SUMMARY
5′-Methylthioadenosine nucleosidase (MTAN) is a bacterial enzyme involved in S-adenosylmethionine-related quorum sensing pathways that induce bacterial pathogenesis factors. Transition state analogues 5′-methylthio- (MT-), 5′-ethylthio- (EtT-) and 5′-butylthio- (BuT-) DADMe-ImmucillinAs are slow-onset, tight-binding inhibitors of Vibrio cholerae MTAN (VcMTAN), with dissociation constants of 73, 70, and 208 pM, respectively. Structural analysis of VcMTAN with BuT-DADMe-ImmucillinA reveals interactions contributing to the high affinity. In V. cholerae cells, these compounds are potent MTAN inhibitors with IC50 values of 27, 31, and 6 nM for MT-, EtT-, and BuT-DADMe-ImmucillinA, disrupting autoinducer production in a dose-dependent manner without affecting growth. MT- and BuT-DADMe-ImmucillinA also inhibit autoinducer-2 production in enterohemorrhagic Escherichia coli O157:H7 with IC50 values of 600, and 125 nM, respectively. BuT-DADMe-ImmucillinA inhibition of autoinducer-2 production in both strains persists for several generations, and causes reduction in biofilm formation. These results support MTAN’s role in quorum sensing, and its potential as target for bacterial anti-infective drug design.
Bacteria communicate to each other by a process known as quorum sensing. When the population density reaches critical levels, they produce and detect signaling molecules known as autoinducers (AIs) to coordinate gene expression and regulate processes beneficial to the microbial communities1. With the growing global threat of multi-drug resistance, nonconventional anti-infective discovery approaches are being explored that are nonlethal to bacteria where the potential to develop resistance is assumed to be less significant. Quorum sensing is an ideal target for bacterial anti-infective design, as many bacterial species use this mechanism to regulate virulence2–5. Several mutant bacterial strains defective in quorum sensing create less potent infections. Quorum sensing-deficient intranasal Streptococcus pneumoniae infections in mouse are less effective at spreading to the lungs or the bloodstream6. In an infant rat Neisseria meningitidis infection model, a quorum sensing-deficient strain is unable to produce viable bacteria in the blood7. These findings, among others, suggest that a number of bacterial infections could be controlled by impeding quorum sensing.
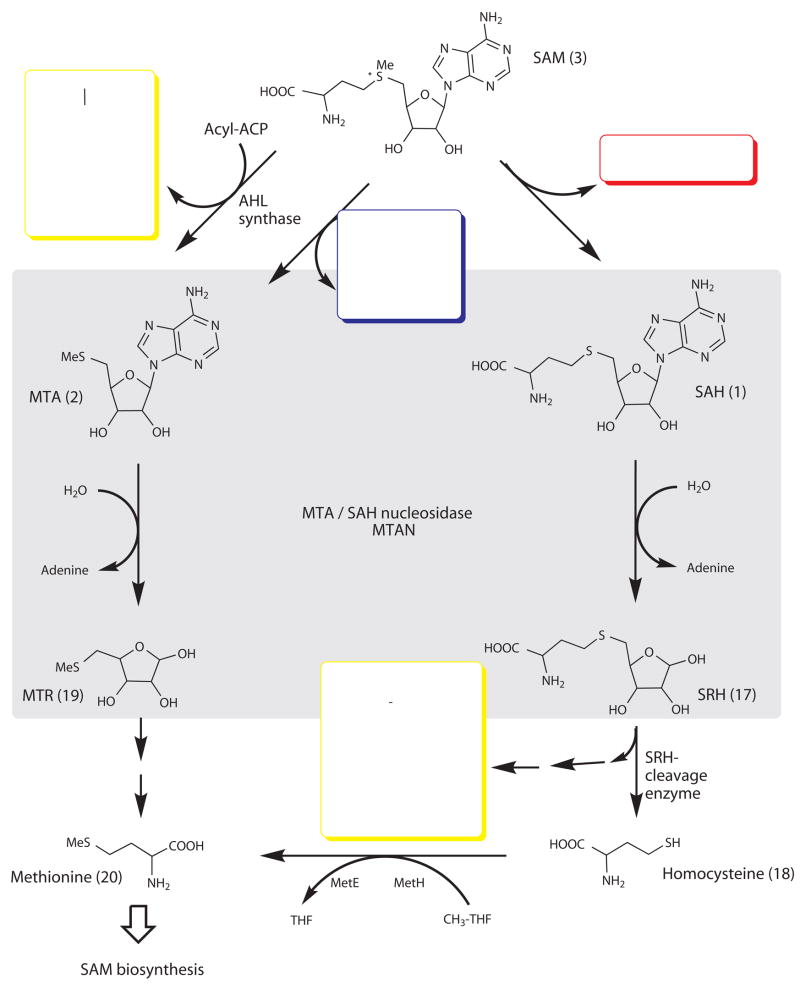
5′-Methylthioadenosine/S-adenosyl homocysteine nucleosidases (MTANs) play a crucial role in maintaining homeostasis in bacteria. MTANs are tightly linked to S-adenosyl methionine pathways that involve methylation reactions yielding S-adenosyl homocysteine (SAH, 1), and polyamine biosynthesis producing methylthioadenosine (MTA, 2) (Fig. 1). MTANs catalyze the hydrolytic deadenylation of MTA and SAH and provide the only known route for their metabolism in bacteria, whose accumulation is expected to inhibit related pathways. In addition, MTANs are directly involved in the biosynthesis of autoinducers. AI-1 and AI-2 are two classes of autoinducers synthesized from S-adenosyl methionine (SAM, 3) (Fig. 1). AI-1 is a family of acyl-homoserine lactones (AHLs, such as hydroxy-butanoyl-L-homoserine lactone, 4) [Au: edit ok, as compound 4 cannot be a class of chemicals?] believed to provide signaling molecules for intra-species communication. AI-2 includes derivatives of 4,5-dihydroxy-2,3-pentanedione (DPD, 5), responsible for inter-species communication. Thus, MTAN inhibition may provide a method of blocking both AI-1 and AI-2 production, and thereby disrupting quorum sensing.
Figure 1.
Role of MTAN in bacterial utilization of S-adenosylmethionine (SAM). The scheme shows the pathways connecting DNA methylation (red box), polyamine synthesis (blue box), autoinducer production (yellow box), and methionine and adenine salvage. AHL synthase catalyzes the transfer of the amino acid moiety of SAM to an acyl acceptor to produce homoserine lactones in the synthesis of AI-1 molecules, and MTA as byproduct. In methyltransferase reactions, SAM produces SAH which is a precursor in the tetrahydrofuran synthesis of AI-2 molecules (shown here as furanosyl boron diester, 14). Blocking MTAN activity is expected to cause accumulation of MTA, resulting in product inhibition of AI-1 production by AHL synthase49. In addition, inhibition of MTAN can directly block the formation of S-ribosylhomocysteine (SRH, 15), the precursor of AI-2. AI-1 and AI-2 are autoinducers used in bacterial quorum sensing, and MTAN offers a means to block formation of these signaling molecules.
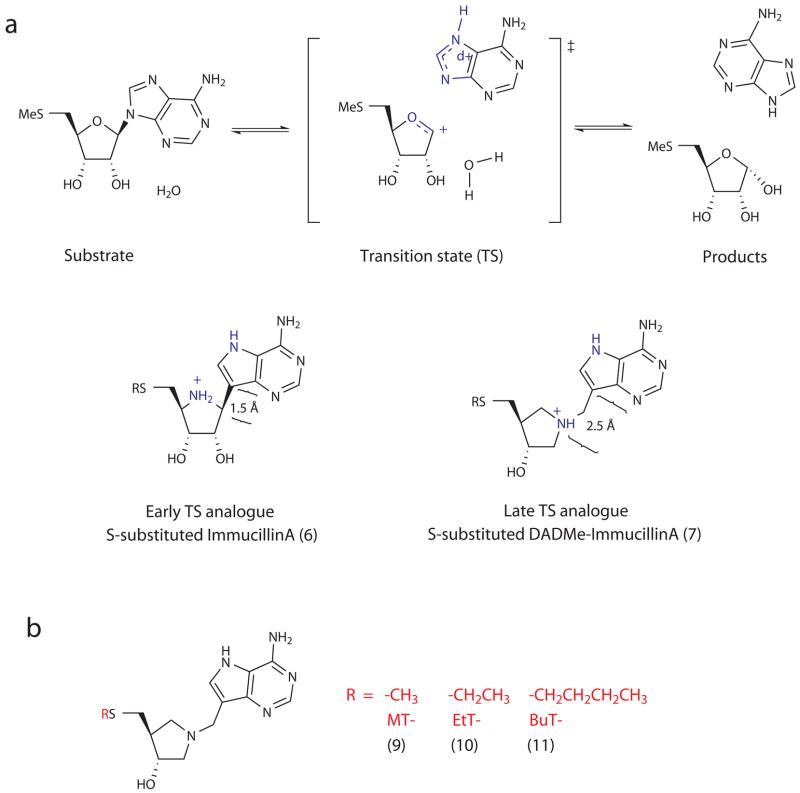
MTAP or 5′-methylthioadenosine phosphorylase is the counterpart to MTAN in humans, and functions similarly in metabolizing MTA but uses phosphate as a nucleophile instead of water. MTAP has been identified as an anticancer target due to its involvement in polyamine biosynthesis, purine and S-adenosylmethionine salvage pathways8,9. The transition state structures of human MTAP as well as MTANs from Escherichia coli (EcMTAN), Streptococcus pneumoniae (SpMTAN), and Neisseria meningitidis (NmMTAN) have been solved using kinetic isotope effects10–13. They all have dissociative SN1 transition states with ribooxacarbenium ion character, which could either be “late” transition states with fully broken N-glycosidic bonds (i.e., C1′-N9 distance of 3.0 Å or greater), or “early” transition states with C1′-N9 distances of 2.0 Å or less (Fig. 2a).
Figure 2.
The reaction catalyzed by MTAN with MTA as substrate. (a, top) shows a dissociative transition state for E. coli with ribooxacarbenium ion character10. Structures of stable analogues for an early dissociative transition state (ImmucillinA), and a late dissociative transition state (DADMe-ImmucillinA) depict differences in bond distances between the adenine leaving group and the ribosyl group, as well as charge localization (a, bottom). (b) Shown is the structure of S-substituted DADMe-ImmucillinA, along with MT-, EtT- and BuT- substituents.
Transition state analysis provides blueprints for the design of stable analogues, which in the study of purine nucleoside phosphorylases, has yielded extremely potent inhibitors currently in clinical trials for autoimmune disease and cancer14,15. The same drug design approach has been extended to MTAP and MTANs10–13. Derivatized ImmucillinA (ImmA) and DADMe-ImmucillinA (DADMe-ImmA) provide two generations of transition state analogues developed for MTAP and MTANs16,17. ImmA derivatives mimic early dissociative transition states, while DADMe-ImmA derivatives resemble late dissociative transition states (Fig. 2a,b). The cationic N1′ of DADMe-ImmA resembles the cationic C1′ of the ribosyl group in late, dissociative transition states. In addition, the methylene group between 9-deazaadenine and the pyrrolidine ring in DADMe-ImmA provides geometric similarity between the adenine leaving group and the ribooxacarbenium site, and the 9-deazaadenine provides chemical stability and mimics the increased pKa at N7 found at the MTAN transition states.
ImmA and DADMe-ImmA derivatives synthesized and tested against MTAP and MTANs exhibit some of the highest affinities ever achieved for noncovalent enzyme-inhibitor interactions18–21. For instance, para-chloro-phenylthio-DADMe-ImmA (6) inhibits purified EcMTAN with a dissociation constant of 47 fM, approaching a Km/Ki ratio of ~108 18. Methylthio-DADMe-ImmA (7) inhibits purified human MTAP with 86 pM affinity, and induces apoptosis in cultured head and neck squamous cell carcinoma cell lines without affecting normal human fibroblast cell lines9. It also suppresses tumor growth in mouse xenografts at doses nontoxic to the animals9. The bioavailability and nontoxic properties of methylthio-DADMe-ImmA make this class of compounds valuable drug candidates. We propose that similar analogues for MTAN may be effective in blocking MTAN activity in cells.
The current work provides in vitro cell characterization of MTAN inhibition and its role in quorum sensing using transition state analogues. Inhibition of Vibrio cholerae MTAN (VcMTAN) activity in recombinant purified enzyme as well as in cell cultures was characterized for the slow-onset, tight-binding DADMe-ImmAs. The effects of these inhibitors on autoinducer production and biofilm formation in pathogenic strains of V. cholerae and E. coli are also described. The results support MTAN’s role in quorum sensing, and indicate that MTAN may be an important target for drug design in anti-infective therapies.
RESULTS
MTAN transition state analogues are picomolar inhibitors of VcMTAN
VcMTAN has substrate specificity for hydrolysis of both MTA and SAH. We obtained a Km of 3 μM for MTA and a kcat of 2 s−1. For SAH, the Km and kcat values were 24 μM, and 0.5 s−1, respectively. With a kcat/Km ratio of 6.6 × 105 M−1s−1 for MTA, VcMTAN’s catalytic efficiency was 60-fold greater than the S. pneumoniae isoform, and 14-fold less than for E. coli MTAN18,20. The transition state analogues 5′-methylthio-DADMe-ImmucillinA (MT-DADMe-ImmA, 7), 5′-ethylthio-DADMe-ImmucillinA (EtT-DADMe-ImmA, 8), and 5′-butylthio-DADMe-ImmucillinA (BuT-DADMe-ImmA, 9) (Fig. 2b) inhibited VcMTAN activity with dissociation constants in the mid-picomolar range (Table 1), compared to E. coli MTAN in the low picomolar, and to S. pneumoniae MTAN in the nanomolar ranges18,20. The same transition state analogues inhibited VcMTAN with an affinity intermediate to that for E. coli and S. pneumoniae MTANs, as predicted by the catalytic enhancement provided by the enzymes. Reaction progress curves in the presence of various concentrations of MT-, EtT-, and BuT-DADMe-ImmA revealed time-dependent, slow-onset inhibition, yielding overall dissociation constants of 73, 70, and 208 pM, respectively (Supplementary Fig. 1a online).
Table 1.
Inhibition constants for purified MTAN activity, cellular MTAN activity, and autoinducer (AI) production determined as described in METHODS.
| R-group | Purified enzyme inhibition Ki*, pM | Cellular MTAN Inhibition IC50, nM | AI Inhibition IC50, nM | |
|---|---|---|---|---|
| BB170 (ai1−ai2+) | BB120 (ai1+ai2+) | |||
| MT- (7) | 73 ± 5 | 27 ± 4 | 0.94 ± 0.13 | 10.5 ± 2.6 |
| EtT- (8) | 70 ± 4 | 31 ± 7 | 11.0 ± 2.0 | 14.0 ± 2.0 |
| BuT- (9) | 208 ± 46 | 6 ± 1 | 1.4 ± 0.3 | 1.0 ± 0.2 |
A method for predicting the transition state structure for MTANs was reported recently, using dissociation constants for known transition state analogues21. This method classifies MTANs as having either early or late dissociative transition states, depending on the ratio of its dissociation constants for 5′-substituted ImmAs and DADMe-ImmAs. Dissociation constants were determined for VcMTAN with methylthio- (10), ethylthio-(11), benzylthio- (12), and para-chloro-phenylthio-ImmucillinA (13) (Supplementary Table 1 online). For the MT-ImmA/DADMe-ImmA inhibitor pair, VcMTAN gave a KImmA/KDADMe-ImmA of 137, indicating a strong preference for the transition state analogue that resembles a late transition state. This analysis predicts a late dissociative transition state for VcMTAN, similar to that of E. coli and S. pneumoniae. In addition to the ImmA dissociation constants being higher than their DADMe-ImmA counterparts, there was no slow onset phase in their inhibition profiles. Thus, the DADMe-ImmA compounds are better mimics of VcMTAN’s transition state, which strongly suggests that it is late and dissociative.
Crystal structure of VcMTAN-BuT-DADMe-ImmA complex
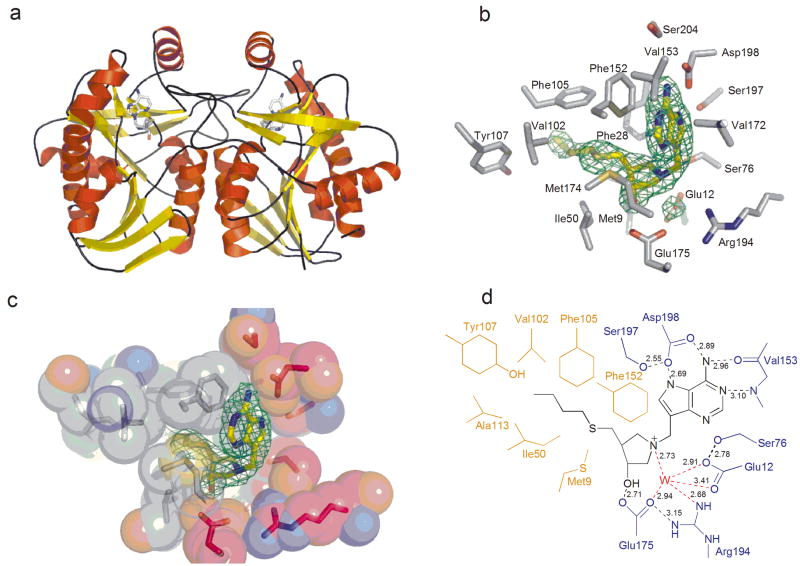
To define the determinants responsible for inhibitor binding, we solved the crystal structure of VcMTAN in complex with BuT-DADMe-ImmA to 2.3 Å resolution (Fig. 3). The final atomic model contained residues 1 – 230 for each monomer of VcMTAN in the asymmetric unit. The largest part of the N-terminal 6-His tag and the last C-terminal residue, 231, were omitted from the structure model due to lack of electron density.
Figure 3.
Crystal structure of VcMTAN in complex with BuT-DADMe-ImmA. (a) Overall structure of the VcMTAN structure showing the asymmetric unit content with the inhibitor BuT-DADMe-ImmA bound in the active sites. (b) The active site of the VcMTAN with a 2Fo − Fc map contoured at 1.2σ surrounding the BuT-DADMe-ImmA inhibitor and the proposed nucleophilic water molecule. (c) Space filling picture of the active site of VcMTAN with BuT-DADMe-ImmA in the active site. Grey represents hydrophobic regions of the protein which interact with hydrophobic parts of the inhibitor. The red color shows parts of the protein that contain charged residues interacting with polar groups of the inhibitor, while green represents loop regions. (d) Schematic drawing of the BuT-DADMe-ImmA inhibitor bound in the active site of VcMTAN showing catalytic contacts.
The VcMTAN structure complexed with BuT-DADMe-ImmA had two monomers in the asymmetric unit related by 2-fold noncrystallographic symmetry which corresponds to the functional dimer (Fig. 3a). Density for the inhibitor in the active site was clearly visible at a σ-level of 5, in maps generated after the first round of refinement (Fig. 3b). The structure of the VcMTAN monomer was a single mixed α/β domain with central twisted nine-stranded mixed β-sheet surrounded by six α-helices (Fig. 3a). Both the monomeric structure and the dimeric form were very similar to the MTAN from E. coli with rms deviations of 0.44 Å comparing the Cα of the two structures although the sequence identity is only 59% 22. The dimer interface involved hydrophobic residues coming from two α-helices and three loop regions from each monomer.
The catalytic site is situated in a pocket formed by residues from β10, a loop between β8 and α4 and a loop contributed by the adjacent subunit (Fig. 3c). The catalytic site can be divided into three parts, the base binding site, the ribose binding site and the 5′-alkylthio-binding site. The purine base contacts Phe152, main chain atoms of Val153, and side chain of Asp198 (Fig. 3d). Phe152 makes hydrophobic stacking interactions with the 9-deazaadenine base of the inhibitor. The carbonyl oxygen of Val153 accepts a hydrogen bond from the N6 amino group (2.96 Å) of adenine while the amide nitrogen of Val153 donates a hydrogen bond to N1 (3.10 Å). The side chain of Asp198 accepts hydrogen bonds from the N6 amino group (2.89 Å) and NH7 (2.69 Å) of the base. Ser197 hydrogen bonds to Oδ2 of Asp198 (2.55 Å) and places the side chain in an orientation favorable for catalysis. The amide nitrogen of Val199 may also orient the Asp198 for catalysis by hydrogen bonding to Oδ1 of the latter.
The pyrrolidine moiety participates in interactions with Met9, Phe208 and Met174 on both sides of this ribosyl mimic. The pyrrolidine moiety shares hydrogen bonds with Glu175 and the proposed catalytic water (WAT3) (Fig. 3d). The Oε1 of Glu175 hydrogen bonds to the 3′-hydroxyl of the pyrrolidine with a distance of 2.71 Å. The protonated N1′ nitrogen of the pyrrolidine donates a hydrogen bond to WAT3 (2.73 Å). WAT3 is further stabilized by several hydrogen bonds with Oε2 of Glu175 (2.94 Å), Oε1 and Oε2 of Glu12 (3.41 and 2.91 Å), and NH1 of Arg194 (2.68A). The side chain of Ser76 is also within hydrogen bond distance to Oε2 of Glu12 (2.78 Å) and is involved in holding Glu12 in place for catalysis.
The 5′-butylthio group is surrounded by hydrophobic residues including Met9, Ile50, Val102, Phe105, Ala113, Phe152, Met174, Tyr107 and Phe208 (Fig. 3c). Both subunits form the catalytic site and Tyr107, Phe105, Ala113 and Val102 reside on the adjacent subunit.
Inhibition of cellular MTAN activity
We cultured V. cholerae N16961 overnight in the presence of the transition state analogues and saw no effect on cell growth as demonstrated by the invariant OD600 at concentrations up to 1 μM, 14,000 times the Ki* value (Supplementary Fig. 1b online). We took the cleared lysates from washed cells and incubated with radiolabeled MTA, and found MTAN activity from cells cultured without inhibitor to be 89 ± 3 pmol/min/OD600 unit, which reflects the variability in the cell density attained by overnight cultures, as well as the amount of active MTAN in extracts. In the presence of the transition state analogues, we saw dose-dependent inhibition of adenine conversion, giving IC50s for the loss of cellular MTAN activity of 27, 31, and 6 nM for MT-, EtT-, and BuT-DADMe-ImmA, respectively (Table 1 and Supplementary Fig. 1c online).
Inhibition of autoinducer production
Under the same conditions used to assay the inhibition of cellular MTAN activity, we measured autoinducer production by V. cholerae N16961 as a function of inhibitors (Table 1). V. cholerae N16961 growth media induced luminescence in quorum sensing V. harveyi reporter strains BB170 and BB120, by a factor of 13.5 (± 4.5), and 2.3 (±1.0), respectively, compared to blank media. BB170 responds to the presence of AI-2 alone, whereas BB120 responds to both AI-1 and AI-2. Marginal induction in BB120 was previously observed for other strains of V. cholerae subjected to the same assay23. It was postulated that in the presence of system 1 (response system for AI-1) in V. harveyi BB120 strain, system 2 (response system for AI-2) is less sensitive to induction23. MTAN inhibitors caused the AI response signal to become progressively weaker as inhibitor concentration increased, and was completely inhibited at 1 μM (Supplementary Fig. 1d online). We obtained IC50 values for suppression of light induction in BB170 of 0.94, 11, and 1.4 nM for MT-, EtT-, and BuT-DADMe-ImmA, whereas in BB120 the IC50s were 10.5, 14, and 1 nM for the same inhibitors (Table 1). Inhibitors alone at concentrations present in AI detection assays, had no effect on light output from the reporter strains, supporting action of the transition state analogues on MTAN of V. cholerae cells for their effect on autoinducer production.
Autoinducer production in MTAN− E. coli
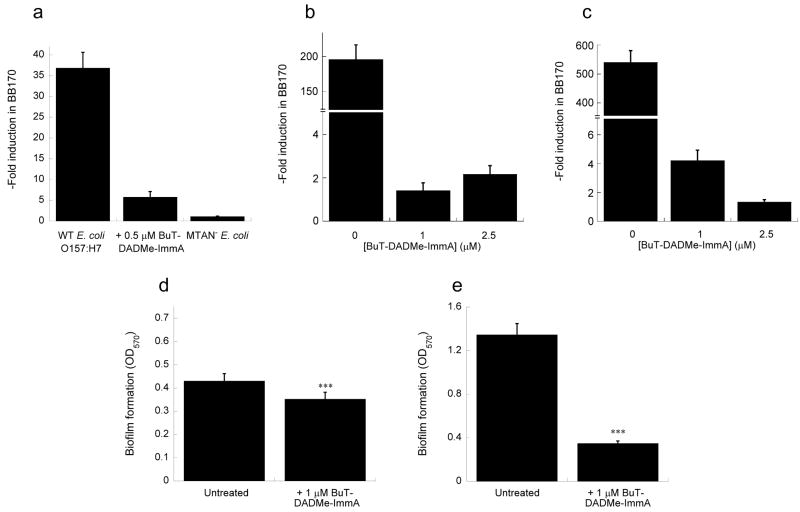
We cultured E. coli O157:H7 and an MTAN knockout strain in the presence of up to 0.5 μM MT- and BuT-DADMe-ImmA and found no growth defects in AB medium (Supplementary Fig. 2a online). AI induction in BB170 reached 37-fold for the wild-type pathogenic strain compared to blank, while administration of MT- and BuT-DADMe-ImmA resulted in a dose-dependent inhibition of AI-2 induction with an IC50 of 600 ± 50 nM, and 125 ± 24 nM, respectively. We found that at only four times the IC50 value for BuT-DADMe-ImmA, induction was reduced to 6-fold (Fig. 4a), while the extent of AI-2 induction for the MTAN knockout was negligible. Genetic interruption of MTAN in E. coli shows that it is not essential for growth although it is important for synthesis of quorum sensing molecules. Wild type E. coli treated with BuT-DADMe-ImmA produced the same phenotype as the MTAN− strain, supporting MTAN as the target for action of the transition state analogue in the cell.
Figure 4.
Effect of BuT-DADMe-ImmA on autoinducer-2 production in pathogenic E. coli and V. cholerae upon short-term and long-term inhibitor treatment, and on static biofilm formation. (a) E. coli O157:H7 ± 0.5 μM BuT-DADMe-ImmA, and an MTAN−strain were grown static in AB medium for >5 generations before assaying the spent medium for autoinducer-2 production. (b) E. coli O157:H7, and (c) V. cholerae N16961 grown shaken in LB for 26 generations, and autoinducer-2 in the spent media was measured. Cultures were prepared in triplicate, and data represent mean values ± s.d. from at least 6 replicates. Biofilm formation studies on (d) E. coli O157:H7, and (e) V. cholerae N16961, ± 1 μM BuT-DADMe-ImmA grown static in LB medium at 25 °C for 24 hours on 96-well format. The observed difference in biofilm formation due to BuT-DADMe-ImmA was statistically significant at t = 5.044, ***p < 0.001, d.f. = 14 for E. coli; and t = 26.689, p < 0.001, d.f. = 14 for V. cholerae.
Persistent suppression of quorum sensing in E. coli and V. cholerae
Pathogenic strains of E. coli and V. cholerae maintained sensitivity to BuT-DADMe-ImmA suppression of autoinducer-2 production for sustained growth cycles. With LB as growth medium, E. coli O157:H7 achieved 195-fold light induction in the V. harveyi BB170 reporter (Fig. 4b). After 26 generations of growth (i.e., 226 expansion of cell number) in the presence of 1 μM or 2.5 μM inhibitor, we saw that light induction in BB170 was suppressed to near basal levels of 1.4- and 2.2- fold, respectively. In V. cholerae N16961, the quorum sensing signal after 26 generations was 540-fold relative to control, and showed dramatic inhibition to 4.2- and 1.3 fold, in the presence of 1 μM or 2.5 μM BuT-DADMe-ImmA (Fig. 4c). Again, we observed that growth of both strains remained uninhibited under these conditions (Supplementary Fig. 2b, c online).
Inhibition of biofilm formation
We used a 96-well plate format for the detection of biofilm formation in both E. coli O157:H7 and V. cholerae N16961. E. coli O157:H7 formed biofilm at the bottom of the plate, whereas V. cholerae N16961 produced biofilm both at the bottom and at the air-liquid interface. Growth of the planktonic cells reached a modest OD600 of 0.5 for E. coli and 0.4 for V. cholerae under static growth conditions and at 25 ° (Supplementary Fig. 2d, e online), while V. cholerae produced three-fold more biofilm than did E. coli.
BuT-DADMe-ImmA (1 μM) did not inhibit growth of cells in the assay, but reduced biofilm production by 18% in E. coli, and 71% in V. cholerae (Fig. 4d, e).
DISCUSSION
The slow-onset, tight-binding inhibition of VcMTAN by MT-, EtT-, and BuT-DADMe-ImmA exhibits some of the highest binding affinities for targets in quorum sensing pathways. Slow onset inhibition is typical for transition state analogues where binding to enzyme equilibrates the protein to a new conformation within seconds to minutes. The enzyme-inhibitor complex is characterized by a slow dissociation rate because of a highly stable inhibited form. Km/Ki values for all three inhibitors are approximately 104, showing strong preference for the transition state analogues over the substrate MTA.
MTANs have dual substrate specificity for MTA and SAH, and are expected to accommodate both methylthio- and homocysteine groups in a manner proportional to their Km values. Transition state analogues that differ only in their 5′-substituents permit direct comparison of VcMTAN’s preference for these groups. MT- and EtT- groups were equally favored at this position, and are also equivalent in blocking quorum sensing in vitro (Table 1). The dissociation constant increases three-fold however, in going from ethyl- to butyl-substituted DADMe-ImmA and suggests modest size specificity within the 5′-binding pocket.
The crystal structure of BuT-DADMe-ImmA in complex with VcMTAN is similar to the crystal structure of EcMTAN in complex with MT-DADMe-ImmA (Supplementary Fig. 3a online)22. The inhibitors in the two structures share a virtual overlap of the 9-deazaadenine and the pyrrolidine ribocation mimic. Similar to EcMTAN, tight binding in the VcMTAN complex is proposed to originate mainly from the conformation adopted by the pyrrolidine group of the inhibitor that allows for the cation at N1′ to be in close proximity to the putative water nucleophile which organizes the geometry of Ser76, Glu12, Arg194, and Glu175 around the catalytic site. The pKa of the N1′ pyrrolidine nitrogen is 8, making it cationic at physiological pH. The DADMe-ImmA inhibitors lack the 2′-hydroxyl moiety of ribosyl groups and allow the presumed catalytic water to be close to the N1′ with a distance of 2.7 Å. This distance was similar to the 2.6 Å in the case of the EcMTAN-MT-DADMe-ImmA structure22. Based on the favorable hydrophobic interactions between the 5′-butylthio group and the hydrophobic pocket in the protein, additional binding affinity would be anticipated relative to MT-DADMe-ImmA. The 3-fold decrease in affinity for BuT- inhibitor relative to MT- inhibitor may correspond to the entropy loss upon binding the flexible butyl group at the catalytic site.
BuT-DADMe-ImmA binds 1000 times stronger to the EcMTAN than to the VcMTAN. Comparisons of the structures overall and the active sites do not reveal obvious explanations for the difference (Supplementary Fig. 3a, b online). The two structures share 59% sequence identity and have almost identical active sites. However, recent studies have demonstrated that residues remote from the active site of purine nucleoside phosphorylase contribute to transition state structure and catalytic efficiency through dynamic motion24. The enhanced catalytic efficiency and inhibitor binding specificity of EcMTAN may also involve the full dynamic architecture of the protein.
Biological effectiveness of MTAN inhibitors in the context of the cell was measured in cell lysates of a virulent strain of Vibrio cholerae (N16961) grown in the presence of inhibitors. Direct measurements of MTAN activity that yielded nanomolar IC50 values for MT-, EtT-, and BuT-DADMe-ImmA demonstrate cell permeability for the inhibitors, most notably in the case of BuT-DADMe-ImmA. Despite having a 3-fold lower affinity with purified VcMTAN, BuT-DADMe-ImmA inhibited cellular VcMTAN activity 5-fold better than its MT-, and EtT- counterparts (Table 1). Although tightly bound in vitro, BuT-DADMe-ImmA inhibition of VcMTAN activity in the cell required a 30-fold increase above the Ki*, suggesting a significant diffusion barrier. With MT-, and EtT-DADMe-ImmA, the diffusion barrier required a gradient close to 500-fold above Ki* to inhibit VcMTAN in growing cells.
Despite the significant diffusion barrier, all three MTAN transition state analogues were potent inhibitors of autoinducer production in V. cholerae N16961, inhibiting quorum sensing induction in both V. harveryi reporter strains.
The role of quorum sensing in enterohemorrhagic E. coli O157:H7 EDL933 has also been extensively studied25–27. It is a highly pathogenic strain of E. coli that causes mortality and morbidity in vulnerable populations by producing Shiga toxins and lesions on intestinal epithelial cells. It uses autoinducers generated at high cell density to modulate concerted biological functions and succeed in host infection.
MT- and BuT-DADMe-ImmA are potent inhibitors of E. coli MTAN with dissociation constants of 2 and 0.3 pM, respectively18. Similar to results from V. cholerae, both MT- and BuT-DADMe-ImmA were able to cross the E. coli cellular membrane and cause nontoxic inhibition of AI-2 production. Interestingly, AI-2 inhibition in E. coli cells was not as efficient as in V. cholerae despite the fact that inhibition of EcMTAN enzyme activity was at least 100-fold stronger than in VcMTAN. A significant barrier to inhibitor permeability in E. coli may explain this discrepancy.
A concern in targeting MTAN to suppress quorum sensing is that overexpression of the quorum sensing pathway (or alternative pathways) might overcome the effect of MTAN inhibitors. Bacterial changes in gene expression in response to cellular signals generally occur rapidly, on the time scale of minutes to a few cell generation times. Serial transfer experiments showed that sensitivity towards BuT-DADMe-ImmA suppression of autoinducer-2 production was maintained in both V. cholerae N16961 and E. coli O157:H7 through progressive passaging of cells. This suggests that inhibition of quorum sensing was not only immediate, it also persisted for several generations.
Biofilm formation is an important bacterial strategy tightly linked to quorum sensing27,28. It is a vital survival mechanism for Vibrio cholerae, in both its infective and noninfective lifestyles29,30. The current model for Vibrio cholerae asserts that at high cell density in the abundance of autoinducers, biofilm formation is impaired by repression of the critical exopolysaccharide regulators HapR and c-di-GMP31,32. While most Vibrio cholerae El Tor strains possess a uniquely inverted quorum sensing mechanism to increase survival and infectivity32,33, the El Tor N16961 strain carries a natural frame-shift mutation in the hapR gene33,34 which abolishes repression on biofilm formation and cholera toxin production33. This makes it conceivable for biofilm formation to be suppressed by BuT-DADMe-ImmA under conditions that also inhibit autoinducer production in this strain.
In E. coli, it has been shown that autoinducer-2 added to cell cultures in microtiter plates increased biofilm formation 30-fold, and that a quorum sensing mutant produced 50% less biofilm than the isogenic wild-type strain35. As in the case of V. cholerae N16961, biofilm formation in E. coli O157:H7 was suppressed by an MTAN inhibitor that disrupts quorum sensing.
Transition state theory has been useful in the development of powerful inhibitors with in vivo effects against target enzymes. MT-, EtT-, and BuT-DADMe-ImmA are transition state analogues of bacterial MTANs and they show high potency in disrupting quorum sensing molecules in pathogenic strains of Vibrio cholerae and Escherichia coli. Streptococcus pneumoniae, Neisseria meningitidis, Klebsiella pneumoniae, Staphylococcus aureus, Helicobacter pylori, are some of the most aggressive human pathogens, and published evidence supports quorum sensing as promoting pathogenesis in these species7,36–40. Each of these bacterial species expresses MTAN and the transition state analogues described here are potent in inhibiting purified MTANs from these sources18,20,21. The potential of inhibiting quorum sensing by targeting MTAN is expected to extend to other pathogens beyond V. cholerae and E. coli.
METHODS
[8-14C]MTA, DADMe-ImmucillinAs
[8-14C]MTA and DADMe-ImmucillinAs were synthesized as described previously9,17
VcMTAN expression and purification
VcMTAN was expressed in E. coli as a His-tagged recombinant protein as described in Supplementary Methods online.
VcMTAN kinetics and inhibition
Kinetic constants were determined by following loss of MTA at 274 nm (Δε274 = 1.6 mM−1cm−1). Reactions were carried out at 25 °C in 100 mM HEPES, pH 7.5, and 50 mM KCl at various MTA concentrations, and initiated by 10 – 12 nM VcMTAN. Inhibition constants (Ki and Ki*) were obtained using a xanthine oxidase-coupled reaction described previously18. Reaction mixtures contained 1 – 2 mM MTA, and various concentrations of MT-, EtT-, and BuT- DADMe-ImmA. Samples were prepared at standard buffer conditions, with ~0.5 units of xanthine oxidase (Sigma), and 12 nM VcMTAN to initiate the reaction at 25 °C, monitored on a UV-Vis spectrophotometer at 293 nm. Inhibition constants were obtained using equation (1) for competitive inhibition using KaleidaGraph 3.6 (Synergy Software):
| (1) |
where νs′ and νs are steady state with and without inhibitor, respectively; Km is the Michaelis constant for MTA; [S] and [I] are the concentrations of MTA and inhibitor, respectively. If the concentration of inhibitor is less than ten times the concentration of enzyme, equation (2) was used for correction:
| (2) |
where I′ is the effective inhibitor concentration; I is the inhibitor concentration used in the assay; ν0′ and ν0 are initial rates with and without inhibitor, respectively; and Et is total MTAN concentration.
Crystallization of the BuT-DADMe-ImmucillinA-MTAN complex
Purified VcMTAN was concentrated to 15 mg mL−1, incubated with 1 mM BuT-DADMe-ImmA, and crystallized using sitting drop vapor diffusion at 18 °C against an 80 μL reservoir containing 0.2 M potassium iodide 20% (w/v) PEG3350, where 1 μL of the protein solution was mixed with 1 μL of the reservoir solution.
Data collection
Crystals were soaked in mother liquor supplemented with 20% glycerol and cooled to −178 °C prior to data collection, which was done subsequently at beamline X29A at the National Synchrotron Lightsource, Brookhaven National Laboratory using an ADSC Quantum 315 detector and 1.10010 Å wavelength. Each frame was exposed for 10 s with an oscillation range of 1°. The HKL2000 suite was used for integration and scaling of the data (Supplementary Table 2 online)41.
Structure determination and refinement
The structure of VcMTAN- BuT-DADMe-ImmA complex was solved by molecular replacement using E. coli MTAN (PDB code 1Z5P. pdb without water) as search model. Molecular replacement with MOLREP, and refinement with REFMAC5 were done using the CCP4i package42–44. COOT was used for molecular modeling45. Clear density was observed in the Fo – Fc maps for the ligands at 3.5σ and were built into the electron density. Majority of the residues (89%) are located in the most favored region of the Ramachandran plot, while the remaining 11% are in the allowed region. Data processing and refinement statistics are summarized in Supplementary Table 2 online. All figures were made using PyMOL46.
Inhibition of cellular MTAN activity
V. cholerae N16961 (ATCC) was grown at 37 °C to stationary phase in LB medium for 16 hours with and without 1 – 1000 nM MT-, EtT-, and BuT-DADMe-ImmA. Cells were washed twice with PBS and lysed with BugBuster Reagent (Novagen). Cleared lysate was incubated with [8-14C]MTA in 50 mM phosphate buffer, pH 7.9, 10 mM KCl at 25 °C for 20 minutes and quenched with perchloric acid to 20% final concentration. Reaction components were separated using reverse-phase HPLC as detailed in Supplementary Methods online, and MTAN activity was evaluated based on 14C-adenine counts.
Autoinducer assay
Autoinducers secreted by V. cholerae N16961 treated with inhibitors were measured using Vibrio harveyi bioluminescence assay47 (described in Supplementary Methods online), where reporter V. harveyi strains produce light in response to autoinducers in V. cholerae spent media. The magnitude of induction is taken as the ratio of light output induced by the V. cholerae filtrate relative to blank, and was plotted against inhibitor concentration, and fitted to equation (3) to obtain the IC50:
| (3) |
where y is the magnitude of induction at inhibitor concentration [I]; y0 is magnitude of induction without inhibitor (untreated sample); c is the maximum difference between treated and untreated sample, and IC50 is the inhibitor concentration representing half maximal induction. Average of at least six replicates was taken, with outliers greater than two standard deviations removed from analysis.
Autoinducer-2 production was measured similarly for enterohemorrhagic E. coli O157:H7 EDL933 (ATCC) grown in autobioinducer medium with 5 – 500 nM MT- and BuT-DADMe-ImmA, as well as for an E. coli MTAN knockout strain without inhibitor treatment.
To evaluate the effects of prolonged incubation with BuT-DADMe-ImmA on the growth and autoinducer production of V. cholerae N16961 and E. coli O157:H7, initial cell culture of these strains was prepared from a 1:100 dilution of overnight seed grown in LB medium at 37 °C, with and without 1 and 2.5 μM BuT-DADMe-ImmA. Cells were grown to stationary phase and aliquots were taken for OD600 and autoinducer-2 assays. Treated and untreated cells were serially diluted from dense cultures into fresh media, and grown under the same conditions of growth and inhibitor concentration for 26 generations. Cultures were prepared in triplicate, and for the bioluminescence assay the average of 6 – 8 replicates was taken.
Biofilm assay
V. cholerae N16961 and E. coli O157:H7 were diluted 1:100 from overnight seed cultures grown in LB medium in sterile, nontreated 96-well plates48. The plates were covered and grown static for 24 hours at 25 °C with and without 1 μM BuT-DADMe-ImmA. After removing the cell suspension and measuring OD600, the plates were rinsed and stained with crystal violet solution for ten minutes. Once the dye was removed and the plate rinsed, the stained biofilm was solubilized using 1:4 acetone: ethanol for E. coli, and DMSO for V. cholerae. The extent of biofilm formation was obtained from the OD570 values, and the average from at least six replicates was reported.
Accession code
The coordinates and structure factors of VcMTAN-BuT-DADMe-ImmA complex are deposited in the protein data bank with accession code 3DP9.
Supplementary Material
Acknowledgments
The authors acknowledge R. H. Furneaux, G. B. Evans, D. H. Lenz, G. F. Painter, and P. C. Tyler of Industrial Research Laboratory, Inc. (Lower Hutt, New Zealand) for supplying the DADMe-Immucillins. M. G. Surette (University of Calgary) for providing Vibrio harveyi strains BB120 and BB170, C. Bradbeer (University of Virginia) for the Escherichia coli MTAN knockout, and NIH Grant GM41916 for funding.
References
- 1.Fuqua WC, Winans SC, Greenberg EP. Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J Bacteriol. 1994;176:269–275. doi: 10.1128/jb.176.2.269-275.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sperandio V. Novel approaches to bacterial infection therapy by interfering with bacteria-to-bacteria signaling. Expert Rev Anti Infect Ther. 2007;5:271–276. doi: 10.1586/14787210.5.2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vendeville A, Winzer K, Heurlier K, Tang CM, Hardie KR. Making ‘sense’ of metabolism: Autoinducer-2, LuxS and pathogenic bacteria. Nat Rev Microbiol. 2005;3:383–396. doi: 10.1038/nrmicro1146. [DOI] [PubMed] [Google Scholar]
- 4.Cegelski L, Marshall GR, Eldridge GR, Hultgren SJ. The biology and future prospects of antivirulence therapies. Nat Rev Microbiol. 2008;6:17–27. doi: 10.1038/nrmicro1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Winzer K, Williams P. Quorum sensing and the regulation of virulence gene expression in pathogenic bacteria. Int J Med Microbiol. 2001;291:131–143. doi: 10.1078/1438-4221-00110. [DOI] [PubMed] [Google Scholar]
- 6.Stroeher UH, Paton AW, Ogunniyi AD, Paton JC. Mutation of luxS of Streptococcus pneumoniae affects virulence in a mouse model. Infect Immun. 2003;71:3206–3212. doi: 10.1128/IAI.71.6.3206-3212.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Winzer K, et al. Role of Neisseria meningitidis luxS in cell-to-cell signaling and bacteremic infection. Infect Immun. 2002;70:2245–2248. doi: 10.1128/IAI.70.4.2245-2248.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harasawa H, et al. Chemotherapy targeting methylthioadenosine phosphorylase (MTAP) deficiency in adult T cell leukemia (ATL) Leukemia. 2002;16:1799–1807. doi: 10.1038/sj.leu.2402570. [DOI] [PubMed] [Google Scholar]
- 9.Basu I, et al. A transition state analogue of 5′-methylthioadenosine phosphorylase induces apoptosis in head and neck cancers. J Biol Chem. 2007;282:21477–21486. doi: 10.1074/jbc.M702287200. [DOI] [PubMed] [Google Scholar]
- 10.Singh V, Lee JE, Nunez S, Howell PL, Schramm VL. Transition state structure of 5′-methylthioadenosine/S-adenosylhomocysteine nucleosidase from Escherichia coli and its similarity to transition state analogues. Biochemistry. 2005;44:11647–11659. doi: 10.1021/bi050863a. [DOI] [PubMed] [Google Scholar]
- 11.Singh V, Schramm VL. Transition-state analysis of S-pneumoniae 5′-methylthioadenosine nucleosidase. J Am Chem Soc. 2007;129:2783–2795. doi: 10.1021/ja065082r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Singh V, Luo M, Brown RL, Norris GE, Schramm VL. Transition-state structure of Neisseria meningitides 5′-methylthioadenosine/S-adenosylhomocysteine nucleosidase. J Am Chem Soc. 2007;129:13831–13833. doi: 10.1021/ja0754204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singh V, Schramm VL. Transition-state structure of human 5′-methylthioadenosine phosphorylase. J Am Chem Soc. 2006;128:14691–14696. doi: 10.1021/ja065419p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Balakrishnan K, Nimmanapalli R, Ravandi F, Keating MJ, Gandhi V. Forodesine, an inhibitor of purine nucleoside phosphorylase, induces apoptosis in chronic lymphocytic leukemia cells. Blood. 2006;108:2392–2398. doi: 10.1182/blood-2006-03-007468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Robak T, Lech-Maranda E, Koerycka A, Robak E. Purine nucleoside analogs as immunosuppressive and antineoplastic agents: mechanism of action and clinical activity. Curr Med Chem. 2006;13:3165–3189. doi: 10.2174/092986706778742918. [DOI] [PubMed] [Google Scholar]
- 16.Evans GB, Furneaux RH, Schramm VL, Singh V, Tyler PC. Targeting the polyamine pathway with transition-state analogue inhibitors of 5′-methylthioadenosine phosphorylase. J Med Chem. 2004;47:3275–3281. doi: 10.1021/jm0306475. [DOI] [PubMed] [Google Scholar]
- 17.Evans GB, et al. Second generation transition state analogue inhibitors of human 5′-methylthioadenosine phosphorylase. J Med Chem. 2005;48:4679–4689. doi: 10.1021/jm050269z. [DOI] [PubMed] [Google Scholar]
- 18.Singh V, et al. Femtomolar transition state analogue inhibitors of 5′-methylthioadenosine/S-adenosylhomocysteine nucleosidase from Escherichia coli. J Biol Chem. 2005;280:18265–18273. doi: 10.1074/jbc.M414472200. [DOI] [PubMed] [Google Scholar]
- 19.Singh V, et al. Picomolar transition state analogue inhibitors of human 5′-methylthioadenosine phosphorylase and X-ray structure with MT-Immucillin-A. Biochemistry. 2004;43:9–18. doi: 10.1021/bi0358420. [DOI] [PubMed] [Google Scholar]
- 20.Singh V, et al. Structure and inhibition of a quorum sensing target from Streptococcus pneumoniae. Biochemistry. 2006;45:12929–12941. doi: 10.1021/bi061184i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gutierrez JA, et al. Picomolar inhibitors as transition-state probes of 5′-methylthioadenosine nucleosidases. ACS Chem Biol. 2007;2:725–734. doi: 10.1021/cb700166z. [DOI] [PubMed] [Google Scholar]
- 22.Lee JE, et al. Structural rationale for the affinity of pico- and femtomolar transition state analogues of Escherichia coli 5′-methylthioadenosine/S-adenosylhomocysteine nucleosidase. J Biol Chem. 2005;280:18274–18282. doi: 10.1074/jbc.M414471200. [DOI] [PubMed] [Google Scholar]
- 23.Bassler BL, Greenberg EP, Stevens AM. Cross-species induction of luminescence in the quorum-sensing bacterium Vibrio harveyi. J Bacteriol. 1997;179:4043–4045. doi: 10.1128/jb.179.12.4043-4045.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saen-Oon S, Ghanem M, Schramm VL, Schwartz SD. Remote mutations and active site dynamics correlate with catalytic properties of purine nucleoside phosphorylase. Biophys J. 2008;94:4078–4088. doi: 10.1529/biophysj.107.121913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Anand SK, Griffiths MW. Quorum sensing and expression of virulence in Escherichia coli O157:H7. Int J Food Microbiol. 2003;85:1–9. doi: 10.1016/s0168-1605(02)00482-8. [DOI] [PubMed] [Google Scholar]
- 26.Sperandio V, Mellies JL, Nguyen W, Shin S, Kaper JB. Quorum sensing controls expression of the type III secretion gene transcription and protein secretion in enterohemorrhagic and enteropathogenic Escherichia coli. Proc Nat Acad Sci USA. 1999;96:15196–15201. doi: 10.1073/pnas.96.26.15196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li J, et al. Quorum sensing in Escherichia coli is signaled by AI-2/LsrR: effects on small RNA and biofilm architecture. J Bacteriol. 2007;189:6011–6020. doi: 10.1128/JB.00014-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Herzberg M, Kaye IK, Peti W, Wood TK. YdgG (TqsA) controls biofilm formation in Escherichia coli K-12 through autoinducer 2 transport. J Bacteriol. 2006;188:587–598. doi: 10.1128/JB.188.2.587-598.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu J, Mekalanos JJ. Quorum sensing-dependent biofilms enhance colonization in Vibrio cholerae. Dev Cell. 2003;5:647–656. doi: 10.1016/s1534-5807(03)00295-8. [DOI] [PubMed] [Google Scholar]
- 30.Matz C, et al. Biofilm formation and phenotypic variation enhance predation-driven persistence of Vibrio cholerae. Proc Nat Acad Sci USA. 2005;102:16819–16824. doi: 10.1073/pnas.0505350102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hammer BK, Bassler BL. Quorum sensing controls biofilm formation in Vibrio cholerae. Mol Microbiol. 2003;50:101–114. doi: 10.1046/j.1365-2958.2003.03688.x. [DOI] [PubMed] [Google Scholar]
- 32.Waters CM, Lu W, Rabinowitz JD, Bassler BL. Quorum sensing controls biofilm formation in Vibrio cholerae through modulation of cyclic di-GMP levels and repression of vpsT. J Bacteriol. 2008;190:2527–2536. doi: 10.1128/JB.01756-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhu J, et al. Quorum-sensing regulators control virulence gene expression in Vibrio cholerae. Proc Natl Acad Sci USA. 2002;99:3129–3134. doi: 10.1073/pnas.052694299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Joelsson A, Liu Z, Zhu J. Genetic and phenotypic diversity of quorum-sensing systems in clinical and environmental isolates of Vibrio cholerae. Infect Immun. 2006;74:1141–1147. doi: 10.1128/IAI.74.2.1141-1147.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gonzalez Barrios AF, et al. Autoinducer 2 controls biofilm formation in Escherichia coli through a novel motility quorum-sensing regulator (MqsR, B3022) J Bacteriol. 2006;188:305–316. doi: 10.1128/JB.188.1.305-316.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Surette MG, Bassler BL. Quorum sensing in Escherichia coli and Salmonella typhimurium. Proc Nat Acad Sci USA. 1998;95:7046–7050. doi: 10.1073/pnas.95.12.7046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dunny GM, Leonard BAB. Cell-cell communication in gram-positive bacteria. Annu Rev Microbiol. 1997;51:527–564. doi: 10.1146/annurev.micro.51.1.527. [DOI] [PubMed] [Google Scholar]
- 38.Balestrino D, Haagensen JAJ, Rich C, Forestier C. Characterization of type 2 quorum sensing in Klebsiella pneumoniae and relationship with biofilm formation. J Bacteriol. 2005;187:2870–2880. doi: 10.1128/JB.187.8.2870-2880.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Joyce EA, et al. LuxS is required for persistent Pneumococcal carriage and expression of virulence and biosynthesis genes. Infect Immun. 2004;72:2964–2975. doi: 10.1128/IAI.72.5.2964-2975.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rader BA, Campagna SR, Semmelhack MF, Bassler BL, Guillemin K. The quorum-sensing molecule autoinducer 2 regulates motility and flagellar morphogenesis in Helicobacter pylori. J Bacteriol. 2007;189:6109–6117. doi: 10.1128/JB.00246-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. In: Carter CWJ, Sweet RM, editors. Methods Enzymol. Vol. 276. Academic Press; New York: 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 42.Potterton E, Briggs P, Turkenburg M, Dodson E. A graphical user interface to the CCP4 program suite. Acta Crystallogr D Biol Crystallogr. 2003;59:1131–1137. doi: 10.1107/s0907444903008126. [DOI] [PubMed] [Google Scholar]
- 43.Vagin A, Teplyakov A. MOLREP: an automated program for molecular replacement. J Appl Cryst. 1997;30:1022–1025. [Google Scholar]
- 44.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 45.Emsley P, Cowtan K. Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 46.DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific; Palo Alto, CA, USA: 2002. [Google Scholar]
- 47.Greenberg EP, Hastings JW, Ulitzur S. Induction of luciferase synthesis in Beneckea harveyi by other marine bacteria. Arch Microbiol. 1979;120:87–91. [Google Scholar]
- 48.O’Toole GA, et al. Genetic approaches to study of biofilms. In: Doyle RJ, editor. Methods Enzymol. Vol. 310. Academic Press; New York: 1999. pp. 91–109. [DOI] [PubMed] [Google Scholar]
- 49.Parsek MR, Val DL, Hanzelka BL, Cronan JE, Greenberg EP. Acyl homoserine-lactone quorum-sensing signal generation. Proc Nat Acad Sci USA. 1999;96:4360–4365. doi: 10.1073/pnas.96.8.4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.