, Huh TL, Park JW. Cytosolic NADP(+)-dependent isocitrate dehydrogenase status modulates oxidative damage to cells. Free Radic Biol Med. 2002;32:1185–1196. doi: 10.1016/s0891-5849(02)00815-8. [DOI] [PubMed] [Google Scholar]
- 17.Ohgaki H, Kleihues P. Epidemiology and etiology of gliomas. Acta Neuropathol. 2005;109:93–108. doi: 10.1007/s00401-005-0991-y. [DOI] [PubMed] [Google Scholar]
- 18.Jennings GT, Minard KI, McAlister-Henn L. Expression and mutagenesis of mammalian cytosolic NADP+-specific isocitrate dehydrogenase. Biochemistry. 1997;36:13743–13747. doi: 10.1021/bi970916r. [DOI] [PubMed] [Google Scholar]
- 19.Xu X, Zhao J, Xu Z, et al. Structures of human cytosolic NADP-dependent isocitrate dehydrogenase reveal a novel self-regulatory mechanism of activity. J Biol Chem. 2004;279:33946–33957. doi: 10.1074/jbc.M404298200. [DOI] [PubMed] [Google Scholar]
- 20.Datta K, Babbar P, Srivastava T, Sinha S, Chattopadhyay P. p53 dependent apoptosis in glioma cell lines in response to hydrogen peroxide induced oxidative stress. Int J Biochem Cell Biol. 2002;34:148–157. doi: 10.1016/s1357-2725(01)00106-6. [DOI] [PubMed] [Google Scholar]
- 21.Sjoblom T, Jones S, Wood LD, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 22.Jones S, Zhang X, Parsons DW, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bleeker FE, Lamba S, Leenstra S, et al. IDH1 mutations at residue p.R132 (IDH1(R132)) occur frequently in high-grade gliomas but not in other solid tumors. Hum Mutat. 2009;30:7–11. doi: 10.1002/humu.20937. [DOI] [PubMed] [Google Scholar]
- 24.Fuxe J, Akusjärvi G, Goike HM, et al. Adenovirus-mediated over-expression of p15INK4B inhibits human glioma cell growth, induces replicative senescence, and inhibits telomerase activity similarly to p16INK4A. Cell Growth Differ. 2000;11:373–384. [PubMed] [Google Scholar]