Abstract
Medulloblastoma is a highly malignant brain tumor that occurs predominantly in children. The molecular pathogenesis of medulloblastoma is under investigation. Previously, we used complementary DNA micro-array analysis to compare patterns of gene expression in medulloblastoma samples versus normal cerebellum. The cytoskeletal protein ezrin was found to be overexpressed in medulloblastoma compared with normal cerebellum, an observation that was further validated by immunohistochemistry and real-time PCR analysis. To assess the role of ezrin in medulloblastoma, we studied ezrin’s role in medulloblastoma migration, invasion, and adhesion. Western blotting and immunofluorescence showed high expression of ezrin in four medulloblastoma cell lines, and ezrin was primarily localized to filopodia. Ezrinspecific small interfering RNA suppressed the formation of filopodia and in vitro migration, invasion, and adhesion. We also used a stably transfected medulloblastoma cell line to study the effect of ezrin overexpression. We showed that high expression of ezrin promotes filopodia formation and in vitro invasion. Finally, athymic mice implanted with ezrin-overexpressing DAOY medulloblastoma cell clones in the cerebellum showed shortened survival compared with controls. These findings suggest that, in addition to other cytoskeletal proteins, ezrin plays an important role in medulloblastoma adhesion, migration, and invasion.
Keywords: cytoskeleton, ezrin, invasion, medulloblastoma, siRNA
Medulloblastoma (MB) is the most common malignant brain tumor to occur in children. Despite advances in surgery, radiation therapy, and chemotherapy, the long-term survival of children is approximately 60%, with significant cognitive and focal neurological deficits caused by both the disease and its treatments.1,2 The major barriers to effective therapy for MB include its invasive behavior and its propensity for cerebrospinal fluid (CSF) dissemination.3 CSF dissemination and leptomeningeal metastasis have been reported in approximately 10%–35% of the cases.4
The molecular biology of MB is beginning to be understood. Perturbed signaling of the central developmental signaling pathways sonic hedgehog homolog (SHH), wingless (WNT/WG), and receptor tyrosine kinase I family ErbB has been implicated in the formation of MB.5–7 Initial cytogenetic and molecular genetic studies of MB identified several nonrandom genetic aberrations that are found on chromosomes 1, 9, 10, 11, 16, and 17 (reviewed by Pietsch et al.8). Loss of chromosome 17p is the most frequent aberration in MB, occurring in up to 50% of the cases and correlating with the occurrence of an isochromosome 17q.9 This suggests the presence of one or more MB-related tumor suppressor gene(s) on chromosome 17p.10,11
Transcriptional profiling of MB has revealed several interesting potential targets in numerous studies.6,12–14 In a previous transcriptional profiling analysis, we identified several genes that were overexpressed in MB compared with normal cerebellum.6 We showed that the cytoskeletal cross-linking protein ezrin was one of the most highly expressed targets in the screen.6 Accordingly, here we describe a study of the role of ezrin in MB.
Ezrin, one of the ERM (ezrin-radixin-moesin) proteins, is involved in the formation of cell-surface processes such as lamellipodia and filopodia and is expressed in a variety of tissues. Ezrin typically concentrates at the apical surface of polarized epithelia.15,16 ERM proteins function to organize specialized cell-membrane domains by linking the actin cytoskeleton to multiple membrane-associated proteins, such as CD44 and adhesion molecules.17 Interestingly, ezrin upregulation has been associated with the proliferation and immortalization of fibroblasts,18 the acquisition of an invasive phenotype in transformed esophageal epithelial cells,19 and endometrial cancer cells.20 In addition, ezrin over-expression enhances the metastatic potential in various tumor types, including osteosarcoma,21,22 rhabdomyosarcoma,23 and carcinomas of the pancreas,24 ovary,25 and endometrium.26
To assess the involvement of ezrin expression in MB, we analyzed ezrin expression in untreated MB cell lines by Western blotting and immunofluorescence. Moreover, using small interfering RNAs (siRNAs) against ezrin, and ezrin-overexpressing MB cell clones, we conducted cellular adhesion, migration, and invasion assays and immunofluorescence and Western blotting methods to elucidate the role of ezrin in MB.
Materials and Methods
Cell Culture
We cultured the permanent human MB cell lines DAOY, ONS76, UW228, and MED8A in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum (FBS), and cell lines D425 and D458 in improved minimal essential medium containing 20% FBS, HEPES, and sodium bicarbonate, maintained at 37°C in a humidified atmosphere (95% air and 5% CO2). D425 and D458 cells were generously provided by Darell Bigner (Duke University, Durham, NC, USA). DAOY, ONS76, UW228, and MED8A cells were a gift of Dolores Dougherty (University of California, San Francisco, San Francisco, CA, USA).
Ezrin siRNA Preparation and Transfection
The ezrin1 siRNA sequences used correspond to bp 312–332 after the start codon of the ezrin gene (5′-AAGCTGGATAAGAAGGTGTCT); and the ezrin2 siRNA sequences correspond to bp 457–477 after the start codon of ezrin (5′-AAGGAATCCTTAGCGAT-GAGA). The 21-bp siRNAs were purchased from Eurogentec S.A. (San Diego, CA, USA). Ezrin siRNA pool (Ez3) was purchased from Dharmacon (Lafayette, CO, USA) and consists of four duplex siRNAs, each independently verified to target the human ezrin open reading frame. Transient transfections of siRNA against ezrin were carried out using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) or DharmaFect transfection reagent (Dharmacon). Cells were plated at 50% confluency in DMEM containing 10% serum without antibiotics. Transfections were carried out 24 h later. The siRNA was diluted in serum-free DMEM. Lipofectamine 2000 was also diluted in serum-free DMEM for 5 min. The two mixtures were combined and incubated for 20 min at room temperature to enable complex formation. Cells were assayed 2 or 3 days after transfection. As a control for off-target effects caused by RNA interference, nontargeting siRNA, designed to have at least four mismatches with human and mouse sequences, was used. Ezrin expression was determined by Western blot analysis using a monoclonal antibody against ezrin.
Ezrin DNA Constructs and MB Cell Transfection
A wild-type human ezrin complementary DNA (cDNA) clone was purchased from the Hospital for Sick Children (Toronto, ON, Canada). The ezrin cDNA was amplified by PCR with the forward primer 5′-GCGGAATTCAC-CGAAACCAATCAATGTC-3′ and the reverse primer 5′-GCGTCTAGATTACAGGGCCTCGAACTC-3′ using Platinum Taq DNA Polymerase High Fidelity (Invitrogen). The fragment obtained was inserted into the plasmid pCRII (Invitrogen) using the TA cloning site and verified by double-strand DNA sequencing. The pFLAG-CMV-2 expression vector (Sigma, St. Louis, MO, USA) was cut at unique EcoRI and XbaI restriction sites, and the cDNA obtained from plasmid pCRII was subcloned into pFLAG-CMV-2 by T4 DNA ligase (Invitrogen). The plasmid was transformed into competent Escherichia coli DH5α cells by heat shock (Invitrogen) before extraction with the Plasmid Midi Kit (Qiagen, Hilden, Germany).
For stable transfections, exponentially growing DAOY cells were seeded 24 h before DNA transfer onto 10-cm2 tissue culture dishes. The cells were cotransfected with pFLAG-CMV-2 with or without the ezrin construct, and with pGK-puro (gift of Jane McGlade, Hospital for Sick Children, Toronto, ON, Canada) using Lipofectamine 2000. Clones from the transfected cells were selected by growth in medium containing 1 μg/ml puromycin (Sigma). Ezrin expression was determined by Western blotting using specific anti-FLAG and anti-ezrin antibodies (see “Western Blotting,” below). The selected MB clones were maintained in DMEM containing 10% fetal bovine serum and 1 μg/ml puromycin for further study.
Microarray Analysis
The Affymetrix Human Genome U133 Plus 2.0 Arrays were used according to the Affymetrix GeneChip Expression Analysis technical manual (Affymetrix, Santa Clara, CA, USA). This platform contains approximately 47,000 human genes. Results of the microarray analysis were scanned using the model 3000 Affymetrix Gene Chip Scanner. Expression analysis was performed using the GeneChip Operating Software according to manufacturer’s specifications. Hybridized Human Genome U133A 2.0 Arrays was scanned using the Affymetrix Microarray Analysis Suite software, version 5.0. Pairwise comparisons were conducted on MB cell lines and MB primary tumor specimens, both compared with normal cerebellum.
Cell Extractions
Cells were harvested and extracted with lysis buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 2 mM EDTA, 1% Triton X-100), with Protease Inhibitor Cocktail tablets (Roche, Mannheim, Germany) and phosphatase inhibitor cocktail tablet II (Calbiochem, La Jolla, CA, USA). The extract was centrifuged, and the supernatant constituted the Triton-soluble fraction. Total cell aliquots were assayed for protein content using the Bio-Rad DC protein assay (Bio-Rad Laboratories, Hercules, CA, USA). Pellets were extracted with sodium dodecyl sulfate (SDS) lysis buffer, boiled for 3 min after sonication, and centrifuged. The supernatant here constituted the Triton-insoluble fraction. To determine the total fraction of CD44 protein, cells were extracted with SDS sample buffer containing Tris (pH 6.8), 20% SDS, glycerol, and β-mercaptoethanol. The extract was boiled for 3 min after sonication. Total and insoluble fractions of cell extracts were used for the detection of CD44.
Western Blotting
Equivalent amounts of protein were loaded onto 8% or 10% (wt/vol) SDS polyacrylamide gels in sample buffer and subjected to electrophoresis. Proteins were then transferred to Immobilon-P polyvinylidene fluoride membranes (Millipore, Bedford, MA, USA), probed with first and second antibodies, and visualized using a chemiluminescence reagent (PerkinElmer Life Sciences Inc., Boston, MA, USA). Dilutions for the first antibodies used in this study were anti-phospho-ezrin (1:500; Cell Signaling, Danvers, MA, USA), anti-ezrin (1:500; Cell Signaling), anti-FLAG (1:500; Sigma), anti-moesin (1:500; Cell Signaling), anti-Flotillin-2 (1:500; BD Biosciences, San Jose, CA, USA), anti-β-actin (1:1,000; Sigma), and anti-CD44 (1:1,000; kindly provided by Dr. Michelle Letarte, the Research Institute, the Hospital for Sick Children, Toronto, ON Canada).27
Immunofluorescence and Confocal Microscopy
The MB cell lines and clones were grown on cover slips, washed three times with phosphate-buffered saline (PBS), and fixed for 10 min with 4% paraformaldehyde. Following three washes with PBS, cells were permeabilized for 5 min with 0.5% Triton X-100. Nonspecific binding was blocked by 1% BSA in PBS for 1 h at room temperature. The primary antibodies were diluted in blocking buffer at the following concentrations: anti-phospho-, ezrin 1:200; antiezrin, 1:200; and anti-CD44, 1:200. Following a 1-h incubation period with the primary antibody, the MB cells were rinsed with PBS and then incubated with fluorescein isothiocyanate–labeled goat antimouse immunoglobulin antibodies or Alexa488 (Molecular Probes, Eugene, OR, USA) diluted 1:200 as secondary antibodies for 30 min. The DNA was stained with 4′,6-diamidino-2-phenylindole (DAPI; Promega, Madison, WI, USA), and filamentous actin was visualized with tetramethyl rhodamine isothiocyanate phalloidin in 1:50 dilution (Molecular Probes). A Zeiss Spinning Disk Confocal microscope (Carl Zeiss Inc., Thornwood, NY, USA) was used to visualize the fluorescence patterns.
Cell Viability
The viability of MB cell lines and clones was assessed using the MTS [3-(4,5-dimethylthiazol-2-yl)-5-(carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt; Promega] assay according to the manufacturer’s instructions. Briefly, 2 × 103 untreated MB cells and cells transfected with ezrin siRNA or controls were suspended in 100 μl culture medium and seeded onto 96-well plates. The MB cells were then incubated at 37°C in a humidified atmosphere in 5% CO2 for 48 h. Each sample was tested in triplicate. MTS and the electron coupling reagent phenazine ethosulfate were then added to MB cells, which were incubated at 37°C for 1 h. The absorbance of the reaction was measured at 520 nm with a Spectrum MAX250 microtiter plate reader (Molecular Devices, Sunnyvale, CA, USA). The percent cell viability was then calculated as (experiment – blank) / (control – blank) × 100%, where the blank was a medium containing MTS solution without cells, and the control was the signal from the wells containing cells with 0.1% ethanol but no serum medium.
Motility Assay
The motility assay was performed using the microliter scale radial monolayer assay as described previously.28 Briefly, 10-well Teflon-coated slides (CSM Inc., Phoenix, AZ, USA) were coated with 10 mg/ml laminin (LN; Sigma) and 0.1% BSA. Cells were then seeded through a cell sedimentation manifold (CSM Inc.) at 3,000 cells/well to establish a circular confluent monolayer at the center of the substrate-coated well. Twenty-four hours after seeding, a circle of best fit that circumscribed the cells was drawn. The cells were allowed to migrate out over a 24-h period, and another circle circumscribing the cells was drawn. Motility results were reported as the change in the diameter of the circumscribed cell population over a 24-h period (mm/h). Measurements were performed using a Leica DM IRE2 inverted microscope (Leica Microsystems, Bannokburn, IL, USA) and digitized using a Spot camera, and image analysis was performed (Scion Image, Frederick, MD, USA).
Invasion Assay
The invasion assay was performed using 24-well BD Bio-coat Matrigel invasion chambers with 8-μm polycarbonate filters (Becton Dickinson, Bedford, MA, USA). The DAOY and ONS76 cell lines transfected with siRNA for ezrin or nontargeting siRNA were seeded onto Matrigel-coated filters, cultured in routine medium, and incubated for 22 h at 37°C in a humidified incubator with 5% CO2. Nonmigratory cells on the upper surface of the filter were removed by wiping with a cotton swab. Invasive cells that penetrated through the pores and migrated to the underside of the membrane were stained with 1% crystal violet solution after fixation with 4% paraformaldehyde in PBS. Cells were counted with a light microscope, and the average cell number was determined.
Adhesion Assay
Adhesion assays were performed as described previously.29 Ninety-six–well plates were coated with 10 μg/ml LN diluted with PBS and 0.1% BSA overnight at 4°C. The unbound sites were then blocked with serum-free medium (PBS containing 0.1% BSA) by incubating the wells for 1 h at 37°C. Single-cell suspensions of MB cells were washed in serum-free medium and resuspended at a concentration of 5 × 105 cells/ml, and 100 μl was added to each well. The plates were incubated for 3 h at 37°C. The plates were washed twice with PBS to remove unbound cells. Adherent cells were fixed with 4% paraformaldehyde for 10 min and 0.5% crystal violet for 20 min. The cells were gently washed, and adherent cells were quantified by measuring the absorbance at 595 nm on a microtiter plate reader.
Tumorigenicity in Athymic Mice
Six-week-old male nu/nu mice were purchased from Charles River Canada Labs (St. Constant, QC, Canada). The animals were housed in groups of five in a vivarium maintained on a light/dark schedule with a constant temperature and humidity. Food and water were available ad libitum. All procedures in mice were reviewed and approved by the Research Ethics Board of the Hospital for Sick Children.
Approximately 1.0 × 105 ezrin-transfected DAOY cells or controls were stereotactically injected into the cerebellum of the athymic mice through a 0.5-mm burr hole in the midline 2 mm posterior to the lambdoid using a rodent head frame (Lab Standard Stereotaxic Instrument, Stoelting, Wood Dale, IL, USA). Mice demonstrating 20% body weight loss or that had difficulty ambulating, feeding, or grooming were sacrificed. The mouse brains and spinal cords were then removed from the calvaria and spine and fixed in 10% formalin before histological examination using hematoxylin and eosin and immunohistochemistry with ezrin antibodies.
Statistical Analysis
Survival curves were generated by the Kaplan-Meier method. The log-rank statistic was used to compare the distributions of survival times. All reported p-values were two-sided and were considered to be statistically significant at <0.05.
Results
Ezrin Is Overexpressed in MB Cell Lines
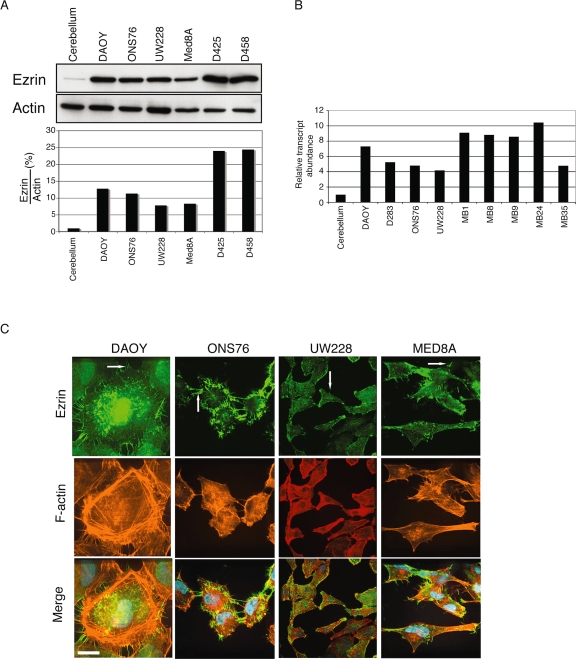
Previously, a transcriptional profiling analysis of pediatric MBs revealed high ezrin expression; therefore, we measured ezrin expression in the normal cerebellum and in multiple MB cell lines (Fig. 1A). Western blotting showed higher expression of ezrin in all MB cell lines than in samples of normal cerebellum. In addition, we compared the ezrin gene expression profile between primary MB samples, MB cell lines, and normal human cerebellum by microarray. We found that ezrin RNA expression was increased relative to normal cerebellum in a subset of primary patient MB samples and in MB cell lines (Fig. 1B). By immunofluorescence confocal microscopy, ezrin (Fig. 1C) is localized to cell-surface membrane extensions and filopodia in MB cells. Previously, ezrin was shown to colocalize with F-actin at filopodia16; although we observed a partial overlap of ezrin and F-actin in MB cells, we found a significant amount of ezrin that did not colocalize with F-actin. We believe this may be due to the aberrant overexpression of ezrin in MB cells, which results in a widespread ubiquitous expression pattern.
Fig. 1.
Ezrin expression in medulloblastoma (MB) cell lines. (A) Top: Ezrin in normal cerebellum and MB cell lines revealed by Western blotting. β-actin was used as a loading control. Ezrin is overexpressed in all MB cell lines compared with normal cerebellum. Bottom: Quantification by densitometry of a representative experiment. (B) Ezrin gene expression analysis of primary patient MB samples (right) and MB cell lines (left) compared with normal human cerebellum using the Affymetrix Human Genome U133 Plus 2.0 Arrays. (C) Distribution of ezrin in DAOY, ONS76, UW228, and MED8A cells. Cells were immunolabeled for ezrin (green) and F-actin (red) to localize endogenous proteins by immunofluorescence. Yellow indicates colocalization of both proteins at filopodia and membrane extensions (arrows). Nuclei were stained using 4’,6-diamidino-2-phenylindole staining (blue). Scale bar, 25 μm.
Ezrin siRNA Inhibits Filopodia Formation
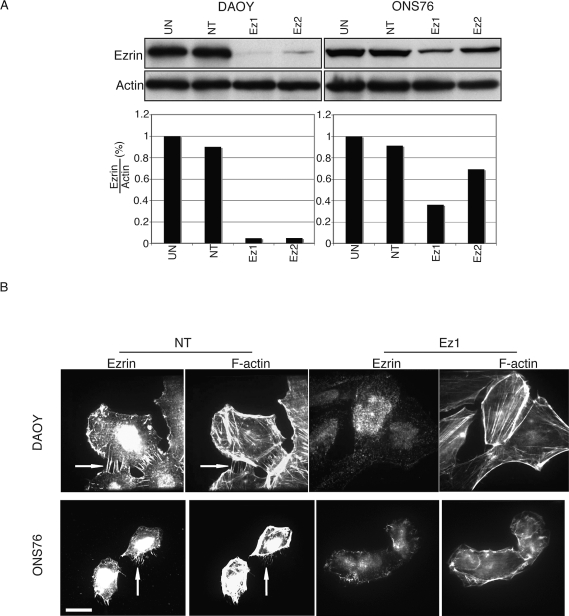
Because ezrin localizes to filopodia and plasma membrane extensions, suggesting a role for ezrin in MB migration, we assessed the effect of ezrin knockdown on MB cell lines. DAOY and ONS76 cell lines were treated with distinct ezrin siRNAs, and Western blot analysis revealed significant reduction of ezrin protein levels (Fig. 2A). By immunofluorescence, both DAOY and ONS76 siRNA-transfected cells showed decreased filopodia formation (arrows, Fig. 2B). Ezrin siRNA treatment of MB cell lines did not substantially alter cell viability compared with controls (see supplementary material, Fig. 1S, A). In addition we assessed whether ezrin siRNA was specific for ezrin by analyzing the expression of the ERM family member moesin by Western blot analysis and found no difference in moesin expression following ezrin knockdown (see supplementary Fig. 2S).
Fig. 2.
Ezrin small interfering RNA (siRNA) depletes endogenous ezrin expression in medulloblastoma cells. (A) Top: Immunoblot analysis of ezrin protein expression in DAOY (left) and ONS76 (right) after ezrin knockdown by siRNA. Bottom: Quantification by densitometry of a representative experiment. Abbreviations: UN, untreated; NT, nontargeting siRNA; Ez1 and Ez2, two different ezrin knockdown sequences used. (B) Morphological change of DAOY and ONS76 cells after transfection with siRNA. After ezrin knockdown, cells were plated on cover slips for immunofluorescence. Cells were labeled with phalloidin for F-actin, and ezrin was labeled with antiezrin antibody by Alexa488-conjugated antimouse antibody. Arrows show filopodia bundles. Scale bar, 25 μm.
Ezrin siRNA Alters MB Adhesion, Migration, and Invasion
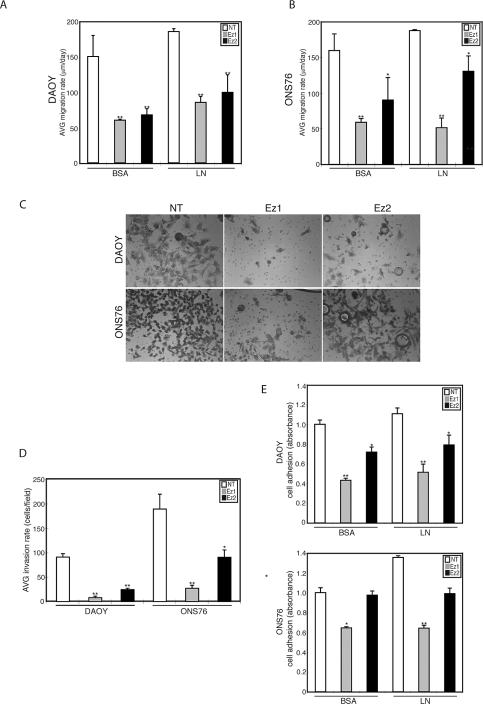
The changes in filopodia formation following siRNA knockdown of ezrin suggest that cell motility functions such as migration and invasion may be affected. Therefore, MB migration was examined using the radial monolayer assay. We show that ezrin knockdown markedly attenuates cell motility in DAOY (Fig. 3A) and ONS76 (Fig. 3B) MB cell lines. In addition, MB cells seeded onto LN as a substrate had slightly higher motility rates than did MB cells seeded onto BSA. Next, we used Boyden chambers to assess in vitro invasion. We observed that ezrin siRNA mediated knockdown reduces MB cell invasion by 60%–80% (Fig. 3C, D). In addition, ezrin knockdown reduces MB cell adhesion of DAOY cells (Fig. 3E). The effect of ezrin knockdown with siRNA-2 (Ez2) on ONS76 adhesion was not as great as that with siRNA-1 (Ez1), likely due to differences in knockdown levels in this cell line (Fig. 2A).
Fig. 3.
Effects of ezrin depletion on medulloblastoma (MB) cell migration, invasion, and adhesion. (A and B) MB cells transfected with ezrin small interfering RNA (siRNA) were seeded through a cell sedimentation manifold to establish a circular confluent monolayer on substratecoated wells (bovine serum albumin [BSA] or laminin [LN]). DAOY (A) and ONS76 (B) cells were incubated for 24 h, and images were used to measure migration. The average (AVG) migration rate was calculated as the change in the diameter of the circle circumscribing the cell population for a 24-h period. (C) Modified Boyden chamber invasion assay after transfection with ezrin siRNA in DAOY and ONS76 MB cell lines. After incubation for 24 h, the cells that migrated through the membrane were stained and representative fields were imaged. Abbreviations: NT, nontargeting siRNA; Ez1 and Ez2, two different ezrin knockdown sequences used. Magnification, ×200. (D) Invasion was quantified by counting cells. Ezrin siRNA significantly inhibited the invasion of MB cell lines. Columns represent the average (AVG) + SEM invasion rate (cells/fields). (E) Adhesion assay for MB cells tested on BSA and LN. Cells were seeded on 96-well plates coated with BSA or LN, incubated for 3 h, fixed with 4% paraformaldehyde, and then stained with crystal violet. The absorbance was measured at 595 nm (mean + SEM). *p < 0.05, **p < 0.01, versus control.
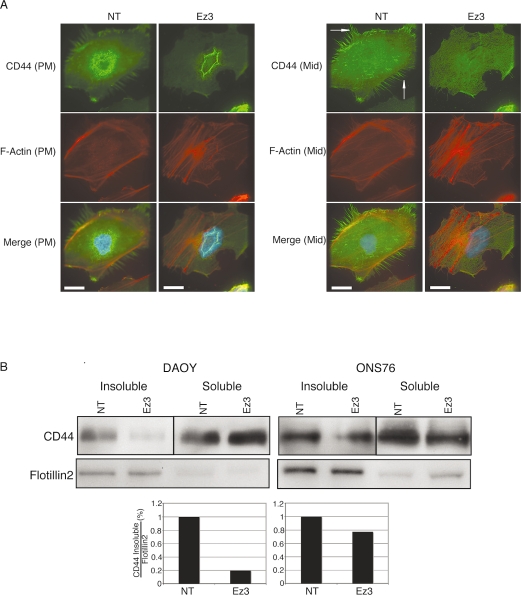
CD44 has previously been identified as an ezrininteracting protein. It is known that the CD44–ezrin complex regulates interactions between the extracellular matrix and cytoskeleton.30–32 Therefore, we examined the effects of ezrin knockdown on CD44 expression and localization. By immunofluorescence, after ezrin siRNA treatment, the localization of CD44 to the filopodia and plasma membrane was diminished compared with controls (Fig. 4A). We then conducted a Western blot analysis using Triton-insoluble and -soluble MB cell lysates to examine the effects of ezrin knockdown on CD44 expression levels. Following ezrin knockdown by siRNA, CD44 levels were decreased in the Triton-insoluble fractions but were unchanged in the soluble fraction compared with controls (Fig. 4B).
Fig. 4.
CD44 expression after transfection with ezrin small interfering RNA (siRNA). (A) Cells were seeded onto bovine serum albumin–coated or laminin-coated cover slips. After fixation, Texas red-X phalloidin and CD44 antibody were used to localize endogenous actin and CD44 by immunofluorescence. Images were acquired by serial focal planes to examine CD44 expression at the plasma membrane (PM, left) or a plane in the middle of the cell (Mid, right). Compared with control cells, ezrin knockdown reduced CD44 expression at filopodia and the plasma membrane in DAOY cells (arrows). Scale bars, 25 μm. (B) Top: Whole-cell lysates and Triton-soluble and -insoluble protein fractions were extracted from DAOY and ONS76 after transfection with ezrin siRNA. Protein (20 μg) was separated with sodium dodecyl sulfate–polyacrylamide gel electrophoresis and blotted onto an Immobilon-P polyvinyl difluoride membrane. CD44 protein was detected using an anti-CD44 antibody in each sample. Flotillin-2, a marker of the insoluble fraction, was used as a loading control. Bottom: Quantification by densitometry of a representative experiment. Abbreviations: NT, nontargeting siRNA; Ez3, ezrin siRNA.
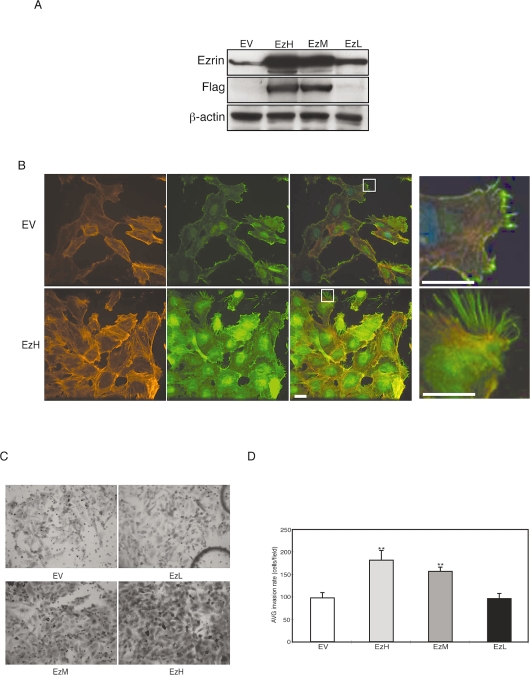
Ezrin Overexpression Increases MB Invasion
We selected three colonies of ezrin-overexpressing DAOY MB clones based on their expression levels: EzH, high protein expression; EzM, intermediate protein expression; and EzL, low protein expression (Fig. 5A). DAOY cells with high expression of FLAG–ezrin exhibited strong cytoplasmic expression of ezrin and long filopodia extensions (Fig. 5B, magnifications at far right). When examined with an in vitro invasion assay described above, DAOY cell clones with high expression of ezrin showed greater invasiveness than did empty vector controls (Fig. 5C, D). These effects were not caused by a change in proliferation (see supplementary Fig. 1S, B).
Fig. 5.
Analysis of ezrin overexpression in medulloblastoma (MB) cell invasion. (A) DAOY cells stably expressing full-length FLAG–ezrin (Flag) or empty vector control (EV) were analyzed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis to assess expression levels. The membranes were probed with anti-ezrin, anti-FLAG, and anti-β-actin and developed with chemiluminescence. Clones identified show high (EzH), intermediate (EzM), or low (EzL) expression of FLAG–ezrin. (B) Empty vector (EV) or FLAG–EzH stable cell lines were immunostained for ezrin (green) and phalloidin (red) and with 4’,6-diamidino-2-phenylindole (blue). The far right panels are enlargements shown by squares in the panels to the left. Scale bars, 25 μm. (C) DAOY cell clones described above were examined by Boyden chamber invasion assay. After a 22-h incubation, the cells that migrated through the membrane were stained and representative fields were imaged. Magnification, ×200. (D) Invasion was quantified by counting cells. Ezrin overexpression correlated with MB invasion rate and is statistically significant. Columns represent the average (AVG) + SEM invasion rate (cells/fields). **p < 0.01, versus control.
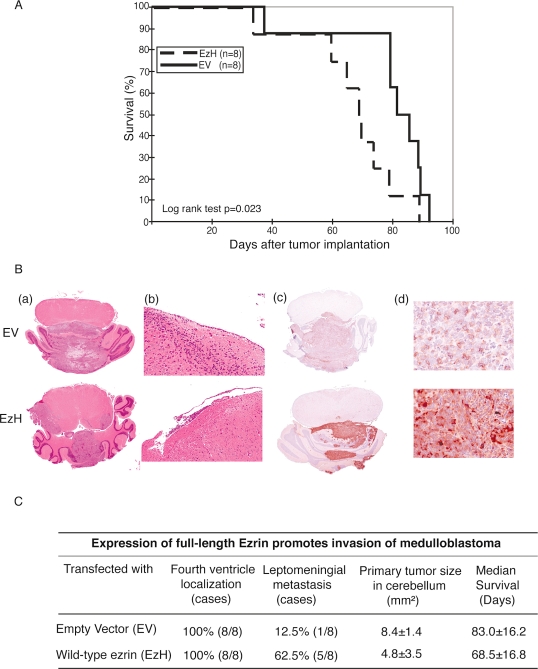
Transfecting DAOY Cells with Ezrin Shortens Survival and Increases Invasiveness
Given that ezrin knockdown reduced the in vitro migration and invasion of MB cells, whereas overexpression of ezrin increased MB invasion, we were interested in examining the in vivo invasiveness of ezrin-overexpressing cells. Inoculating FLAG–EzH DAOY MB cell clones and empty vector controls into the cerebellum of athymic mice resulted in a statistically significant increase in survival among mice with tumor cells injected with empty vector control (median survival, 83 days, vs. 68.5 days with EzH MB cells, p < 0.05; Fig. 6A). Histopathological analysis of the brains of mice with MB cells transfected with empty vector control or EzH showed intracerebellar tumors in both cases; however, only the EzH-derived tumor overexpressed ezrin (Fig. 6B, c and d). Interestingly, five of eight mice (62.7%) in the ezrin high-expressing group (EzH) exhibited leptomeningeal metastasis, compared with one of eight (12.5%) for the empty vector control cells (Fig. 6C).
Fig. 6.
(A) Kaplan-Meier survival analysis for nude mice with intracerebellar human medulloblastoma (MB) cells transfected with either empty vector (EV) or a vector encoding full-length ezrin (EzH). The EV control mice lived longer than did the EzH mice (log-rank test, p < 0.05). The median survival of the EV group was 83 days, compared with 68.5 days in the EzH group. (B) Whole-mount photomicrographs from mouse brains depicting size and dissemination of MB cells. Mice were injected with empty vector (EV) or full-length ezrin (EzH), and the sections were stained with hematoxylin and eosin (a and b) and processed for immunohistochemistry with antiezrin (c and d). EzH-expressing MB cells showed greater leptomeningeal dissemination (b and c) than did EV controls. Original magnifications: ×1 for a and c; ×10 for b; ×40 for d. (C) Analysis of in vivo xenograft implant localization and invasion.
Discussion
Previous studies have shown that ezrin is expressed in a variety of epithelial and neuroepithelial neoplasms.24,26,33–35 In human brain tumors, ezrin expression has been reported in astrocytomas but not in oligodendrogliomas.34 Tynninen et al.36 studied a series of human gliomas of varying grades of malignancy using tissue microarray profiling and showed that high ezrin expression correlates with shorter recurrence period and shorter overall patient survival. Similarly, Park et al.,6 using transcriptional profiling of human MB cell lines and specimens, have shown that ezrin is one of several genes upregulated in MB compared with its expression in the normal cerebellum.6 This finding, together with our previous interest in cytoskeletal protein–matrix interactions in human brain tumors, encouraged us to examine the role of ezrin in MB. Here, we show that ezrin is expressed in a series of MB cell lines. Furthermore, in MB cells, ezrin is phosphorylated at threonine 567, a phosphorylation site that is required for cytoskeletal rearrangement and oncogene-induced transformation.37 Finally, in the present study, we have manipulated ezrin expression by siRNA knockdown and by overexpression using gene transfection techniques and identified a role for ezrin in filopodial formation, invasion, and leptomeningeal dissemination.
Experimental models have shown a direct relationship between ezrin expression and tumor migration. In an endometrial cancer model, Ohtani et al.20 demonstrated that neoplastic cell migration was inhibited by ezrin antisense oligonucleotides. Wick et al.38 showed that glioma cell invasion could be blocked using dominant negative ezrin expression constructs. Our data support these studies in that siRNA for ezrin blocked migration and invasion of MB cells. The mechanism by which ezrin exerts its effects on cell adhesion is not entirely clear. However, Poullet et al.39 showed that ezrin interacts with focal adhesion kinase and induces its activation independently of cell–matrix adhesion.
The immunocytochemical localization of ezrin has been the subject of several studies. A cytoplasmic localization of ezrin occurs in a variety of human tumors, including anaplastic astrocytomas34 and carcinoma of the ovary,40 prostate,41 and lung,35 but not in the normal tissues from which these tumors originate. A redistribution of ezrin from the cytosol to microvilli and membrane ruffles has been demonstrated in a variety of cancer cell lines in response to various growth factors, including epidermal growth factor and hepatocyte growth factor/scatter factor.42,43 The redistribution of ezrin into cell membrane structures such as cell ruffles and filopodia has been implicated as a possible mechanism for enhanced tumor invasion and migration in vitro.25 Our data show that in MB cells ezrin localizes to both filopodia and cytoplasm. Furthermore, siRNA for ezrin decreased the number of filopodia in DAOY and ONS76 MB cells.
Ezrin likely exerts its cellular effects through its interaction with several protein-binding partners. As one example, CD44 was the first cell-surface protein to be identified with which ezrin and the ERM proteins interact. Hyaluronan (HA) is the principal ligand of CD44, but other CD44 ligands include collagen, fibronectin, LN, and chondroitin sulfate.30,44 Annabi et al.45 suggested that membrane type 1 matrix metalloproteinase (MT1-MMP)-mediated CD44 cell-surface cleavage may play a critical role in efficient cell detachment from HA and could consequently promote glioma cell migration and invasion. Moreover, CD44 is a multifunctional cell-surface adhesion molecule involved in cell–cell and cell–matrix interactions, as well as cell migration.30–32 It has been shown that the cytoplasmic domain of CD44 binds directly to the amino terminal of ERM proteins.46,47 Parker et al.48 reported that 93.5% MB tumors are positive for CD44 using flow cytometry. To elucidate the relationship between CD44 and ezrin expression in MB, we used Triton X-100–extracted MB lysates from ezrin siRNA–treated MB cultures to examine CD44 protein levels and found a decrease in the insoluble lysate fraction, but not in the soluble fraction. These findings suggest that treatment of MB cells with ezrin siRNA destabilizes the interaction of CD44 and ezrin and modulates CD44 protein stability and/or localization, which may affect cytoskeletal structure and the invasiveness of MB cells.
In addition to ezrin knockdown by siRNA, we were interested in examining the effects of ezrin overexpression in MB cell lines stably transfected with a full-length ezrin cDNA. Prior reports have shown that overexpression of ezrin increased carcinoma cell invasion in vitro.49 Our present studies showed that MB cells stably over-expressing ezrin expression had higher invasion rates than did controls. While the mechanism by which ezrin overexpression leads to enhanced invasion and migration is not understood, a previous study by MacDonald et al.50 has shown that activation of the Ras/mitogen-activated protein (MAP) kinase pathway may be involved in MB dissemination. Our data are of interest because they show that athymic mice implanted in the cerebellum with ezrin-overexpressing MB cells have shortened survivals and exhibit a propensity to leptomeningeal dissemination compared with controls. However, our data are in contrast to the work of Elliott et al.,49 who showed that overexpression of ezrin per se is not sufficient to induce metastasis of breast cancer in a subcutaneous tumor model, although a dominant-negative aminoterminal ezrin mutant diminished breast cancer metastasis, suggesting that multiple pathways are likely involved in the metastatic cascade.
In conclusion, we show for the first time that ezrin plays a role in MB motility, invasion, and dissemination. Therefore, ezrin may be considered along with other cytoskeletal proteins, such as actin, fascin, tubulin, and rho-GTPases, that are becoming increasingly more important to study, analyze, and target in the context of the invasiveness of human brain tumors.
Acknowledgments
This work was supported by grants from the Canadian Institutes of Health Research (grant MOP-74610), Pediatric Brain Tumor Foundation, Wiley and Laurie Berman Funds for Brain Tumor Research, and b.r.a.i.n.child. J.T.R. is a recipient of a Canadian Institutes of Health Research Scientist Award.
References
- 1.Louis DN, Pomeroy SL, Cairncross JG. Focus on central nervous system neoplasia. Cancer Cell. 2002;1:125–128. doi: 10.1016/s1535-6108(02)00040-5. [DOI] [PubMed] [Google Scholar]
- 2.Tarbell NJ, Loeffler JS, Silver B, et al. The change in patterns of relapse in medulloblastoma. Cancer. 1991;68:1600–1604. doi: 10.1002/1097-0142(19911001)68:7<1600::aid-cncr2820680722>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 3.Rutka JT. Medulloblastoma. Clin Neurosurg. 1997;44:571–585. [PubMed] [Google Scholar]
- 4.David KM, Casey AT, Hayward RD, Harkness WF, Phipps K, Wade AM. Medulloblastoma: Is the 5-year survival rate improving? A review of 80 cases from a single institution. J Neurosurg. 1997;86:13–21. doi: 10.3171/jns.1997.86.1.0013. [DOI] [PubMed] [Google Scholar]
- 5.Buhren J, Christoph AH, Buslei R, Albrecht S, Wiestler OD, Pietsch T. Expression of the neurotrophin receptor p75NTR in medulloblastomas is correlated with distinct histological and clinical features: Evidence for a medulloblastoma subtype derived from the external granule cell layer. J Neuropathol Exp Neurol. 2000;59:229–240. doi: 10.1093/jnen/59.3.229. [DOI] [PubMed] [Google Scholar]
- 6.Park PC, Taylor MD, Mainprize TG, et al. Transcriptional profiling of medulloblastoma in children. J Neurosurg. 2003;99:534–541. doi: 10.3171/jns.2003.99.3.0534. [DOI] [PubMed] [Google Scholar]
- 7.Yokota N, Mainprize TG, Taylor MD, et al. Identification of differentially expressed and developmentally regulated genes in medulloblastoma using suppression subtraction hybridization. Oncogene. 2004;23:3444–3453. doi: 10.1038/sj.onc.1207475. [DOI] [PubMed] [Google Scholar]
- 8.Pietsch T, Taylor MD, Rutka JT. Molecular pathogenesis of childhood brain tumors. J Neurooncol. 2004;70:203–215. doi: 10.1007/s11060-004-2750-7. [DOI] [PubMed] [Google Scholar]
- 9.Biegel JA, Lloreta J, Ariza A, et al. Isochromosome 17q in primitive neuroectodermal tumors of the central nervous system. Genes Chromosomes Cancer. 1989;1:139–147. doi: 10.1002/gcc.2870010206. [DOI] [PubMed] [Google Scholar]
- 10.Cogen PH, Daneshvar L, Metzger AK, Duyk G, Edwards MS, Sheffield VC. Involvement of multiple chromosome 17p loci in medulloblastoma tumorigenesis. Am J Hum Genet. 1992;50:584–589. [PMC free article] [PubMed] [Google Scholar]
- 11.Steichen-Gersdorf E, Baumgartner M, Kreczy A, Maier H, Fink FM. Deletion mapping on chromosome 17p in medulloblastoma. Br J Cancer. 1997;76:1284–1287. doi: 10.1038/bjc.1997.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lindsey JC, Lusher ME, Anderton JA, et al. Identification of tumourspecific epigenetic events in medulloblastoma development by hypermethylation profiling. Carcinogenesis. 2004;25:661–668. doi: 10.1093/carcin/bgh055. [DOI] [PubMed] [Google Scholar]
- 13.Oliver TG, Grasfeder LL, Carroll AL, et al. Transcriptional profiling of the sonic hedgehog response: A critical role for N-myc in proliferation of neuronal precursors. Proc Natl Acad Sci U S A. 2003;100:7331–7336. doi: 10.1073/pnas.0832317100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pomeroy SL, Tamayo P, Gaasenbeek M, et al. Prediction of central nervous system embryonal tumour outcome based on gene expression. Nature. 2002;415:436–442. doi: 10.1038/415436a. [DOI] [PubMed] [Google Scholar]
- 15.Berryman M, Franck Z, Bretscher A. Ezrin is concentrated in the apical microvilli of a wide variety of epithelial cells whereas moesin is found primarily in endothelial cells. J Cell Sci. 1993;105(4):1025–1043. doi: 10.1242/jcs.105.4.1025. [DOI] [PubMed] [Google Scholar]
- 16.Bretscher A. Regulation of cortical structure by the ezrin-radixin-moesin protein family. Curr Opin Cell Biol. 1999;11:109–116. doi: 10.1016/s0955-0674(99)80013-1. [DOI] [PubMed] [Google Scholar]
- 17.Tsukita S, Yonemura S. ERM (ezrin/radixin/moesin) family: From cytoskeleton to signal transduction. Curr Opin Cell Biol. 1997;9:70–75. doi: 10.1016/s0955-0674(97)80154-8. [DOI] [PubMed] [Google Scholar]
- 18.Kaul SC, Mitsui Y, Komatsu Y, Reddel RR, Wadhwa R. A highly expressed 81 kDa protein in immortalized mouse fibroblast: Its proliferative function and identity with ezrin. Oncogene. 1996;13:1231–1237. [PubMed] [Google Scholar]
- 19.Shen ZY, Xu LY, Chen MH, et al. Upregulated expression of Ezrin and invasive phenotype in malignantly transformed esophageal epithelial cells. World J Gastroenterol. 2003;9:1182–1186. doi: 10.3748/wjg.v9.i6.1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ohtani K, Sakamoto H, Rutherford T, Chen Z, Satoh K, Naftolin F. Ezrin, a membrane-cytoskeletal linking protein, is involved in the process of invasion of endometrial cancer cells. Cancer Lett. 1999;147:31–38. doi: 10.1016/s0304-3835(99)00272-4. [DOI] [PubMed] [Google Scholar]
- 21.Khanna C, Khan J, Nguyen P, et al. Metastasis-associated differences in gene expression in a murine model of osteosarcoma. Cancer Res. 2001;61:3750–3759. [PubMed] [Google Scholar]
- 22.Khanna C, Wan X, Bose S, et al. The membrane-cytoskeleton linker ezrin is necessary for osteosarcoma metastasis. Nat Med. 2004;10:182–186. doi: 10.1038/nm982. [DOI] [PubMed] [Google Scholar]
- 23.Yu Y, Khan J, Khanna C, Helman L, Meltzer PS, Merlino G. Expression profiling identifies the cytoskeletal organizer ezrin and the developmental homeoprotein Six-1 as key metastatic regulators. Nat Med. 2004;10:175–181. doi: 10.1038/nm966. [DOI] [PubMed] [Google Scholar]
- 24.Akisawa N, Nishimori I, Iwamura T, Onishi S, Hollingsworth MA. High levels of ezrin expressed by human pancreatic adenocarcinoma cell lines with high metastatic potential. Biochem Biophys Res Commun. 1999;258:395–400. doi: 10.1006/bbrc.1999.0653. [DOI] [PubMed] [Google Scholar]
- 25.Chen Z, Fadiel A, Feng Y, Ohtani K, Rutherford T, Naftolin F. Ovarian epithelial carcinoma tyrosine phosphorylation, cell proliferation, and ezrin translocation are stimulated by interleukin 1alpha and epidermal growth factor. Cancer. 2001;92:3068–3075. doi: 10.1002/1097-0142(20011215)92:12<3068::aid-cncr10149>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 26.Ohtani K, Sakamoto H, Rutherford T, et al. Ezrin, a membrane-cytoskeletal linking protein, is highly expressed in atypical endometrial hyperplasia and uterine endometrioid adenocarcinoma. Cancer Lett. 2002;179:79–86. doi: 10.1016/s0304-3835(01)00857-6. [DOI] [PubMed] [Google Scholar]
- 27.Letarte M. Human p85 glycoprotein bears three distinct epitopes defined by several monoclonal antibodies. Mol Immunol. 1986;23:639–644. doi: 10.1016/0161-5890(86)90101-x. [DOI] [PubMed] [Google Scholar]
- 28.Berens ME, Rief MD, Loo MA, Giese A. The role of extracellular matrix in human astrocytoma migration and proliferation studied in a micro-liter scale assay. Clin Exp Metastasis. 1994;12:405–415. doi: 10.1007/BF01755884. [DOI] [PubMed] [Google Scholar]
- 29.Hyun Hwang J, Smith CA, Salhia B, Rutka JT. The role of fascin in the migration and invasiveness of malignant glioma cells. Neoplasia. 2008;10:149–159. doi: 10.1593/neo.07909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Naor D, Sionov RV, Ish-Shalom D. CD44: Structure, function, and association with the malignant process. Adv Cancer Res. 1997;71:241–319. doi: 10.1016/s0065-230x(08)60101-3. [DOI] [PubMed] [Google Scholar]
- 31.Rudzki Z, Jothy S. CD44 and the adhesion of neoplastic cells. Mol Pathol. 1997;50:57–71. doi: 10.1136/mp.50.2.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sneath RJ, Mangham DC. The normal structure and function of CD44 and its role in neoplasia. Mol Pathol. 1998;51:191–200. doi: 10.1136/mp.51.4.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bohling T, Turunen O, Jääskeläinen J, et al. Ezrin expression in stromal cells of capillary hemangioblastoma. An immunohistochemical survey of brain tumors. Am J Pathol. 1996;148:367–373. [PMC free article] [PubMed] [Google Scholar]
- 34.Geiger KD, Stoldt P, Schlote W, Derouiche A. Ezrin immunoreactivity is associated with increasing malignancy of astrocytic tumors but is absent in oligodendrogliomas. Am J Pathol. 2000;157:1785–1793. doi: 10.1016/S0002-9440(10)64816-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tokunou M, Niki T, Saitoh Y, Imamura H, Sakamoto M, Hirohashi S. Altered expression of the ERM proteins in lung adenocarcinoma. Lab Invest. 2000;80:1643–1650. doi: 10.1038/labinvest.3780174. [DOI] [PubMed] [Google Scholar]
- 36.Tynninen O, Carpen O, Jääskeläinen J, Paavonen T, Paetau A. Ezrin expression in tissue microarray of primary and recurrent gliomas. Neuropathol Appl Neurobiol. 2004;30:472–477. doi: 10.1111/j.1365-2990.2004.00562.x. [DOI] [PubMed] [Google Scholar]
- 37.Tran Quang C, Gautreau A, Arpin M, Treisman R. Ezrin function is required for ROCK-mediated fibroblast transformation by the Net and Dbl oncogenes. EMBO J. 2000;19:4565–4576. doi: 10.1093/emboj/19.17.4565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wick W, Grimmel C, Wild-Bode C, Platten M, Arpin M, Weller M. Ezrin-dependent promotion of glioma cell clonogenicity, motility, and invasion mediated by BCL-2 and transforming growth factor-beta2. J Neurosci. 2001;21:3360–3368. doi: 10.1523/JNEUROSCI.21-10-03360.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Poullet P, Gautreau A, Kadaré G, Girault JA, Louvard D, Arpin M. Ezrin interacts with focal adhesion kinase and induces its activation independently of cell-matrix adhesion. J Biol Chem. 2001;276:37686–37691. doi: 10.1074/jbc.M106175200. [DOI] [PubMed] [Google Scholar]
- 40.Moilanen J, Lassus H, Leminen A, Vaheri A, Bützow R, Carpén O. Ezrin immunoreactivity in relation to survival in serous ovarian carcinoma patients. Gynecol Oncol. 2003;90:273–281. doi: 10.1016/s0090-8258(03)00262-2. [DOI] [PubMed] [Google Scholar]
- 41.Pang ST, Fang X, Valdman A, et al. Expression of ezrin in prostatic intraepithelial neoplasia. Urology. 2004;63:609–612. doi: 10.1016/j.urology.2003.09.068. [DOI] [PubMed] [Google Scholar]
- 42.Bretscher A. Rapid phosphorylation and reorganization of ezrin and spectrin accompany morphological changes induced in A-431 cells by epidermal growth factor. J Cell Biol. 1989;108:921–930. doi: 10.1083/jcb.108.3.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jiang WG, Hiscox S, Singhrao SK, et al. Induction of tyrosine phosphorylation and translocation of ezrin by hepatocyte growth factor/scatter factor. Biochem Biophys Res Commun. 1995;217:1062–1069. doi: 10.1006/bbrc.1995.2877. [DOI] [PubMed] [Google Scholar]
- 44.Gunthert U, Schwärzler C, Wittig B, et al. Functional involvement of CD44, a family of cell adhesion molecules, in immune responses, tumour progression and haematopoiesis. Adv Exp Med Biol. 1998;451:43–49. doi: 10.1007/978-1-4615-5357-1_7. [DOI] [PubMed] [Google Scholar]
- 45.Annabi B, Naud E, Lee YT, Eliopoulos N, Galipeau J. Vascular progenitors derived from murine bone marrow stromal cells are regulated by fibroblast growth factor and are avidly recruited by vascularizing tumors. J Cell Biochem. 2004;91:1146–1158. doi: 10.1002/jcb.10763. [DOI] [PubMed] [Google Scholar]
- 46.Hirao M, Sato N, Kondo T, et al. Regulation mechanism of ERM (ezrin/radixin/moesin) protein/plasma membrane association: Possible involvement of phosphatidylinositol turnover and Rho-dependent signaling pathway. J Cell Biol. 1996;135:37–51. doi: 10.1083/jcb.135.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yonemura S, Hirao M, Doi Y, et al. Ezrin/radixin/moesin (ERM) proteins bind to a positively charged amino acid cluster in the juxtamembrane cytoplasmic domain of CD44, CD43, and ICAM-2. J Cell Biol. 1998;140:885–895. doi: 10.1083/jcb.140.4.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Parker K, Pilkington GJ. Morphological, immunocytochemical and flow cytometric in vitro characterisation of a surface-adherent medulloblastoma. Anticancer Res. 2005;25:3855–3863. [PubMed] [Google Scholar]
- 49.Elliott BE, Meens JA, SenGupta SK, Louvard D, Arpin M. The membrane cytoskeletal crosslinker ezrin is required for metastasis of breast carcinoma cells. Breast Cancer Res. 2005;7:R365–R373. doi: 10.1186/bcr1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.MacDonald TJ, Brown KM, LaFleur B, et al. Expression profiling of medulloblastoma: PDGFRA and the RAS/MAPK pathway as therapeutic targets for metastatic disease. Nat Genet. 2001;29:143–152. doi: 10.1038/ng731. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.