Abstract
One of the hallmark pathologies of Alzheimer’s disease (AD) is amyloid plaque deposition. Plaques appear hypointense on T2- and T2*-weighted MR images probably due to the presence of endogenous iron, but no quantitative comparison of various imaging techniques has been reported. We estimated the T1, T2, T2*, and proton density values of cortical plaques and normal cortical tissue and analyzed the plaque contrast generated by a collection of T2-, T2*-, and susceptibility-weighted imaging (SWI) methods in ex vivo transgenic mouse specimens. The proton density and T1 values were similar for both cortical plaques and normal cortical tissue. The T2 and T2* values were similar in cortical plaques, which indicates that the iron content of cortical plaques may not be as large as previously thought. Ex vivo plaque contrast was increased compared to a previously reported spin echo sequence by summing multiple echoes and by performing SWI; however, gradient echo and susceptibility weighted imaging was found to be impractical for in vivo imaging due to susceptibility interface-related signal loss in the cortex.
Keywords: MR microscopy, Alzheimer’s disease, magnetic resonance imaging, magnetic resonance micro imaging, transgenic mice, susceptibility weighted imaging
INTRODUCTION
Amyloid precursor protein (APP) is a normal transmembrane protein found in cells throughout the body. One of the products of proteolytic degradation of APP in the brain is amyloid β peptide (Aβ). Disordered amyloid metabolism is believed to be central in the pathogenesis of Alzheimer’s disease (AD). One of the principal defining pathologies of AD is amyloid plaque deposition (1), the reduction of which has been identified as a major therapeutic objective (2). Murine models of AD have been created by inserting one or more human mutations into the mouse genome (3,4). These transgenic mice display extensive plaque formation, whereas plaques are not found in wild-type mice. Transgenic mouse models of Alzheimer’s disease allow controlled study of this phenomenon and enable testing of anti-amyloid interventions that might be useful in humans.
Benveniste et al. first demonstrated the ability of MRI to visualize amyloid plaques in ex vivo human brain specimens (5). Subsequent works by multiple groups have investigated imaging plaques in transgenic mice without exogenous contrast agents. Plaques have been visualized ex vivo using spin-echo (SE) (6,7) and fast spin-echo (FSE) (8) techniques, both being T2-weighted, and gradient-echo (GRE) techniques (9). In vivo plaque detection has been performed using a wider array of techniques, including T1ρ-weighting (10), macroscopic T2 mapping (11), FSE imaging (12), SE imaging (6,13), and gradient-echo (GRE) imaging (9,14).
It has been postulated that the inherent contrast of plaques is due to the accumulation of iron (6,9,14). Iron reduces both T2 and T2* in and near the plaque causing the plaque to present as a hypointense region. Since each of the previously mentioned studies were conducted with differing mouse models, ages, imaging parameters and experimental design, it is difficult to compare them in order to determine the ideal plaque imaging method. Furthermore, no work has investigated the ability of susceptibility-weighted imaging (SWI) to increase plaque contrast. Naturally occurring iron in the plaque should cause a phase shift in the plaque voxel, which could provide increased contrast in SWI images (15,16). In this work we quantitatively compare the ability of various imaging methods to visualize cortical plaques ex vivo. We used ex vivo brain specimens of transgenic mice because a study of this scope would be impractical in vivo due to prohibitively long scan times. Ex vivo specimens were used to estimate T2, T2*, and T1, from which optimal parameters could be determined for nine different imaging sequences: spin-echo (SE), fast spin-echo (FSE), multiple spin-echo (mSE), multiple asymmetric spin-echo (mASE), gradient-echo (GRE), multiple gradient-echo (mGRE), mASE with SWI (mASE-SWI), GRE with SWI (GRE-SWI), and mGRE with SWI (mGRE-SWI). The parameters and experimental setup were based on the previously reported spin-echo sequence used to image individual plaques in vivo and ex vivo (6,13). Although relaxation values will differ between in vivo and ex vivo specimens, the ex vivo comparison provides a starting point for future in vivo studies. After ex vivo comparison, a double transgenic amyloid precursor protein/presenillin 1 (APP/PS1) (3) mouse model was imaged to subjectively compare the gradient-echo, spin-echo and asymmetric spin-echo sequences.
METHODS
Ex vivo experiments were performed on the fixed brain specimens of four 9-month-old APP/PS1 transgenic mice (3), one 22-month-old APP transgenic mouse, and two 4-month-old wild-type mice. In order to obtain fixed ex vivo brain specimens the animals were perfused transcardially with PBS and fixed with neutral buffered 10% formalin after an overdose with sodium pentobarbital (200 mg/kg, i.p.). The brain was removed, fixed further in formalin overnight, and equilibrated in 0.1 M sodium phosphate, pH 7.4, for 24 h. The brain was embedded vertically in 2% agar in a 15-mm outer diameter glass tube for ex vivo MRI. After imaging the ex vivo brains were sectioned and stained for Thio-S and iron using the same protocol as described by Jack et al. (13). In vivo experiments were performed on a 12-month-old APP/PS1 transgenic mouse. The mouse was anesthetized using 1.0–1.5% isoflurane and O2/N2O and positioned in a custom-built cradle for head immobilization during imaging. Body temperature was monitored by a rectal probe and maintained with a hot water circulating system. Experimental protocols were approved by the Institutional Animal Care and Use Committees of both the University of Minnesota and the Mayo Clinic in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
MRI experiments were performed with a Unity Inova spectrometer (Varian, Palo Alto, CA) interfaced to a 9.4-T/31-cm horizontal bore magnet equipped with actively shielded gradients capable of 450 mT/m in a rise time of 200 µs (Resonance Research, Inc., Billerica, MA). RF transmission and reception was performed with a quadrature surface coil consisting of 1-cm diameter loops.
The relevant relaxation time constants of plaques and normal cortical tissue were estimated ex vivo in order to give guidance in choosing echo times and repetition times for the various sequences. The T1 value of cortical plaques and normal cortical tissue was estimated in the fixed brains of two 9-month-old APP/PS1 transgenic mice; the T2 and T2* values, the proton density, and the frequency shift of cortical plaques and cortical tissue were estimated in the fixed brains of four 9-month-old APP/PS1 transgenic mice. Normal cortical tissue was defined as a region within the cortex with homogenous intensity. In any region of interest there is likely to be numerous plaques too small to be detected by MRI. Since this is necessarily the background tissue for identifying large plaques, it has been defined as normal cortical tissue. The density of small plaques in normal appearing regions will depend on the age and strain of the mouse; therefore, the results may differ for different mouse populations.
The T1 values of normal cortical tissue and cortical plaques were estimated using the saturation recovery method with the SE sequence (6). Images were acquired with a resolution of 60 × 60 × 120 µm3; 29 different repetition time (TR) values ranging from 50 ms to 12000 ms; echo time (TE) = 24 ms; spectral width (sw) = 30 kHz; x, y, and z matrices of 256 × 96 × 32 with a field-of-view (FOV) of 15.36 × 5.76 × 3.84 mm3; total imaging time was 60 hr.
The T2 values of normal cortical tissue and cortical plaques were estimated using the SE sequence with a single localizing 180° pulse in the z direction. The 180° localizing pulse in the y direction was removed to allow for the acquisition of shorter echo times for a more reliable T2 estimation. Images were acquired with a resolution of 60 × 60 × 120 µm3; TR = 2000 ms; 10 different TE values ranging from 6.88 ms to 75.88 ms; sw = 60 kHz; x, y, and z matrices of 256 × 192 × 32 with a FOV of 15.36 × 11.52 × 3.84 mm3; total imaging time was 3.4 hr. The spectral width was increased for the T2 estimations to attain a shorter echo time.
The T2* values of normal cortical tissue and cortical plaques were measured using the GRE sequence. Images were acquired with a resolution of 60 × 60 × 120 µm3; TR = 659 ms; 10 different TE values ranging from 2.58 ms to 75.88 ms; sw = 30 kHz; x, y, and z matrices of 256 × 256 × 32 with a FOV of 15.36 × 15.36 × 3.84 mm3.
The relaxation parameter estimation was a two-stage analysis. The first stage consisted of fitting the relaxation curves for each individual pixel identified as a cortical plaque or normal appearing cortical tissue using a non-linear least squares algorithm. In the second stage the weighted average of the individual relaxation parameter estimates was calculated. The weight given to each parameter estimate was inversely proportional to the standard error of the estimate. To estimate the frequency offset of the plaques, a phase map was generated at each echo time in the GRE sequence to determine the effective frequency offset of the plaques relative to the normal cortical tissue. The average phase of the plaques was fit to a line to estimate the average frequency offset. It should be noted that the estimated T2 value may not correspond to the actual T2 value because the data was acquired with an echo train. Since this study is concerned with the contrast produced by echo trains, the T2 value estimated with an echo train is the more useful value in this context.
The next portion of the study was designed to compare the ability of different imaging methods to generate plaque contrast against normal appearing cortical tissue. The pulse sequence diagrams for the sequences are shown in Fig. 1. The spin echo sequences (SE, mSE, mASE, and mASE-SWI) were all based on the previously reported SE sequence (6). The multiple-echo sequences (mSE, mASE, and mGRE) generated an echo train with no phase encoding performed between the echoes. In this way, the set of phase-encoded echoes collected at each TE produced a fully encoded image that was processed independently. The images were summed in absolute value mode to increase the signal-to-noise ratio (SNR). The receiver gain was equal for all of the echoes. The GRE based sequences are shown in Fig. 1a. The GRE sequence acquired only one echo, while the mGRE sequence acquired 8 echoes. The echoes were acquired with unipolar readout gradients to minimize the variation from image to image. The SE sequences are shown in Fig. 1b. The spin echo asymmetry is indicated by Δτ. The SE sequence acquired one echo with Δτ = 0, the mSE sequence acquired 5 echoes with Δτ = 0, and the mASE sequence acquired 5 echoes with Δτ = 8 ms.
Figure 1.
Pulse sequence diagrams. (a) The gradient echo sequences use an hyperbolic secant pulse to excite a slab then perform 3D imaging on that slab. The GRE sequence acquires a single gradient echo, and the mGRE sequence acquires 8 echoes. The readout gradients are unipolar to minimize distortions between the echoes. (b) The spin echo sequences are fully adiabatic sequences to minimize flip angle variation across the object. The sequence has a spin echo asymmetry equal to Δτ, which is zero for the SE and mSE sequences.
The parameters for all sequences were designed to give the same resolution (60 × 60 × 120 µm3) and total acquisition time (1 hr 30 min). The echo times for all sequences were chosen to give optimal plaque contrast based on the results of the relaxation estimates. The SE sequence parameters were: TR = 1758 ms; TE = 43 ms; sw = 30 kHz; x, y, and z matrices of 256 × 96 × 32 with a FOV of 15.36 × 5.76 × 3.84 mm3. The mSE sequence parameters were: TR = 1758 ms; TE = 28, 43, 58, 73, 88 ms; sw = 30 kHz; x, y, and z matrices of 256 × 96 × 32 with a FOV of 15.36 × 5.76 × 3.84 mm3. The mASE sequence parameters were TR = 1758 ms; TE = 30, 45, 60, 75, 90 ms; Δτ = 8 ms; sw = 30 kHz; x, y, and z matrices of 256 × 96 × 32 with a FOV of 15.36 × 5.76 × 3.84 mm3. The FSE sequence parameters were TR = 1758 ms; TE = 45, 60 ms; echo train length = 2; Nex = 2; sw = 30 kHz; x, y, and z matrices of 256 × 96 × 32 with a FOV of 15.36 × 5.76 × 3.84 mm3. The GRE sequence parameters were TR = 659 ms; TE = 39 ms; sw = 30 kHz; x, y, and z matrices of 256 × 256 × 32 with a FOV of 15.36 × 15.36 × 3.84 mm3. The mGRE sequence parameters were TR = 659 ms; TE = 15, 23, 31, 39, 47, 55, 63, 71 ms; sw = 30 kHz; x, y, and z matrices of 256 × 256 × 32 with a FOV of 15.36 × 15.36 × 3.84 mm3. The GRE sequences required a larger FOV in y because there was no refocusing pulse to select a slab in the y direction. To generate the SWI images, a phase map was generated using a filtered k-space to remove the sources of slowly varying phase. The phase map was mapped to a phase mask and then multiplied into the magnitude image 5 times. In vivo GRE, SE, and mASE images were acquired with the same parameters given above, except the GRE sequence had TE = 23 ms.
The sequences were compared by manually identifying plaques common to all nine image sets. After zero-filling by a factor of two in the in-plane directions, a 5 × 5 pixel ROI was drawn around each plaque. Another 5 × 5 pixel ROI was drawn in a region of normal appearing cortical tissue near the plaque. The plaque contrast-to-noise ratio (CNR) was calculated as,
| (1) |
where St is the mean of the signal in the tissue ROI, Sp is the mean of the signal in the plaque ROI, σi is the variance of the ith pixel in the plaque ROI, and N is the total number of pixels in the plaque ROI. Eq. 1 is adapted from the visibility equation derived by Constable and Henkelman (17). The average CNR was calculated across all five brain specimens for each sequence. A total of 427 plaques were counted in the five specimens.
In order to correlate histologic sections with MR images, images of the individual histologic sections were combined to create a digitized 3D volume of the specimen. In order to match the through-plane resolution of the histologic and MR images, appropriate multiples of the 30-µm histologic sections were used. The digitized histologic volume was then spatially matched to the ex vivo MRI volume using an image analysis software package (18). A linked cursor system in the image analysis software program was then used to identify amyloid plaques common to the three spatially registered digitized datasets: ex vivo MRI, and in vitro Thio-S staining, and in vitro iron staining.
RESULTS
The calculated relaxation parameters are shown in Table 1. The optimal echo times were calculated using the equation
| (2) |
where T2t is the T2 or T2* value for normal cortical tissue, and T2p is the T2 or T2* value for cortical plaques. The maximal T2 contrast occured at TE = 42.1 ms; the maximal T2* contrast occured at TE = 38.8 ms. The average phase of the plaques relative to the normal tissue was fit to a line to arrive at a frequency shift of −0.7 Hz.
Table 1.
Estimated values of the relaxation parameters of cortical plaques and normal cortical tissue in ex vivo APP/PS1 brain specimens.
| Cortical Plaques | Normal Cortical Tissue | |
|---|---|---|
| Proton density (a.u.) | 1.00 ± 0.10 | 1.042 ± 0.081 |
| T1 (s) | 1.51 ± 0.07 | 1.64 ± 0.03 |
| T2 (ms) | 32.1 ± 2.2 | 56.6 ± 1.3 |
| T2* (ms) | 31.1 ± 1.6 | 49.4 ± 3.0 |
| Frequency shift (Hz) | −0.7 ± 0.2 | 0.0 ± 0.1 |
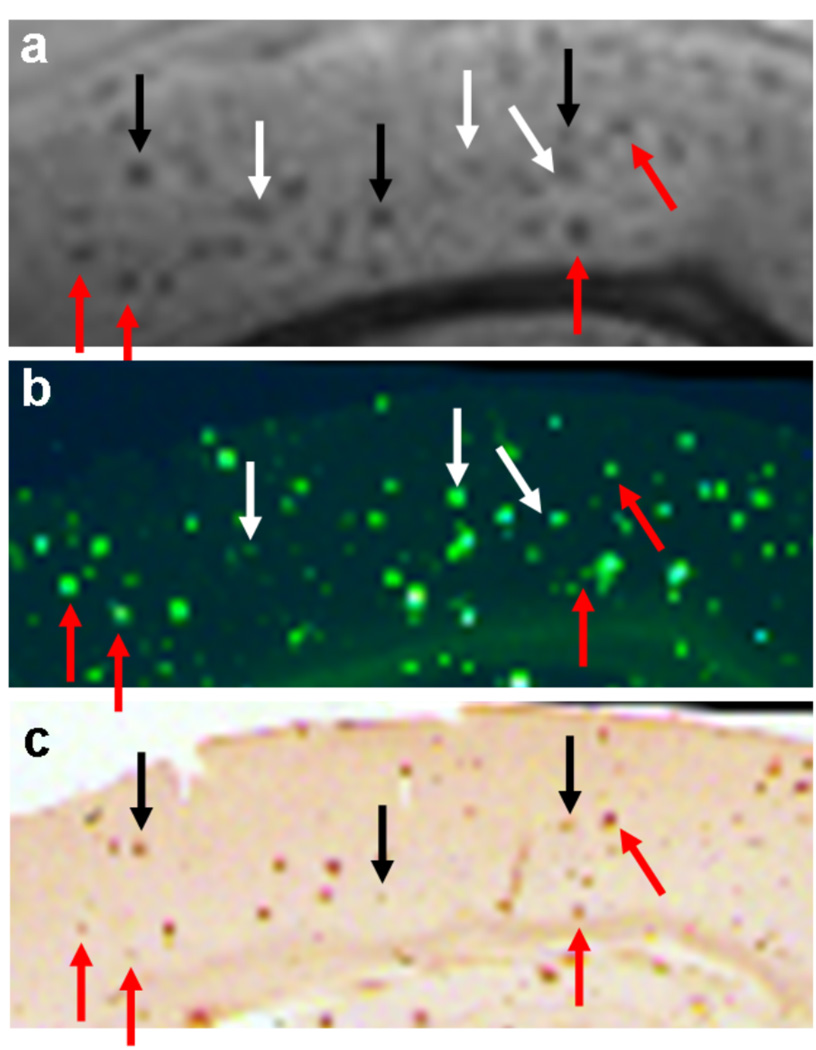
Fig. 2 shows representative ex vivo images from the three major classes of sequences: T2*-, T2-, and susceptibility-weighted. Fig. 3 shows the same magnified region for all nine sequences. The average CNR of all counted plaques is shown for each imaging method. The images are ordered by increasing CNR (defined in Eq. 1). The multi-echo imaging sequences (designated by a lower-case ‘m’) all produced increased plaque CNR compared to their single echo counterparts. Similarly, all SWI imaging sequences produced increased plaque CNR compared to the magnitude-only counterparts. Fig. 4 shows the correlation between the mGRE sequence, Thio-S and iron staining. Red arrows indicate plaques that were common to all three modalities, black arrows indicate plaques that were present in the MRI and iron staining, and white arrows indicate plaques that were present in MRI and Thio-S staining. Fig. 5 shows representative ex vivo images of a wild-type mouse brain using the three major classes of sequences. It can be seen that none of the imaging methods introduce hypointense regions that could be falsely identified as plaques.
Figure 2.
Representative images from T2-, T2*-, and susceptibility-weighted imaging sequences in a 22-month-old ex vivo APP brain specimen. (a) An image from the mSE sequence (T2-weighted). (b) An image from the mGRE sequence (T2*-weighted). (c) An image from the mGRE-SWI sequence (susceptibility-weighted).
Figure 3.
Magnified images from all nine sequences in a 9-month-old ex vivo APP/PS1 brain. The images are ordered by increasing plaque CNR (defined in Eq. 1). The multi-echo imaging sequences (designated by a lower-case ‘m’) all produced increased plaque CNR compared to their single echo counterparts. Similarly, all SWI imaging sequences produced increased plaque CNR compared to the magnitude-only counterparts.
Figure 4.
Magnified images from the mGRE sequence (a), Thio-S staining (b), and iron staining (c). Red arrows point to plaques that were common to all three images. Black arrows point to plaques that were common to MRI and iron staining. White arrows point to plaques that were common to MRI and Thio-S staining.
Figure 5.
Representative images from T2-, T2*-, and susceptibility-weighted imaging sequences in a 4-month-old ex vivo wild-type brain specimen. (a) An image from the mSE sequence (T2-weighted). (b) An image from the GRE sequences (T2*-weighted). (c) An image from the mGRE-SWI sequence (susceptibility-weighted). It can be seen that none of the imaging methods introduce hypointense regions that could be falsely identified as plaques.
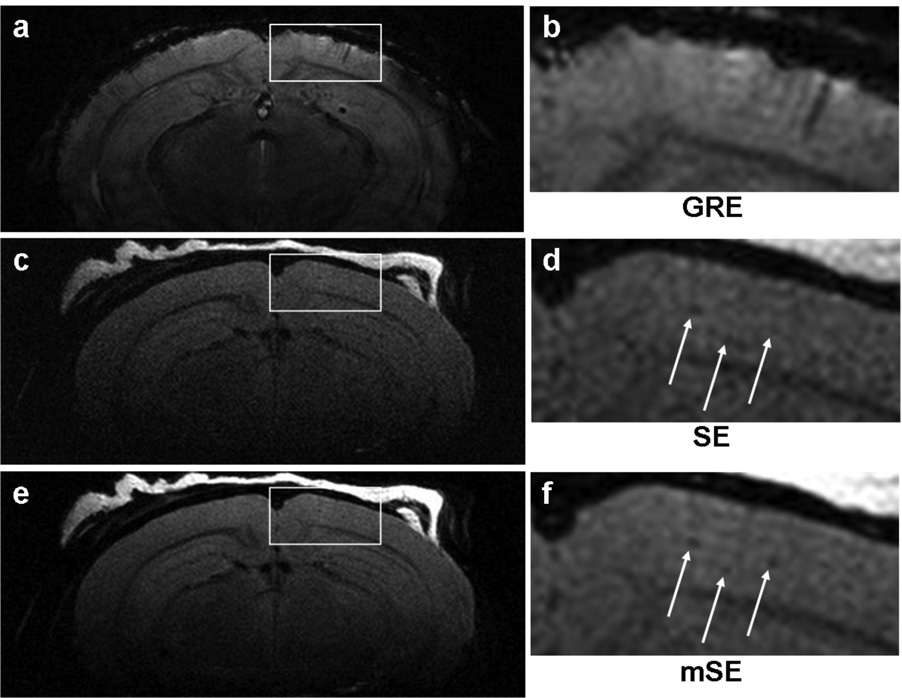
Fig. 6a shows a typical image from a GRE sequence in vivo. The echo time of this image was 23 ms, which is less than the optimal echo time for plaque contrast of 39.3 ms as estimated ex vivo. Although the optimal echo time will differ in vivo, it is unlikely to be significantly different from what was estimated ex vivo. The results were similar for manual shimming as well as automated shimming using FASTMAP (19). The severe susceptibility artifacts at the cortical interface at long echo times were judged to severely limit the utility of GRE- and SWI-based sequences for in vivo imaging. Fig. 6b shows a typical image from a SE sequence in vivo, and Fig. 6c shows a typical image from a mASE sequence in vivo with 8 ms of spin echo asymmetry. It was found that 8 ms was the maximal T2* weighting that could be applied without susceptibility-induced signal loss in the cortex.
Figure 6.
Representative in vivo images from the GRE, SE, and mASE sequences in a 12-month-old APP/PS1 mouse. (a) The GRE image showed cortical signal loss due to the susceptibility mismatch interfaces at TE = 23 ms. Sinuses inferior to the image border also caused a rippling artifact in the image. Since plaque contrast is maximal near 40 ms, this effect makes in vivo plaque imaging with GRE-based sequences impractical. (b) The SE sequence that was previously demonstrated to visualize plaques in vivo. (c) The mASE sequence provides increased SNR and image quality compared to the SE sequence. A quantitative comparison of the SE and mASE sequence in vivo is beyond the scope of this study, but it will be needed in the future.
DISCUSSION
The fact that plaques contain iron has led to the assumption that T2*-weighting would be superior to T2-weighting in creating plaque contrast. The T2 and T2* estimates presented here are surprisingly similar, which may indicate that the iron concentration in the plaques is not as high as previously thought (20–22). The iron and Thio-S staining indicate that different individual plaques can have a wide range of fibrillar amyloid and iron concentrations, which could result in a range of T2 and T2* values. The values given here are an average for all plaques identified. A more comprehensive study is needed to see if there is a correlation between the concentrations of iron, amyloid and T2 in individual plaques. The similar T2 and T2* values may explain why previous studies have not succeeded in resolving cortical plaques with GRE sequences (14,23). The echo times in these studies were all less than 15 ms, which is not an optimal echo time according to the T2* estimates presented here. These studies took place at 7T and 11.7 T, respectively, so the T2* values will differ slightly. However, an echo time of 15 ms is likely too short to generate significant contrast at those field strengths. Other studies have used T2-weighted methods to visualize plaques, which our current results indicate will give similar contrast to T2*-weighted methods for a given echo time. The frequency shift of the plaques, presumably caused by the naturally occurring iron content of plaques, is small, but long gradient echo times can be used to generate phase contrast due to this frequency shift.
The first conclusion drawn from the ex vivo comparison was that for a given acquisition time, a FSE technique is inferior to a summed echo technique. It is postulated that this is due to the blurring caused by FSE techniques (24). The ex vivo results also indicate that T2*-weighted images provide superior plaque CNR compared to T2-weighted images, and SWI images provide the greatest CNR. The similar T1 values between plaques and normal tissue indicate that the differing TR between the SE- and GRE-based sequences is not a factor in plaque contrast. The T2 and T2* decay curves indicate that the contrast for a single gradient or spin echo will not be significantly different, but the gradient echo sequences gave consistently higher CNR. This could be due to a slight blooming effect in the gradient echo images. An increase in the apparent size of the plaques will decrease the partial volume effect and increase contrast. Furthermore, the multi-gradient echo sequences allow one to sum more echoes in the high contrast echo time region, which results in a greater overall CNR. Although the frequency shift of the plaques is less than 1 Hz, the phase accumulated at long echo times is still large enough to give a large increase in plaque CNR through SWI. Therefore, when performing ex vivo imaging, the mGRE-SWI sequence will provide the greatest plaque CNR for a given scan time. It should be noted that the plaque darkness in the images may not correlate exactly with the calculated CNR in Fig. 3. The plaque darkness is determined by the difference between the plaque signal and the background signal without regard to the uncertainty in the value of the plaque signal. That variance can be significantly different between the sequences; therefore, the plaques may appear darker in one sequence despite the fact that the CNR is reduced. In this work the calculated CNR value was assumed to be a more suitable measure of plaque visibility than simple visual inspection. The lack of hypointense pixels in the ex vivo images of the wild-type mouse brain indicate that the T2*- and susceptibility-weighting do not introduce structures into the image that could be erroneously identified as plaques.
It is apparent in Fig. 3 that the CNR of some plaques increases dramatically in moving from a SE sequence to a GRE or SWI sequence, while the CNR of other plaques appears to change minimally. Qualitative analysis of the iron and Thio-S stains do not present a predictable pattern as to which plaques will have increased CNR and which will not. It is likely dependent on plaque size, iron content and amyloid content. Previous studies by El Tannir El Tayara et al. have shown that the T2 value of cortical tissue containing plaques is not related to amyloid load (20,25). This indicates that the relaxation mechanisms involved are not solely dependent on the amount of iron or amyloid alone. They are more likely a complex interplay between the amount and morphology of the iron and the amyloid as well as other factors yet to be studied.
The in vivo GRE images indicate that the air-to-tissue and/or the fat-to-skull interfaces cause large enough susceptibility artifacts to make GRE- and SWI-based methods impractical for in vivo visualization of cortical plaques. GRE imaging techniques have been used to visualize striatal plaques in vivo (9,14,26); however, most molecular biology studies focus on cortical and hippocampal plaques in these animal models of human AD. The longest gradient echo time that could be reached without areas of severe signal loss in the cortex was less than 10 ms, but the T2* contrast is not large enough to visualize plaque until the echo time is greater than 20 ms. Similarly, the phase accumulated by the plaques is too small to make SWI imaging feasible at echo times shorter than 20 ms. Thus, the in vivo techniques for cortical plaque imaging are optimally performed with spin-echo based sequences. A mASE sequence with a small amount of spin echo asymmetry could increase plaque contrast without inducing excessive susceptibility artifacts. Future studies will be needed to validate the ability of the mASE to resolve individual plaques in vivo. Although SWI appears to be impractical for in vivo mouse imaging, it may be suitable for in vivo human imaging due to the increased distance from the air/tissue interface to the region of interest.
Because plaques are on the order of the voxel size, the CNR of plaques is directly related to the ability to detect them. We believe this ex vivo study provides a valuable comparison of a large group of imaging sequences in their ability to generate plaque contrast. Based on the findings presented here, future in vivo studies on mouse models of Alzheimer’s disease can make educated pulse sequence design choices.
Acknowledgments
Grant Sponsors: NIH grants R01 AG22035, P41 RR008079, and P30 NS057091, the Minnesota Partnership for Biotechnology and Medical Genomics, and the W.M. Keck Foundation
REFERENCES
- 1.Hardy J, Selkoe D. The amyloid hypothesis of Alzheimer's disease: Progress and problems on the road to therapeutics. Science. 2002;297:353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 2.Schenk D, Barbour R, Dunn W, Gordon G, Grajeda H, Guido T, Hu K, Huang J, Johnson-Wood K, Khan K, Kholodenko D, Lee M, Liao Z, Liebergurg I, Motter R, Mutter L, Soriano F, Shopp G, Vasquez N, Vandevert C, Walker S, Wogulis M, Yednock T, Games D, Seugert P. Immunization with amyloid-beta attenuates Alzheimer's-disease-like pathology in the PDAPP mouse. Nature. 1999;400:173–177. doi: 10.1038/22124. [DOI] [PubMed] [Google Scholar]
- 3.Holcomb L, Gordon M, McGowan E, Yu X, Benkovic S, Jantzen P, Wright K, Saad I, Mueller R, Morgan D, Sanders S, Zehr C, O’Campo K, Hardy J, Prada C, Eckman C, Younkin S, Hsiao KKD. Accelerated Alzheimer-type phenotype in transgenic mice carrying both mutant amyloid precursor protein and presenilin 1 transgenes. Nat Med. 1998;4:97–100. doi: 10.1038/nm0198-097. [DOI] [PubMed] [Google Scholar]
- 4.Hsiao K, Chapman P, Nilsen S, Eckman C, Harigaya Y, Younkin D, Yang F, Cole G. Correlative memory deficits, Aβ elevation, and amyloid plaques in transgenic mice. Science. 1996;274:99–102. doi: 10.1126/science.274.5284.99. [DOI] [PubMed] [Google Scholar]
- 5.Benveniste H, Einstein G, Kim K, Hulette C, Johnson G. Detection of neuritic plaques in Alzheimer's disease by magnetic resonance microscopy. Proc Natl Acad Sci. 1999;96:14079–14084. doi: 10.1073/pnas.96.24.14079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jack CR, Garwood M, Wengenack TM, Borowski BJ, Curran GL, Lin J, Adriany G, Grohn OH, Grimm R, Poduslo JF. In vivo visualization of Alzheimer's amyloid plaques by magnetic resonance imaging in transgenic mice without a contrast agent. Magn Reson Med. 2004;52:1263–1271. doi: 10.1002/mrm.20266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang J, Yarowsky P, Gordon MN, Di Carlo G, Munireddy S, van Zijl PC, Mori S. Detection of amyloid plaques in mouse models of Alzheimer's disease by magnetic resonance imaging. Magn Reson Med. 2004;51:452–457. doi: 10.1002/mrm.10730. [DOI] [PubMed] [Google Scholar]
- 8.Lee S-P, Falangola MF, Nixon RA, Duff K, Helpern JA. Visualization of β-amyloid plaques in a transgenic mouse model of Alzheimer's disease using MR microscopy without contrast reagents. Magn Reson Med. 2004;52:538–544. doi: 10.1002/mrm.20196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Faber C, Zahneisen B, Tippmann F, Schroeder A, Fahrenholz F. Gradient-echo and CRAZED imaging for minute detection of Alzheimer plaques in an APPV717I × ADAM10-dn mouse model. Magn Reson Med. 2007;57:696–703. doi: 10.1002/mrm.21201. [DOI] [PubMed] [Google Scholar]
- 10.Borthakur A, Gur T, Wheaton AJ, Corbo M, Trojanowski JQ, Lee VM, Reddy R. In vivo measurement of plaque burden in a mouse model of Alzheimer's disease. J Magn Reson Imaging. 2006;24:1011–1017. doi: 10.1002/jmri.20751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Helpern J, Lee S, Falangola M, Dyakin V, Bogart A, Ardekani B, Duff K, Branch C, Wisniewski T, de Leon M, Wolf O, O’Shea J, Nixon R. MRI assessment of neuropathology in a transgenic mouse model of Alzheimer's disease. Magn Reson Med. 2004;51:794–798. doi: 10.1002/mrm.20038. [DOI] [PubMed] [Google Scholar]
- 12.Braakman N, Matysik J, van Duinen SG, Verbeek F, Schliebs R, de Groot HJ, Alia A. Longitudinal assessment of Alzheimer's β-amyloid plaque development in transgenic mice monitored by in vivo magnetic resonance microimaging. J Magn Reson Imaging. 2006;24:530–536. doi: 10.1002/jmri.20675. [DOI] [PubMed] [Google Scholar]
- 13.Jack CR, Wengenack TM, Reyes DA, Garwood M, Curran GL, Borowski BJ, Lin J, Preboske GM, Holasek SS, Adriany G, Poduslo JF. In vivo magnetic resonance microimaging of individual amyloid plaques in Alzheimer's transgenic mice. J Neurosci. 2005;25(43):10041–10048. doi: 10.1523/JNEUROSCI.2588-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vanhoutte G, Dewachter I, Borghgraef P, Van Leuven F, Van der Linden A. Noninvasive in vivo MRI detection of neuritic plaques associated with iron in APP[V717I] transgenic mice, a model for Alzheimer's disease. Magn Reson Med. 2005;53:607–613. doi: 10.1002/mrm.20385. [DOI] [PubMed] [Google Scholar]
- 15.Haacke EM, Ayaz M, Khan A, Manova ES, Krishnamurthy B, Gollapalli L, Cuilla C, Kim I, Petersen F, Kirsch W. Establishing a baseline phase behavior in magnetic resonance imaging to determine normal vs. abnormal iron content in the brain. J Magn Reson Imaging. 2007;26:256–264. doi: 10.1002/jmri.22987. [DOI] [PubMed] [Google Scholar]
- 16.Haacke E, Xu Y, Cheng Y-CN, Reichenbach JR. Susceptibility Weighted Imaging (SWI) Magn Reson Med. 2004;52:612–618. doi: 10.1002/mrm.20198. [DOI] [PubMed] [Google Scholar]
- 17.Constable RT, Henkelman RM. Contrast, resolution, and detectability in MR imaging. J Comput Assist Tomogr. 1991;15(2):297–303. doi: 10.1097/00004728-199103000-00021. [DOI] [PubMed] [Google Scholar]
- 18.Robb R. A software system for interactive and quantitative analysis of biomedical images. In: Hohne K, Fuchs H, Pizer S, editors. 3D imaging in medicine. Berlin: Springer-Verlag; 1990. pp. 333–361. [Google Scholar]
- 19.Gruetter R. Automatic, localized in vivo adjustment of all 1st-order and 2nd-order shim coils. Magn Reson Med. 1993;29(6):804–811. doi: 10.1002/mrm.1910290613. [DOI] [PubMed] [Google Scholar]
- 20.El Tannir El Tayara N, Delatour B, Le Cudennec C, Guegan M, Volk A, Dhenain M. Age-related evolution of amyloid burden, iron load, and MR relaxation times in a transgenic mouse model of Alzheimer's disease. Neurobiol Dis. 2006;22:199–208. doi: 10.1016/j.nbd.2005.10.013. [DOI] [PubMed] [Google Scholar]
- 21.Falangola MF, Lee S-P, Nixon RA, Duff K, Helpern JA. Histological co-localization of iron in Aβ plaques of PS/APP transgenic mice. Neurochem Res. 2005;30(2):201–205. doi: 10.1007/s11064-004-2442-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meadowcroft M, Connor J, Smith M, Yang Q. Micro MR imaging of beta-amyloid plaques and co-registration with iron deposition and histological analysis in both human Alzheimer's disease and APP/PS1 transgenic mice. Toronto, Ontario, Canada. In Proceedings of the 16th Annual Meeting of ISMRM; 2008. p. 256. [Google Scholar]
- 23.Dhenain M, Privat N, Duyckaerts C, Jacobs R. Senile plaques do not induce susceptibility effects in T2*-weighted MR microscopic images. NMR Biomed. 2002;15:197–203. doi: 10.1002/nbm.760. [DOI] [PubMed] [Google Scholar]
- 24.Constable RT, Gore JC. The loss of small objects in variable TE imaging: Implications for FSE, RARE, and EPI. Magn Reson Med. 1992;28:9–24. doi: 10.1002/mrm.1910280103. [DOI] [PubMed] [Google Scholar]
- 25.El Tannir El Tayara N, Volk A, Dhenain M, Delatour B. Transverse relaxation time reflects brain amyloidosis in young APP/PS1 transgenic mice. Magn Reson Med. 2007;58:179–184. doi: 10.1002/mrm.21266. [DOI] [PubMed] [Google Scholar]
- 26.Dhenain M, El Tannir El Tayara N, Wu T-D, Guegan M, Volk A, Quintana C, Delatour B. Characterization of in vivo MRI detectable thalamic amyloid plaques from APP/PS1 mice. Neurobiol Aging. doi: 10.1016/j.neurobiolaging.2007.05.018. In Press. [DOI] [PubMed] [Google Scholar]