Abstract
Chromosome instability (CIN) is a hallmark of many tumors and correlates with the presence of extra centrosomes1-4. However, a direct mechanistic link between extra centrosomes and CIN has not been established. It has been proposed that extra centrosomes generate CIN by promoting multipolar anaphase, a highly abnormal division that produces 3 or more aneuploid daughter cells. Here, we use long-term live-cell imaging to demonstrate that cells with multiple centrosomes rarely undergo multipolar cell divisions, and the progeny of these divisions are typically inviable. Thus, multipolar divisions cannot explain observed rates of CIN. By contrast, we observe that CIN cells with extra centrosomes routinely undergo bipolar cell divisions, but display a significantly elevated frequency of lagging chromosomes during anaphase. To define the mechanism underlying this mitotic defect, we generated cells that differ only in their centrosome number. We demonstrate that extra centrosomes alone are sufficient to promote chromosome missegregation during bipolar cell division. These segregation errors are a consequence of cells passing through a transient ‘multipolar spindle intermediate’ in which merotelic kinetochore-microtubule attachment errors accumulate prior to centrosome clustering and anaphase. These findings provide a direct mechanistic link between extra centrosomes and CIN, two common characteristics of solid tumors. We propose that this mechanism may be a common underlying cause of CIN in human cancer.
Keywords: syntelic, tetraploid, aneuploid, mitosis
A conspicuous feature of many tumor cells is an elevated rate of gain or loss of whole chromosomes, a phenomenon referred to as chromosome instability1. Cells with CIN missegregate chromosomes 10-100 times more frequently than non-transformed or chromosomally stable diploid cancers cells. CIN is thus a major source of aneuploidy 1, 5 and has important implications not only for tumor initiation, where aneuploidy can play a causal role 6, but also for tumor cell evolution, where elevated rates of chromosome missegregation may enable clonal expansion of cells with proliferative advantages, metastatic potential, or chemoresistance 5, 7, 8. Despite its importance, the mechanisms leading to CIN in most cancers are not defined.
One proposed mechanism underlying CIN is through extra centrosome-mediated multipolar spindle assembly followed by asymmetric chromosome segregation resulting in massive aneuploidy 3. Correlative support for this idea comes from the fact that extra centrosomes and multipolar mitotic figures are common in CIN cancers, yet rare in chromosomally stable tumors 9-12. However, an obvious paradox arises when considering such multipolar cell division as an underlying mechanism of CIN: aneuploidy compromises cell fitness 6, 13, 14 and therefore massive aneuploidy following multipolar cell division would likely compromise viability 3, 15-17. To date, neither the frequency of multipolar divisions nor the fate of the resulting progeny has been systematically characterized in cancer cells.
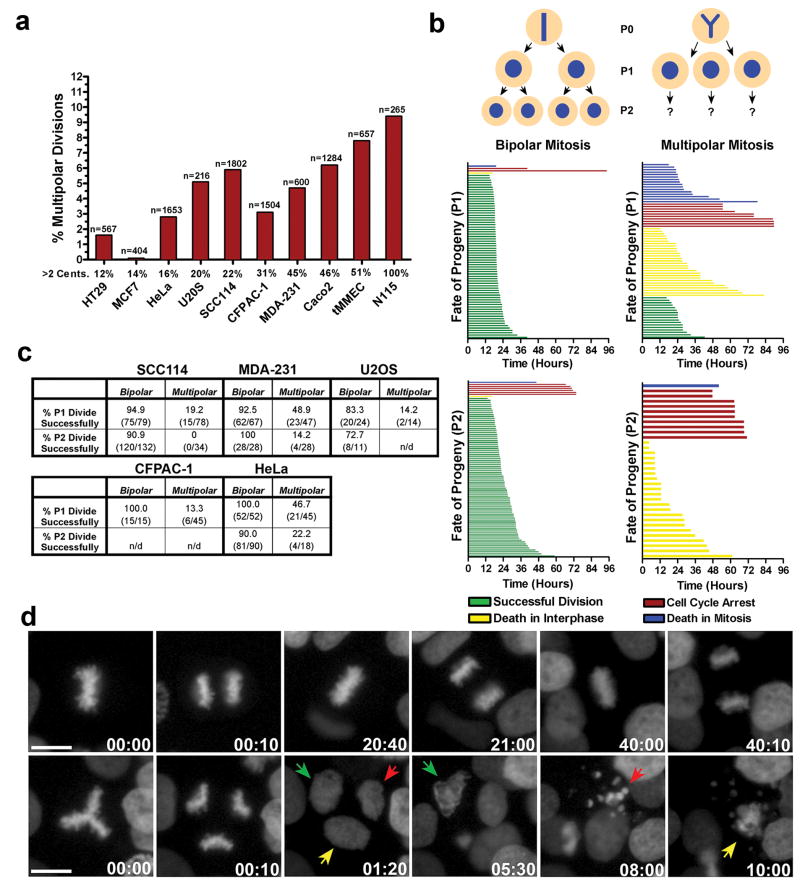
To directly visualize the relationship between multipolar cell division and cell viability, we generated a variety of cancer cell lines from different tissues of origin that stably express the chromosome marker H2B-GFP and performed long-term live cell imaging. The percentage of cells that harbored extra centrosomes varied significantly, from ∼12% in HT-29 human colon cancer cells to 100% in mouse neuroblastoma N1E-115 cells (Fig.1a). Nevertheless, our analysis clearly indicated that the fraction of cells undergoing multipolar cell division, defined by the segregation of chromosomes to 3 or more poles during anaphase, was always markedly less than the fraction of cells possessing extra centrosomes (Fig.1a). Thus, as expected from previous work, clustering of supernumerary centrosomes into two poles is an efficient mechanism that limits multipolar cell division 3, 15-19.
Figure 1. Multipolar cell divisions are rare and the progeny are typically inviable.
a) The percentage of different cancer cells that undergo multipolar cell division based on live-cell imaging (n= number of cell divisions). b) A representative cell fate analysis from SCC114 cells. Individual cell fates of progeny from both bipolar (left column) and multipolar (right column) cell divisions (represented by single colored lines) are shown. c) Percentage of progeny from bipolar and multipolar cell divisions that undergo successful cell division. d) Still frames from the imaging experiment represented in b showing a representative SCC114 cell undergoing several rounds of bipolar cell division (top row), or a single multipolar cell division (bottom row). Colored arrows track the fate of the three progeny from the multipolar cell division (Supp. Movie 1). Time, hours:minutes. Scale bar, 10 μm.
We determined the fate of cells that underwent a multipolar anaphase. We observed that although multipolar anaphase often produced 3 or more mononucleated daughter cells, cytokinesis failure along one or both division planes to produce binucleate or polynucleate progeny was also common (Supp. Fig. 1a). Both mono- and polynucleated progeny (P1) of spontaneously arising multipolar divisions were then tracked over a 4-day period and classified as: 1) Undergoing mitotic cell death with a multipolar spindle configuration; 2) Dying during the subsequent interphase; 3) Undergoing cell-cycle arrest; or 4) Successfully completing a second round of cell division to generate P2 progeny (Fig. 1b, c). Strikingly, we found that the vast majority of progeny (P1) of multipolar cells died or arrested, regardless of tissue of origin or whether the cells were mono- or polynucleated (Fig. 1b, c, d, Supp. Fig. 1a, b, Supp. Movie 1). Moreover, when rare P1 progeny from multipolar divisions completed a second round of mitosis to generate P2 progeny, even fewer of the resulting daughter cells were capable of further division (Fig. 1b, c). Finally, we observed that progeny from multipolar divisions were usually inviable even if they were born from binucleated, presumably tetraploid, mother cells. This suggests that doubling the chromosome content does not efficiently buffer the deleterious effects of massive aneuploidy that results from multipolar anaphase (Supp. Fig. 2).
Taken together, these data provide two reasons why multipolar cell division alone cannot explain the high rates of chromosome missegregation in CIN cells. First, the frequencies of multipolar division are not high enough to account for the observed rates of chromosome missegregation in these cell lines: For example, whereas MCF-7 and HT-29 cells missegregate chromosomes on average once every 2 or 5 divisions, respectively 1, 13, they each undergo multipolar anaphase only once every 50 or more divisions. Second, the failure of progeny to continue proliferating reveals that multipolar mitosis cannot give rise to persistently unstable cells.
We therefore considered other mechanisms by which multiple centrosomes might generate CIN. Recent work has revealed that merotely, a type of error in which single kinetochores attach to microtubules emanating from different poles 20, 21, is common in CIN cells 13. Merotelic attachments are particularly dangerous because they are poorly sensed by the spindle assembly checkpoint and, if not corrected, may give rise to lagging chromosomes during anaphase that can lead to missegregation events 20-23. However, the cause of merotely in CIN cancers is unknown.
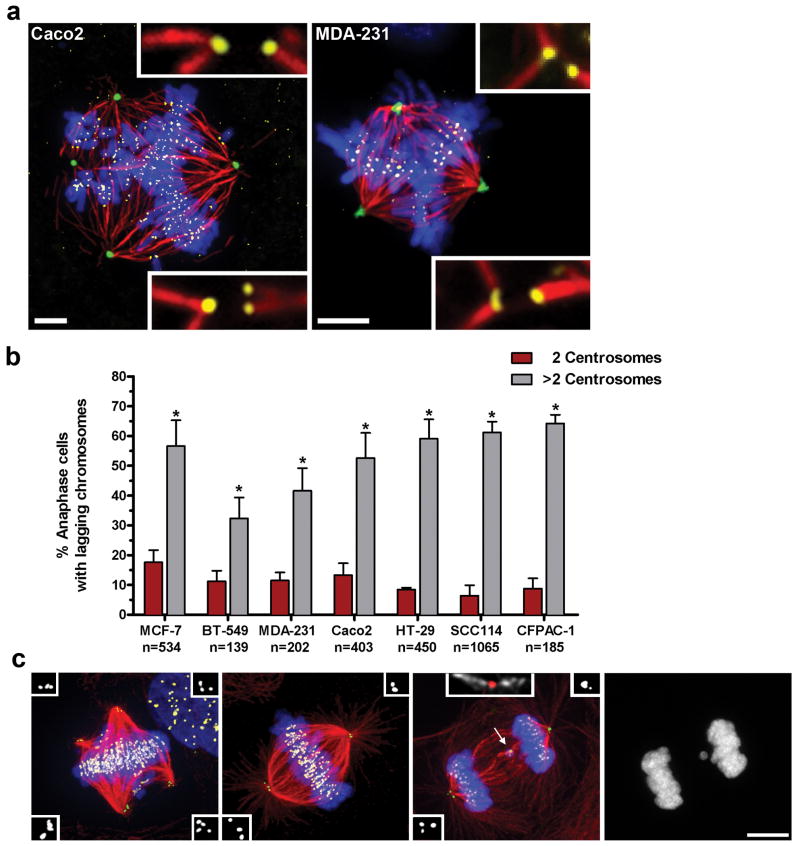
We hypothesized that cells with extra centrosomes pass through transient multipolar intermediates prior to centrosome clustering and that the geometry of such intermediates predisposes to merotelic attachments. Supporting this idea, we found that CIN cells with extra centrosomes spend a majority of mitosis in a multipolar configuration prior to centrosome clustering (Supp. Fig. 3), and, furthermore, we directly observed numerous merotelic attachments within multipolar spindles by high-resolution microscopy (Fig. 2a, Supp. Fig. 8). Moreover, as observed in tetraploid PtK1 cells 24, 25, both the frequency and number of lagging chromosomes are dramatically elevated in rare cells that undergo multipolar anaphase, indicating that merotelic attachments are enriched within the multipolar configuration (Supp. Fig. 4, Supp. Movie 2).
Figure 2. Extra centrosomes correlate with increases in lagging chromosomes.
a) Multipolar Caco2 and MDA-231 cells stained for pericentrin (green), microtubules (red), chromosomes (blue), and kinetochores (yellow). Merotelic attachments are shown in insets. Scale bars, 5 μm. b) The percentage of cancer cells with 2 (red) or >2 (grey) centrosomes that exhibit one or more lagging chromosomes during bipolar anaphase (n=number of anaphase cells counted; error bars represent mean ± SE from at least 4 independent experiments; asterisks denote P-values < 0.02 and are derived from an unpaired two-tailed t-test. c) Representative MCF-7 cells with extra centrosomes during prometaphase (multipolar spindle intermediate), metaphase (bipolar spindle with clustered centrosomes), and anaphase (with clustered centrosomes), stained for centrioles (green, and inset white), microtubules (red), chromosomes (blue), and centromeres (yellow). Arrow indicates a lagging chromosome caused by merotelic attachment, in which microtubules emanating from both poles attach to a single kinetochore (inset; microtubules white, centromere red). A detailed description of the criteria used for scoring lagging chromosomes can be found in Supp. Fig. 7. Scale bar, 10 μm.
As a further test of the hypothesis, a panel of CIN cell lines was analyzed by fixed-cell imaging and scored for both lagging chromosomes (markers for merotelic attachment) and centrosome number during mid-anaphase (Fig. 2b, c). The presence of extra centrosomes during bipolar anaphase correlated with a significant increase (3-10 fold) in the frequency of lagging chromosomes in every cell line examined, consistent with the idea that extra centrosomes increase the formation of merotelic attachments that can persist through anaphase (Fig. 2b, c).
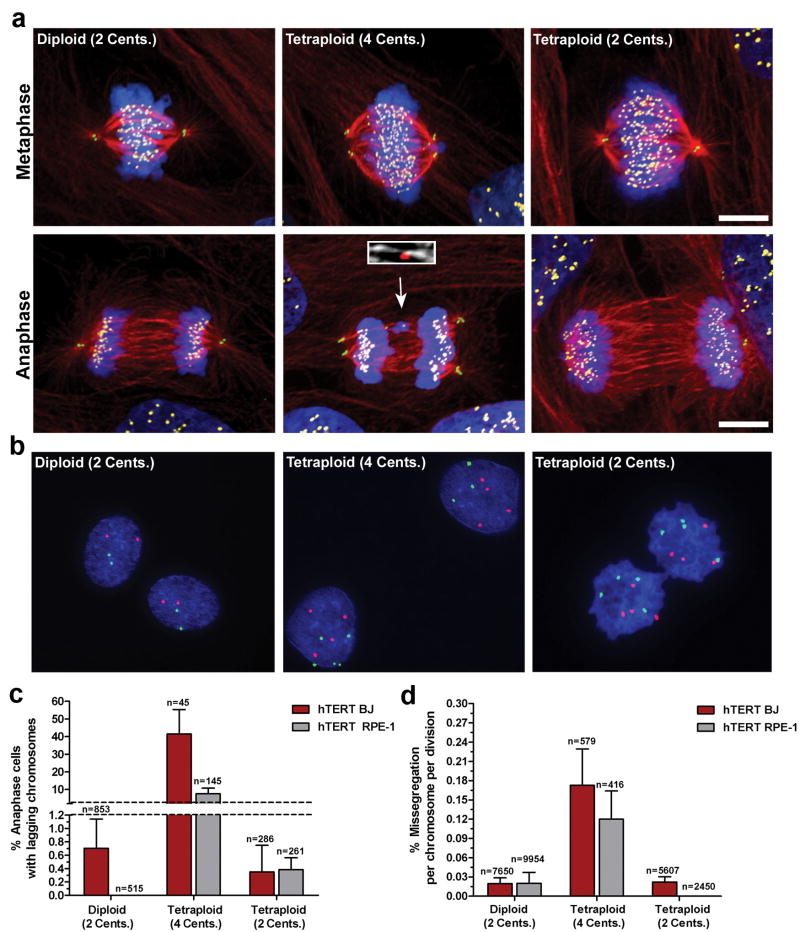
We next tested whether extra centrosomes alone are sufficient to promote chromosome missegregation during bipolar cell division. A significant technical challenge for studying the consequences of centrosome amplification has been the difficulty in obtaining genetically matched cells that do or do not contain extra centrosomes. We circumvented this problem in two ways. First, we generated tetraploid cells with extra centrosomes by using cytochalasin D to inhibit cytokinesis in chromosomally stable non-transformed telomerase immortalized human BJ and RPE1 cell lines. Since non-transformed cells are prone to cell-cycle arrest after tetraploidization, we transiently knocked down p53 by siRNA to facilitate passage of tetraploids through mitosis. Like cancer cells, tetraploid cells clustered their extra centrosomes into two poles during the subsequent mitosis and displayed a significantly increased frequency of anaphase lagging chromosomes during bipolar anaphase relative to matched diploids that were also exposed to cytochalasin and depleted of p53 (Fig. 3a, c).
Figure 3. Extra centrosomes promote chromosome missegregation.
a) Human hTERT BJ fibroblasts (Diploid, 2 centrosomes; Tetraploid, 4 centrosomes; Tetraploid, 2 centrosomes) during metaphase and anaphase stained for centrioles (green), microtubules (red), chromosomes (blue), and centromeres (yellow). Arrow indicates a lagging chromosome caused by merotelic attachment (inset; microtubules white, centromere red). b) FISH using centromeric probes specific for chromosomes 6 (green) and 8 (red) in hTERT-BJ fibroblasts. c) Percentage of hTERT BJ (red) and hTERT RPE-1 (grey) cells that exhibit one or more lagging chromosomes during bipolar anaphase (n=number of anaphases counted). d) Missegregation frequency per chromosome per division in hTERT BJ (red) and hTERT RPE-1 (grey) cells (n=number of cell divisions counted). Error bars represent mean ± SE from at least 4 independent experiments. Scale bar, 10 μm.
Interestingly, both tetraploid BJ and RPE-1 cells spontaneously lost their extra centrosomes after passage in culture (Fig. 3a and data not shown). We therefore used sequential FACS sorting to isolate pure populations of tetraploid cells with a normal complement of centrosomes. Fluorescence in situ hybridization (FISH) and karyotyping demonstrated that these cells contained a tetraploid complement of chromosomes (Fig. 3b, Supp. Fig. 5). Consequently, this procedure generated pure populations of tetraploids with only 2 centrosomes during mitosis and allowed us to compare the rate of lagging chromosomes in tetraploid cells possessing 2 or >2 centrosomes. Strikingly, the loss of extra centrosomes was accompanied by a decrease in the fraction of cells with lagging chromosomes to a level observed in diploid cells (Fig. 3a, c). These findings strongly suggest that the increased rate of lagging chromosomes in newly generated tetraploid cells is due to extra centrosomes rather than a duplicated genome.
To determine if the observed increases in lagging chromosomes in cells containing extra-centrosomes leads to chromosome missegregation, we used anaphase/telophase FISH to measure the rate of chromosome missegregation in BJ fibroblasts and RPE-1 cells (Fig. 3b, d). To ensure that cells with extra centrosomes passed through a bipolar mitosis, FISH signals were only scored in anaphase or telophase cells in which the daughters each possessed a single nucleus. The missegregation rates per chromosome in diploid, newly generated tetraploid, and late-passage tetraploid cells closely mirrored the results obtained by scoring lagging chromosomes as tetraploids with >2 centrosomes showed missegregation rates ∼6-8 fold higher than diploids or tetraploids with 2 centrosomes (Fig. 3d). These rates correspond to 1 chromosome missegregation for every ∼6 divisions in tetraploid BJ fibroblasts with >2 centrosomes, compared to 1 chromosome missegregation for every ∼50 divisions in tetraploids with 2 centrosomes. Thus, extra centrosomes promote chromosome missegregation even after cells cluster centrosomes to assemble bipolar spindles.
Although the above data suggest that the majority of missegregation events can be explained by merotelic attachments and lagging chromosomes, some missegregation events in cells with extra centrosomes may, at low frequency, arise by other mechanisms. For example, we occasionally observed single chromosomes bi-orienting between two inefficiently clustered centrosomes even after all other chromosomes had aligned at the metaphase plate (see Fig 3a, top row, middle panel). Presumably, these bi-oriented polar chromosomes could be under tension, satisfy the SAC, and thus segregate both sisters to a single daughter upon anaphase entry 4. Indeed, in tetraploid RPE1 cells, we did identify a single such example by live-cell imaging (Supp. Fig. 6, Supp. Movie 3). However, this mechanism does not occur frequently enough to contribute significantly to chromosome missegregation in the extra-centrosomal cells we examined: We did not observe a single such bi-oriented chromosome during anaphase in any of our fixed-cell samples, and only a very minor fraction of the ∼8000 CIN cell divisions we imaged by H2B-GFP showed chromosomes at the poles during anaphase onset, consistent with previous imaging analyses 13, 26.
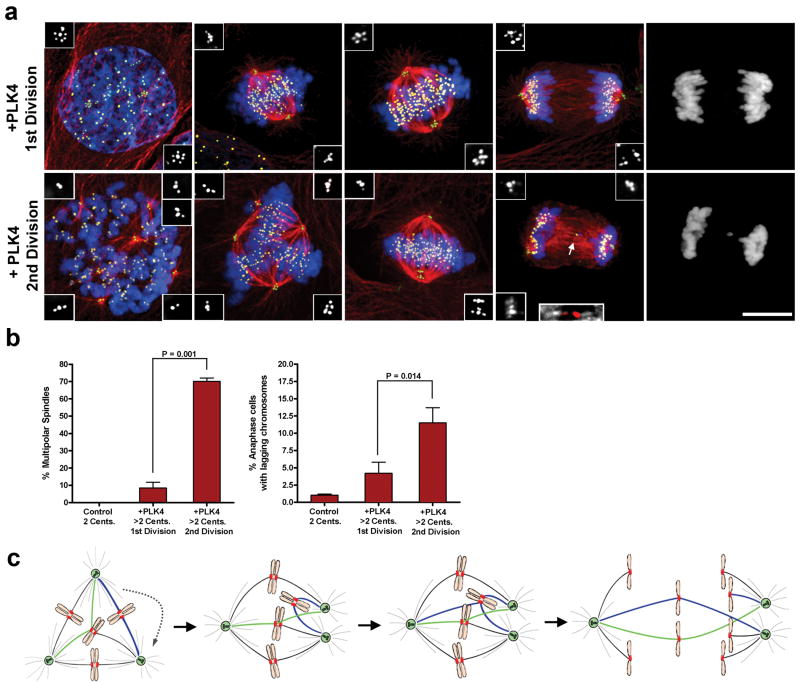
Finally, we designed an experiment where, in otherwise genetically identical cells, we could directly test the hypothesis that transient multipolar spindle intermediates generate anaphase lagging chromosomes. We recognized that this could be accomplished by monitoring mitosis over 2 generations after induction of PLK4, a kinase that regulates centriole replication and whose overexpression has previously been shown to cause centriole amplification 27. To do this, we used an U2OS osteosarcoma cell line in which PLK4 expression is regulated by a doxycycline-inducible promoter 27. After a 15 hr. induction of PLK4, cells contained two ‘rosettes’ of overduplicated centrioles, each comprised of a mother centriole surrounded by numerous daughter centrioles 27 (Fig. 4a). Importantly, because the extra centrioles assembled before mitosis, they remained engaged and functioned as single units to enable bipolar spindle assembly without a multipolar spindle intermediate (Fig. 4a, b). In these cells, despite centriole amplification, the frequency of lagging chromosomes was similar to that observed in control cells with 2 centrosomes (Fig. 4b). By contrast, in the second cell cycle after centriole overduplication, supernumerary centrioles disengaged prior to duplication 28 and multipolar intermediates were common in the mitosis that followed (Fig. 4a, b). In these cells, a marked increase in the frequency of lagging chromosomes was observed following centrosome clustering and the onset of bipolar anaphase (Fig. 4b). Thus, extra centrosomes force cells to pass through a multipolar spindle intermediate and thereby enhance the formation of merotelic attachments and lagging chromosomes.
Figure 4. ‘Multipolar spindle intermediates’ promote merotelic attachment.
a) U2OS cells undergoing their 1st or 2nd mitotic division following PLK4 overexpression stained for centrioles (green, inset white), microtubules (red), chromosomes (blue), and centromeres (yellow). Arrow indicates a lagging chromosome caused by merotelic attachment (inset; microtubules white, centromere red). b) The percentage of mitotic cells exhibiting multipolar spindles or lagging chromosomes for each condition. Error bars represent the mean ± SE from 5 independent experiments; P-value derived from paired two-tailed t-test. Scale Bar, 10 μm. c) Extra centrosomes promote merotelic attachment (green microtubules) by altering spindle geometry. In addition, syntelic attachments (blue microtubules) also accumulate upon centrosome clustering and may promote further enhancement of merotely. Unresolved merotelic attachments can give rise to lagging chromosomes at anaphase.
We propose that extra centrosomes generate CIN primarily by promoting merotelic kinetochore-microtubule attachments. This is due to the unique spindle geometry that occurs when cells resolve transient multipolar intermediates into bipolar spindles, in a manner broadly similar to what occurs during spindle assembly after nocodazole or monastrol washout 20, 23 (Fig. 4c). Moreover, syntelic attachments (both sister kinetochores attached to the same pole) are also expected to be enriched when chromosomes bi-orient between two centrosomes that eventually cluster (Fig. 4c), and these syntelic attachments may promote further merotely (Supp. Movie 4) 21, 23. Many of these merotelic attachment errors are corrected prior to anaphase by mechanisms that involve the release or destabilization of inappropriately attached microtubules 20, 21, 29; this can be inferred by the dramatic decrease in the frequency and number of lagging chromosomes observed in cells that enter mitosis from a multipolar configuration rather than a bipolar configuration (Supp. Fig. 4c), or by directly quantifying the number of merotelic attachments before and after centrosome clustering (Supp. Fig. 8). However, the overall increase in the number of initial merotelic attachments during the multipolar spindle intermediate reduces the likelihood that all errors will be corrected prior to anaphase onset, thereby causing a net increase in the frequency of lagging chromosomes and chromosome missegregation errors 13.
In sum, we have demonstrated that extra centrosomes are not simply innocent bystanders in CIN cells; rather, their presence directly promotes chromosome missegregation that may then facilitate the evolution of more malignant phenotypes. This finding clarifies the longstanding correlation between centrosome amplification and CIN, and provides one simple and unifying explanation for the observed high rates of merotely in CIN cancers. Previously, a variety of genetic mutations have been implicated as causes of CIN, but these defects appear to be relatively infrequent 30. By contrast, extra centrosomes are prevalent among CIN cells, and we suggest that the mechanism described here is a common contributor to chromosomal instability in human cancer.
Methods Summary
All cell lines were maintained at 37°C with 5% CO2 atmosphere. Immunofluorescence microscopy was performed as previously described 17. Fixed cell images were collected by confocal immunofluorescence on a Yokogawa CSU-X1 spinning disk confocal mounted on a Nikon Ti-E inverted microscope (Nikon Instruments). Live-cell imaging was performed using a TE2000-E2 inverted Nikon microscope equipped with the Nikon Perfect Focus system enclosed within a temperature and CO2-controlled environment that maintained an atmosphere of 37°C and 3-5% humidified CO2. Sequential FACS sorting of tetraploids with 8c DNA content was used to generate tetraploid cells with two centrosomes. Detailed descriptions of FISH, karyotyping, imaging, cell lines, culture conditions, and antibodies used in this study can be found in supplementary methods.
Supplementary Material
Acknowledgments
We would like to thank D. Compton, R. King, D. Livingston, A. Manning, T. Rapoport, J. Shah, T. Stukenberg, and members of the Pellman lab for comments/discussion of the manuscript; L. Cameron of the Confocal and Light Microscopy Core Facility at the Dana-Farber Cancer Institute; Donna Neuberg for help with statistical analysis; C. King for help with the model figure; J. Iwasa for creating the animations; and L. Moreau and S. Thompson for technical advice on chromosome spreads and FISH analysis. Reagents were kindly provided by D. Compton, S. Gollin, R. King, A. Khodjakov, E. Nigg, and G.Wahl. N. Ganem is a fellow of the Leukemia and Lymphoma Society and D. Pellman is an HHMI Investigator. This work was supported by NIH grant GM083299. D.P wishes to dedicate this manuscript to the memory of H. Hanafusa.
Footnotes
The authors declare no competing financial interests.
References
- 1.Lengauer C, Kinzler KW, Vogelstein B. Genetic instability in colorectal cancers. Nature. 1997;386:623–7. doi: 10.1038/386623a0. [DOI] [PubMed] [Google Scholar]
- 2.D'Assoro AB, Lingle WL, Salisbury JL. Centrosome amplification and the development of cancer. Oncogene. 2002;21:6146–53. doi: 10.1038/sj.onc.1205772. [DOI] [PubMed] [Google Scholar]
- 3.Nigg EA. Centrosome aberrations: cause or consequence of cancer progression? Nat Rev Cancer. 2002;2:815–25. doi: 10.1038/nrc924. [DOI] [PubMed] [Google Scholar]
- 4.Sluder G, Nordberg JJ. The good, the bad and the ugly: the practical consequences of centrosome amplification. Curr Opin Cell Biol. 2004;16:49–54. doi: 10.1016/j.ceb.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 5.Rajagopalan H, Lengauer C. Aneuploidy and cancer. Nature. 2004;432:338–41. doi: 10.1038/nature03099. [DOI] [PubMed] [Google Scholar]
- 6.Weaver BA, Silk AD, Montagna C, Verdier-Pinard P, Cleveland DW. Aneuploidy acts both oncogenically and as a tumor suppressor. Cancer Cell. 2007;11:25–36. doi: 10.1016/j.ccr.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 7.Gao C, et al. Chromosome instability, chromosome transcriptome, and clonal evolution of tumor cell populations. Proc Natl Acad Sci U S A. 2007;104:8995–9000. doi: 10.1073/pnas.0700631104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194:23–8. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- 9.Ghadimi BM, et al. Centrosome amplification and instability occurs exclusively in aneuploid, but not in diploid colorectal cancer cell lines, and correlates with numerical chromosomal aberrations. Genes Chromosomes Cancer. 2000;27:183–90. [PMC free article] [PubMed] [Google Scholar]
- 10.Lingle WL, et al. Centrosome amplification drives chromosomal instability in breast tumor development. Proc Natl Acad Sci U S A. 2002;99:1978–83. doi: 10.1073/pnas.032479999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pihan GA, et al. Centrosome defects can account for cellular and genetic changes that characterize prostate cancer progression. Cancer Res. 2001;61:2212–9. [PubMed] [Google Scholar]
- 12.Sato N, et al. Correlation between centrosome abnormalities and chromosomal instability in human pancreatic cancer cells. Cancer Genet Cytogenet. 2001;126:13–9. doi: 10.1016/s0165-4608(00)00384-8. [DOI] [PubMed] [Google Scholar]
- 13.Thompson SL, Compton DA. Examining the link between chromosomal instability and aneuploidy in human cells. J Cell Biol. 2008;180:665–72. doi: 10.1083/jcb.200712029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Torres EM, Williams BR, Amon A. Aneuploidy: cells losing their balance. Genetics. 2008;179:737–46. doi: 10.1534/genetics.108.090878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brinkley BR. Managing the centrosome numbers game: from chaos to stability in cancer cell division. Trends Cell Biol. 2001;11:18–21. doi: 10.1016/s0962-8924(00)01872-9. [DOI] [PubMed] [Google Scholar]
- 16.Basto R, et al. Centrosome amplification can initiate tumorigenesis in flies. Cell. 2008;133:1032–42. doi: 10.1016/j.cell.2008.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kwon M, et al. Mechanisms to suppress multipolar divisions in cancer cells with extra centrosomes. Genes Dev. 2008;22:2189–203. doi: 10.1101/gad.1700908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Quintyne NJ, Reing JE, Hoffelder DR, Gollin SM, Saunders WS. Spindle multipolarity is prevented by centrosomal clustering. Science. 2005;307:127–9. doi: 10.1126/science.1104905. [DOI] [PubMed] [Google Scholar]
- 19.Yang Z, Loncarek J, Khodjakov A, Rieder CL. Extra centrosomes and/or chromosomes prolong mitosis in human cells. Nat Cell Biol. 2008;10:748–51. doi: 10.1038/ncb1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cimini D. Merotelic kinetochore orientation, aneuploidy, and cancer. Biochim Biophys Acta. 2008;1786:32–40. doi: 10.1016/j.bbcan.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 21.Salmon ED, Cimini D, Cameron LA, DeLuca JG. Merotelic kinetochores in mammalian tissue cells. Philos Trans R Soc Lond B Biol Sci. 2005;360:553–68. doi: 10.1098/rstb.2004.1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cimini D, et al. Merotelic kinetochore orientation is a major mechanism of aneuploidy in mitotic mammalian tissue cells. J Cell Biol. 2001;153:517–27. doi: 10.1083/jcb.153.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cimini D, Moree B, Canman JC, Salmon ED. Merotelic kinetochore orientation occurs frequently during early mitosis in mammalian tissue cells and error correction is achieved by two different mechanisms. J Cell Sci. 2003;116:4213–25. doi: 10.1242/jcs.00716. [DOI] [PubMed] [Google Scholar]
- 24.Heneen WK. Kinetochores and microtubules in multipolar mitosis and chromosome orientation. Exp Cell Res. 1975;91:57–62. doi: 10.1016/0014-4827(75)90140-8. [DOI] [PubMed] [Google Scholar]
- 25.Sluder G, Thompson EA, Miller FJ, Hayes J, Rieder CL. The checkpoint control for anaphase onset does not monitor excess numbers of spindle poles or bipolar spindle symmetry. J Cell Sci. 1997;110(Pt 4):421–9. doi: 10.1242/jcs.110.4.421. [DOI] [PubMed] [Google Scholar]
- 26.Gascoigne KE, Taylor SS. Cancer cells display profound intra- and interline variation following prolonged exposure to antimitotic drugs. Cancer Cell. 2008;14:111–22. doi: 10.1016/j.ccr.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 27.Kleylein-Sohn J, et al. Plk4-induced centriole biogenesis in human cells. Dev Cell. 2007;13:190–202. doi: 10.1016/j.devcel.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 28.Tsou MF, Stearns T. Mechanism limiting centrosome duplication to once per cell cycle. Nature. 2006;442:947–51. doi: 10.1038/nature04985. [DOI] [PubMed] [Google Scholar]
- 29.Bakhoum SF, Thompson SL, Manning AL, Compton DA. Genome stability is ensured by temporal control of kinetochore-microtubule dynamics. Nat Cell Biol. 2008 doi: 10.1038/ncb1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chandhok NS, Pellman D. A little CIN may cost a lot: revisiting aneuploidy and cancer. Curr Opin Genet Dev. 2009;19:74–81. doi: 10.1016/j.gde.2008.12.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.