Abstract
The O6-methylguanine-DNA methyltransferase gene (MGMT) is methylated in several cancers, including gliomas. However, the functional role of cysteine-phosphate-guanine (CpG) island (CGI) methylation in MGMT silencing is still controversial. The aim of this study was to investigate whether MGMT CGI methylation correlates inversely with RNA expression of MGMT in glioblastomas and to determine the CpG region whose methylation best reflects the level of expression. The methylation level of CpG sites that are potentially related to expression was investigated in 54 glioblastomas by pyrosequencing, a highly quantitative method, and analyzed with respect to their MGMT mRNA expression status. Three groups of patients were identified according to the methylation pattern of all 52 analyzed CpG sites. Overall, an 85% rate of concordance was observed between methylation and expression (p < 0.0001). When analyzing each CpG separately, six CpG sites were highly correlated with expression (p < 0.0001), and two CpG regions could be used as surrogate markers for RNA expression in 81.5% of the patients. This study indicates that there is good statistical agreement between MGMT methylation and expression, and that some CpG regions better reflect MGMT expression than do others. However, if transcriptional repression is the key mechanism in explaining the higher chemosensitivity of MGMT-methylated tumors, a substantial rate of discordance should lead clinicians to be cautious when deciding on a therapeutic strategy based on MGMT methylation status alone.
Keywords: CpG island methylation, expression, glioblastomas, MGMT
Epigenetic alterations such as histone modification and DNA methylation have been associated with tumor formation and progression. There is an increasing interest in aberrant promoter DNA methylation, as this mechanism is related to transcriptional repression and is involved in the disruption of key cellular pathways. Among the genes with a promoter cysteine-phosphate-guanine (CpG) island (CGI) susceptible to DNA methylation, the O6-methylguanine-DNA methyltransferase gene (MGMT) is one of the best studied. The currently investigated CpG sites were indeed found to be methylated in several cancers, notably in gliomas.1 The study of MGMT promoter methylation status is of particular interest as MGMT encodes a DNA repair protein that removes alkylating lesions, thereby providing resistance to alkylating chemotherapeutic agents such as temozolomide. A methylated MGMT promoter might thus reflect a “chemosensitivity state” to alkylating agents, as DNA methylation normally leads to transcriptional repression.
Hypermethylation of the MGMT promoter was found to be of favorable prognostic value in patients with glioblastomas (GBM) or low-grade gliomas when treated with the alkylating agent temozolomide, with or without additional radiotherapy.2–6 However, the role of 5′ CGI hypermethylation in MGMT gene silencing is still controversial. Several studies using cell lines have reported a relationship between MGMT gene expression and promoter methylation.7–10 One study in glioma cell lines even demonstrated a graded relationship between methylation and expression.7 Many studies in primary human gliomas take for granted that MGMT DNA methylation is directly linked to MGMT transcriptional repression and therefore often focus on measuring MGMT DNA methylation as a marker of chemosensitivity to alkylating agents. However, to our knowledge, only one of the studies investigating both MGMT DNA methylation and MGMT expression (immunohistochemistry or mRNA) in gliomas actually showed a mutually exclusive presence of methylation or expression in a small series of eight human primary gliomas.1 Other investigations showed only a partial correlation between these two measures11–13 or no correlation at all.14,15 Notably, it was shown that MGMT DNA methylation and MGMT protein expression (immunohistochemistry) cannot be used interchangeably to predict survival for patients with malignant gliomas.11 Another study of anaplastic gliomas showed a correlation between MGMT protein expression and survival but not between MGMT DNA methylation and survival.14 The observed discordances between these measures point out the necessity of clarifying the exact relationship between MGMT CGI methylation and MGMT expression.
The MGMT promoter contains a 777-bp CGI with 97 CpG sites. Studies of cell lines showed that differences in methylation levels were located within two large regions,9,10 one of which contains the region that is most commonly investigated by the methylation-specific PCR assay (MSP).1 The MSP region includes nine CpG sites that partially cover the first noncoding exon and the minimal enhancer. However, it is not known if these nine CpG sites are the sites that best reflect the status of expression. On the other hand, according to Pieper et al.,16 the changes in methylation between an MGMT-expressing and nonexpressing cell line are focused in four CpG sites rather than being diffusely and uniformly distributed throughout the CGI. Two of these were approximately 130 nucleotides downstream of the transcription start site (TSS) and included the region currently investigated by MSP, while the two others were approximately 200 nucleotides upstream of the TSS.16 Taking these data together, it remains uncertain whether there are specific CpG sites, which are not necessarily located side by side, that influence the MGMT promoter activity level.
The aim of this study was to investigate whether MGMT CGI methylation reflects the expression of the gene in human GBM and to determine the CpG region whose methylation correlates best with the level of expression in primary tumors. Previously used assays assessing the methylation status of the MGMT CGI have provided mostly qualitative results or low-resolution quantitative results and considered the methylation state of only about 10 CpG sites. To investigate which CpG region is of interest, we quantitatively analyzed the methylation levels of 60 CpG sites for 54 GBM tumors by pyrosequencing, a high-throughput and reliable method,17,18 and correlated the results with the MGMT expression level of the tumors.
Materials and Methods
Patients
Patients were selected from our database containing clinical information regarding patients with a primary brain tumor that had been seen in our department since 2003. Inclusion criteria were (1) 18 or more years of age at onset, (2) histological diagnosis of GBM according to the WHO classification, (3) availability of cryopreserved tumor material, and (4) written informed consent for molecular analysis.
Samples and Bisulfite Treatment
DNA from frozen tumors and nonneoplastic brain tissues (18 from epilepsy patients, 1 cerebellar gray cortex, 1 cerebellar white matter, and 4 from amyotrophic lateral sclerosis patients) was extracted using a standard protocol (QIAmp DNA Mini Kit; Qiagen, Courtaboeuf, France). We bisulfite-converted 300 ng of DNA using the Gold DNA Methylation Kit (Zymo Research, Orange, CA, USA) according to the manufacturer’s instructions.
Pyrosequencing
Quantitative DNA methylation analysis of the bisulfite-treated DNA was performed by pyrosequencing or—in the case of several sequencing primers—by serial pyrosequencing.18 Primers for PCR amplification, pyrosequencing, and primer extension reactions were purchased from Biotez (Buch, Germany). Regions of interest were amplified using 30 ng of bisulfite-treated human genomic DNA and 5–7.5 pmol of forward and reverse primer, one of which was biotinylated. Sequences of oligonucleotides for PCR amplification and pyrosequencing are given in Table 1. Reaction conditions were 1× HotStar Taq buffer supplemented with 1.6 mM MgCl2, 200 mM deoxyribonucleotide triphosphates, and 2.0 U HotStar Taq polymerase (Qiagen) in a 25-μl volume. The PCR program consisted of a 15-min denaturing step at 95°C followed by 50 cycles of 30 s at 95°C, 30 s at the respective annealing temperatures (Table 1), and 20 s at 72°C, with a final 5-min extension at 72°C. Then, 3–5 μl of the amplification products was incubated with 2 μl streptavidin-coated Sepharose beads (GE Health-care, Uppsala, Sweden) in 68 μl binding buffer (10 mM Tris, 2 M NaCl, 1 mM EDTA, 0.1% Tween 20; pH 7.6, adjusted with 1 M HCl). The template strands were purified and rendered single-stranded on a Pyrosequencing workstation (Pyrosequencing AB, Uppsala, Sweden). Beads were released into 12 μl annealing buffer (20 mM Tris, 2 mM Mg-acetate; pH 7.6, adjusted with 4 M acetic acid) containing 4 pmol of the respective sequencing primer (Table 1), and sequencing primers were annealed to the target by incubation at 80°C for 2 min. Quantitative DNA methylation analysis was carried out on a PSQ 96MD system with the PyroGold SQA Reagent Kit (Pyrosequencing AB), and results were analyzed using the Q-CpG software (version 1.0.9, Pyrosequencing AB).
Table 1.
Nucleotide positions, sequences of primers used for pyrosequencing reactions for the respective analyses, and annealing temperature for the respective PCR amplifications
| Nucleotide Position | PCR Forward Primer | PCR Reverse Primer | PCR Temperature (°C) | Sequencing Primer | No. CpGs |
|---|---|---|---|---|---|
| −510 to −303 | 5′-GGTTTGGGGGTTT TTGATTAG | 5′-Biotin-CCTTTTCCTATC ACAAAAATAATCC | 60°C | 5′-YGGTWTTAGGAG GGGAG | 14 |
| 5′-GTAGGATAGGGATT TTTATTAA | 13 | ||||
| −288 to +3 | 5′-TAAATTAAGGTATAG AGTTTTAGG | 5′-Biotin-AAAACCTAAAAA AAACAAAAAAAC | 58°C | 5′-AGGAAGTTGGGAAGG | 10 |
| 5′-TTTGTATAGGTAGAAG GGTTA | 11 | ||||
| −20 to +178 | 5′-GTTTTTTTGTTTTTTT TAGGTTTT | 5′-Biotin-AAACRACCCAA ACACTCACC | 60°C | 5′-GGTTTYGTTTYGTTT TAGATT | 9 |
| 5′-AGGATATGTTGGGATAGTT | 11 | ||||
| +75 to +332 | 5′-GGATATGTTGGGATAGTT | 5′-Biotin-ACAACACCTA AAAAACACTTAAAAC | 58°C | 5′-GTTTTGAYGTTYGTAGGTT | 9 |
Nucleotide numbering starts at the transcription start site.
Expression Analysis
Total RNA was isolated using an RNA extraction kit (RNeasy Lipid Tissue Mini Kit, Qiagen) according to the manufacturer’s instructions. RNA quality was assessed using an Agilent 2100 Bioanalyser (Agilent Technologies, Massy, France). cDNA was prepared from each RNA sample (1 μg) using a combination of random primers (Promega, Lyon, France) and Moloney murine leukemia virus reverse transcriptase (Invitrogen, Cergy Pontoise, France). Real-time PCR was carried out in a 25-μl volume containing 5 μl of 20-fold diluted cDNA, 600 nM of each primer, and 12.5 μl of 2× SYBR Green buffer (ABgene, Courtaboeuf, France). Values were normalized to the expression levels of the housekeeping gene ALAS (5-aminolevulinate synthase). In parallel, the same experiments were performed on five nontumoral brain samples, also to normalize quantitative PCR values using the 2ΔΔCT method.19 Oligonucleotides used are as follows: MGMT forward primer, 5′-CCTGGCTGAATGCCTATTTC-3′; MGMT reverse primer, 5′-GATGAGGATGGGGACAGGATT-3′; ALAS forward primer, 5′-TGCAGTCCTCAGCGCAGT-3′; and ALAS reverse primer, 5′-TGGCCCCAACTTCCA-TCAT-3′.
Statistical Analysis
Statistical analysis was performed using the XLSTAT software (Addinsoft, Paris, France). The Mann-Whitney test was used to determine the relationship between methylation, as a quantitative variable, and expression, as a qualitative variable (low expression versus high expression relative to the average expression of eight controls). To analyze the methylation percentage as a qualitative variable (methylated vs. unmethylated), a cutoff value based on the methylation of the controls was chosen and defined as follows: cutoff = methylation average of controls (from CpGs x to y) + 2*STDEVP, where STDEVP is the standard deviation of controls from CpGs x to y. The chi-square test was used to determine the repartition between methylation and expression as qualitative variables. Two-sided p-values less than 0.05 were considered significant. The following variables were investigated: MGMT gene expression, MGMT promoter methylation status, and age. Hierarchical clustering was based on the degree of methylation (as a percentage) for each. Average linkage (unweighted pair-group method using arithmetic averages [UPGMA]) was used with squared Euclidean distance as an interval measure.
Results
MGMT CGI Methylation of GBM
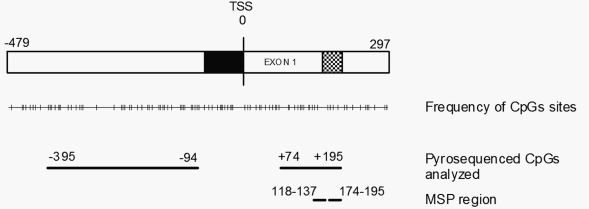
The degree of CpG methylation in the MGMT CGI was investigated for 54 patients with GBM and for 24 nontumoral brain tissues. We analyzed a total of 68 CpG sites located in the promoter region of MGMT and extending into the first noncoding exon (CpGs at nucleotides −452 to +195 relative to the TSS). Eight CpG sites situated at −452 to −399 relative to the TSS were highly methylated for both tumoral and nontumoral brain samples, as determined by pyrosequencing, suggesting that these CpG sites are situated outside of the promoter region that is susceptible to de novo methylation. This region was therefore excluded from further analysis. Moreover, analysis of glioma cell lines and tumors by cloning and sequencing showed that eight CpG sites situated at −90 to +69 corresponded to a methylation-free region in all samples (data not shown). Since this result indicates that this region has no impact on expression, we excluded it from our analysis. In total, 52 CpGs of the MGMT CGI that were potentially correlated with expression were analyzed further. Thirty-five of these CpG sites were upstream and 17 were downstream of the TSS (Fig. 1).
Fig. 1.
A 777-bp O6-methylguanine-DNA methyltransferase gene (MGMT) cysteine-phosphate-guanine (CpG) island with 97 CpG sites, including the minimal promoter, first noncoding exon, and minimal enhancer, shown as black, white, and hatched bars, respectively. Nucleotides are numbered with respect to the transcription start site (TSS). The 52 CpG sites analyzed by pyrosequencing are indicated; 35 were upstream of the TSS, and 17 were downstream of the TSS. The methylation-specific PCR assay (MSP) region (118–137 and 174–195 for methylated primers) currently investigated is also indicated.
Three Groups of Patients Were Distinguished by Their Methylation Pattern among the 52 Analyzed CpG Sites
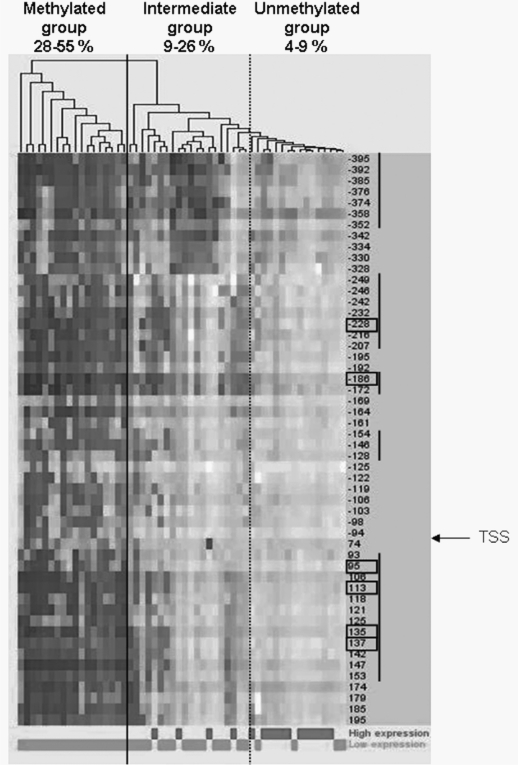
Hierarchical clustering of the methylation profiles of the 52 CpG sites provided strong evidence of two distinct patient classes (Fig. 2). The first class grouped 18 patients (33%) with considerable methylation at almost all of the CpG sites tested (average range, 28%–55% methylation: group 1, methylated). The second class grouped the 36 other patients (67%) that had a lower global methylation level (average range, 4%–26%). This second class could be further separated into two subgroups, one showing a nonhomogeneous methylation pattern along the CGI (range average, 9%–26%: group 2, intermediate) with elevated methylation at only some CpG sites, and the other almost completely unmethylated (average range, 4%–9%: group 3, unmethylated). When integrated into the hierarchical clustering model, the control brain samples (average range, 6%–11%) clustered with the second class, between the intermediate group and the unmethylated group. Methylation patterns were found to be homogeneous between control brain samples along the CpG sites tested.
Fig. 2.
Hierarchical clustering of the 54 glioblastomas for the 52 cysteine-phosphate-guanine (CpG) sites analyzed by pyrosequencing. The darkness of the gray represents the methylation level at each CpG. CpG numbering starts at the CpG just before the transcription start site (TSS). A vertical solid line separates the two distinct patient classes provided by the hierarchical clustering. The vertical dashed line separates the intermediate and unmethylated groups. Methylation range averages for the 52 analyzed CpG sites are indicated for each group. At the bottom, the expression level of the O6-methylguanine-DNA methyltransferase gene is also indicated for each patient as a qualitative variable.
MGMT CGI Methylation and MGMT Gene Expression
MGMT gene expression was determined for all patients and for eight control brain samples. The MGMT expression level was relatively homogeneous among controls. Thirty-eight patients (70%) expressed lower levels of MGMT transcripts relative to the average expression of the controls, and 16 (30%) expressed higher levels of MGMT.
MGMT Expression Analysis of Methylated, Intermediate, and Unmethylated Groups
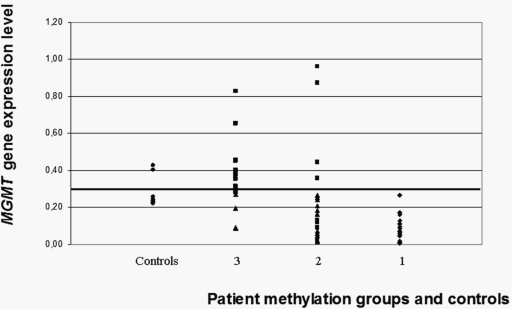
The three groups of patients (group 1, methylated; group 2, intermediate; group 3, unmethylated) were examined with regard to their expression level (low vs. high; Fig. 3). All patients in group 1 expressed a low level of MGMT. The specificity was lower for the other two groups. Twelve of 16 patients (75%) from group 3 expressed a high level of MGMT. Similarly, this relationship was not exclusive for patients of group 2, although a majority of them expressed a low level of MGMT (16 of 20, 80%). The relationship between methylation and expression as qualitative variables was statistically significant among the three groups of patients (p < 0.0001).
Fig. 3.
Distribution of O6-methylguanine-DNA methyltransferase gene (MGMT) expression levels for methylated (group 1), intermediate (group 2), unmethylated (group 3), and control groups (based on the analysis of all 52 cysteine-phosphate-guanine [CpG] sites). MGMT expression average of controls, which represent the cutoff value for low and high expressing groups, is indicated by a horizontal bar.
We also investigated whether the degree of expression within the group of low-expressing or high-expressing patients was different according to their methylation profile. Markedly, no differences were found in the expression level among the low-expressing patients in groups 2 and 3, or among the high-expressing patients in the same groups. The minimum methylation level observed within low-expressing patients of group 2 was, on average, 9% for all 52 CpG sites analyzed. It thus appears that this level of methylation might be sufficient to suppress MGMT gene expression.
MGMT Expression Analysis for the Entire CGI and for Each of the 52 CpG Sites
Despite the fact that the relationship between MGMT expression and the three groups of patients, methylated (group 1), intermediate (group 2), and unmethylated (group 3), was unquestionable, it was nonexclusive.
Methylation of the entire set of 52 CpG sites and MGMT expression were analyzed as qualitative variables (Table 2). The methylation status for each patient was determined according to the cutoff defined in “Materials and Methods.” For all 52 CpG sites, the cutoff was 10.65%. The majority of the methylated patients expressed a low level of MGMT (33 of 36, 91.6%), and the majority of the unmethylated patients expressed a high level of MGMT (13 of 18, 72.2%). Overall, concordant results were observed in 85% of cases.
Table 2.
Comparison of the repartition between methylation and expression for analyzed CpG regions and CpG sites
| Expression |
Unmethylated (n) |
Methylated (n) |
p-Value |
Concordant Results |
|||
|---|---|---|---|---|---|---|---|
| High | Low | High | Low | n | % | ||
| All 52 CpGs | 13 | 5 | 3 | 33 | p < 0.0001 | 46 | 85 |
| CpG −228 | 14 | 8 | 2 | 30 | p < 0.0001 | 44 | 81.5 |
| CpG −186 | 12 | 6 | 4 | 32 | p < 0.0001 | 44 | 81.5 |
| CpG +95 | 12 | 5 | 4 | 33 | p < 0.0001 | 45 | 83 |
| CpG +113 | 13 | 6 | 3 | 32 | p < 0.0001 | 45 | 83 |
| CpG +135 | 13 | 6 | 3 | 32 | p < 0.0001 | 45 | 83 |
| CpG +137 | 13 | 7 | 3 | 31 | p < 0.0001 | 44 | 81.5 |
| CpGs −395 to −352 | 14 | 10 | 2 | 28 | p < 0.0001 | 42 | 78 |
| CpGs −249 to −207 | 15 | 13 | 1 | 25 | p < 0.0001 | 40 | 74 |
| CpGs −186 to −172 | 12 | 6 | 4 | 32 | p < 0.0001 | 44 | 81.5 |
| CpGs −154 to −128 | 16 | 16 | 0 | 22 | p < 0.0001 | 38 | 70 |
| CpGs +93 to +153 | 13 | 7 | 3 | 31 | p < 0.0001 | 44 | 81.5 |
| MSP region | 12 | 11 | 4 | 27 | p = 0.0017 | 39 | 72 |
Abbreviations: CpG, cysteine-phosphate-guanine; MSP, methylation-specific PCR assay.
We then examined whether a specific CpG site could best reflect the expression of MGMT. We analyzed each CpG separately to identify discriminating CpG sites. MGMT expression and MGMT methylation were considered as qualitative and quantitative variables, respectively. We found that all CpG sites were significantly related to expression. We thus analyzed both methylation and expression as qualitative variables for each CpG alone to determine the specificity. CpGs −228, −186, 95, 113, 135, and 137 were those that best correlated with expression, with 81.5%–83% concordance (Table 2; Fig. 2).
We also aimed to identify a CpG region as a marker of expression since reducing the number of analyzed CpG sites could therefore facilitate clinical study setting. For this, we selected the CpG sites most correlated with expression, using methylation as a quantitative variable (CpGs at −395 to −352, −249 to −207, −186 to −172, −154 to −128, and +93 to +153; p ≤ 0.0006) and analyzed them with both methylation and expression as qualitative variables (Table 2). The most concordant results were observed for CpGs −186 to −172 and CpGs +93 to +153 with 81.5% concordance. The other CpG regions showed a larger percentage of discordant results due to an elevated percentage of unmethylated patients with low expression, up to 50% for CpGs −154 to −128.
In addition, we separately analyzed the region that is commonly studied by MSP (CpGs at +118 to +137 and at +174 to +195 for the methylated primers; Fig. 1). Fifty-seven percent of patients were methylated. Although the majority of methylated patients expressed a low level of MGMT (27 of 31, 87%), unmethylated patients were found to express either a high (12 of 23) or low (11 of 23) level of MGMT. Overall, 28% discordant results were observed, demonstrating the analytical superiority of the novel regions identified in our study.
Discussion
This study demonstrates that MGMT CGI methylation correlates inversely with MGMT gene expression, but also points out that the relationship between methylation and expression is not absolute.
In the 54 analyzed GBM tumors, the methylation pattern of the 52 CpG sites studied was found to be homogeneous (group 1, methylated, or group 3, unmethylated) in 63% of patients, whereas an intermediate methylation pattern, with some CpG sites being methylated and others not, was found in 37% of patients (group 2). Methylation of the whole 52 CpG sites correlated with expression with an 85% rate of concordance. To identify small CpG regions that best reflect MGMT gene expression, notably for a clinical study setting, we analyzed each CpG separately. Six isolated CpG sites (CpGs −228, −186, +95, +113, +135, and +137) were of interest, as well as two CpG regions (−186 to −172, and +93 to +153), each with a minimum of 81.5% of concordant results between methylation and expression.
The four CpG candidates (−228, −186, +125, and +137) that could be linked to expression according to Pieper et al.16 were among those with the most concordant results (80%–81.5%) in our study. The region commonly investigated by MSP was not among the regions that best correlated with expression, although we found a rate of methylation (57%) similar to previous reports (45%–68%).3,4,11,14,20
Promoter CGIs can present a differential pattern of methylation along the CGI, and some CpG sites may be more important than others with regard to expression.21 In the case of MGMT, we identified five distinct regions associated with gene silencing, two of them reflecting MGMT expression better than the others. These regions bracketed a relatively methylation-free region of the CGI, which contained the minimal promoter, the TSS, and the first CpG sites of the noncoding exon. These results reinforce the fact that MGMT silencing is influenced by methylation of sites that are distant from the TSS.9,10,16 Therefore, it seems that core regions do not necessarily include the TSS and the minimal promoter.
We examined whether transcription factor binding sites were present within particular CpG regions that correlated best with expression. Putative transcription factor binding sites were described initially by Harris et al.22 when they cloned and sequenced the promoter region of MGMT. Proposed activator protein-1 (AP1) and AP2 binding sites are located upstream of the CGI, and the majority of specificity protein 1 (Sp1) binding sites are within the methylation-free region. Remarkably, one CpG region associated with MGMT expression (−249 to −207) harbors an Sp1 binding site overlapping CpG sites −246 and −242. However, neither these latter CpG sites nor the region −249 to −207 was among those that best reflect the MGMT expression. Very few reports have been dedicated to a functional analysis of potential MGMT transcription factors. Glucocorticoid-responsive element and nuclear factor κB (NF-κB) transcription factor binding sites were reported23,24 but are not located at specific hypermethylated CpG regions associated with MGMT expression. The NF-κB transcription factor binding sites are nonetheless located just upstream of the CpG region +93 to +153.
The relationship between methylation and expression could be confirmed for 85% of the patients, at best. Other epigenetic mechanisms could explain the fact that some patients were unmethylated for the MGMT CGI but expressed a low level of MGMT, such as histone modifications.25 It is possible that MGMT DNA methylation plays only an indirect role in the regulation of MGMT expression.16 Indeed, it has been suggested that DNA methylation in cancer could be a secondary process to an initial dramatic change in expression.26,27 MGMT gene silencing observed in nonmethylated patients could also be the result of some genetic alterations. The MGMT gene is located in 10q26, a region that is frequently heterozygously lost in GBM.28 However, no relationship was found between the expression of MGMT transcripts and the loss of 10q. The presence of mutations within the body of the gene has not been extensively explored. Only one report described MGMT gene mutations in 10 of 40 patients with esophageal cancer,29 but whether these mutations affect enzymatic function or lead to modification of the expression level was not investigated. In contrast, some patients with a methylated MGMT CGI still expressed high levels of MGMT. A recent study in cell lines and primary gliomas found that MGMT expression correlates with NF-κB activation, whereas no relationship was found between MGMT methylation and MGMT expression.24 Furthermore, in HEK293 cells, overexpression of NF-κB/p65 increased MGMT expression despite the fact that the MGMT promoter was methylated in these cells.
Many retrospective clinical studies investigated either MGMT expression or MGMT methylation to determine the prognostic impact of MGMT. Both biomarkers have shown a prognostic association between MGMT status and outcome. Whereas MGMT protein levels seem to be largely controlled transcriptionally and not translationally or posttranslationally,30 MGMT methylation does not always reflect gene expression as demonstrated in our and other studies, and it seems difficult to use one measure interchangeably with the other.11,12,31 To date, MGMT promoter methylation status seems to be a robust prognostic predictor in GBM patients treated with alkylating chemotherapy.4 Indeed, Gorlia et al.32 recently recommended stratifying patients according to MGMT promoter methylation in future GBM trials without further indication about MGMT expression. Hegi and colleagues showed that treatment with temozolomide led to a clear survival benefit in GBM patients with a methylated MGMT promoter,4 whereas no association was found between MGMT expression as determined by immunohistochemistry and patient survival in the same population.31 Conversely, Brell et al.14 observed a prognostic impact of MGMT expression as determined by immunohistochemistry in 72 anaplastic glioma patients who received chemotherapy. This impact was not confirmed when analyzing MGMT promoter methylation status as a marker. These discrepancies do not seem related to intratumoral heterogeneity since MGMT methylation and MGMT expression appear to be a global tumor phenomenon.15,33 Rather, it may be explained by observer variability relative to the immunohistochemistry technique31 in addition to population dissimilarities, grade of malignancy, and size of each cohort. Unfortunately, in our study, we could not determine which of the two biomarkers is clinically more relevant due to the limited number and short follow-up of our cohort. However, if transcriptional repression is the main mechanism explaining the higher chemosensitivity of MGMT-methylated tumors, our data suggest that clinical decisions based on MGMT methylation status alone could leave behind a significant minority of unmethylated patients with low expression of MGMT who are potentially sensitive to treatment. Conversely, some methylated patients with high expression of MGMT could also be inadequately included.
To conclude, pyrosequencing appears to be a good and reliable technique to evaluate MGMT methylation status. However, the best MGMT methylation assay remains unsettled,13,30–32 and prospective comparisons of available assays are still needed. Furthermore, careful comparisons between methylation and expression studies should also be performed in future prospective clinical trials to determine which information best predicts sensitivity to alkylating agents.
Acknowledgments
S.E. was supported by the Ligue Nationale Contre le Cancer. This study was supported by the Ligue Nationale Contre le Cancer, the Fédération pour la Recherche sur le Cerveau, and the Association pour la Recherche sur le Cancer.
References
- 1.Esteller M, Hamilton SR, Burger PC, Baylin SB, Herman JG. Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is a common event in primary human neoplasia. Cancer Res. 1999;59:793–797. [PubMed] [Google Scholar]
- 2.Esteller M, Garcia-Foncillas J, Andion E, et al. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med. 2000;343:1350–1354. doi: 10.1056/NEJM200011093431901. [DOI] [PubMed] [Google Scholar]
- 3.Hegi ME, Diserens AC, Godard S, et al. Clinical trial substantiates the predictive value of O-6-methylguanine-DNA methyltransferase promoter methylation in glioblastoma patients treated with temozolomide. Clin Cancer Res. 2004;10:1871–1874. doi: 10.1158/1078-0432.ccr-03-0384. [DOI] [PubMed] [Google Scholar]
- 4.Hegi ME, Diserens AC, Gorlia T, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352:997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 5.Criniere E, Kaloshi G, Laigle-Donadey F, et al. MGMT prognostic impact on glioblastoma is dependent on therapeutic modalities. J Neurooncol. 2007;83:173–179. doi: 10.1007/s11060-006-9320-0. [DOI] [PubMed] [Google Scholar]
- 6.Everhard S, Kaloshi G, Criniere E, et al. MGMT methylation: a marker of response to temozolomide in low-grade gliomas. Ann Neurol. 2006;60:740–743. doi: 10.1002/ana.21044. [DOI] [PubMed] [Google Scholar]
- 7.Costello JF, Futscher BW, Tano K, Graunke DM, Pieper RO. Graded methylation in the promoter and body of the O6-methylguanine DNA methyltransferase (MGMT) gene correlates with MGMT expression in human glioma cells. J Biol Chem. 1994;269:17228–17237. [PubMed] [Google Scholar]
- 8.Costello JF, Futscher BW, Kroes RA, Pieper RO. Methylation-related chromatin structure is associated with exclusion of transcription factors from and suppressed expression of the O-6-methylguanine DNA methyltransferase gene in human glioma cell lines. Mol Cell Biol. 1994;14:6515–6521. doi: 10.1128/mcb.14.10.6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Watts GS, Pieper RO, Costello JF, Peng YM, Dalton WS, Futscher BW. Methylation of discrete regions of the O6-methylguanine DNA methyltransferase (MGMT) CpG island is associated with heterochromatinization of the MGMT transcription start site and silencing of the gene. Mol Cell Biol. 1997;17:5612–5619. doi: 10.1128/mcb.17.9.5612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qian XC, Brent TP. Methylation hot spots in the 5’ flanking region denote silencing of the O6-methylguanine-DNA methyltransferase gene. Cancer Res. 1997;57:3672–3677. [PubMed] [Google Scholar]
- 11.Maxwell JA, Johnson SP, Quinn JA, et al. Quantitative analysis of O6-alkylguanine-DNA alkyltransferase in malignant glioma. Mol Cancer Ther. 2006;5:2531–2539. doi: 10.1158/1535-7163.MCT-06-0106. [DOI] [PubMed] [Google Scholar]
- 12.Mollemann M, Wolter M, Felsberg J, Collins VP, Reifenberger G. Frequent promoter hypermethylation and low expression of the MGMT gene in oligodendroglial tumors. Int J Cancer. 2005;113:379–385. doi: 10.1002/ijc.20575. [DOI] [PubMed] [Google Scholar]
- 13.Jeuken JW, Cornelissen SJ, Vriezen M, et al. MS-MLPA: an attractive alternative laboratory assay for robust, reliable, and semiquantitative detection of MGMT promoter hypermethylation in gliomas. Lab Invest. 2007;87:1055–1065. doi: 10.1038/labinvest.3700664. [DOI] [PubMed] [Google Scholar]
- 14.Brell M, Tortosa A, Verger E, et al. Prognostic significance of O6-methylguanine-DNA methyltransferase determined by promoter hypermethylation and immunohistochemical expression in anaplastic gliomas. Clin Cancer Res. 2005;11:5167–5174. doi: 10.1158/1078-0432.CCR-05-0230. [DOI] [PubMed] [Google Scholar]
- 15.Grasbon-Frodl EM, Kreth FW, Ruiter M, et al. Intratumoral homogeneity of MGMT promoter hypermethylation as demonstrated in serial stereotactic specimens from anaplastic astrocytomas and glioblastomas. Int J Cancer. 2007;121:2458–2464. doi: 10.1002/ijc.23020. [DOI] [PubMed] [Google Scholar]
- 16.Pieper RO, Patel S, Ting SA, Futscher BW, Costello JF. Methylation of CpG island transcription factor binding sites is unnecessary for aberrant silencing of the human MGMT gene. J Biol Chem. 1996;271:13916–13924. doi: 10.1074/jbc.271.23.13916. [DOI] [PubMed] [Google Scholar]
- 17.Dupont JM, Tost J, Jammes H, Gut IG. De novo quantitative bisulfite sequencing using the pyrosequencing technology. Anal Biochem. 2004;333:119–127. doi: 10.1016/j.ab.2004.05.007. [DOI] [PubMed] [Google Scholar]
- 18.Tost J, Gut IG. DNA methylation analysis by pyrosequencing. Nat Protoc. 2007;2:2265–2275. doi: 10.1038/nprot.2007.314. [DOI] [PubMed] [Google Scholar]
- 19.Auger N, Thillet J, Wanherdrick K, et al. Genetic alterations associated with acquired temozolomide resistance in SNB-19, a human glioma cell line. Mol Cancer Ther. 2006;5:2182–2192. doi: 10.1158/1535-7163.MCT-05-0428. [DOI] [PubMed] [Google Scholar]
- 20.Blanc JL, Wager M, Guilhot J, et al. Correlation of clinical features and methylation status of MGMT gene promoter in glioblastomas. J Neurooncol. 2004;68:275–283. doi: 10.1023/b:neon.0000033385.37098.85. [DOI] [PubMed] [Google Scholar]
- 21.Ushijima T. Detection and interpretation of altered methylation patterns in cancer cells. Nat Rev Cancer. 2005;5:223–231. doi: 10.1038/nrc1571. [DOI] [PubMed] [Google Scholar]
- 22.Harris LC, Potter PM, Tano K, Shiota S, Mitra S, Brent TP. Characterization of the promoter region of the human O6-methylguanine-DNA methyltransferase gene. Nucleic Acids Res. 1991;19:6163–6167. doi: 10.1093/nar/19.22.6163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Biswas T, Ramana CV, Srinivasan G, et al. Activation of human O6-methylguanine-DNA methyltransferase gene by glucocorticoid hormone. Oncogene. 1999;18:525–532. doi: 10.1038/sj.onc.1202320. [DOI] [PubMed] [Google Scholar]
- 24.Lavon I, Fuchs D, Zrihan D, et al. Novel mechanism whereby nuclear factor kappaB mediates DNA damage repair through regulation of O(6)-methylguanine-DNA-methyltransferase. Cancer Res. 2007;67:8952–8959. doi: 10.1158/0008-5472.CAN-06-3820. [DOI] [PubMed] [Google Scholar]
- 25.Zhao W, Soejima H, Higashimoto K, et al. The essential role of histone H3 Lys9 di-methylation and MeCP2 binding in MGMT silencing with poor DNA methylation of the promoter CpG island. J Biochem (Tokyo) 2005;137:431–440. doi: 10.1093/jb/mvi048. [DOI] [PubMed] [Google Scholar]
- 26.Turker MS. Gene silencing in mammalian cells and the spread of DNA methylation. Oncogene. 2002;21:5388–5393. doi: 10.1038/sj.onc.1205599. [DOI] [PubMed] [Google Scholar]
- 27.Clark SJ, Melki J. DNA methylation and gene silencing in cancer: which is the guilty party? Oncogene. 2002;21:5380–5387. doi: 10.1038/sj.onc.1205598. [DOI] [PubMed] [Google Scholar]
- 28.Behin A, Hoang-Xuan K, Carpentier AF, Delattre JY. Primary brain tumours in adults. Lancet. 2003;361:323–331. doi: 10.1016/S0140-6736(03)12328-8. [DOI] [PubMed] [Google Scholar]
- 29.Wang L, Zhu D, Zhang C, et al. Mutations of O6-methylguanine-DNA methyltransferase gene in esophageal cancer tissues from Northern China. Int J Cancer. 1997;71:719–723. doi: 10.1002/(sici)1097-0215(19970529)71:5<719::aid-ijc5>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 30.Pieper RO. Understanding and manipulating O6-methylguanine-DNA methyltransferase expression. Pharmacol Ther. 1997;74:285–297. doi: 10.1016/s0163-7258(97)00003-x. [DOI] [PubMed] [Google Scholar]
- 31.Preusser M, Janzer RC, Felsberg J, et al. Anti-O6-methylguaninemethyltransferase (MGMT) immunohistochemistry in glioblastoma multiforme: observer variability and lack of association with patient survival impede its use as clinical biomarker. Brain Pathol. 2008;18:520–532. doi: 10.1111/j.1750-3639.2008.00153.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gorlia T, van den Bent MJ, Hegi ME, et al. Nomograms for predicting survival of patients with newly diagnosed glioblastoma: prognostic factor analysis of EORTC and NCIC trial 26981-22981/CE.3. Lancet Oncol. 2008;9:29–38. doi: 10.1016/S1470-2045(07)70384-4. [DOI] [PubMed] [Google Scholar]
- 33.Parkinson JF, Wheeler HR, Clarkson A, et al. Variation of O(6)-methylguanine-DNA methyltransferase (MGMT) promoter methylation in serial samples in glioblastoma. J Neurooncol. 2008;87:71–78. doi: 10.1007/s11060-007-9486-0. [DOI] [PubMed] [Google Scholar]