Abstract
The Kruppel-like Factor (KLF) family of zinc-finger transcription factors are critical regulators of cell differentiation, phenotypic modulation and physiologic function. An emerging body of evidence implicates an important role for these factors in cardiovascular biology, however, the role of KLFs in muscle biology is only beginning to be understood. This article reviews the published data describing the role of KLFs in cardiomyocytes, smooth muscle cells, and skeletal muscle and highlights the importance of these factors in cardiovascular development, physiology and disease pathobiology.
Keywords: KLF, kruppel, cardiomyocyte, heart, smooth muscle, myocyte, skeletal, transcription
Introduction
Cardiovascular disease remains one of the most prevalent causes of morbidity and mortality worldwide [1] and a better understanding of the molecular underpinnings of disease pathogenesis will be invaluable for therapeutic advancement. The heart and vasculature are comprised of two important muscle cell types that play central roles in the development of cardiovascular disease – the cardiomyocyte and the vascular smooth muscle cell (SMC). In addition, phenotypic changes in skeletal muscle that occur with vascular insufficiency or chronic heart failure are becoming increasingly recognized as significant component of disease pathobiology [2–4]. As such, dissecting the molecular mechanisms that drive dysfunction in these cell types represents fertile ground for the development of novel therapies [5].
A number of cytosolic signaling pathways involved in development and remodeling of the heart, vasculature and periphery have been elucidated in cardiomyocytes [6], SMC [7] and skeletal muscle [8]. Importantly, many of these pathways converge on a critical set of transcription factors that can serve as signal integrators and produce alterations in gene expression, cellular identity, proliferative state, and physiologic function [4, 9–14]. Of these transcriptional regulators, the Kruppel-Like Factor (KLF) family of zinc-finger transcription factors has recently been a subject of intense investigation in human health and disease [12, 15–17]. This review will focus on the emerging role of the KLF family of transcription factors in the biology of cardiomyocytes, vascular smooth muscle cells and skeletal muscle.
Kruppel-Like Factors (KLFs)
KLFs are a subfamily of the zinc-finger class of DNA-binding transcriptional regulators. Several features distinguish KLFs from the larger family of zinc-finger transcription factors. First, KLFs contain three Cysteine2/Histidine2 containing zinc fingers located at the extreme C-terminus of the protein [18, 19]. Second, KLFs share a highly conserved seven residue sequence between zinc fingers, TGEKP(Y/F)X [20]. Third, most KLFs are able to bind consensus sequences such as the CACCC element or GT box and such DNA-binding specificity is dictated by three critical residues within each zinc finger [19, 20]. In contrast to the zinc-finger regions, the non-DNA-binding regions are highly divergent, modulate transactivation and transrepression, and often mediate protein-protein interactions.
The name “Kruppel-like factor” is derived from the shared homology of these proteins to the DNA-binding domains of the Drosophila protein kruppel (a German word meaning “cripple”). Kruppel is an early developmental fly gene whose role in body patterning was described in the pioneering work of Nusslein-Volhard and Weischaus who observed that Drosophila embryos deficient in kruppel died as a result of abnormal thoracic and abdominal segmentation and thus appeared “crippled” [21–24]. The first mammalian kruppel homolog, termed erythroid Kruppel-like factor (EKLF/KLF1), was identified in 1993 as a factor specifically expressed in the red blood cell lineage [25]. Gene targeting approaches have demonstrated an essential role for EKLF/KLF1 in β-globin gene induction and erythrocyte development [8, 26]. Since the identification of EKLF/KLF1, a total of 17 mammalian KLFs have been identified (designated KLF1 through KLF17, based on their chronologic order of identification) and are known to be expressed in a broad range of cell types. These factors have been shown to have important roles in a diverse array of cellular processes including cardiac remodeling [27, 28], hematopoiesis [8, 12, 26], angiogenesis [29], neoplasia [15, 30], flow-mediated endothelial gene expression [31, 32], gluconeogenesis [33], monocyte activation [34, 35], and determination of pluripotent stem cell fate [36]. In this review, we will discuss the published studies that implicate KLFs as important regulators of cardiac, VSMC and skeletal muscle biology (summarized in Table 1).
Table 1.
Summary of published findings pertaining to Kruppel-like Factors in Muscle Biology.
| FUNCTION / OBSERVATION | REFERENCES | ||
|---|---|---|---|
| HEART | |||
| KLF5 |
|
[28] | |
| KLF15 |
|
[27, 40] | |
| KLF13 |
|
[37, 47, 50] | |
| KLF10 |
|
[38] | |
| SMOOTH MUSCLE | |||
| KLF5 |
|
[17, 28, 71–74, 76, 77] | |
| KLF4 |
|
[85, 86] | |
| KLF15 |
|
[93] | |
| KLF13 |
|
[91] | |
| SKELETAL MUSCLE | |||
| KLF15 |
|
[40, 96] | |
| KLF13 |
|
[47] | |
| KLF6 |
|
[95] |
KLFs in the Heart
Despite significant progress in the understanding of KLFs in human health and disease over the past decade, the expression and function of KLFs in the heart is only beginning to be elucidated. To date, there are four published reports describing the function of KLFs in the heart – KLF5 in cardiac fibroblasts [28], KLF15 (postnatal) [27] and KLF13 (embryonic) [37] in cardiomyocytes, and a brief report describing a cardiac phenotype in systemic KLF10-null mice [38]. Figure 1 highlights some of these published findings pertaining to KLFs in the myocardium.
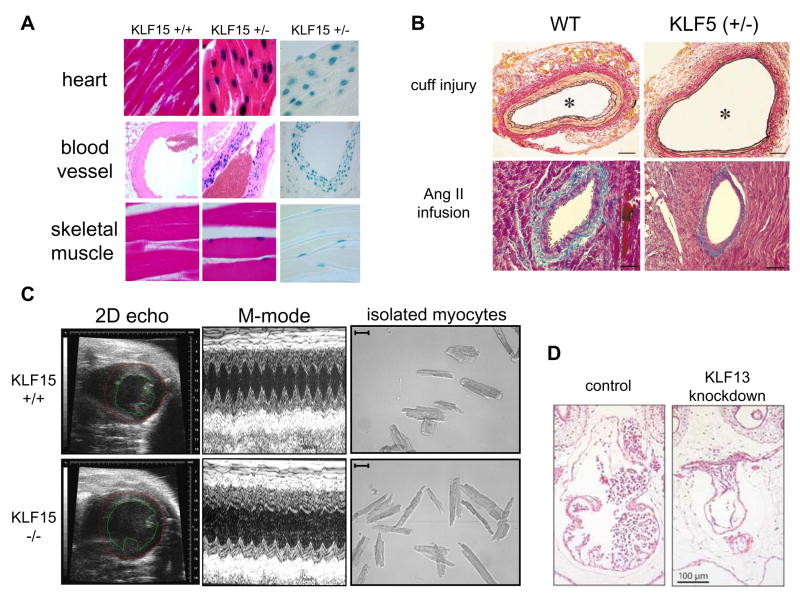
Figure 1.
Kruppel-Like Factors are Important Regulators of Muscle Biology in the Cardiovascular System. (A) KLF15 is expressed in cardiomyocytes, SMC and skeletal muscle [27, 40]. KLF15 (+/−) mice were generated by targeting the native mouse KLF15 gene with a lacZ cassette containing a nuclear localization signal. Histologic sections from KLF15 (+/+) vs. (+/−) tissues are shown. KLF15 (+/−) tissues are shown both with and without eosin counterstaining. (Adapted with permission from PNAS and JBC) (B) KLF5 (+/−) mice have reduced neointimal formation in response to vascular injury (top) and reduced perivascular fibrosis in response to angiotensin II infusion (bottom) [28]. (Adapted with permission from Nature Medicine). (C) KLF15 (−/−) develop eccentric hypertrophy, severe LV dysfunction, and eccentric myocyte enlargement with pressure overload [27]. (Adapted with permission from PNAS). (D) KLF13 knockdown in Xenopus embryos causes atrial septal abnormalities and defects in ventricular trabeculation [37]. (Adapted with permission from EMBO Journal)
KLF5
KLF5 (also known as BTEB2/IKLF) was first identified from a human placental library using a homology screening strategy [39]. An appreciation for the importance of this factor in cardiac biology has been elegantly developed principally through studies in the laboratory of Dr. Ryozo Nagai [28]. As will be described later in this review, KLF5 has also been implicated as a central regulator of vascular smooth muscle cell differentiation, growth and gene expression. In the heart, the published evidence to date suggests that KLF5 is expressed principally in cardiac fibroblasts while expression is undetectable in cardiomyocytes. In primary cultured cardiac fibroblasts, KLF5 expression was shown to be upregulated with angiotensin II stimulation in a parallel fashion to PDGF-A (a well-described angiotensin target gene in cardiac fibroblasts). From a mechanistic standpoint, angiotensin-induced induction of PDGF-A was shown to be dependent on KLF5 recruitment to the PDGF-A promoter.
In an effort to study the role of KLF5 in cardiovascular biology in vivo, the mouse KLF5 gene was targeted systemically [28]. Complete absence of KLF5 is embryonic lethal by embryonic day E8.5 though the precise developmental defect in these mice has not been well characterized. However, KLF5(+/−) mice are viable into adulthood and partial deficiency of KLF5 in vivo resulted in an attenuated hypertrophic response to angiotensin II infusion as assessed by cardiac mass, wall thickness, cardiac fibrosis, and PDGF-A expression [28]. In addition, angiotensin-induced expression of TGF-β and collagen type IV were also attenuated in KLF5(+/−) hearts suggesting that induction of these important angiotensin effectors may also be KLF5 dependent. Interestingly, these investigators also found that KLF5 can physically interact with the retinoic acid receptor-alpha (RARα) and their data raises the possibility that RARα activation modulates KLF5-dependent myocardial remodeling. Taken together, these data implicate KLF5 expression in cardiac fibroblasts plays an important role in cardiac remodeling [28].
KLF15
As KLF5 was principally expressed in cardiac fibroblasts, the possibility that KLFs were expressed in cardiomyocytes and had important roles in cardiomyocyte biology remained unanswered. Our group identified KLF15 as being expressed in cardiomyocytes [40] and has recently provided evidence that this factor functions as a negative regulator of hypertrophic remodeling thus demonstrating a novel role for this family of transcription factors in postnatal cardiomyocyte function [27]. In addition, we have shown that KLF15 is also expressed in primary cardiac fibroblasts [27], however, the significance of this finding has not been completely elucidated. Figure 1A shows β-galactosidase staining of tissues from adult KLF15(+/−) mice (in which a nuclear localized lacZ gene was “knocked in” to the KLF15 locus by homologous recombination) and clearly illustrates robust KLF15 expression in all three muscle types.
KLF15 has an intriguing expression pattern in the heart – it is not detectable during embryonic development and is expressed only at very low levels in the early postnatal period. However, KLF15 expression is robustly upregulated by 30 days postnatally, at a time when genes such as ANF, BNP and cyclin-A [40] are downregulated. Furthermore, KLF15 expression is dramatically reduced with pressure overload hypertrophy in murine models as well as in human subjects with valvular aortic stenosis. [27, 40] KLF15 expression is also reduced by pharmacologic agonists known to induce cardiomyocyte hypertrophy such as phenylephrine and endothelin-1 [27]. Finally, a recent report detailing expression profiles in cultured myocytes treated with hydrogen-peroxide to induce oxidative stress revealed over 50% reduction in KLF15 expression [41] although the physiologic relevance of this finding is not known.
To understand the role of KLF15 in cardiac biology, we undertook gain and loss of function studies [27]. Overexpression of KLF15 in neonatal rat ventricular myocytes potently inhibits three cardinal features of hypertrophic remodeling (cell growth, protein synthesis and hypertrophic gene expression) under both basal and stimulated conditions. To study the role of KLF15 deficiency in vivo we targeted the mouse KLF15 gene to create systemic KLF15(−/−) mice. These KLF15(−/−) mice are viable, fertile and do not have cardiac hypertrophy at baseline (8–12 weeks age). Despite their seemingly “normal” baseline cardiac phenotype, these animals are exquisitely sensitive to stress and develop a severe form of eccentric hypertrophic remodeling in response to pressure overload characterized by cavity enlargement, depressed systolic function, and exaggerated fetal gene expression (Figure 1C). The morphologic changes in KLF15(−/−) hearts are likely due to a primary cardiomyocyte defect as isolated myocytes from failing hearts are eccentrically hypertrophied (large and elongated). There was no significant apoptotic activity in these hearts after one week of aortic constriction, suggesting that apoptotic cell dropout was not contributing to the observed phenotype.
From a mechanistic standpoint, KLF15 can potently inhibit the transcriptional activity of MEF2 and GATA4 [27] – two well-characterized transcriptional activators of hypertrophic remodeling that can function synergistically in the myocardium [42, 43]. The primary downstream mechanism by which MEF2 and GATA4 carry out their pro-hypertrophic effects is via enhanced DNA binding. Consistent with this observation, KLF15 appears to be able to inhibit the ability of these factors to bind target promoters although the molecular basis for this inhibitory effect remains to be elucidated.
Interestingly, there are striking similarities between the phenotypes of KLF15-deficient mice and mice with transgenic overexpression of either MEF2 or GATA4 in the myocardium. Transgenic overexpression of MEF2A and MEF2C leads to a dose-dependent dilated cardiomyopathy that is dramatically worsened with pressure overload [44]. Cardiomyocytes from these MEF2-transgenic hearts have large and elongated myocytes [44] similar to those observed with KLF15-deficiency [27]. Transgenic overexpression of GATA4 in the heart at high levels results in a severe cardiomyopathy and premature death [45]. Even modest GATA4 overexpression results in a spontaneous cardiomyopathy characterized by increased heart mass and hypertrophic gene expression [45]. Given that MEF2 and GATA4 factors can cooperate and serve as integrators for multiple upstream signaling pathways, it is not surprising that heightened activity of these two factors leads to a marked decompensation in KLF15(−/−) mice in response to stress. The inhibition of MEF function by KLF15 may also have implications for postnatal cardiac development. Previous studies by the Molkentin and colleagues [46] showed an increase in MEF2 binding and activity in the postnatal period. In this context, the induction of KLF15 expression in the heart after birth constitutes an intriguing expression pattern. It is interesting to speculate that KLF15 may serve as a transcriptional “brake” on excessive MEF2 activity during the transition from neonatal to adult myocardium [27]. Further studies in our laboratory are actively underway to further dissect the precise function and regulation of KLF15 in the cardiomyocyte and cardiac fibroblast – both in the postnatal period and in response to various hypertrophic stimuli.
KLF13
KLF13 (also known as FKLF-2/BTEB3) has previously been reported to have restricted expression in erythroid cells, T-lymphocytes, heart and skeletal muscle [47, 48]. Low-level detection of KLF13 transcripts has been detected by RT-PCR in other adult mouse tissues [49] although the full significance of this finding is unclear. In the mouse embryo, there is robust developmental expression of KLF13 in the heart, cephalic mesenchyme, dermis, and epithelial layers of the gut and urinary bladder [50]. There are a number of reports describing the important role of KLF13 in the transcriptional regulation of erythroid gene expression [51, 52] and its critical role in the induction of RANTES expression in activated T-lymphocytes [48, 53].
A role for KLF13 in the embryonic myocardium was recently elucidated by the Nemer laboratory in elegant studies of BNP gene regulation and Xenopus development [37]. A proximal BNP promoter that is sufficient for maximal cardiac transcription has been previously defined by this group [54]. Interestingly, there is an evolutionarily conserved KLF consensus site (CACCC) in this proximal BNP promoter that is flanked by nearby GATA sites and is essential for promoter activity. Based on this finding, the investigators hypothesized that a KLF family member may be important in regulation of BNP expression. Indeed, KLF13 was shown to be expressed in cardiac myocytes (atria > ventricles), and that its ability to bind the CACCC element in the proximal BNP promoter was critical for transactivation. KLF13 was also shown to activate multiple cardiac promoters (BNP, ANF, β-MHC, α-cardiac actin) in a synergistic fashion with GATA4. In addition, KLF13 was shown to physically interact with GATA4 and detailed structure-function analyses mapped the site of this interaction to the N-terminal zinc finger of GATA4 [37].
To determine the role of KLF13 in heart development, its expression pattern was determined in the mouse embryonic heart. Cardiac expression of KLF13 was first detected at E9.5 with subsequent expression in the developing atrial myocardium, ventricular trabeculae, AV cushions and the truncus arteriosus. Postnatally, KLF13 was substantially downregulated in the heart with expression predominantly restricted to the valves and interventricular septum. Given this expression pattern, the role of KLF13 in cardiac development was further explored in elegant studies using KLF13 knockdown in the Xenopus embryo. Embryos with KLF13 deficiency developed atrial septal defects and ventricular hypotrabeculation (Figure 1D) similar to that seen in humans or mice with hypomorphic GATA4 alleles [11]. This hypoplastic phenotype was not associated with increased apoptosis and raises the possibility that KLF13 may be involved in regulating cardioblast proliferation. Interestingly, these defects could be rescued with overexpression of GATA4 in a dose-dependent manner, suggesting that KLF13 and GATA4 are indeed mutual cofactors in heart development in vivo. These findings raise the intriguing possibility that KLF13 may be a novel candidate gene for human congenital heart disease. The function of KLF13 in the mammalian heart or any potential role in postnatal cardiac function has not been reported to date.
KLF10
In contrast to KLF5,15, and 13 there is very little known about the role of KLF10/TIEG1 in the heart. This transcription factor was first described in osteoblasts as a “TGF-β Inducible Early Gene” and thus carried the name (TIEG-1)[55]. Subsequent studies from the laboratory or Dr. Thomas Spelsberg have revealed an important role for this factor in the regulation of bone mineralization [56], osteoclast differentiation [56] and epithelial proliferation [57, 58]. KLF10 can modulate Smad signaling in osteoblasts and mice with systemic KLF10 deficiency are osteopenic [59] and have impaired tendon healing in response to mechanical injury [60].
Since its first description, KLF10/TIEG1 was known to have low-level expression from whole-heart RNA [55]. However, its precise cellular expression within the heart (myocytes vs. fibroblasts) and functional role in the myocardium remain poorly understood. A recent report by the Spelsberg laboratory described the cardiac phenotype of systemic KLF10(−/−) mice [38]. It is not known whether KLF10 is expressed in the developing heart and embryonic defects have not been described in KLF10(−/−) mice. However, male KLF10(−/−) mice (but not females) examined at 16 months age have spontaneous pathologic cardiac hypertrophy with increased heart mass, wall thickness, tissue fibrosis and myocyte disarray with preservation of LV systolic function. The exact timing of this hypertrophic remodeling is not well characterized, although no significant cardiac fibrosis was observed in 4 month-old KLF10(−/−) mice. Expression profiling in these hypertrophic KLF10(−/−) hearts revealed that KLF10 may regulate the expression of Pituitary Tumor Transforming Gene (Pttg1), however the functional significance of this finding has yet to be elucidated. In future studies, it will be important to determine the cellular expression pattern of KLF10 in the heart, particularly whether it is expresses in cardiomyocytes and/or cardiac fibroblasts. As TGFβ is a critical mediator of cardiac fibrosis and angiotensin-mediated cardiac remodeling [61], further studies are necessary to more precisely define the regulation and transcriptional function of KLF10 in the heart.
Important Questions for KLFs in the Heart
From the published studies described in this review, it is clear that KLFs play an important role in cardiac development, maturation and postnatal function. However, several important questions remain regarding the role of this family of factors in cardiac biology. It is likely that other KLFs are expressed in the heart and have significant functions. The timing and relative expression for all of the KLFs expressed in the myocardium must be determined. In addition, there are multiple cell types found within the heart (cardiomyocyte, fibroblast, endothelial cell, vascular smooth muscle cell, immune cells) and expression profile of these cardiac KLFs within each cell type must be explored. For example KLF5 is expressed in cardiac fibroblasts and VSMC [28], while KLF15 is expressed in all three muscle types [27, 40]. We are actively exploring the relative function of KLF15 in the cardiomyocyte vs. the cardiac fibroblast, as both tissue play significant roles in myocardial remodeling and the response to angiotensin II. It is likely that conditional gene targeting and inducible transgenesis, where feasible, will be necessary to dissect the precise in vivo roles of these factors in cardiac biology. The overlapping and divergent function of KLFs expressed within the same cell type (e.g. KLF13 and KLF15) is also a critical area of investigation. For example, both KLF13 and KLF15 can modulate GATA4 activity; however the former can activate multiple cardiac promoters in a synergistic fashion with GATA4 [37] while the latter can potently repress GATA-mediated induction of the these promoters [27]. These data raise the intriguing possibility that KLF15 may function as a postnatal repressor, in part, via antagonism of KLF13 at critical promoters and that this crosstalk may involve competition for GATA4 binding. Furthermore, the possibility that KLFs directly regulate the expression of each other in the heart is another important area on inquiry as such regulation has been demonstrated in other tissues [62]. In fact, the broad aspects of regulation of the KLF expression in the myocardium is poorly understood and will provide invaluable insights into their tissue-specific functions. Finally, the mechanism of KLF action must be explored within the context of the multiprotein transcriptional complexes that operate in the myocardium. It will be critical to define how each KLF interacts with other important DNA-binding proteins, chromatin remodeling complexes, nuclear enzymes and transport complexes, and RNA-modifying factors within the nucleus.
KLFs in Smooth Muscle Biology
Vascular smooth muscle cells (VSMC) are a critical cellular constituent of the blood vessel wall. Indeed, these cells exhibit remarkable plasticity and can readily change phenotype in response to a diverse array of external stimuli including mechanical injury, growth factors and oxidative stress [63]. As VSMC mature, they are relatively quiescent, proliferating at a low rate and exhibiting a differentiated, contractile phenotype [64]. These properties allow the VSMC to carry out its primary function under physiologic conditions - regulating vessel tone by contraction and relaxation. The VSMC is also capable of a multitude of other functions including vessel repair. In response to vascular injury, VSMC undergo a dramatic change in phenotype — from that of a quiescent / differentiated cell to one that dedifferentiates, proliferates, migrates, re-expresses fetal isoforms of contractile proteins and elaborates extracellular matrix [64–67]. These features contribute to the development of occlusive vascular diseases such as atherosclerosis, restenosis, and transplant arteriopathy [68, 69]. Despite its importance, the molecular mechanisms that control VSMC development and differentiation have yet to be fully elucidated [70]. Recent investigations have defined an important role for KLFs in the regulation of VSMC biology and a summary these published findings are provided in Table 1.
KLF5
Multiple lines of evidence strongly implicate KLF5 as an important regulator of SMC gene expression and function. Initial insights can be gleaned from the expression pattern of this factor in the embryonic and adult vasculature. KLF5 expression is robust in fetal but not adult SMC. However, expression can be re-induced in adult SMCs after vascular injury or with angiotensin II treatment. [71–73] Importantly, KLF5 expression in injured human vessels is most abundant in neointimal SMCs and the development of restenosis is associated with increased neointimal expression of KLF5. Finally, pro-survival (e.g. survivin) [74] or pro-proliferative pathways (e.g. mitogen-activated protein kinases) [75] that are activated following vessel injury can, in turn, augment KLF5 expression. These key observations suggest that KLF5 may be an important regulator of the SMCs response to vascular injury.
The most cogent evidence implicating KLF5 in the SMC injury response is derived from loss-of-function experiments [28]. As stated previously, complete absence of KLF5 is embryonically lethal while KLF5(+/−) mice are viable and fertile. In contrast to wild-type mice, KLF5(+/−) mice exhibit a marked reduction in neointima formation and SMC proliferation following vascular injury (Figure 1B, top). Furthermore, in response to angiotensin II infusion, KLF5 (+/−) mice also demonstrate reduced perivascular fibrosis suggesting an important role in adventitial remodeling following vessel injury (Figure 1B, bottom). Direct evidence that KLF5 can influence gene expression and cellular behavior is derived from overexpression studies. Forced overexpression of KLF5 can promote cellular proliferation [76, 77] and can induce expression of a number of factors that are well-known to influence the vascular injury response such as PDGF-A/B, PAI-1, iNOS, and the VEGF receptors [72]. Thus, through increases in cellular proliferation and alterations in gene expression, KLF5 can critically regulate vascular remodeling.
Given the importance of KLF5 in cardiovascular biology, the mechanisms regulating its activity has been the subject of intense interest. While several pathways have been described, a particularly intriguing mechanism was revealed by Shindo and colleagues who demonstrated that KLF5 interacts with retinoic acid receptors (RAR) [28]. Importantly, these investigators found that an RARα agonist suppresses KLF5 function whereas an RARα antagonist activates KLF5. These studies suggest that the interaction of KLF5 with RARα maybe important in vascular remodeling and that manipulation of this interaction may provide a novel means of regulating this process. Indeed, Fujiu and colleagues have recently demonstrated that Am80, a synthetic RARα that has been successfully used in the treatment of promyelocytic leukemia, inhibits the transcriptional activity of KLF5. Furthermore, oral administration of Am80 significantly blunts the development of in-stent neointimal proliferation in a rabbit stent model [78].
KLF5 has also been shown to interact with factors best known for their role in chromatin remodeling. For example, KLF5 activity is increased via its interaction with the coactivator and acetylase p300 [76]. Importantly, this augmentation of KLF5 function is, at least in part, mediated by acetylation of KLF5 itself. Two factors that can inhibit KLF5 activity have also been identified — the myeloid leukemia associated oncoprotein SET (identified as a KLF-interactor by MALDI-TOF mass spectroscopy) and HDAC1 [76]. Previous studies suggest that SET serves as a histone chaperone that can help displace and/or assemble nucleosomal histones [79]. SET was shown to negatively regulate the ability of KLF5 to bind DNA, transactivate target promoters and exert its pro-proliferative effects. Similarly, HDAC1 can also interact with KLF5 and inhibit its ability to bind DNA though the mechanistic basis for the inhibitory effects of SET and HDAC are distinct. In the case of SET, this is thought to occur by noncatalytic inhibition of p300-meidated KLF5 acetylation. In contrast, the inhibitory effect of HDAC1 is thought to occur through direct interaction with KLF5 and interference of the p300-KLF5 activation complex. Indeed, these data highlight that potent factors such as KLF5 have multiple layers of regulation in the nucleus.
KLF4
Kruppel-like factor 4 was first identified as being highly expressed in epithelial cells and gain/loss of function studies in vivo have confirmed a critical role for this factor in epithelial cell differentiation [80–82]. Subsequent work has implicated KLF4 as being important in a broad range of cellular processes such as endothelial pro-inflammatory activation [83], macrophage gene expression [35], tumor cell development [30] and stem cell biology [36, 84].
Studies from several laboratories have also identified KLF4 in smooth muscle cells. Like KLF5, KLF4 expression is very low under basal conditions. However, in rodent models of vascular injury, KLF4 expression is rapidly induced achieving peak levels by 1–2 hours and returning to baseline by 24 hours. In addition, potent growth factors that regulate smooth muscle cell proliferation, differentiation, and extracellular matrix formation such as PDGF-bb and TGFβ1 can regulate KLF4 expression [85, 86]. Using siRNA approaches, studies from the Owens’ laboratory suggest that KLF4 mediates, at least in part, the repressive effects of PDGF-bb on SMC gene expression. This effect may occur as a consequence of KLF4 mediated repression of myocardin – a critical transcriptional coactivator involved in SMC differentiation [85, 87–90]. In contrast to PDGF-bb, TGFβ1 is known to induce SMC gene expression. Although KLF4 can also be induced by TGFβ1, in this setting it is has been shown to repress the TGFβ1-dependent increase in SMC differentiation markers such as α-SMC actin and SM22α. Taken together, these studies suggest that KLF4 may promote the de-differentiated phenotype seen in response to vascular injury. However, while these in vitro studies have provided critical insights into KLF4 function, the role of this factor in SMC biology in vivo has not been elucidated.
KLF13
Martin and colleagues demonstrated that KLF13, also known as basic transcription element-binding protein (BTEB 3), activates the minimal promoter for the smooth muscle specific SM22α gene [91]. However, further studies are required to further delineate the role of this factor in SMC biology.
KLF15
KLF15 was identified by our group and others as being highly expressed in several tissues including adipose, liver, kidney, and all three muscle types [40, 92]. Recent studies from our group and others implicate KLF15 as an important regulator of cardiac hypertrophy [27], glucose homeostasis [33] and adipocyte differentiation [33, 40]. With respect to SMCs, KLF15 is expression is robust at baseline but strongly attenuated following vascular injury or in response to pro-proliferative stimuli [93]. This is in marked contrast to the induction of KLF4 and KLF5 seen with vascular injury and indicates that KLF15 may have a distinct role in SMC biology. In contrast to the pro-proliferative effects of KLF5, overexpression of KLF15 in cultured SMC potently inhibits their proliferation. Consistent with this observation, mice deficient in KLF15 exhibit an exaggerated neointimal response to vascular injury. These studies implicate KLF15 as an important negative regulator of SMC proliferation [93]. Furthermore, the opposing effects of KLF5 and KLF15 in SMC biology suggests that the balance of these two factors may be an important determinant of the vascular injury response.
KLFs in Skeletal Muscle
Despite the initial description of the Drosophila kruppel protein as a critical determinant of myogenic fate in the developing fly [94], the role of KLFs in skeletal muscle is significantly less developed than in cardiac or smooth muscle. Although a number of KLFs have been identified as expressed in developing or mature skeletal muscle (KLF6 [95], KLF13 [47], KLF15 [40]), there are very few reports describing their regulation and potential roles in this tissue (Table 1) [40, 96]. Our group has demonstrated that KLF15 is robustly expressed in skeletal muscle where it regulates expression of the glucose transporter GLUT4. Recently, we have reported that KLF15(−/−) mice have severe fasting hypoglycemia which is caused by a defect in the ability of critical amino acids to enter the gluconeogenetic pathway in the liver [33] though the significance of this metabolic block in the heart or skeletal muscle has not been fully determined. KLF15 has also been shown to regulate the fasting-induced transcriptional activation of mitochondrial acetyl-CoA sythetase-2 in skeletal muscle [96]. Indeed, a deeper understanding of the expression, regulation and functional significance of KLFs in skeletal muscle has important implications for understanding the molecular pathways perturbed in diseases such as diabetes and metabolic syndrome, congestive heart failure, and peripheral arterial insufficiency [2–4].
Future Directions
The KLFs are emerging as important regulators of muscle biology with great pertinence to cardiovascular disease. Further investigation is needed to better define the spatiotemporal expression of the entire family of KLFs across these muscle tissues both developmentally and postnatally. In addition, the mechanisms by which the KLFs themselves are regulated are not well characterized. While there are multiple KLFs expressed in each tissue, their divergent versus redundant roles must be elucidated as the timing and relative expression of these potent transcription factors is a critical determinant of their physiologic function. As such, conditional gene targeting and inducible transgenesis will likely be necessary to dissect the precise roles of these factors in muscle biology in vivo. Defining target genes that are both induced and repressed by KLFs in each tissue type using pathway-based analysis will provide invaluable insights into the broad cellular processes that are modulated by each factor. Most importantly, a deeper understanding of the precise molecular mechanisms by which these regulators activate and repress gene expression (e.g. post-translational modification, domain mapping studies, identification of interaction partners, modulation of chromatin remodeling) will be of fundamental importance not only for cardiovascular biology, but also for a global understanding of gene expression and cell differentiation.
Nonstandard Abbreviations
- KLF
Kruppel-like factor
- PDGF
platelet derived growth factor
- TGFβ
transforming growth factor β
- MEF
myocyte enhancer factor
- HDAC
histone deacetylase
- MALDI-TOF
matrix-assisted laser desorption ionization / time-of-flight
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jain MK, Ridker PM. Anti-inflammatory effects of statins: clinical evidence and basic mechanisms. Nat Rev Drug Discov. 2005;4:977–87. doi: 10.1038/nrd1901. [DOI] [PubMed] [Google Scholar]
- 2.Ventura-Clapier R, Mettauer B, Bigard X. Beneficial effects of endurance training on cardiac and skeletal muscle energy metabolism in heart failure. Cardiovasc Res. 2007;73:10–8. doi: 10.1016/j.cardiores.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 3.Libera LD, Vescovo G. Muscle wastage in chronic heart failure, between apoptosis, catabolism and altered anabolism: a chimaeric view of inflammation? Curr Opin Clin Nutr Metab Care. 2004;7:435–41. doi: 10.1097/01.mco.0000134374.24181.5b. [DOI] [PubMed] [Google Scholar]
- 4.Finck BN, Kelly DP. PGC-1 coactivators: inducible regulators of energy metabolism in health and disease. J Clin Invest. 2006;116:615–22. doi: 10.1172/JCI27794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McKinsey TA, Olson EN. Toward transcriptional therapies for the failing heart: chemical screens to modulate genes. J Clin Invest. 2005;115:538–46. doi: 10.1172/JCI24144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heineke J, Molkentin JD. Regulation of cardiac hypertrophy by intracellular signalling pathways. Nat Rev Mol Cell Biol. 2006;7:589–600. doi: 10.1038/nrm1983. [DOI] [PubMed] [Google Scholar]
- 7.Gerthoffer WT. Mechanisms of vascular smooth muscle cell migration. Circ Res. 2007;100:607–21. doi: 10.1161/01.RES.0000258492.96097.47. [DOI] [PubMed] [Google Scholar]
- 8.Nuez B, Michalovich D, Bygrave A, Ploemacher R, Grosveld F. Defective haematopoiesis in fetal liver resulting from inactivation of the EKLF gene. Nature. 1995;375:316–8. doi: 10.1038/375316a0. [DOI] [PubMed] [Google Scholar]
- 9.Akazawa H, Komuro I. Roles of cardiac transcription factors in cardiac hypertrophy. Circ Res. 2003;92:1079–88. doi: 10.1161/01.RES.0000072977.86706.23. [DOI] [PubMed] [Google Scholar]
- 10.Adhikari N, Charles N, Lehmann U, Hall JL. Transcription factor and kinase-mediated signaling in atherosclerosis and vascular injury. Curr Atheroscler Rep. 2006;8:252–60. doi: 10.1007/s11883-006-0081-1. [DOI] [PubMed] [Google Scholar]
- 11.Epstein JA, Parmacek MS. Recent advances in cardiac development with therapeutic implications for adult cardiovascular disease. Circulation. 2005;112:592–7. doi: 10.1161/CIRCULATIONAHA.104.479857. [DOI] [PubMed] [Google Scholar]
- 12.Perry C, Soreq H. Transcriptional regulation of erythropoiesis. Fine tuning of combinatorial multi-domain elements. Eur J Biochem. 2002;269:3607–18. doi: 10.1046/j.1432-1033.2002.02999.x. [DOI] [PubMed] [Google Scholar]
- 13.Oettgen P. Regulation of vascular inflammation and remodeling by ETS factors. Circ Res. 2006;99:1159–66. doi: 10.1161/01.RES.0000251056.85990.db. [DOI] [PubMed] [Google Scholar]
- 14.Puigserver P, Spiegelman BM. Peroxisome proliferator-activated receptor-gamma coactivator 1 alpha (PGC-1 alpha): transcriptional coactivator and metabolic regulator. Endocr Rev. 2003;24:78–90. doi: 10.1210/er.2002-0012. [DOI] [PubMed] [Google Scholar]
- 15.Wei D, Kanai M, Huang S, Xie K. Emerging role of KLF4 in human gastrointestinal cancer. Carcinogenesis. 2006;27:23–31. doi: 10.1093/carcin/bgi243. [DOI] [PubMed] [Google Scholar]
- 16.Feinberg MW, Lin Z, Fisch S, Jain MK. An emerging role for Kruppel-like factors in vascular biology. Trends Cardiovasc Med. 2004;14:241–6. doi: 10.1016/j.tcm.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 17.Suzuki T, Aizawa K, Matsumura T, Nagai R. Vascular implications of the Kruppel-like family of transcription factors. Arterioscler Thromb Vasc Biol. 2005;25:1135–41. doi: 10.1161/01.ATV.0000165656.65359.23. [DOI] [PubMed] [Google Scholar]
- 18.Turner J, Crossley M. Basic Kruppel-like factor functions within a network of interacting haematopoietic transcription factors. Int J Biochem Cell Biol. 1999;31:1169–74. doi: 10.1016/s1357-2725(99)00067-9. [DOI] [PubMed] [Google Scholar]
- 19.Bieker JJ. Isolation, genomic structure, and expression of human erythroid Kruppel-like factor (EKLF) DNA Cell Biol. 1996;15:347–52. doi: 10.1089/dna.1996.15.347. [DOI] [PubMed] [Google Scholar]
- 20.Dang DT, Zhao W, Mahatan CS, Geiman DE, Yang VW. Opposing effects of Kruppel-like factor 4 (gut-enriched Kruppel-like factor) and Kruppel-like factor 5 (intestinal-enriched Kruppel-like factor) on the promoter of the Kruppel-like factor 4 gene. Nucleic Acids Res. 2002;30:2736–41. doi: 10.1093/nar/gkf400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nusslein-Volhard C, Wieschaus E. Mutations affecting segment number and polarity in Drosophila. Nature. 1980;287:795–801. doi: 10.1038/287795a0. [DOI] [PubMed] [Google Scholar]
- 22.Jackle H, Rosenberg UB, Preiss A, Seifert E, Knipple DC, Kienlin A, Lehmann R. Molecular analysis of Kruppel, a segmentation gene of Drosophila melanogaster. Cold Spring Harb Symp Quant Biol. 1985;50:465–73. doi: 10.1101/sqb.1985.050.01.058. [DOI] [PubMed] [Google Scholar]
- 23.Zuo P, Stanojevic D, Colgan J, Han K, Levine M, Manley JL. Activation and repression of transcription by the gap proteins hunchback and Kruppel in cultured Drosophila cells. Genes Dev. 1991;5:254–64. doi: 10.1101/gad.5.2.254. [DOI] [PubMed] [Google Scholar]
- 24.Gloor H. Arch. Julius-Klaus-Stift VererbForsch. 1950;25:38–44. [Google Scholar]
- 25.Miller IJ, Bieker JJ. A novel, erythroid cell-specific murine transcription factor that binds to the CACCC element and is related to the Kruppel family of nuclear proteins. Mol Cell Biol. 1993;13:2776–86. doi: 10.1128/mcb.13.5.2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perkins AC, Sharpe AH, Orkin SH. Lethal beta-thalassaemia in mice lacking the erythroid CACCC-transcription factor EKLF. Nature. 1995;375:318–22. doi: 10.1038/375318a0. [DOI] [PubMed] [Google Scholar]
- 27.Fisch S, Gray S, Heymans S, Haldar SM, Wang B, Pfister O, Cui L, Kumar A, Lin Z, Sen-Banerjee S, Das H, Petersen CA, Mende U, Burleigh BA, Zhu Y, Liao R, Jain MK. Kruppel-Like Factor 15 is a Novel Regulator of Cardiomyocyte Hypertrophy. Proc Nat Acad Sci. 2007 doi: 10.1073/pnas.0701981104. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shindo T, Manabe I, Fukushima Y, Tobe K, Aizawa K, Miyamoto S, Kawai-Kowase K, Moriyama N, Imai Y, Kawakami H, Nishimatsu H, Ishikawa T, Suzuki T, Morita H, Maemura K, Sata M, Hirata Y, Komukai M, Kagechika H, Kadowaki T, Kurabayashi M, Nagai R. Kruppel-like zinc-finger transcription factor KLF5/BTEB2 is a target for angiotensin II signaling and an essential regulator of cardiovascular remodeling. Nat Med. 2002;8:856–63. doi: 10.1038/nm738. [DOI] [PubMed] [Google Scholar]
- 29.Bhattacharya R, Senbanerjee S, Lin Z, Mir S, Hamik A, Wang P, Mukherjee P, Mukhopadhyay D, Jain MK. Inhibition of vascular permeability factor/vascular endothelial growth factor-mediated angiogenesis by the Kruppel-like factor KLF2. J Biol Chem. 2005;280:28848–51. doi: 10.1074/jbc.C500200200. [DOI] [PubMed] [Google Scholar]
- 30.Rowland BD, Bernards R, Peeper DS. The KLF4 tumour suppressor is a transcriptional repressor of p53 that acts as a context-dependent oncogene. Nat Cell Biol. 2005;7:1074–82. doi: 10.1038/ncb1314. [DOI] [PubMed] [Google Scholar]
- 31.SenBanerjee S, Lin Z, Atkins GB, Greif DM, Rao RM, Kumar A, Feinberg MW, Chen Z, Simon DI, Luscinskas FW, Michel TM, Gimbrone MA, Jr, Garcia-Cardena G, Jain MK. KLF2 Is a novel transcriptional regulator of endothelial proinflammatory activation. J Exp Med. 2004;199:1305–15. doi: 10.1084/jem.20031132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parmar KM, Larman HB, Dai G, Zhang Y, Wang ET, Moorthy SN, Kratz JR, Lin Z, Jain MK, Gimbrone MA, Jr, Garcia-Cardena G. Integration of flow-dependent endothelial phenotypes by Kruppel-like factor 2. J Clin Invest. 2006;116:49–58. doi: 10.1172/JCI24787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gray S, Wang B, Orihuela Y, Hong E, Fisch S, Haldar S, Cline G, Kim J, Peroni O, Kahn B, Jain M. Regualtion of Gluconeogenesis by Kruppel-like Factor 15. Cell Metabolism. 2007 doi: 10.1016/j.cmet.2007.03.002. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Das H, Kumar A, Lin Z, Patino WD, Hwang PM, Feinberg MW, Majumder PK, Jain MK. Kruppel-like factor 2 (KLF2) regulates proinflammatory activation of monocytes. Proc Natl Acad Sci U S A. 2006;103:6653–8. doi: 10.1073/pnas.0508235103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Feinberg MW, Cao Z, Wara AK, Lebedeva MA, Senbanerjee S, Jain MK. Kruppel-like factor 4 is a mediator of proinflammatory signaling in macrophages. J Biol Chem. 2005;280:38247–58. doi: 10.1074/jbc.M509378200. [DOI] [PubMed] [Google Scholar]
- 36.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–76. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 37.Lavallee G, Andelfinger G, Nadeau M, Lefebvre C, Nemer G, Horb ME, Nemer M. The Kruppel-like transcription factor KLF13 is a novel regulator of heart development. Embo J. 2006;25:5201–13. doi: 10.1038/sj.emboj.7601379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rajamannan NM, Subramaniam M, Abraham TP, Vasile VC, Ackerman MJ, Monroe DG, Chew TL, Spelsberg TC. TGFbeta inducible early gene-1 (TIEG1) and cardiac hypertrophy: Discovery and characterization of a novel signaling pathway. J Cell Biochem. 2007;100:315–25. doi: 10.1002/jcb.21049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sogawa K, Imataka H, Yamasaki Y, Kusume H, Abe H, Fujii-Kuriyama Y. cDNA cloning and transcriptional properties of a novel GC box-binding protein, BTEB2. Nucleic Acids Res. 1993;21:1527–32. doi: 10.1093/nar/21.7.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gray S, Feinberg MW, Hull S, Kuo CT, Watanabe M, Sen-Banerjee S, DePina A, Haspel R, Jain MK. The Kruppel-like factor KLF15 regulates the insulin-sensitive glucose transporter GLUT4. J Biol Chem. 2002;277:34322–8. doi: 10.1074/jbc.M201304200. [DOI] [PubMed] [Google Scholar]
- 41.Clerk A, Kemp TJ, Zoumpoulidou G, Sugden PH. Cardiac myocyte gene expression profiling during H2O2-induced apoptosis. Physiol Genomics. 2006 doi: 10.1152/physiolgenomics.00168.2006. [DOI] [PubMed] [Google Scholar]
- 42.Czubryt MP, Olson EN. Balancing contractility and energy production: the role of myocyte enhancer factor 2 (MEF2) in cardiac hypertrophy. Recent Prog Horm Res. 2004;59:105–24. doi: 10.1210/rp.59.1.105. [DOI] [PubMed] [Google Scholar]
- 43.Pikkarainen S, Tokola H, Kerkela R, Ruskoaho H. GATA transcription factors in the developing and adult heart. Cardiovasc Res. 2004;63:196–207. doi: 10.1016/j.cardiores.2004.03.025. [DOI] [PubMed] [Google Scholar]
- 44.Xu J, Gong NL, Bodi I, Aronow BJ, Backx PH, Molkentin JD. Myocyte enhancer factors 2A and 2C induce dilated cardiomyopathy in transgenic mice. J Biol Chem. 2006;281:9152–62. doi: 10.1074/jbc.M510217200. [DOI] [PubMed] [Google Scholar]
- 45.Liang Q, De Windt LJ, Witt SA, Kimball TR, Markham BE, Molkentin JD. The transcription factors GATA4 and GATA6 regulate cardiomyocyte hypertrophy in vitro and in vivo. J Biol Chem. 2001;276:30245–53. doi: 10.1074/jbc.M102174200. [DOI] [PubMed] [Google Scholar]
- 46.Molkentin JD, Markham BE. Myocyte-specific enhancer-binding factor (MEF-2) regulates alpha-cardiac myosin heavy chain gene expression in vitro and in vivo. J Biol Chem. 1993;268:19512–20. [PubMed] [Google Scholar]
- 47.Asano H, Li XS, Stamatoyannopoulos G. FKLF-2: a novel Kruppel-like transcriptional factor that activates globin and other erythroid lineage genes. Blood. 2000;95:3578–84. [PubMed] [Google Scholar]
- 48.Song A, Chen YF, Thamatrakoln K, Storm TA, Krensky AM. RFLAT-1: a new zinc finger transcription factor that activates RANTES gene expression in T lymphocytes. Immunity. 1999;10:93–103. doi: 10.1016/s1074-7613(00)80010-2. [DOI] [PubMed] [Google Scholar]
- 49.Scohy S, Gabant P, Van Reeth T, Hertveldt V, Dreze PL, Van Vooren P, Riviere M, Szpirer J, Szpirer C. Identification of KLF13 and KLF14 (SP6), novel members of the SP/XKLF transcription factor family. Genomics. 2000;70:93–101. doi: 10.1006/geno.2000.6362. [DOI] [PubMed] [Google Scholar]
- 50.Martin KM, Metcalfe JC, Kemp PR. Expression of Klf9 and Klf13 in mouse development. Mech Dev. 2001;103:149–51. doi: 10.1016/s0925-4773(01)00343-4. [DOI] [PubMed] [Google Scholar]
- 51.Feng D, Kan YW. The binding of the ubiquitous transcription factor Sp1 at the locus control region represses the expression of beta-like globin genes. Proc Natl Acad Sci U S A. 2005;102:9896–900. doi: 10.1073/pnas.0502041102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang P, Basu P, Redmond LC, Morris PE, Rupon JW, Ginder GD, Lloyd JA. A functional screen for Kruppel-like factors that regulate the human gamma-globin gene through the CACCC promoter element. Blood Cells Mol Dis. 2005;35:227–35. doi: 10.1016/j.bcmd.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 53.Ahn YT, Huang B, McPherson L, Clayberger C, Krensky AM. Dynamic interplay of transcriptional machinery and chromatin regulates “late” expression of the chemokine RANTES in T lymphocytes. Mol Cell Biol. 2007;27:253–66. doi: 10.1128/MCB.01071-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Grepin C, Dagnino L, Robitaille L, Haberstroh L, Antakly T, Nemer M. A hormone-encoding gene identifies a pathway for cardiac but not skeletal muscle gene transcription. Mol Cell Biol. 1994;14:3115–29. doi: 10.1128/mcb.14.5.3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Subramaniam M, Harris SA, Oursler MJ, Rasmussen K, Riggs BL, Spelsberg TC. Identification of a novel TGF-beta-regulated gene encoding a putative zinc finger protein in human osteoblasts. Nucleic Acids Res. 1995;23:4907–12. doi: 10.1093/nar/23.23.4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Subramaniam M, Gorny G, Johnsen SA, Monroe DG, Evans GL, Fraser DG, Rickard DJ, Rasmussen K, van Deursen JM, Turner RT, Oursler MJ, Spelsberg TC. TIEG1 null mouse-derived osteoblasts are defective in mineralization and in support of osteoclast differentiation in vitro. Mol Cell Biol. 2005;25:1191–9. doi: 10.1128/MCB.25.3.1191-1199.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Subramaniam M, Hefferan TE, Tau K, Peus D, Pittelkow M, Jalal S, Riggs BL, Roche P, Spelsberg TC. Tissue, cell type, and breast cancer stage-specific expression of a TGF-beta inducible early transcription factor gene. J Cell Biochem. 1998;68:226–36. [PubMed] [Google Scholar]
- 58.Tachibana I, Imoto M, Adjei PN, Gores GJ, Subramaniam M, Spelsberg TC, Urrutia R. Overexpression of the TGFbeta-regulated zinc finger encoding gene, TIEG, induces apoptosis in pancreatic epithelial cells. J Clin Invest. 1997;99:2365–74. doi: 10.1172/JCI119418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bensamoun SF, Hawse JR, Subramaniam M, Ilharreborde B, Bassillais A, Benhamou CL, Fraser DG, Oursler MJ, Amadio PC, An KN, Spelsberg TC. TGFbeta inducible early gene-1 knockout mice display defects in bone strength and microarchitecture. Bone. 2006;39:1244–51. doi: 10.1016/j.bone.2006.05.021. [DOI] [PubMed] [Google Scholar]
- 60.Tsubone T, Moran SL, Subramaniam M, Amadio PC, Spelsberg TC, An KN. Effect of TGF-beta inducible early gene deficiency on flexor tendon healing. J Orthop Res. 2006;24:569–75. doi: 10.1002/jor.20101. [DOI] [PubMed] [Google Scholar]
- 61.Rosenkranz S. TGF-beta1 and angiotensin networking in cardiac remodeling. Cardiovasc Res. 2004;63:423–32. doi: 10.1016/j.cardiores.2004.04.030. [DOI] [PubMed] [Google Scholar]
- 62.Funnell AP, Maloney CA, Thompson LJ, Keys J, Tallack M, Perkins AC, Crossley M. Erythroid kruppel-like factor directly activates the basic kruppel-like factor gene in erythroid cells. Mol Cell Biol. 2007;27:2777–90. doi: 10.1128/MCB.01658-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Davis CA, Haberland M, Arnold MA, Sutherland LB, McDonald OG, Richardson JA, Childs G, Harris S, Owens GK, Olson EN. PRISM/PRDM6, a transcriptional repressor that promotes the proliferative gene program in smooth muscle cells. Mol Cell Biol. 2006;26:2626–36. doi: 10.1128/MCB.26.7.2626-2636.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Owens GK, Kumar MS, Wamhoff BR. Molecular regulation of vascular smooth muscle cell differentiation in development and disease. Physiol Rev. 2004;84:767–801. doi: 10.1152/physrev.00041.2003. [DOI] [PubMed] [Google Scholar]
- 65.Ross R. The pathogenesis of atherosclerosis: a perspective for the 1990s. Nature. 1993;362:801–9. doi: 10.1038/362801a0. [DOI] [PubMed] [Google Scholar]
- 66.Ross R. Atherosclerosis--an inflammatory disease [see comments] New England Journal of Medicine. 1999;340:115–26. doi: 10.1056/NEJM199901143400207. [DOI] [PubMed] [Google Scholar]
- 67.Pislaru S, Simari RD. The translation of transcription. Circ Res. 2005;97:1083–4. doi: 10.1161/01.RES.0000194573.70503.b9. [DOI] [PubMed] [Google Scholar]
- 68.Jawien A, Bowen-Pope DF, Lindner V, Schwartz SM, Clowes AW. Platelet-derived growth factor promotes smooth muscle migration and intimal thickening in a rat model of balloon angioplasty. J Clin Invest. 1992;89:507–11. doi: 10.1172/JCI115613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ferns GA, Raines EW, Sprugel KH, Motani AS, Reidy MA, Ross R. Inhibition of neointimal smooth muscle accumulation after angioplasty by an antibody to PDGF. Science. 1991;253:1129–32. doi: 10.1126/science.1653454. [DOI] [PubMed] [Google Scholar]
- 70.Wei J, Gorman TE, Liu X, Ith B, Tseng A, Chen Z, Simon DI, Layne MD, Yet SF. Increased neointima formation in cysteine-rich protein 2-deficient mice in response to vascular injury. Circ Res. 2005;97:1323–31. doi: 10.1161/01.RES.0000194331.76925.5c. [DOI] [PubMed] [Google Scholar]
- 71.Hoshino Y, Kurabayashi M, Kanda T, Hasegawa A, Sakamoto H, Okamoto E, Kowase K, Watanabe N, Manabe I, Suzuki T, Nakano A, Takase S, Wilcox JN, Nagai R. Regulated expression of the BTEB2 transcription factor in vascular smooth muscle cells: analysis of developmental and pathological expression profiles shows implications as a predictive factor for restenosis. Circulation. 2000;102:2528–34. doi: 10.1161/01.cir.102.20.2528. [DOI] [PubMed] [Google Scholar]
- 72.Nagai R, Suzuki T, Aizawa K, Shindo T, Manabe I. Significance of the transcription factor KLF5 in cardiovascular remodeling. J Thromb Haemost. 2005;3:1569–76. doi: 10.1111/j.1538-7836.2005.01366.x. [DOI] [PubMed] [Google Scholar]
- 73.Sakamoto H, Sakamaki T, Kanda T, Hoshino Y, Sawada Y, Sato M, Sato H, Oyama Y, Nakano A, Takase S, Hasegawa A, Nagai R, Kurabayashi M. Smooth muscle cell outgrowth from coronary atherectomy specimens in vitro is associated with less time to restenosis and expression of a key Transcription factor KLF5/BTEB2. Cardiology. 2003;100:80–5. doi: 10.1159/000073043. [DOI] [PubMed] [Google Scholar]
- 74.Bafford R, Sui XX, Wang G, Conte M. Angiotensin II and tumor necrosis factor-alpha upregulate survivin and Kruppel-like factor 5 in smooth muscle cells: Potential relevance to vein graft hyperplasia. Surgery. 2006;140:289–96. doi: 10.1016/j.surg.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 75.Kawai-Kowase K, Kurabayashi M, Hoshino Y, Ohyama Y, Nagai R. Transcriptional activation of the zinc finger transcription factor BTEB2 gene by Egr-1 through mitogen-activated protein kinase pathways in vascular smooth muscle cells. Circ Res. 1999;85:787–95. doi: 10.1161/01.res.85.9.787. [DOI] [PubMed] [Google Scholar]
- 76.Miyamoto S, Suzuki T, Muto S, Aizawa K, Kimura A, Mizuno Y, Nagino T, Imai Y, Adachi N, Horikoshi M, Nagai R. Positive and negative regulation of the cardiovascular transcription factor KLF5 by p300 and the oncogenic regulator SET through interaction and acetylation on the DNA-binding domain. Mol Cell Biol. 2003;23:8528–41. doi: 10.1128/MCB.23.23.8528-8541.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sun R, Chen X, Yang VW. Intestinal-enriched Kruppel-like factor (Kruppel-like factor 5) is a positive regulator of cellular proliferation. J Biol Chem. 2001;276:6897–900. doi: 10.1074/jbc.C000870200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fujiu K, Manabe I, Ishihara A, Oishi Y, Iwata H, Nishimura G, Shindo T, Maemura K, Kagechika H, Shudo K, Nagai R. Synthetic retinoid Am80 suppresses smooth muscle phenotypic modulation and in-stent neointima formation by inhibiting KLF5. Circ Res. 2005;97:1132–41. doi: 10.1161/01.RES.0000190613.22565.13. [DOI] [PubMed] [Google Scholar]
- 79.Seo SB, McNamara P, Heo S, Turner A, Lane WS, Chakravarti D. Regulation of histone acetylation and transcription by INHAT, a human cellular complex containing the set oncoprotein. Cell. 2001;104:119–30. doi: 10.1016/s0092-8674(01)00196-9. [DOI] [PubMed] [Google Scholar]
- 80.Dai X, Segre JA. Transcriptional control of epidermal specification and differentiation. Curr Opin Genet Dev. 2004;14:485–91. doi: 10.1016/j.gde.2004.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jaubert J, Cheng J, Segre JA. Ectopic expression of kruppel like factor 4 (Klf4) accelerates formation of the epidermal permeability barrier. Development. 2003;130:2767–77. doi: 10.1242/dev.00477. [DOI] [PubMed] [Google Scholar]
- 82.Segre JA, Bauer C, Fuchs E. Klf4 is a transcription factor required for establishing the barrier function of the skin. Nat Genet. 1999;22:356–60. doi: 10.1038/11926. [DOI] [PubMed] [Google Scholar]
- 83.Hamik AL, Kumar Z, Balcells A, Sinha M, Katz Sumita, Feinberg J, Gerzsten M, Edelman RE, Jain MK. Kruppel-like Factor 4 Regualtes Endothelial Inflammation. Journal of Biological Chemistry. 2007 doi: 10.1074/jbc.M700078200. in press. [DOI] [PubMed] [Google Scholar]
- 84.Nakatake Y, Fukui N, Iwamatsu Y, Masui S, Takahashi K, Yagi R, Yagi K, Miyazaki J, Matoba R, Ko MS, Niwa H. Klf4 cooperates with Oct3/4 and Sox2 to activate the Lefty1 core promoter in embryonic stem cells. Mol Cell Biol. 2006;26:7772–82. doi: 10.1128/MCB.00468-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Liu Y, Sinha S, McDonald OG, Shang Y, Hoofnagle MH, Owens GK. Kruppel-like factor 4 abrogates myocardin-induced activation of smooth muscle gene expression. J Biol Chem. 2005;280:9719–27. doi: 10.1074/jbc.M412862200. [DOI] [PubMed] [Google Scholar]
- 86.King KE, Iyemere VP, Weissberg PL, Shanahan CM. Kruppel-like factor 4 (KLF4/GKLF) is a target of bone morphogenetic proteins and transforming growth factor beta 1 in the regulation of vascular smooth muscle cell phenotype. J Biol Chem. 2003;278:11661–9. doi: 10.1074/jbc.M211337200. [DOI] [PubMed] [Google Scholar]
- 87.Parmacek MS. Myocardin-related transcription factors: critical coactivators regulating cardiovascular development and adaptation. Circ Res. 2007;100:633–44. doi: 10.1161/01.RES.0000259563.61091.e8. [DOI] [PubMed] [Google Scholar]
- 88.Du KL, Ip HS, Li J, Chen M, Dandre F, Yu W, Lu MM, Owens GK, Parmacek MS. Myocardin is a critical serum response factor cofactor in the transcriptional program regulating smooth muscle cell differentiation. Mol Cell Biol. 2003;23:2425–37. doi: 10.1128/MCB.23.7.2425-2437.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pipes GC, Creemers EE, Olson EN. The myocardin family of transcriptional coactivators: versatile regulators of cell growth, migration, and myogenesis. Genes Dev. 2006;20:1545–56. doi: 10.1101/gad.1428006. [DOI] [PubMed] [Google Scholar]
- 90.Wang Z, Wang DZ, Pipes GC, Olson EN. Myocardin is a master regulator of smooth muscle gene expression. Proc Natl Acad Sci U S A. 2003;100:7129–34. doi: 10.1073/pnas.1232341100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Martin KM, Ellis PD, Metcalfe JC, Kemp PR. Selective modulation of the SM22alpha promoter by the binding of BTEB3 (basal transcription element-binding protein 3) to TGGG repeats. Biochem J. 2003;375:457–63. doi: 10.1042/BJ20030870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Uchida S, Tanaka Y, Ito H, Saitoh-Ohara F, Inazawa J, Yokoyama KK, Sasaki S, Marumo F. Transcriptional regulation of the CLC-K1 promoter by myc-associated zinc finger protein and kidney-enriched Kruppel-like factor, a novel zinc finger repressor. Mol Cell Biol. 2000;20:7319–31. doi: 10.1128/mcb.20.19.7319-7331.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gra SW, Croce B, Sakuma K, Chen M, Fisch Z, Haldar S, Simon S, Jain DIMK. Targeting of KLF15 reveals a critical role in the vascular smooth muscle cells response to injury (Abstract) AHA Scientific Sessions. 2006 [Google Scholar]
- 94.Ruiz-Gomez M, Romani S, Hartmann C, Jackle H, Bate M. Specific muscle identities are regulated by Kruppel during Drosophila embryogenesis. Development. 1997;124:3407–14. doi: 10.1242/dev.124.17.3407. [DOI] [PubMed] [Google Scholar]
- 95.Matsumoto N, Laub F, Aldabe R, Zhang W, Ramirez F, Yoshida T, Terada M. Cloning the cDNA for a new human zinc finger protein defines a group of closely related Kruppel-like transcription factors. J Biol Chem. 1998;273:28229–37. doi: 10.1074/jbc.273.43.28229. [DOI] [PubMed] [Google Scholar]
- 96.Yamamoto J, Ikeda Y, Iguchi H, Fujino T, Tanaka T, Asaba H, Iwasaki S, Ioka RX, Kaneko IW, Magoori K, Takahashi S, Mori T, Sakaue H, Kodama T, Yanagisawa M, Yamamoto TT, Ito S, Sakai J. A Kruppel-like factor KLF15 contributes fasting-induced transcriptional activation of mitochondrial acetyl-CoA synthetase gene AceCS2. J Biol Chem. 2004;279:16954–62. doi: 10.1074/jbc.M312079200. [DOI] [PubMed] [Google Scholar]