Abstract
Repression of the NF-κB pathway has been extensively researched due to its pivotal role in inflammation. We investigated the potential of the aryl hydrocarbon receptor (AHR) to suppress NF-κB regulated gene expression, especially acute phase genes, such as serum amyloid A (Saa). Using AHR mutants, it was determined that nuclear translocation and heterodimerization with ARNT are essential, but DNA-binding is not involved in AHR-mediated Saa repression. A number of AHR ligands were capable of repressing saa3 expression. AHR activation leads to a decrease in RELA and C/EBP/β recruitment to and histone acetylation at Saa3 gene promoter. A battery of acute-phase genes (e.g. C-reactive protein and haptoglobin) induced by cytokine exposure was repressed by AHR activation in mouse hepatocytes. Dietary exposure to an AHR ligand represses cytokine induced acute-phase response in liver. Use of a human liver-derived cell line revealed similar repression of Saa mRNA levels and secreted protein. Repression of AHR expression also enhanced Saa induction in response to cytokines, suggesting that AHR is capable of constitutively repressing Saa gene expression. These results establish a role for AHR in inflammatory signaling within the liver, presenting a new therapeutic opportunity, and signify AHR’s ability to function in a DNA-independent manner.
Keywords: Ah receptor, Serum amyloid, TCDD, inflammation, acute phase response, liver
The acute phase response (APR) represents a major adaptive physiological first-line reaction to potentially deleterious environmental stresses including infection, inflammation, chemical stress and neoplastic growth. Homeostatic disruption by such factors initiates the APR, primarily within the liver, resulting in a complex but highly coordinated change in the pattern of hepatic gene expression. The stimulation and repression of a subset of predominantly secreted hepatic factors known as the positive and negative acute phase proteins (APP) respectively, signals to the body the need to respond to a perceived stress. Numerous proteins have been classified as belonging to the APP, including; plasminogen, fibrinogen, C-reactive protein (CRP), α1-antitrypsin, α2-macroglobulin, and serum amyloid A (SAA)1. Many APP are pleiotropic in nature and generally serve to modulate the immune system and metabolic processes to counteract a perceived stress. However, persistent pathophysiological conditions such as cancer or autoimmune diseases (e.g. rheumatoid arthritis) can lead to chronic stress and sustained APR induction with subsequent deleterious effects on immune signaling, catabolism, cachexia and amyloidosis. Consequently, clarification of the transcriptional regulation of specific acute phase proteins and the potential to modulate their expression has obvious clinical benefits.
Induction of the APR is principally driven through cytokine signaling and activation of transcription factors such as, nuclear factor-κB (NF-κB); signal transducer and activator of transcription-3 (STAT3) and CCAAT-enhancer binding protein β (C/EBPβ or NF-IL6). The transcriptional activity of NF-κB can be influenced by numerous factors, both such as post-translational modification of NF-κB subunits, or through cross talk with other transcription factors (e.g. nuclear receptors)2. Recently, evidence has been provided implicating the aryl hydrocarbon receptor (AHR) or dioxin receptor as a modulator of NF-κB activity3. AHR, a ligand-activated transcription factor belonging to the basic helix-loop-helix PAS protein family has an established role in xenobiotic metabolism, driving the expression of detoxification enzymes, CYP1A1 being a prime example. AHR adheres to the paradigm of a ligand-activated transcription factor; ligand binding promotes the dissociation of AHR from a cytoplasmic chaperone complex thus facilitating nuclear translocation of AHR4. Nuclear AHR readily forms a heterodimer with AHR-nuclear translocator (ARNT) thus forming a competent transcription factor capable of binding cognate DNA dioxin response elements (DRE) and stimulating the expression of AHR target genes5. Recent evidence suggests that AHR activity is not restricted solely to xenobiotic metabolism but may also exert modulatory effects upon diverse cellular processes through the phenomenon of receptor cross-talk6.
Here, evidence is presented highlighting the potential of AHR to negatively regulate the transcriptional activity of factors that regulate the inflammation response with particular emphasis upon suppression of acute phase protein expression in the liver. Utilizing a murine in vivo model in conjunction with mouse and human cell culture systems we demonstrate, the capacity of ligand activated AHR to directly attenuate cytokine-mediated induction of the APR component SAA mRNA and protein, as well as other APR genes. Furthermore, we demonstrate that this attenuation occurs in the absence of direct interaction of AHR and its cognate response element. This represents the first report documenting the ability of AHR to repress inflammatory signaling in a non-traditional fashion and highlights the potential of AHR as a target for the therapeutic management of inflammatory disease.
MATERIALS AND METHODS
Reagents and Mice
Anti-RELA antibody (sc-372) and anti-C/EBPβ (sc-150) were purchased from Santa Cruz Biotechnology Inc. and anti-acetyl-histone H4 (Lys5) antibody was purchased from Upstate. Affinity-purified anti-AHR rabbit polyclonal antibody was obtained from BioMol. Recombinant interleukins were purchased from PeproTech Inc. Hepa1c1c7 and Huh7 cells were obtained from American Type Culture Collection and Curt Omiecinski (Penn State University), respectively. C57BL6/J were obtained from Jackson Laboratory, while Ahrfx/fxCreAlb and Ahr−/− mice were gifts from Chris Bradfield (University of Wisconsin, Madison).
AHR Ligand Dietary Exposure
Female C57BL6/J and Ahr−/− mice, 10–12 weeks old mice with mean weights 19.8 ± 0.7 g and 16.9 ± 1.4 g respectively, were used in this study. Mice were maintained on a standard 12 h light/dark cycle with ad libitum access to standard chow and water. Mice were given ad libitum access to purified AIN93M (Dyets, Inc.) or purified diet supplemented with 0.4 g/kg β-NF for 18 h (overnight). The next day mice were given intra-peritoneal injections of vehicle (phosphate-buffered saline) or 10 µg/kg murine IL1B/IL6 as indicated. 4 h post- i.p. injection mice were sacrificed by asphyxiation with CO2 and hepatic tissue harvested.
Cell Culture
Hepa1c1c7 and Huh7 established cell-lines were cultured in α-minimum essential medium with 8% fetal bovine serum (FBS). SV-40 virus immortalized mAHR−/− hepatocytes were maintained in αMEM, 10 nM dexamethasone and 4% FBS at 34°C. The cells were grown in the absence of dexamethasone during experiments. Primary bone marrow (BM) cells were isolated from lower limb bones of 8–12 week old C57BL/6J mice, BM cells were cultured overnight in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 8% FBS and penicillin/streptomycin. Non-adherent cells were centrifuged and plated in DMEM supplemented with 10 ng/ml granulocyte-monocyte colony stimulating factor and 2 mM glutamine. Half the volume of medium was replaced every day for 4 days prior to treatment.
Primary Hepatocyte Isolation
Primary murine hepatocytes were isolated by the in situ two-step perfusion method from mice7. Hepatocytes were maintained in culture media (Hepatozyme-SFM (Invitrogen)/2.5% DMSO/10 nM dexamethasone/100 IU/ml penicillin and 100 µg/ml streptomycin). Cells were cultivated for 5 d before treatment.
Expression Constructs
Plasmid constructs pcDNA3-mAhR, pcDNA3-ARNT-HA, pEYFPmAhR, pCI-XAP2, pGudLuc 6.1 were generated previously. The mAhR mutant constructs, pcDNA3-mAhR ΔH1 (Δ43–51) and pEYFP-mAhR ΔH1 were generated using loop-out mutagenesis with a QuickChange site-directed mutagenesis kit (Stratagene, La Jolla, CA).
Transient Transfections and Luciferase Assays
SV-40 virus immortalized mAHR−/− hepatocytes8 were transfected using Lipofectamine 2000 according to the manufacturer’s protocol.
Antibodies and Protein Blot Analysis
AHR was detected using mouse monoclonal antibody RPT1 (Affinity Bioreagents). Primary antibodies were detected with a biotinylated rabbit anti-mouse antibodies (Jackson Immunoresearch). Biotinylated secondary antibodies were detected using either 125I-streptavidin (Amersham Biosciences) or ECL.
RNA Isolation and Real-Time PCR
RNA was isolated from cells with TRI® reagent (Sigma) and reverse transcribed with a High Capacity cDNA Archive kit (Applied Biosystems). Quantitative real-time PCR was performed on iQ systems (BioRad) using iQ SYBR Green master mix (BioRad), according to the manufacturer’s protocol. Expression values of genes of interest were normalized to that of ribosomal protein L13a (RpL13a) or β-actin. The sequence of the primers used in real-time PCR are listed in Table S1.
Chromatin Immunoprecipitation Assay
Chromatin immunoprecipitation assays (ChIP) were performed as described previously9. Briefly, cells cultured in 150 mm culture dishes were crosslinked with 1% formaldehyde for 8 min at 37°C and sonicated using a Branson Sonifier 450 to generate 500 – 700 bp fragments. The sonicated lysate was diluted to two A260 units and 1 ml of this lysate was subjected to immunoprecipitation with the appropriate antibodies given above and protein A sepharose. The level of enrichment of promoter fragments was determined by PCR or real-time PCR. The Saa3 primers used were 5′-GCGCAATCTGGGGAAAGAAGATGT and 5′-TGAGTGGCTTCTGTCCTTTGCTGA (forward and reverse, respectively); for Saa2, 5′-TACTACACCCCAGAAGATTGCCAC and 5′-AGGTGAGAGGAGGCAGGCATTTAT.
siRNA Transfections
Repression of AHR expression was performed with siRNA oligos purchased from Dharmacon RNAi Technologies. Approximately 60% confluent Huh7 cells were transfected with 120 nM scrambled or anti-AHR oligos using Dharmafect-1 transfection reagent following manufacturer’s protocol. Culture medium was changed after 24 h and the cells were allowed to recover for an additional 12 h before treatment.
ELISA
Huh7 cells were treated with 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) and interleukins for 10 h or 24 h under serum-free conditions. Culture media was analyzed for SAA protein levels using a human SAA ELISA kit purchased from Anogen (Yes Biotech Laboratories Ltd.) according to the manufacturer’s protocol. The level of SAA in mouse serum was determined using a mouse SAA ELISA kit (Immunology Consultants Lab, Inc. Newberg, OR). Statistical comparison of treatments was performed using the Student’s t-test (α=0.05).
DNA Microarray Analysis
RNA was isolated from 106 sorted cells using TRI® reagent and further purified with RNeasy® columns. RNA integrity was confirmed by Bioanalyzer (Applied Biosystems). Samples were then hybridized to Affy mouse 2.0A genome chips. Labeling, hybridization and washing were performed at the microarray core facility, the Pennsylvania State University. Data was processed and significantly altered genes were identified using GeneChip Operating Software (GCOS). Genes were declared as increased or decreased in AHR and DNA-binding mutant AHR (A78D-AHR) transfected cells, as compared to control transfections.
RESULTS
Identification of DNA-binding Independent Effects Mediated by the AHR
The established mechanism of AHR function involves binding of the AHR-ARNT heterodimer to DNA bearing the DRE sequence. In order to investigate the possibility of a DNA-binding independent manner of AHR function, simian virus 40 (SV40) immortalized AHR null mouse hepatocytes were transfected with either the AHR or DNA-binding defective mutant (A78D) AHR expressing plasmid or control vector. The A78D-AHR mutant has previously been shown to bind ligand, translocate to the nucleus and heterodimerize with ARNT, yet it fails to bind DNA at its response element10. Green fluorescent protein (GFP) expressing plasmid was cotransfected along with AHR plasmids in a ratio of 1:3 and a high level of transfection efficiency was obtained (Fig. S1a). GFP expressing transfected cells were subjected to FACS and RNA and protein were isolated. AHR expression was confirmed by Western blot (Fig. 1a) and the RNA was used for microarray experiments. Single samples were analyzed on individual DNA microarrays. Data analysis was targeted to identify the subset of genes altered in WT-AHR as well as A78D-AHR transfected cells but not in the control. Multiple APR genes were identified to be repressed by the wild-type and DNA-binding defective mutant AHR (Table 1). Maximum repression was observed for Saa3 mRNA, which was selected for further analysis.
Figure 1.
Functional dissociation of the AHR function involved in Saa3 repression. (a) Western blot analysis of WT-AHR and A78D-AHR protein expression in SV40 immortalized AHR-null mouse hepatocytes transfected with a combination of GFP and WT-AHR/A78D-AHR/control vector in a ratio of 1:3, using Lipofectamine2000 transfection reagent. Cells were sorted for GFP expression using FACS analysis. (b) Schematic representation of murine WT-AHR domains and the deletion/mutation (arrowheads) for each AHR mutant. (c) and (d) Real-time PCR on RNA isolated from SV40 immortalized AHR-null hepatocytes transfected with WT-AHR or various AHR mutants for 24 h. (e) Western blot analysis of the WT-AHR and AHR mutants expressed in experiment depicted in panels c and d. WT, wild-type; A78D, DNA-binding mutant; ΔH1, heterodimerization mutant; K14A, nuclear localization mutant. Data represent mean induction ± SEM (n = 3/treatment group) and were analyzed by one-way ANOVA to determine significance (*P < 0.05).
Table 1.
Genes regulated by transiently expressed A78D-AHR or WT-AHR in immortalized AHR-null murine hepatocytes compared to control transfected cells.
 |
 |
 |
|
|---|---|---|---|
| aryl-hydrocarbon receptor | Ahr | 2.6 | 2.6 |
| trimethyllysine hydroxylase, epsilon | Tmlhe | 2.1 | 2.7 |
| LIM domain containing preferred translocation partner in lipoma | Lpp | 1.7 | 2.2 |
| acidic (leucine-rich) nuclear phosphoprotein 32 family, member A | Anp32a | 1.7 | 1.8 |
| denticleless homolog (Drosophila) | Dtl | 1.6 | 1.6 |
| DNA segment, Chr 9, ERATO Doi 306, expressed | D9Ertd306e | 1.5 | 1.6 |
| PREDICTED: LIM domain only 7 [Mus musculus], mRNA sequence | Lmo7 | 1.5 | 1.7 |
| germ cell-less homolog (Drosophila) | Gcl | 1.4 | 1.6 |
| zinc finger protein 207 | Zfp207 | 1.4 | 1.6 |
| WD repeat domain 26 | Wdr26 | 1.4 | 1.6 |
| protein tyrosine phosphatase, receptor type, J | Ptprj | 1.4 | 1.7 |
| RIKEN cDNA 3300001H21 gene | 3300001H21R ik | 0.7 | 0.6 |
| zinc finger protein 179 | Zfp179 | 0.7 | 0.6 |
| Ceruloplasmin | Cp | 0.7 | 0.5 |
| procollagen, type VI, alpha 1 | Col6a1 | 0.7 | 0.6 |
| matrilin 2 | Matn2 | 0.7 | 0.6 |
| interferon, alpha-inducible protein 27 | Ifi27 | 0.7 | 0.5 |
| vanin 3 | Vnn3 | 0.7 | 0.6 |
| Transferring | Trf | 0.7 | 0.6 |
| procollagen, type VI, alpha 2 | Col6a2 | 0.7 | 0.6 |
| procollagen, type VI, alpha 2 | Col6a2 | 0.7 | 0.6 |
| myxovirus (influenza virus) resistance 1 | Mx1 | 0.7 | 0.6 |
| complement component factor h | Cfh | 0.7 | 0.6 |
| complement component 1, s subcomponent | C1s | 0.7 | 0.5 |
| slit homolog 3 (Drosophila) | Slit3 | 0.7 | 0.6 |
| Kruppel-like factor 10 | Klf10 | 0.7 | 0.6 |
| lipocalin 2 | Lcn2 | 0.6 | 0.6 |
| lipopolysaccharide binding protein | Lbp | 0.6 | 0.5 |
| serine (or cysteine) peptidase inhibitor, clade A, member 3M | Serpina3m | 0.6 | 0.6 |
| proteoglycan 4 (megakaryocyte stimulating factor, articular superficial zone protein) | Prg4 | 0.6 | 0.5 |
| complement component 3 | C3 | 0.6 | 0.5 |
| FBJ osteosarcoma oncogene | Fos | 0.6 | 0.4 |
| serine (or cysteine) peptidase inhibitor, clade A, member 3N | Serpina3n | 0.5 | 0.5 |
| STEAP family member 4 | Steap4 | 0.5 | 0.4 |
| serine (or cysteine) peptidase inhibitor, clade A, member 3G | Serpina3g | 0.5 | 0.6 |
| serum amyloid A 3 | Saa3 | 0.4 | 0.4 |
Generation and Characterization of an AHR Heterodimerization Mutant
It is important to examine whether heterodimerization of AHR and ARNT is essential for the DNA-independent effects of AHR. An AHR-heterodimerization mutant (ΔH1-AHR) was constructed by deleting the DNA sequence that encodes for amino acids 43 – 51, which encompass helix-1 of the helix-loop-helix domain (Fig. 1b). Immunoprecipitation of HA-tagged ARNT failed to co-precipitate the ΔH1-AHR, demonstrating the inability of this mutant to heterodimerize with ARNT in contrast with WT-AHR (Fig. S2a). Consequently, ΔH1-AHR was unable to drive the expression of DRE-driven luciferase in Cos-1 cells (Fig. S2b) as well as that of endogenous Cyp1a1 mRNA in BP-8 cells, a rat hepatoma-derived cell-line deficient in AHR (Fig. S2c). However, a photoaffinity ligand binding assay demonstrated that ΔH1-AHR was still capable of binding ligand as efficiently as WT-AHR (Fig. S2d). TCDD treatment of yellow fluorescent protein (YFP)-tagged ΔH1-AHR and WT-AHR transfected Cos-1 cells demonstrated that ΔH1-AHR is capable of translocating into the nucleus upon ligand activation (Fig. S2e). Thus, the ΔH1-AHR is selectively deficient in its ability to heterodimerize with ARNT and drive classical DRE-dependent gene expression.
Saa3 Repression Requires AHR Heterodimerization and Nuclear Translocation
The conventional AHR pathway requires nuclear translocation and heterodimerization with ARNT. A previously described K14A-AHR mutant incapable of translocating to the nucleus11 and the ΔH1-AHR mutant (Fig. 1b), were expressed in SV40-immortalized AHR null cells along with GFP-plasmid. Saa3 expression was repressed by the WT-AHR and A78D-AHR, but not by K14A-AHR and ΔH1-AHR (Fig. 1c). Thus, DNA-binding independent gene regulation by AHR appears to require nuclear translocation and heterodimerization. As expected, Cyp1a1 – the prototypic AHR target gene, was induced in the cells transfected with WT-AHR, but not with A78D-AHR or ΔH1-AHR (Fig 1d). K14A-AHR minimally induced Cyp1a1; however, this may be due to overexpression. It should be noted that activation of AHR by TCDD did not alter the extent of Saa3 repression in SV40-immortalized AHR null hepatocytes (data not shown).
Saa3 Repression Under Different Experimental Conditions
The AHR-mediated Saa3 repression observed in immortalized AHR null cells was confirmed in other model systems. Saa3 transcription was induced in Hepa1c1c7 cells, a mouse hepatoma derived cell-line, by treatment with different pro-inflammatory cytokines such as interleukin-1β (IL1B), interleukin-6 (IL6), tumor necrosis factor-α (TNFA) or a combination of IL1B and IL6 (Fig. 2a and 2b). Activated AHR repressed the induction of Saa3 by ~50% for each cytokine treatment. AHR ligand dependent repression of Saa3 induction by AHR may provide an interesting therapeutic approach for chronic inflammatory diseases. However, it is essential to confirm that Saa3 repression is not limited to only high doses of an AHR ligand. To this end, repression of cytokine-induced Saa3 was studied with decreasing doses of TCDD in Hepa1c1c7 cells. TCDD was able to effectively repress Saa3 even at the lowest dose tested (200 pM) (Fig. 2e). Also, Hepa1c1c7 cells were treated with different AHR ligands to determine if Saa3 repression was a TCDD-specific effect. The established AHR ligands, benzo[a]pyrene (B[a]P), β-naphthoflavone (β-NF), α-naphthoflavone (α-NF) and M50354 were all able to repress Saa3 induction (Fig. 2c). M50354 is a recently described AHR agonist compound capable of attenuating atopic allergic responses12. Interestingly, all ligands tested were effective at repressing Saa3, while TCDD was far more effective at inducing CYP1A1 (Fig. 3d). These results would support the concept that an AHR ligand may be found that is highly selective in eliciting a gene repression response.
Figure 2.
AHR directly represses Saa3 transcription in a dose-responsive and ligand-dependent manner. (a and b) AHR represses Saa3 induction by various cytokines. Hepa1c1c7 cells were treated with 10 nM TCDD or vehicle control. After 30 min, cells were treated with 2 ng/ml IL1B, IL6, TNFA or a combination of IL1B and IL6 for an additional 6 h. (c and d) Various classes of AHR ligands can suppress Saa3. Hep1c1c7 cells were treated with different AHR ligands at the described doses for 30 min prior to interleukin (IL1B + IL6, 2 ng/ml each) treatment. Saa3 mRNA (c) and Cyp1a1 mRNA (d) were measured by real-time PCR. (e and f) Analysis of TCDD dose-response of AHR-mediated Saa3 repression. Hepa1c1c7 cells treated with increasing doses of TCDD (0.2 nM to 10 nM) for 30 min prior to interleukin (IL1B + IL6, 2ng/ml each) treatment. (e) Closed triangles represent repression of IL-induced Saa3 mRNA by various doses of TCDD, as determined by real-time PCR. Closed squares represent uninduced Saa3 mRNA levels, as a control. (f) TCDD-driven Cyp1a1 mRNA induction, as measured by real-time PCR. (g) AHR-mediated Saa3 repression is due to direct transcriptional inhibition. Real-time PCR on RNA from Hepa1c1c7 cells treated first with (black bars), or without (open bars), the translational inhibitor – cycloheximide (10 µg/ml) for 30 min, then with TCDD (10 nM) for 30 min and finally with TNFA for 6 h. Data represent mean induction ± SEM (n = 3/treatment group) and were analyzed to determine significance (*P < 0.05).
Figure 3.
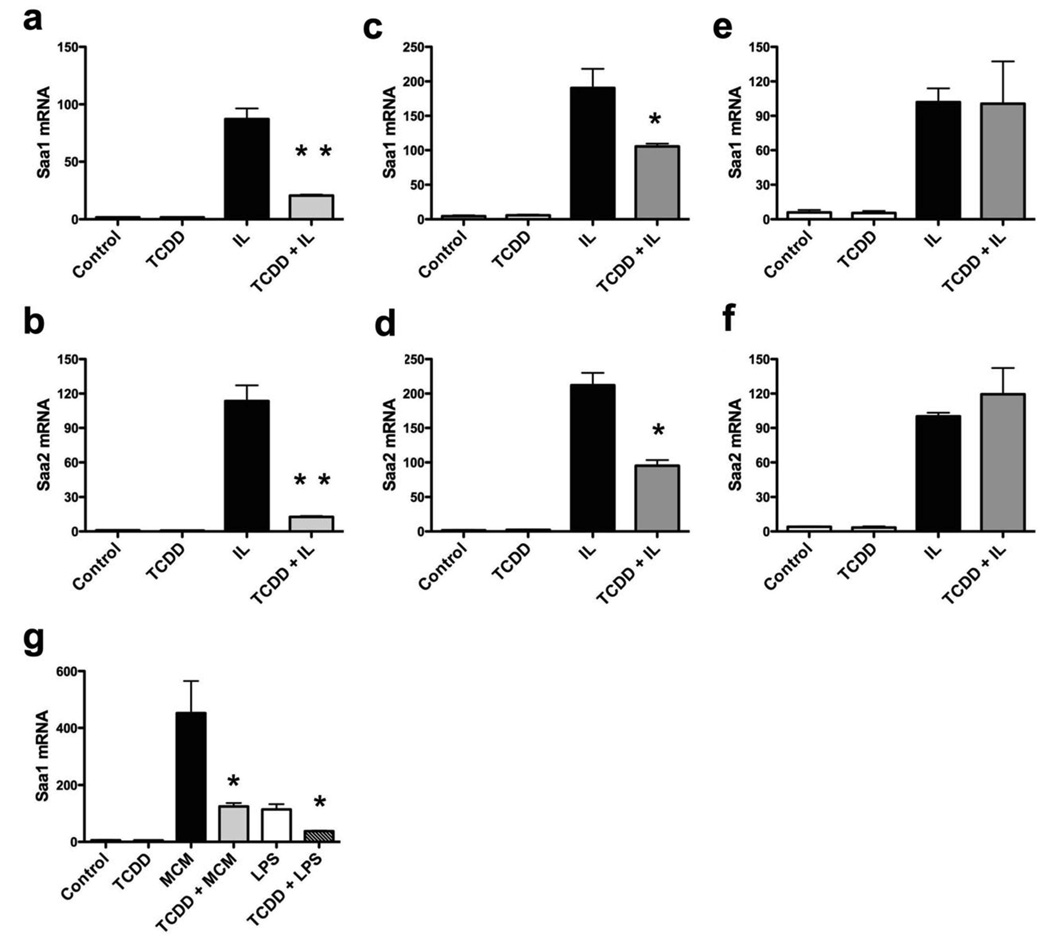
AHR activation represses gene expression of several Saa-family members. (a and b) Real-time PCR on RNA from TCDD (10 nM, 30 min) followed by interleukin (2 ng/ ml each of IL1B and IL6) treated Hepa1c1c7 cells. Effect of AHR activation on induction of other SAA family members, Saa1 (a) and Saa2 (b), was determined. Data represents the mean +/− S.D. of triplicate measurements. Experiment was repeated thrice with similar results. (c and d) Real-time PCR on RNA from primary mouse hepatocytes treated with TCDD (10 nM for 30 min) followed by interleukin (2 ng/ ml each of IL1B and IL6) for 24 h. Prior to treatment, cells were transferred to α-MEM with 1 mg/ml bovine serum albumin for 24 h. Repression of Saa1 (c) and Saa2 (d) mRNA was measured. (e and f) Real-time PCR measurement of Saa1 (e) and Saa2 (f) mRNA, as described above, in AHR-deficient primary mouse hepatocytes isolated from Ahrfx/fxCreAlb mice. (g) Primary murine bone marrow cells were cultured to promote differentiation into macrophages, as outlined in the text. After a 3 d LPS challenge, the conditioned media was collected from macrophage containing plates (MCM–macrophage conditioned media) and used to treat Hepa1c1c7 cells for 6 h following TCDD (10 nM, 30 min) pre-treatment. ‘LPS’ refers to LPS-spiked media that was maintained under similar culture conditions in the absence of any cells, and thus was devoid of any cytokines secreted by macrophages. Saa1 mRNA levels were determined by real-time PCR. Data represent mean induction ± SEM (n = 3/treatment group) and were analyzed to determine significance (*P < 0.05; **P < 0.01).
AHR Mediated Saa3 Repression is a Direct Transcriptional Effect
In order to ascertain that AHR directly effects the transcription of Saa3, Hepa1c1c7 cells were pretreated with cycloheximide, a translation inhibitor. Though cycloheximide treatment elevated the constitutive level of Saa3 expression, it did not alter its repression by AHR activation (Fig. 2g). This indicates that AHR-mediated Saa3 repression is a direct effect and not secondary to changes in the expression of another protein. Gene repression can be mediated by a decrease in transcription rate or by alteration of mRNA stability. After challenging with TCDD and ILB/IL6, Hepa1c1c7 cells were treated with Actinomycin D, a transcription inhibitor, and the level of Saa3 mRNA was assessed over 4 h. The decay rate of Saa3 mRNA was not significantly altered by AHR activation over the time period examined (data not shown).
AHR Activation Represses Other Saa Family Member Genes
All members of the SAA family are upregulated simultaneously in an acute phase response. Hence, we examined the effect of AHR activation on the expression of Saa1 and Saa2 in Hepa1c1c7 cells (Fig. 3a and 3b). Cytokine-mediated induction of both Saa1 and Saa2 was repressed by AHR activation by 75 and 85 percent, respectively. This is significant as Saa1 and Saa2 are the major hepatic serum amyloid isoforms. Interestingly, Saa1 and Saa2 did not appear to be repressed in the previous microarray results from WT-AHR or A78D-AHR transfected SV40-immortalized mouse hepatocytes, the reason for which is not clear.
Saa Repression is AHR Dependent
AHR-deficient or AHR-expressing primary hepatocytes were isolated from Ahrfx/fxCreAlb (hepatocyte-specific conditional AHR null)13 or C57BL6/J mice respectively. Saa transcription was highly induced in these cells by IL1B/IL6 treatment. TCDD was able to restrict the induction of Saa1 and Saa2 in AHR-expressing (Fig. 3c and 3d), but not in AHR-deficient, hepatocytes (Fig. 3e and 3f). This, along with the observation that AHR transfection in AHR null cells is required for suppressing Saa3 (Fig. 1c), clearly establishes that AHR is essential for Saa repression by TCDD.
AHR Can Repress Saa Induction Mediated by Complex Inflammatory Medium
Different cytokines can have counter-regulatory effects on various aspects of an inflammatory response. Thus, it is possible that IL1B and IL6 mediated Saa induction might not simulate the exact response obtained with a combination of cytokines, as expected in an inflammatory response in vivo. To confirm the ability of AHR to repress Saa induction under such circumstances, primary bone-marrow cells were isolated from C57BL6J mice and were cultured to promote differentiation into macrophages. Following a three-day LPS challenge, the conditioned culture medium was collected from the macrophages and used to treat Hepa1c1c7 cells. To differentiate the effect of secreted cytokines from those of LPS, LPS-containing culture medium was incubated in the absence of macrophages and used as a control. AHR-activation was able to repress the induction of Saa1 in response to the macrophage-conditioned-media or LPS alone (Fig. 3g). Similar results were obtained examining Saa2 repression by TCDD (data not shown). This demonstrates AHR’s ability to repress Saa induction under a physiologically attainable concentration/combination of cytokines.
Mechanistic Insights into AHR-mediated Saa Repression
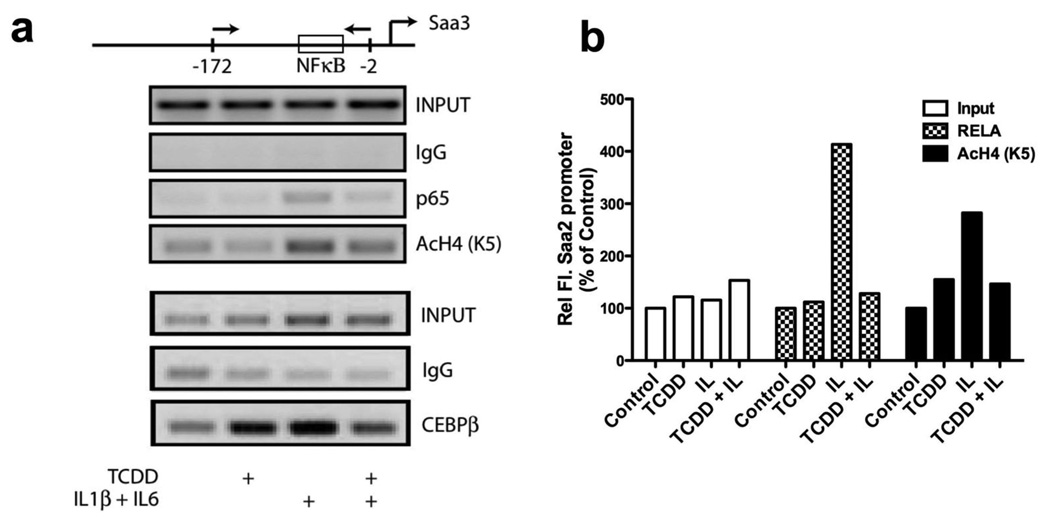
The fact that AHR directly represses Saa3 and that the K14A-AHR (nuclear localization) mutant failed to repress Saa3 induction, suggests that AHR likely affects the formation of a transcription complex within the nucleus and perhaps at the Saa promoters. Chromatin immunoprecipitation (ChIP) assays in Hepa1c1c7 cells demonstrate that activated AHR appears to reduce the presence of the RELA (p65) subunit of NF-κB at the Saa3 and Saa2 promoters in response to interleukin treatment (Fig. 4a, 4b). In addition, C/EBPβ presence on the SAA3 promoter after cytokine treatment was greatly inhibited after TCDD co-treatment (Fig. 4a). AHR has previously been shown to physically interact with RELA14, which might then contribute to preventing RELA recruitment to Saa promoters in response to IL1B/IL6 treatment. Chromatin immunoprecipitation with an acetylated-histone 4 antibody demonstrated that AHR activation also reduced histone acetylation at Saa2 and Saa3 promoters (Fig. 4a, 4b).
Figure 4.
AHR directly represses Saa transcription. (a and b) ChIP assay to determine the effect of AHR activation on Saa2 and Saa3 promoters. Hepa1c1c7 cells were treated with TCDD (10 nM for 30 min) prior to interleukin treatment (2 ng/ml each of IL1B and IL6 for 20 min). Immunoprecipitation was performed with antibodies for RELA and acetylated histones (K5). Changes at the Saa3 promoter were assessed by PCR (a), while changes at Saa2 promoter was analyzed by real-time PCR (b). Data represents one of three independent experiments.
AHR Mediated Suppression Extends to Other APR Genes
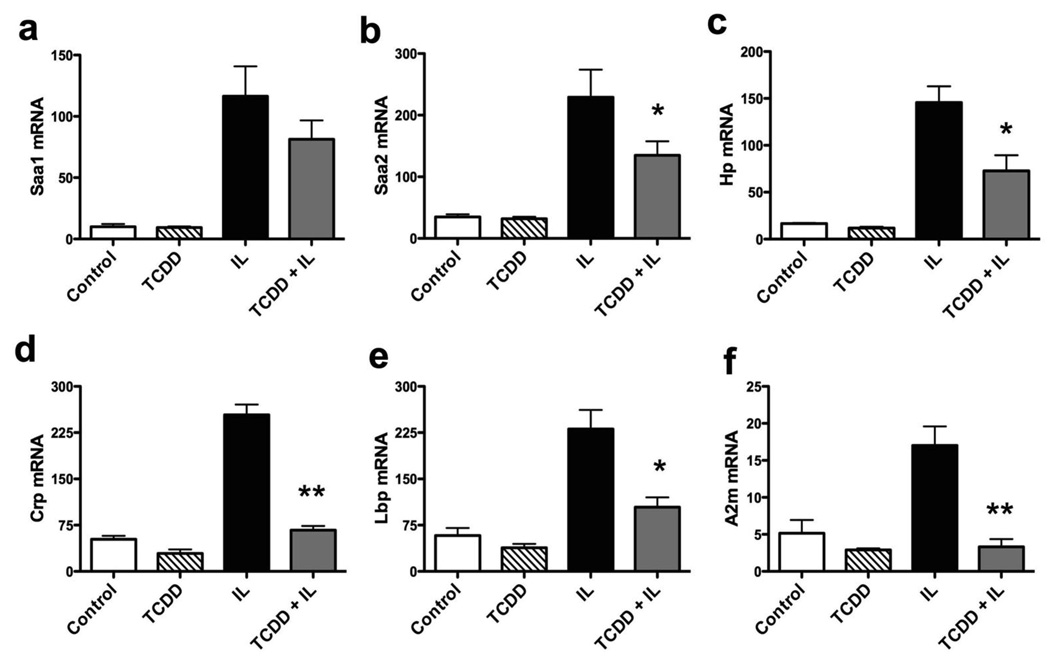
After confirming the repression of Saa1 and Saa2 in primary mouse hepatocytes, expression of other acute phase response genes was also examined by real-time PCR (Fig. 5a–f). AHR activation was able to repress induction of a number of acute phase genes; including, C-reactive protein (CRP), LPS-binding protein (LBP), haptoglobin, alpha-2-macroglobulin, and alpha-1-acid glycoprotein-1. This suggests that AHR represses the acute-phase response through a common transcriptional regulatory mechanism.
Figure 5.
AHR activation represses a battery of APR genes. Real-time PCR on RNA from primary mouse hepatocytes treated with TCDD (10 nM, 30 min) followed by interleukin (2 ng/ml each of IL1B and IL6) for 24 h. Repression of various APR genes was assayed. CRP, C-reactive protein; LBP, LPS-binding protein; A2m, alpha-2-macroglobulin; Hp, haptoglobin. Data represent mean induction ± SEM (n = 3/treatment group) and were analyzed to determine significance (*P < 0.05; **P < 0.01).
AHR Activation In Vivo Leads to Attenuation of Cytokine Mediated Acute Phase Response
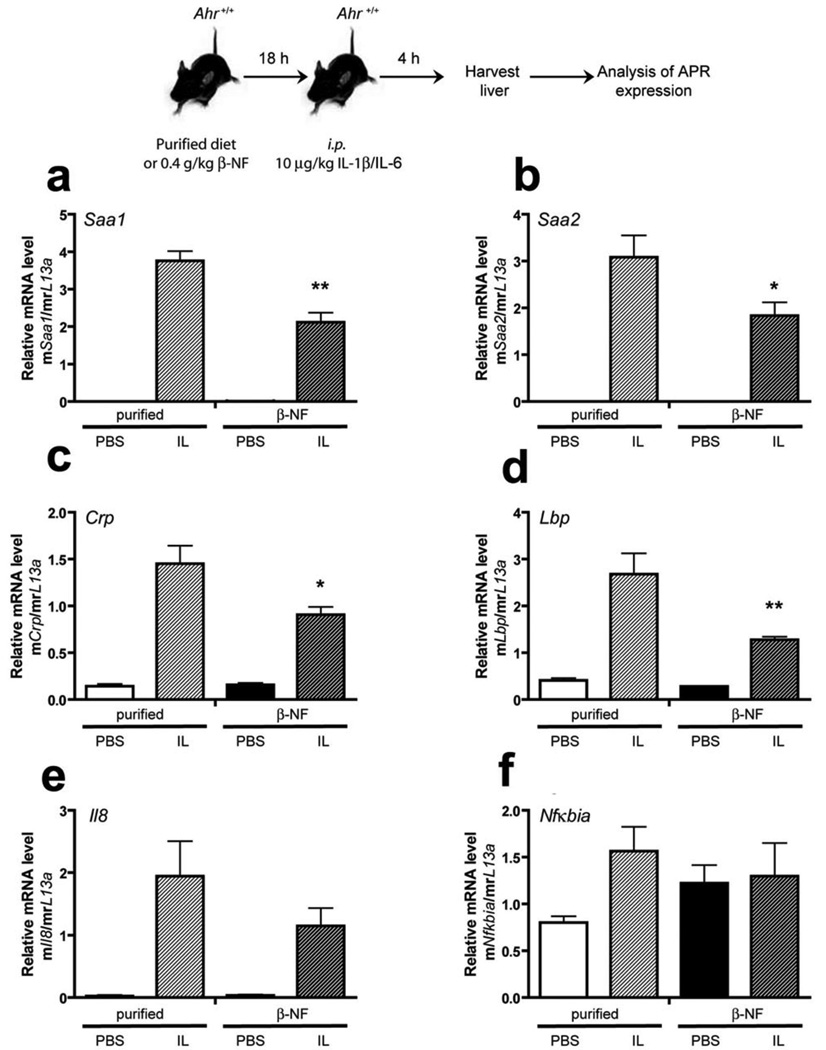
In order to test whether activation of the AHR leads to repression of cytokine-mediated acute-phase response, we reasoned that continual exposure to an AHR ligand would be more effective, thus a dietary route of exposure was chosen. After a preliminary dose-response experiment with dietary β-NF indicated that 0.4 g of β-NF/kg yielded about 10% of the Cyp1a1 inducibility observed at higher doses of β-NF in mice, BNF concentration was chosen for subsequent experiments (data not shown). C57BL6/J mice were fed a semi-purified control diet or a diet containing 0.4 g of β-NF/kg overnight. Mice were then i.p. injected with IL1B and IL-6, and after 5 h liver RNA was isolated. The level of Cyp1a2 mRNA induction was measured in C57BL6/J and Ahr-null mouse liver in the presence and absence of IL1B/IL-6 as a measure of AHR activation (Fig. S4). These results revealed that β-NF induced Cyp1A2 mRNA in C57BL6/J mice indicating that dietary β-NF effectively induced AHR activity. Real-time PCR analysis revealed that Saa1, Saa2 and Crp mRNA induction after cytokine treatment were all significantly reduced by the presence of an AHR ligand (Fig. 6). In contrast, dietary β-NF had no effect on cytokine-mediated acute-phase gene expression in Ahr-null mice, indicating that the observed repression in C57BL6/J mice is AHR-mediated (Fig 3S). Interestingly, β-NF treatment of Ahr-null mice leads to an increase in cytokine-mediated induction of certain acute-phase response genes (e.g. Lpb). As expected, IL1B/IL6 exposure in mice leads to the induction of Il-8 and Nfkbia mRNA in liver, two genes regulated by inflammatory signaling (Fig. 6e–f). In addition, β-NF failed to significantly influence the level of induction of Il-8 and Nfkbia (commonly known as IκBα) mRNA, suggesting that the AHR’s ability to alter inflammatory gene regulation is gene context specific. However, Il8 mRNA levels did show a non-statistically significant repressed response to β-NF exposure.
Figure 6.
β-NF attenuates cytokine-mediated APR expression in vivo. Female C57B6/J mice were given ad libitum access to purified chow ± 0.4 g/kg β-NF for 18 h. After 18 h mice were given i.p. injection of vehicle (PBS) or 10 mg/kg murine IL1B/IL6 as indicated. 4 h post- i.p. injection mice were sacrificed and hepatic tissue harvested for RNA isolation and subsequent quantitative PCR analysis of APR and pro-inflammatory genes Saa1, Saa2, Crp, Lbp, Il-8 and Ikb (Nfkbia) (a–f). Data represent mean induction ± SEM (n = 3/treatment group) and were analyzed by one-way ANOVA to determine significance (*P < 0.05; **P < 0.01).
AHR Mediated Repression of SAA is Observed in Human Huh7 Cells
Another important question to address is whether human cells would elicit a similar response to AHR activation in the presence of cytokines. SAA3 is not expressed in human liver15, while SAA1 and SAA2 have a very high sequence similarity, thus making it difficult to design unique primer-set for detecting SAA2. Hence, SAA1 mRNA levels were monitored to assess the effect of AHR activation on the acute phase response. Huh7 cells, a human hepatocarcinoma derived cell line, were treated with vehicle or TCDD to activate the AHR, followed by treatment with human IL1B and IL6. Under these conditions, activation of AHR repressed SAA1 mRNA induction by 75% (Fig. 7a). Changes in the level of secreted SAA protein were determined by ELISA and were found to mimic changes in mRNA (Fig. 7b). Since it is not possible to differentiate between different SAA family members by ELISA, this repression of SAA reflects the changes in the levels of all secreted SAA family members.
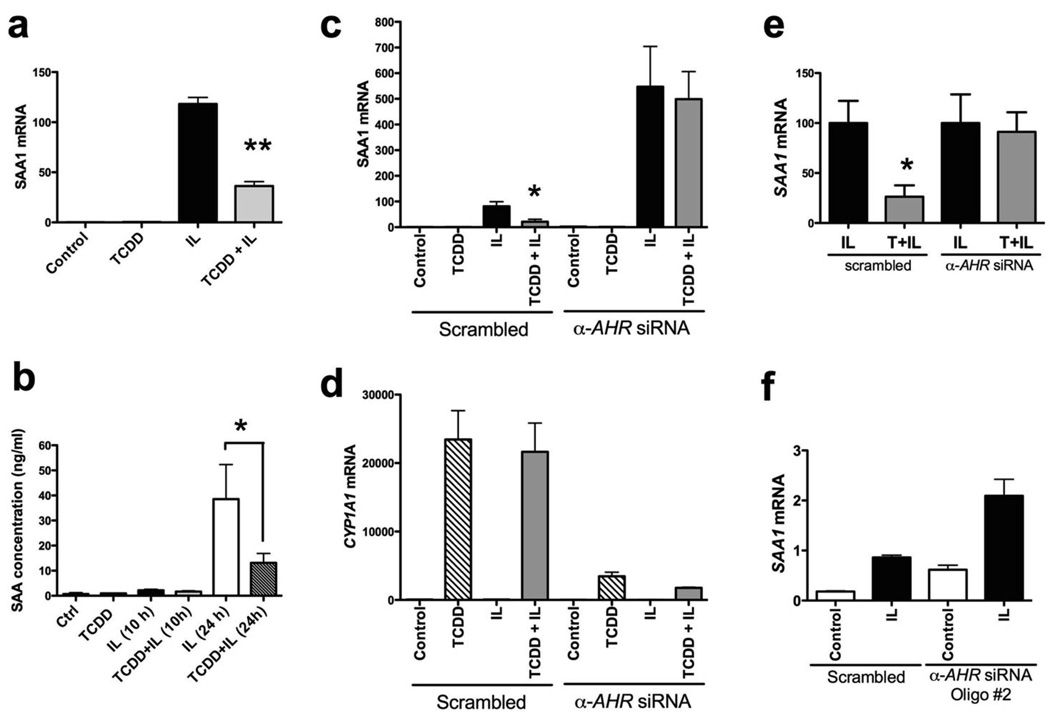
Figure 7.
AHR-mediated repression of SAA in human cells. (a) Real-time on RNA from Huh7 cells treated with TCDD (10 nM, 30 min) followed by human interleukins (2 ng/ml each of IL1B and IL6) for 6 h. Human SAA1 mRNA abundance was assayed. (b) ELISA to quantify SAA protein secreted by Huh7 cells, treated for 10 h or 24 h with TCDD and interleukins. Just prior to treatment, cells were transferred to serum-free medium. (c and d) siRNA-driven AHR knock-down in Huh7 cells. 36 h after siRNA transfection, cells were treated with TCDD and interleukins, as in (A). SAA1 mRNA (c) and CYP1A1 (d) mRNA levels were determined by real-time PCR. (e) An alternate representation of data from (c). SAA1 induction, upon interleukin exposure of scrambled and anti-AHR siRNA transfected cells, was scaled to 100 units. (f) Real-time PCR to assess SAA1 mRNA induction in Huh7 cells electroporated with a different anti-AHR siRNA oligo (Dharmafect). Data represent mean induction ± SEM (n = 3/treatment group) and were analyzed to determine significance (*P < 0.05; **P < 0.01).
Although TCDD exerts its effects almost exclusively through the AHR, we wished to confirm that the observed TCDD-mediated Saa1 repression in human cells is indeed AHR dependent. Repression of AHR expression in Huh7 cells was accomplished using AHR siRNA oligonucleotides. As expected, diminished AHR expression resulted in a loss of SAA1 repression with TCDD treatment (Fig.7c and 7e). AHR repression was verified by the loss of its ability to induce a transcriptional target gene, CYP1A1 (Fig. 7d). Interestingly, the loss of AHR resulted in an enhanced induction of SAA1 with IL1B/IL6 treatment (Fig. 7c). In order to confirm that this was not an off-target effect of the AHR siRNA oligo sequence, a second anti-AHR siRNA oligo was transfected into Huh7 cells by electroporation. Loss of AHR expression blocked TCDD-mediated repression of cytokine induced SAA1 expression (Fig. 7e). Repression of AHR expression by this second AHR siRNA also resulted in enhanced SAA1 induction by IL1B/IL6 treatment (Fig. 7f). This suggests that AHR may function to constitutively suppress the level of SAA1 transcription.
Whether AHR-mediated repression of inflammatory genes is a universal phenomenon or a promoter specific effect was examined. Two known NF-κB regulated genes, Il-8 and Nfkbia were induced by IL1B/IL6 and both remained unaffected upon co-treatment with TCDD (data not shown). Thus, AHR-mediated repression of cytokine-mediated induction of acute phase gene expression is context specific and does not appear to occur at every target gene regulated by NFκB.
DISCUSSION
A dysregulated inflammatory/immune response appears to be the underlying cause of many diseases, such as multiple sclerosis, asthma, lupus and rheumatoid arthritis. An acute phase response dominates the initial reaction to perceived insults and commences a series of biochemical and neuroendocrine changes that facilitate mounting an inflammatory/immune response. Acute phase proteins largely expressed by the liver serve various tasks in this process. However, persistent activation of the APR has its own perils. Elevated CRP is associated with increased cardiovascular risk and has been proposed to be a better clinical marker of atherosclerosis and related events than lipid levels16. SAA is an apolipoprotein for high-density lipoproteins (HDL) and influences cholesterol metabolism leading to enhanced inflammation. Conversely, constant elevation of SAA, and even alpha-2-macroglobulin, leads to extracellular amyloid plaques that interfere with organ function and underlie the pathology of diseases such as Alzheimer’s17. In about 5% of rheumatoid arthritis patients AA amyloid deposition occurs, which can then lead to renal dysfunction and other adverse complications18. Therapies that can selectively reduce circulating SAA levels should be useful in managing amyloidosis in these patients.
Recent studies have demonstrated that SAA can contribute to systemic inflammation19. For example, SAA has been shown to serve as a ligand that activates TLR2, RAGE and FPRL1 receptors leading to an enhanced inflammatory response20–22. Collectively, signaling through these receptors can lead to activation of MAPKs, p42/44, JNK, and p38 kinases, as well as the transcription factors AP-1 and NFκB. The ability of SAA to activate FPRL1 receptor in synoviocytes can result in synovial hyperplasia as well as endothelial angiogenesis, two common phenotypes observed in rheumatoid arthritis22. Interestingly, elevated levels of the AHR have been observed in synovial tissue of rheumatoid arthritis patients, suggesting another possible target tissue for modulating AHR activity23.
Transcription of SAA and most other APPs, are primarily regulated by NF-κB, NF-IL6 (C/EBP-β) and STAT324. The inhibitory effects of the glucocorticoid receptor (GR), and estrogen receptor (ER) on NF-κB induced gene transcription has received wide attention25. NF-κB signaling allows multiple levels of regulation, which have been utilized by NRs to interact with this pathway. Cytokines engage distinct receptors on the cell surface, leading to recruitment and activation of a family of adaptor proteins through various post-translational modifications. Eventually, signaling from different cytokine receptors converges on phosphorylation-dependent activation of the IKK complex (IκB-kinase complex), which in turn releases NF-κB to translocate into the nucleus. Activated AHR may inhibit any of these cell-surface receptors or the immediate downstream cytoplasmic signaling to repress NF-κB activity. However, in the context of APR gene regulation, AHR effectively repressed Saa3 mRNA when induced separately by IL1B, IL6 and TNFA. Also, K14A-AHR the nuclear localization mutant, was unable to repress Saa3 mRNA induction. This demonstrates that AHR-mediated NF-κB suppression is not due to an effect on upstream cytokine signaling, but is primarily a nuclear phenomenon.
In ChIP assay, activation of AHR diminished cytokine-induced association of the RELA subunit of NF-κB with its response elements in Saa2 and Saa3 promoters. Other groups have previously demonstrated the ability of AHR to physically interact with RELA14. While a direct physical interaction between the two proteins might certainly explain the reduction in RELA recruitment to Saa promoters, it cannot be the sole mechanism for AHR-NF-κB cross-talk, because AHR activation is unable to universally repress NF-κB driven gene expression (Fig. 7G and 7H). Also, we did not observe a significant reduction in RELA or p50 protein levels upon AHR activation (data not shown). Yet another possible mechanism could be the ability of the AHR to interact with RelB and bind to RelB-like response elements, as has been observed in U937 macrophages26. However, we have been unable to detect the AHR at the SAA3 promoter by ChIP analysis.
The degree of APR repression observed in mice in this study is not as marked as that observed in cell culture experiments. There may be many reasons for this observation, although we believe that treatment of mice with an AHR agonist will have a dual effect on inflammatory signaling. There are two basic mechanisms that can modulate inflammatory signaling that probably are cell-type specific; the first is the ability of the AHR/ARNT heterodimer to repress gene transcription, which is the subject of this report. The second is the ability of various AHR ligands to enhance inflammatory signaling, apparently through a DRE-mediated mechanism. For example, IL1B and TCDD co-treatment of MCF-7 cells leads to a syngeristic induction of Il6 expression27. The use of an AHR ligand that can effectively dissociate the repressive effects from the DRE-driven transactivation effects should tilt the overall AHR-mediated responses towards anti-inflammatory effects. Exposure of Hepa 1 cells to the partial agonist α-naphthoflavone reveals that this compound is effective at repressing cytokine-mediated SAA3 induction yet is a relatively poor inducer of DRE-driven transcriptional activity (Fig. 3C). This result would support the concept that a highly selective AHR ligand can be identified that would yield anti-inflammatory activity.
Ligand-activated receptors have multiple domains that impart different functionalities, such as DNA-binding, ligand-binding, dimerization and co-regulator recruitment. However, depending on the manner of activation and the physiological context, the functionalities of soluble receptors can be dissociated from their biological roles, as in the case of GR28 and ER29. Here, we demonstrate for the first time that DNA-binding is not essential for AHR-mediated repressive effects on NF-κB transactivation, while heterodimerization with ARNT and nuclear translocation are required. Aside from xenobiotic metabolizing enzyme induction, this is also the first report identifying a functional molecular role for AHR in a biological process, and not just regulation of individual genes. The data presented in this report demonstrate the functional interplay between AHR and inflammatory signaling pathways to regulate the expression of multiple APR genes, an important aspect of the hepatic inflammatory response. This report identifies a novel physiological function performed by the AHR in murine as well as human systems. AHR-mediated transcriptional repression is not conducted in the classical DRE-dependent fashion, but most likely involves multiple protein-protein interaction mechanisms. The fact that dietary exposure to the AHR ligand β-NF effectively represses cytokine-mediated APR in liver, as seen in figure 6 underscores the possibility of utilizing the AHR as a therapeutic target for treatment of inflammatory/autoimmune disease. However, in order to therapeutically utilize the ability of AHR to function as a repressor of APR, and possibly other inflammatory phenomena, it is necessary to identify selective ligands that would not also induce xenobiotic metabolism.
Supplementary Material
This paper contains supplemental information that is available on the Laboratory Investigation web site.
ACKNOWLEDGEMENTS
We thank Dr. Chris Bradfield for the Ahrfx/fxCreAlb and Ahr−/− mice. This work was supported by Public Health Service grant ES04869 from the National Institute of Environmental Health Sciences.
ABBREVIATIONS
- APR
acute phase response
- AHR
aryl hydrocarbon receptor
- ARNT
AHR-nuclear translocator
- β-NF
β-naphthoflavone
- IL1B
interleukin 1β
- SAA
serum amyloid A
- TCDD
2,3,7,8-tetrachlorodibenzo-p-dioxin
REFERENCES
- 1.Ramadori G, Christ B. Cytokines and the hepatic acute-phase response. Semin Liver Dis. 1999;19:141–155. doi: 10.1055/s-2007-1007106. [DOI] [PubMed] [Google Scholar]
- 2.Pascual G, Glass CK. Nuclear receptors versus inflammation: mechanisms of transrepression. Trends Endocrinol Metab. 2006;17:321–327. doi: 10.1016/j.tem.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 3.Tian Y, Rabson AB, Gallo MA. Ah receptor and NF-kappaB interactions: mechanisms and physiological implications. Chem Biol Interact. 2002;141(1–2):97–115. doi: 10.1016/s0009-2797(02)00068-6. [DOI] [PubMed] [Google Scholar]
- 4.Schmidt JV, Bradfield CA. Ah receptor signaling pathways. Annu Rev Cell Dev Biol. 1996;12:55–89. doi: 10.1146/annurev.cellbio.12.1.55. [DOI] [PubMed] [Google Scholar]
- 5.Bacsi SG, Reisz-Porszasz S, Hankinson O. Orientation of the heterodimeric aryl hydrocarbon (dioxin) receptor complex on its asymmetric DNA recognition sequence. Mol Pharmacol. 1995;47:432–438. [PubMed] [Google Scholar]
- 6.Barouki R, Coumoul X, Fernandez-Salguero PM. The aryl hydrocarbon receptor, more than a xenobiotic-interacting protein. FEBS Lett. 2007;581:3608–3615. doi: 10.1016/j.febslet.2007.03.046. [DOI] [PubMed] [Google Scholar]
- 7.Madden CR, Finegold MJ, Slagle BL. Expression of hepatitis B virus X protein does not alter the accumulation of spontaneous mutations in transgenic mice. J Virol. 2000;74:5266–5272. doi: 10.1128/jvi.74.11.5266-5272.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murray IA, Reen RK, Leathery N, et al. Evidence that ligand binding is a key determinant of Ah receptor-mediated transcriptional activity. Arch Biochem Biophys. 2005;442:59–71. doi: 10.1016/j.abb.2005.07.014. [DOI] [PubMed] [Google Scholar]
- 9.Spencer VA, Sun JM, Li L, et al. Chromatin immunoprecipitation: a tool for studying histone acetylation and transcription factor binding. Methods. 2003;31:67–75. doi: 10.1016/s1046-2023(03)00089-6. [DOI] [PubMed] [Google Scholar]
- 10.Levine SL, Petrulis JR, Dubil A, et al. A tetratricopeptide repeat half-site in the aryl hydrocarbon receptor is important for DNA binding and trans-activation potential. Mol Pharmacol. 2000;58:1517–1524. doi: 10.1124/mol.58.6.1517. [DOI] [PubMed] [Google Scholar]
- 11.Petrulis JR, Kusnadi A, Ramadoss P, et al. The hsp90 Co-chaperone XAP2 alters importin beta recognition of the bipartite nuclear localization signal of the Ah receptor and represses transcriptional activity. J Biol Chem. 2003;278:2677–2685. doi: 10.1074/jbc.M209331200. [DOI] [PubMed] [Google Scholar]
- 12.Morales JL, Krzeminski J, Amin S, et al. Characterization of the antiallergic drugs 3-[2-(2-phenylethyl) benzoimidazole-4-yl]-3-hydroxypropanoic acid and ethyl 3-hydroxy-3-[2-(2-phenylethyl)benzoimidazol-4-yl]propanoate as full aryl hydrocarbon receptor agonists. Chem Res Toxicol. 2008;21:472–482. doi: 10.1021/tx700350v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walisser JA, Glover E, Pande K, et al. Aryl hydrocarbon receptor-dependent liver development and hepatotoxicity are mediated by different cell types. Proc Natl Acad Sci U S A. 2005;102:17858–17863. doi: 10.1073/pnas.0504757102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tian Y, Ke S, Denison MS, et al. Ah receptor and NF-kappaB interactions, a potential mechanism for dioxin toxicity. J Biol Chem. 1999;274:510–515. doi: 10.1074/jbc.274.1.510. [DOI] [PubMed] [Google Scholar]
- 15.Kluve-Beckerman B, Drumm ML, Benson MD. Nonexpression of the human serum amyloid A three (SAA3) gene. DNA Cell Biol. 1991;10:651–661. doi: 10.1089/dna.1991.10.651. [DOI] [PubMed] [Google Scholar]
- 16.Danesh J, Wheeler JG, Hirschfield GM, et al. C-reactive protein and other circulating markers of inflammation in the prediction of coronary heart disease. N Engl J Med. 2004;350:1387–1397. doi: 10.1056/NEJMoa032804. [DOI] [PubMed] [Google Scholar]
- 17.Veerhuis R, Boshuizen RS, Familian A. Amyloid associated proteins in Alzheimer's and prion disease. Curr Drug Targets CNS Neurol Disord. 2005;4:235–248. doi: 10.2174/1568007054038184. [DOI] [PubMed] [Google Scholar]
- 18.Gillmore JD, Lovat LB, Persey MR, et al. Amyloid load and clinical outcome in AA amyloidosis in relation to circulating concentration of serum amyloid A protein. Lancet. 2001;358:24–29. doi: 10.1016/S0140-6736(00)05252-1. [DOI] [PubMed] [Google Scholar]
- 19.Malle E, Sodin-Semrl S, Kovacevic A, Serum amyloid A. An acute-phase protein involved in tumour pathogenesis. Cell Mol Life Sci. 2008 doi: 10.1007/s00018-008-8321-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cheng N, He R, Tian J, et al. Cutting edge: TLR2 is a functional receptor for acute-phase serum amyloid A. J Immunol. 2008;181:22–26. doi: 10.4049/jimmunol.181.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Okamoto H, Katagiri Y, Kiire A, et al. Serum amyloid A activates nuclear factor-kappaB in rheumatoid synovial fibroblasts through binding to receptor of advanced glycation end-products. J Rheumatol. 2008;35:752–756. [PubMed] [Google Scholar]
- 22.Lee MS, Yoo SA, Cho CS, et al. Serum amyloid A binding to formyl peptide receptor-like 1 induces synovial hyperplasia and angiogenesis. J Immunol. 2006;177:5585–5594. doi: 10.4049/jimmunol.177.8.5585. [DOI] [PubMed] [Google Scholar]
- 23.Kobayashi S, Okamoto H, Iwamoto T, et al. A role for the aryl hydrocarbon receptor and the dioxin TCDD in rheumatoid arthritis. Rheumatology (Oxford) 2008;47:1317–1322. doi: 10.1093/rheumatology/ken259. [DOI] [PubMed] [Google Scholar]
- 24.Shimizu H, Yamamoto K. NF-kappa B and C/EBP transcription factor families synergistically function in mouse serum amyloid A gene expression induced by inflammatory cytokines. Gene. 1994;149:305–310. doi: 10.1016/0378-1119(94)90166-x. [DOI] [PubMed] [Google Scholar]
- 25.Glass CK, Ogawa S. Combinatorial roles of nuclear receptors in inflammation and immunity. Nat Rev Immunol. 2006;6:44–55. doi: 10.1038/nri1748. [DOI] [PubMed] [Google Scholar]
- 26.Vogel FA, Matsumura F. A new cross-talk between the aryl hydrocarbon receptor and RelB, a member of the NFkappaB family. Biochem. Pharmacol. Epub. 2008 doi: 10.1016/j.bcp.2008.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hollingshead BD, Beischlag TV, Dinatale BC, et al. Inflammatory signaling and aryl hydrocarbon receptor mediate synergistic induction of interleukin 6 in MCF-7 cells. Cancer Res. 2008;68:3609–3617. doi: 10.1158/0008-5472.CAN-07-6168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reichardt HM, Kaestner KH, Tuckermann J, et al. DNA binding of the glucocorticoid receptor is not essential for survival. Cell. 1998;93:531–541. doi: 10.1016/s0092-8674(00)81183-6. [DOI] [PubMed] [Google Scholar]
- 29.Valentine JE, Kalkhoven E, White R, et al. Mutations in the estrogen receptor ligand binding domain discriminate between hormone-dependent transactivation and transrepression. J Biol Chem. 2000;275:25322–25329. doi: 10.1074/jbc.M002497200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This paper contains supplemental information that is available on the Laboratory Investigation web site.