Abstract
Aligning lipid bilayers in nanoporous anodized aluminum oxide (AAO) is a new method to help study membrane proteins by electron paramagnetic resonance (EPR) and solid-state nuclear magnetic resonance (NMR) spectroscopic methods. The ability to maintain hydration, sample stability, and compartmentalization over long periods of time, and to easily change solvent composition are major advantages of this new method. To date, 1,2-dimyristoyl-sn-glycero-3-phosphatidylcholine (DMPC) has been the only phospholipid used for membrane protein studies with AAO substrates. The different properties of lipids with varying chain lengths require modified sample preparation procedures to achieve well formed bilayers within the lining of the AAO substrates. For the first time, the current study presents a simple methodology to incorporate large quantities of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphatidylcholine (POPC), DMPC, and 1,2-dipalmitoyl-3-sn-phosphatidylcholine (DPPC) phospholipids inside AAO substrate nanopores of varying sizes. 2H and 31P solid-state NMR were used to confirm the alignment of each lipid and compare the efficiency of alignment. This study is the first step in standardizing the use of AAO substrates as a tool in NMR and EPR and will be useful for future structural studies of membrane proteins. Additionally, the solid-state NMR data suggest possible applications of nanoporous aluminum oxide in future vesicle fusion studies.
Keywords: solid-state nuclear magnetic resonance spectroscopy, lipid bilayer, nanoporous aluminum oxide, nanotube arrays, bilayer nanotubes
1. Introduction
Solid-state nuclear magnetic resonance (NMR) spectroscopy is a widely used tool to study membrane proteins in their natural lipid environment that could not otherwise be probed using x-ray crystallography. A variety of pertinent information about integral membrane proteins can be obtained in aligned lipid bilayers [1, 2]. The incorporation of a protein into an aligned lipid bilayer reveals unique structural information such as peptide tilt angle and lipid chain dynamics [3]. 2H NMR spectroscopy of deuterated acyl chains of lipids is a powerful approach to monitor the dynamic properties inside lipid bilayers. [4]. Phosphorus NMR spectroscopy is a valuable technique for studying the alignment and degree of formation of lipid bilayers. A broad axially symmetric 31P NMR powder spectrum is obtained from unoriented lipid bilayers and consists of a σ ⊥ downfield and σ|| upfield. A bilayer that is aligned such that the membrane normal is perpendicular with respect to the static magnetic field, B0, will yield a sharp peak at σ ⊥ [5]. Using a combination of powder and aligned bilayers formation in AAO substrate provides a complete picture of both the surface and interior dynamics of the lipid bilayers.
The two most commonly used methods to align lipid bilayers are mechanical alignment on glass plates and magnetically aligned phospholipids bilayers (bicelles). Glass plate alignment is achieved by sandwiching lipids between thin glass plates and then hydrating the sample in a temperature controlled humidity chamber [6]. Drawbacks to this very common method include low sample stability which limits the length of sample viability; thus, the length of maximal NMR experimentation time, and a low maximum hydration achievable which affects the creation of a realistic cellular membrane mimicking environment. Aligned bicelles are prepared by mixing long and short chain phospholipids such as DMPC and DHPC (1,2-dihexanoyl-sn-glycero-3-phosphocholine) under optimized experimental conditions [7, 8]. Drawbacks to the bicelle alignment procedure include the inability to perform experiments requiring the removal or replacement of solvent after the bicelle sample is created, low sample stability of non ether-linked lipids after being heated, the inability of some lipids (such as POPC) to align in the magnetic field of the NMR spectrometer, and the limited range of alignment temperatures for a given bicelle composition [3].
Recently, nanoporous anodized aluminum oxide substrates have been used to align bilayers for use in NMR and electron paramagnetic resonance (EPR) spectroscopy [9–11]. The lipids were shown to self assemble in such a way that their bilayer normals are all perpendicular to the static magnetic field when the magnetic field is parallel to the long axis of the aluminum oxide pores. This alignment indicates that the bilayers form rings that line the inside of the pores (much like an onion) (see Figure 1 in Reference [10]). A structural study of lipid nanotube arrays by Gaede et al. demonstrated that the outermost lipid layer of each pore was most likely covalently attached to the substrate while the other lipid components were washed away by extrusion [12]. In addition to adhering to the surface, a model that the bilayer exists in a shape similar to the Golgi apparatus with gentle curves was suggested in this comprehensive biophysical study of POPC phospholipids in AAO substrate [12].
Since the discovery that bilayers could be aligned in AAO substrates, membrane proteins have been successfully incorporated into the AAO substrate lipid system for investigation with NMR and EPR spectroscopy [3, 10, 11, 13]. The surface area of exposed lipids in the nanopores is comparable to a glass plate preparation, but there are many advantages of using the AAO substrate system for protein alignment in a lipid bilayer [13]. These advantages include a long shelf life of hydrated sample (months), the ability to rapidly dehydrate and rehydrate with full sample viability, constant access to the bilayer solvent to add or remove it by flowing new solvents in, and the ability of samples to be frozen and thawed with no sample degradation.
The future development of AAO substrates as a platform for performing microarray analysis including small molecules screens and protein-protein interaction screens is very promising [14]. The compartmentalized nature of the AAO substrate in combination with its robust stability to sample degradation even when storing proteins in lipid poises it as a natural candidate for this growing field. Difficult to study membrane proteins like ion channels or metabotropic receptors involved in vesicle exo and endocytosis could be tested for interactions with small molecule drugs in a high throughput environment. New protein-protein interactions between a vesicle containing protein and a protein in a lipid filled nanopore could elucidate new integral membrane protein interactions and cellular signaling/recognition pathways [15]. Recently, fluorescence experiments have shown that protein-protein interactions can be monitored between an immobilized protein inside AAO substrate without lipid [14]. This first proof of principle experiment lays the groundwork for the logical extension of creating membrane bound protein microarrays and biosensors. The current study lays a foundation for standardized lipid bilayer preparations that could be used for such microarray analyses.
One major difficulty in the creation of AAO substrate bilayer samples is sample preparation in which more than one layer of lipid is present. To date, most studies of AAO substrates and lipids have used an extruder or some device to wash through the pores of the AAO substrate after lipid deposition or as a method to deposit the lipid. The high first lipid layer retention of the AAO substrate allows for a single easily deposited first layer. However, for NMR spectroscopy more than one lipid bilayer is desired.
In the current study, we discuss a straightforward method of forming lipid bilayers in AAO substrates. This sample procedure is very fast, easy to repeat, and reveals that more than one pore size and lipid composition may be aligned using the same technique with minor modifications. As biologically relevant lipid systems are varied and their choice often affect the protein structure being studied, applying the AAO substrate lipid alignment technique to new lipids will help future users of this technique [16].
2. Results and Discussion
In order to establish a consistent and reliable AAO lipid bilayer substrate sample preparation method, the viability of samples was assessed using 31P and 2H solid-state NMR spectroscopy. The most important difference between this sample preparation method and those used in many other procedures found in the literature is that there is no extruding or washing buffer through the pores that would remove all but one layer of lipid [12]. After publication of the first membrane protein incorporated into aligned bilayers embedded in AAO substrates by the Lorigan lab (Reference [11]), multiple requests for a detailed sample procedure were received. This current study addresses those requests by providing a simple sample preparation procedure to yield efficient and reproducible aligned lipid nanotube arrays optimized for maximum NMR signal intensity (see material and methods).
There are three important considerations when preparing an AAO lipid substrate sample; amount of buffer used, heating time, and degree of homogenization before lipid deposition. First, the amount of buffer used to homogenize each lipid was found to be particularly important. Each lipid was homogenized in different amounts of buffer that varied slightly from sample to sample. It may be a consequence of the need for a critical density of lipid vesicles to start vesicle fusion on a substrate that exact vesicle water ratios were needed. Indeed, the current investigation lends support to this study of vesicle fusion by Keller et al. which suggests the need for a critical density before vesicle fusion to a substrate [17]. The variability in buffer needed from sample to sample depends upon the condition in which the lipids dry. For example, clumps of lipid require additional buffer to be added after one or two freeze thaw cycles, while thin layers require only the initial amount of buffer. The additional buffer to the clumped lipids enhances homogenization of the lipids.
Heating time during both the initial lipid deposition step and the washing step were also varied depending upon phospholipid type. Presumably, the increase in heating times, which were needed with increased chain length, was caused by differences in lipid phase transition and viscosity. The higher phase transition temperature of the longer chain lipids indicate that more heat is needed to help the phospholipids go into the more fluid Lα phase in which the increased fluidity might help fuse the phospholipids into large lipid bilayers. Thus, it could be that more heat is needed with the longer lipid chains to help them form bilayers more completely. Alternatively, the added heat could help the longer chain phospholipids reduce their viscosity such that they could better flow into the pores of the AAO substrates [18]. In either case, the amount of heating time is a crucial factor that varies with chain length and must be considered when making AAO substrates with different types of phospholipids.
Third, the degree of homogenization of the sample is a variable parameter that plays a pivotal role in sample preparation. The simplest explanation of this observation is that there is a range of optimal sizes of vesicles for proper AAO substrate formation. Particularly, if too much sonication is performed, a fast moving component in the 31P NMR spectra (an isotropic peak) will appear revealing the presence of vesicles or micelles that have not been properly deposited into the AAO substrates (data not shown). This fast moving component most likely originates from small lipid vesicles or micelles that cannot form bilayers in the AAO substrate and float in excess buffer after having been fractionated too much by sonication. If the sample is not homogenized enough such that there are still small clumps of lipid, it can be concluded that not all of the lipids are small enough to fit in the pores.
Figure 1 demonstrates the alignment of each lipid bilayer system in 200 nm, 100 nm, and 20 nm pore diameter AAO substrates with 31P NMR spectroscopy. Each 31P NMR spectrum clearly shows one peak at σ ⊥ relative to the crushed AAO substrate powder pattern spectra for each phospholipid. Improper hydration caused a broadening of the peak and any improperly created plates caused the appearance of a shoulder to the left of the peak indicating the presence of more powder components. The smallest diameter pore has a slightly shifted σ ⊥ from the other pore diameters (σ ⊥ of 20 nm pore size is 2.4 ppm upfield of 100 nm and 200 nm pores). This small pore diameter σ ⊥ change was confirmed by comparing its powder spectrum shift (data not shown). One possible explanation is that the lipid headgroups in the smaller pore size may have a preferred curvature between the substrate and the lipid bilayer such that the smaller pore size stabilizes the lipids less. This lower stability might have caused an increased motion; thus, shrinking the 31P chemical shift anisotropy (CSA) and shifting the 31P σ ⊥ peak.
Figure 1.

31P NMR spectra of (A) POPC, (B) DMPC, and (C) DPPC bilayers in AAO substrates. Each lipid was tested in three different substrate condition; 200 nm and 100 nm AAO substrate pore size (solid line), 20 nm pore size (dashed), and crushed 200 nm AAO substrate (dotted line). For clarity, only one solid line is shown for the 200 nm and 100 nm AAO substrate because they are identical in lineshape.
Figure 2 compares the 31P NMR spectra of unoriented POPC lipid bilayers incorporated into AAO substrates (σ11=−19.8 ppm, σ22=−16.7 ppm, and σ33=31.4 ppm) and the powder multi-lamellar vesicle (MLV) (σ11=−15.8 ppm, σ22=−12.2 ppm, and σ33=31.8 ppm) of POPC bilayers. The AAO substrate is clearly shifted to the left when compared to the MLV sample. Considering the increased stability associated with being in a solid support environment, it is unlikely that the decrease in chemical shift of AAO is caused by a higher degree of motion of the bilayers. Films of aluminum oxide formed anodically from aluminum in a electrolytic conditions which contain many anions [19]. Specifically, in phosphate solutions, many PO3− anions gather on the outer parts of the film. It has been shown that charged anions affect the absolute value of the CSA by decreasing it (shifting the σ ⊥ peak to the left) [20, 21]. We propose that charges remaining from the preparation of the AAO substrate interact with the lipid bilayer and cause a slight shift in the 31P NMR spectrum of AAO substrate bound lipids, not an increase in motional dynamics.
Figure 2.
31P NMR spectra of POPC formed in crushed AAO substrates (solid line), aligned AAO substrate (dotted line), and multilamellar vesicles (dashed line).
Additionally, slight asymmetry is observed in the 31P aligned spectra of Figure 2 on the baseline. This might have arise from the gentle Golgi like curvature proposed by Gaede et al. [12]. The non-perfect perpendicular alignment of the bilayer normals at periodic points in the nanopore may have caused a small amount of non σ ⊥ components exhibited as an asymmetry in the peak.
An alternative explanation of the 31P shift of the 20 nm aligned peak to the left seen in Figure 1 follows from the conclusion that charges from the substrate affect the 31P CSA. It could be that there were more charges in the smaller pore diameter of the AAO substrate that were artifacts of aluminum oxide creation or that an increased proximity of the charges to the lipid bilayer due to the smaller pore volume lead to an increased anion effect on the CSA of the 20 nm pore AAO substrate. The observation that the 20 nm pore size has a different effect on the lipids has been seen in a much different context that might be caused by a similar mechanism. Specifically, polymophonuclear leukocytes (a neutrophil involved in immune response) were activated differently and their pseudopodia extended further when exposed to 20 nm pore size AAO substrates than the 200 nm pore diameter AAO substrate [22]. Thus, the mechanism or mechanisms that caused different headgroup dynamics in the phospholipids in AAO substrates such as charge distribution may also play a role in the use of alumina ceramics for biomedical devices and other cellular interfaces with nanoporous aluminum oxide.
Figure 3 shows 2H NMR spectra for each pore size (200 nm, 100 nm, and 20 nm) and each lipid (DPPC, DMPC, and POPC). Temperature profiles from 20 °C to 65 °C support normal phase temperature behavior and no out of the ordinary temperature affects (temperature series data not shown). This supports a recent DSC study indicating that the phase behavior of phospholipids in AAO substrates is not altered when compared to other bilayer preparation methods [23]. The size of the pore diameter did not affect the acyl chain order nor was there any major affect on the peak linewidths. Additionally, quadrupolar splittings (approximately 24 kHz) and hence molecular order in all lipids were comparable to glass plate preparation of aligned lipid bilayers further indicating that the acyl chains are not disturbed in the AAO substrate. The knowledge that regardless of the substrate pore size, the interior of the bilayer is unperturbed by the AAO substrate is important to have confirmed for future protein incorporations into AAO substrate. Thus, integral membrane proteins will not likely be disturbed by the presence of the AAO substrate.
Figure 3.
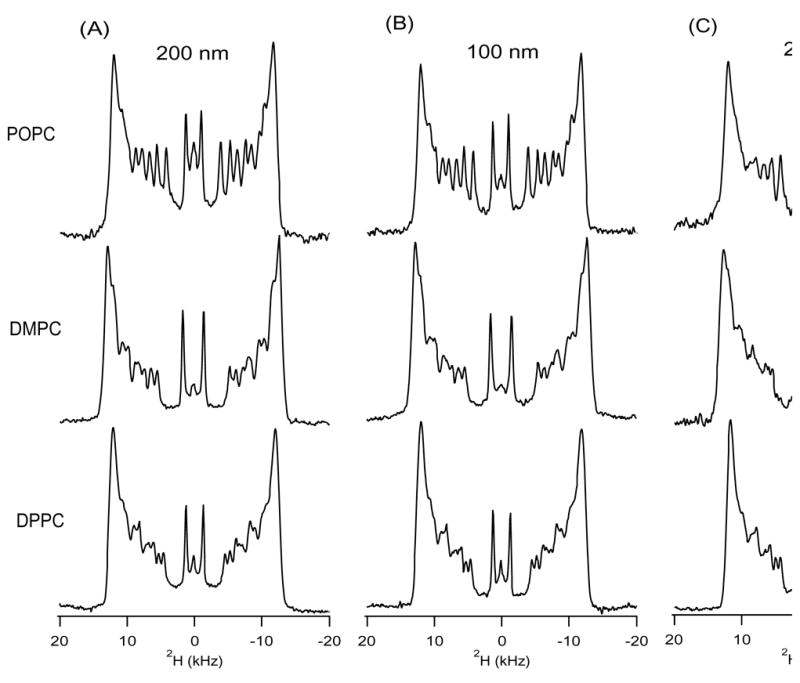
2H NMR spectra of fully acyl chain deuterated POPC (top), DMPC (middle), and DPPC (bottom). Different sizes of AAO substrate pores were tested; (A) 200 nm, (B) 100 nm, and (C) 20 nm. POPC and DMPC spectra were taken at 37 °C and the DPPC spectrum was taken at 50 °C.
However, a decrease in signal to noise is observed with decreasing pore size. This is most easily observed in the comparison of POPC lipids in the 200 nm pore substrate with POPC lipids in the 20 nm pore diameter substrate. Each plate has approximately an equal total pore volume when the pore density and size are considered (109, 1010, and 1011 pores/cm2 for 200 nm, 100 nm, and 20 nm diameter pores respectively). Therefore, the decrease in signal to noise is most likely caused by less sample forming bilayers confined within the inside of the pores. This suggests the possibility that lipid vesicle fusion is hindered in the small pores size when the homogenized lipid samples are deposited.
Two possible differences in pore diameter may have caused such a change in vesicle fusion; radius of bilayer curvature and charge interactions. Radius of curvature of a bilayer is a crucial but not well determined requirement for proper vesicle fusion [24, 25]. Changes in the radius of bilayer curvature caused by different pore diameters could be the source of the signal to noise differential associated with decreased pore diameter. Alternatively, the difference in charge distribution in the AAO substrate in the small nanopores versus the large nanopores may play an important role as van der Waal interactions are important determinants of vesicle attachment to solid substrates [26]. It is more likely that radius of curvature is responsible because the first layer of the bilayer is known to strongly attach to the substrate [12].
Furthermore, DMPC lipids exhibited a greater signal to noise ratio when compared to POPC lipids and DPPC lipids. It may be that the higher viscosity of the longer chain lipids caused them to stay in the pore during washing, forcing them to form bilayers in the heated NMR environment whereas the less viscous POPC lipid allowed non-fused lipid to flow out during the preparation of 20 nm plates (Reference [18] and personal observations). Another possibility is that the shorter chain lipids better adhere to the smaller pore and formed more uniform pore layers inhibiting any trapping of vesicles inside the pore that might fuse later. In either scenario, this circumstance should be considered when choosing a phospholipid to use for solid-state NMR experiments using AAO substrates.
In summary, 2H and 31P NMR spectroscopy demonstrated the proper perpendicular alignment of POPC, DMPC, and DPPC using the procedure standardized in the methods section. Sharp single 31P peaks at the σ ⊥ of the bilayers confirmed the presence of a uniform distribution of bilayers. Acyl chain dynamics demonstrated that the pore size does not affect the inner core of the bilayer. This is important for future integral membrane protein studies using an AAO substrate lipid system because changes in acyl chain dynamics may introduce artifacts in protein experiments. Three new phenomena were observed while optimizing the sample preparation procedure in this study. First, the σ ⊥ shift of 31P of 20 nm pores versus 200 nm pores indicates that the headgroups are affected differently by pore size, an important factor to be considered in the design of experiments to study surface membrane proteins. Second, the 31P σ ⊥ shift of the lipids in AAO substrates with respect to the MLVs shows the importance of charges created in the AAO substrate manufacturing and with further study may provide a mechanism of attachment of the lipids to the substrate as suggested in Gaede et al. [12]. Finally, the decreased S/N ratio in the 2H NMR spectra as the chain length decreased demonstrated that vesicle binding and fusion is dependent upon the lipid type (properties such as viscosity may be primary factors) and the radius of curvature of the inner bilayer. This study provides a detailed procedure for forming aligned POPC, DMPC, and DPPC lipid bilayers in varying pore size AAO substrates in addition to presenting new factors to consider when using these nanotube arrays for membrane protein studies. Finally, the current study has also paved the way for future experimentation that could use the different lipids presented here and different pore diameters to explore the curvature and lipid length effects on vesicle fusion events, an essential part of biology [25].
3. Experimental
3.1 Materials
DMPC (1,2-dimyristoyl-sn-glycero-3-phosphatidylcholine), DPPC (1,2-dipalmitoyl-sn-glycero-3-phophatidylcholine), and POPC (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphatidylcholine) were purchased from Avanti Polar Lipids (Alabaster, AL). All phospholipids were dissolved in chloroform and stored at −20 °C prior to use. TRIS (tris[hydroxymethyl]aminomethane) was purchased from Sigma/Aldrich. Deuterium-depleted water, deuterated d31-POPC, d54-DMPC, and d62-DPPC were purchased from Isotec (Miamisburg, OH). AAO substrate was purchased from Whatman (England). A small suctioning vaccum tool was purchased from Laboratory Supplies Co. Inc. (Hicksville, NY).
3.2 Buffer, lipid, and AAO substrate preparation
Each phospholipid required a slightly different sample preparation procedure. Approximately 1 mg of lipid was deposited on each AAO substrate plate by the end of the sample preparation. The drying procedure was common to all lipids. 1 mL of 20 mg/mL dissolved lipid was aliquoted in a 12mm × 75mm test tube. A constant stream of N2 gas was applied into the test tube such that the lipid dried in the lowest part of the test tube. Care was taken to not trap chloroform below a layer of dried lipid at the bottom of the test tube. Small amounts of leftover, trapped chloroform created poor samples. Tilting and rotating the test tube while drying helped to prevent chloroform trapping. The samples were left under N2 gas for one hour before being placed in a vacuum dessiccator overnight.
A TRIS buffer (pH=7, 10 mM) was prepared using deuterium depleted water and adjusted to pH=7. 150–180 μL buffer (11.1%–13.3% [w/v]), 185–195 μL (10.3%–10.8% [w/v]) buffer, and 210–230 μL (8.7%–9.5% [w/v]) buffer was added to the POPC, DMPC, and DPPC lipid samples, respectively. Each sample was vortexed for two minutes followed by two freeze thaw cycles in which hot tap water was used to warm each sample after freezing with liquid nitrogen. This procedure was repeated 3–6 times until the sample was completely homogeneous (composition was smooth and milky white). Depending upon how the lipid dried, 5–10 minutes of sonication helped homogenize the samples instead of vortexing during one or two freeze thaw cycles. Additionally, to homogenize the viscous DPPC samples, 210 μL of buffer was first added and if needed later, after a freeze thaw cycle, 10 μL buffer aliquots were added until fully homogenized. 20 5mm × 8mm rectangular pieces of AAO substrate were cut with razor blades. Uniformity in substrate size was found to be critical to maintaining sample hydration later. All plates were placed on a plastic Petrie dish for the rest of the sample preparation.
3.3 Lipid deposition on AAO substrate and wiping of excess lipid
In a 40 °C oven, the cut AAO substrates were pre-warmed. Uniformity during the following process is critical to successful sample preparation. If one plate is not prepared properly (too dry, lipid clogging the pores, lipid not wiped off well enough, etc.), the sample will have significant non-aligned portions when observed with 31P or 2H solid-state NMR spectroscopy. Consequently, the following sample preparation tips help achieve maximal lipid incorporation into the pores with no dry plates, no pores blocked by lipids, and no lipids outside of the pores.
With a pipette, two 5 μL aliquots of freshly homogenized lipid were added to each substrate close enough to each other to touch and mix. The pipette tip was used to touch the AAO substrate below the droplets so that the surface tension would break and water would flow all over the AAO substrate (the substrate looks wet and clear after water had infused it). All 20 plates were then put into a 40 °C oven for 4, 5, and 6–7 minutes for POPC, DMPC, and DPPC respectively. This is one of the two crucial steps for good bilayer formation. The heat of the oven helps form the bilayers and lets them flow easily into the pores of the substrate. The humidity of the environment dictates the exact time needed to heat each lipid. The exact times of drying varied up to one minute from day to day depending upon the exact environment humidity and temperature. The ideal time to remove the plates is right before any lipid dries on the substrate but not too soon that the sample is still glistening from being too wet.
Immediately after removal from the oven, the excess lipid not inside the pores was wiped gently with a folded kimwipe. The best samples were obtained when, after wiping, the spot on each substrate where the drops of lipid were initially placed were clearly still wet while the surrounding substrate areas were dry.
Because of the high phase transition temperature (45 °C) of DPPC it was necessary to do extra preparatory steps in this part of the sample preparation. This included keeping the homogenized lipid in a hot water bath while depositing it and placing the Petrie dish holding the substrate on top of a beaker of steaming water to constantly heat the substrates. This step increased the phospholipid viscosity so that the DPPC flows easily into the pores. The Petrie dish with steam was used for the duration of the DPPC preparation. Additionally, the higher temperature for DPPC was needed during all steps of sample preparation and during the NMR experiments due to the higher phase transition temperature of DPPC when compared to DMPC and POPC [27].
3.4 Washing the substrate and stacking the plates
Immediately after wiping the excess lipid from the top of each substrate, 7 μL of deuterium depleted water was added to the center of each plate. The pipette was touched to the substrate through the water droplet as was done before during lipid deposition to make sure that the water fully hydrated each substrate. The hydrated substrate was then heated in the 40 °C oven for 2 minutes, 2 minutes, and 3 minutes for POPC, DMPC, and DPPC respectively. The purpose of this heating is to again help the bilayers form properly in the pores. Also note that these times may very up to 30 seconds depending upon environmental conditions such as temperature and humidity.
After removal from the oven, each plate still glistened from excess water. Using the corner of a kimwipe, each sample was very quickly dabbed to remove almost all excess water. This second step is crucial to the creation of well-aligned lipid bilayers. If the plates were not uniformly wet after slightly dabbing, then they were never uniformly hydrated when they were stacked. Within seconds after dabbing, the plates began to dry to the point when they lose the water suctioned between them and the Petrie dish and turn from visibly wet (clear, not glistening) to only slightly clear but distinguishable still by the fact that where the original drops of lipid were placed is wetter than the rest of the plate. This state disappeared and the plate dried within 20 seconds and it was in this time that the plate was picked up with a small suctioning vacuum tool and placed on comparably sized glass plate on parafilm. If the dabbing and water application was uniform, then all the plates went into this hydration state within one or two minutes allowing for a constant stacking of plates such that the hydration of the previous stacked substrate would be preserved in between two other hydrated plates. After stacking, one 3 μL aliquot of deuterium depleted water was placed on the top substrate followed by another glass plate. The stack of 20 plates was then wrapped tightly with parafilm. Finally, any plates that did not dry properly, that stuck too much, or were out of the ordinary for any reason (usually 2–3 of 20 plates) were never used.
3.5 Powder sample and multi lamellar vesicle preparation
The powder AAO lipid samples were prepared by crushing the stacked AAO substrates after the above preparation procedure with a mortar and pestle. Parafilm was used to contain the unoriented lipid sample. Multilamellar vesicles were prepared by placing the homogenized lipid mixtures inside a small bag that was used inside the same flat coil solid-state NMR probe for consistency.
3.6 Solid-state NMR Spectroscopy
All NMR experiments were conducted on a 11.7 T Bruker AVANCE 500 MHz wide bore solid-state NMR spectrometer. All spectra were taken with a Bruker double resonance flat coil solid-state NMR probe of dimension 8.5 mm × 14 mm. The resonance frequencies used for 2H and 31P, 1H were 76.8 MHz and 202.4 MHz, and 500.1 MHz respectively. The 2H NMR spectra were recorded using a standard quadrupole-echo pulse sequence (3.0 μs 90° pulses, and a 0.4 s recycle delay). For the 2H NMR data, 8000 scans were taken and the free induction decay was processed using 100 Hz of exponential line broadening. The 31P NMR spectra were recorded with 6.5 dB 1H decoupling using a 7.0 μs 90° pulse and a 4 s recycle delay. For the 31P NMR data, 1024 scans were averaged and processed with 100 Hz of exponential line broadening. H3PO4 (85%) was used to reference all 31P chemical shifts to 0 ppm. Topspin software (Bruker Biospin, Billerica MA) was used for data analysis.
Figure 4.

Diagram representation of the sample preparation procedure; Step 1-dry lipid in test tube, Step 2-dissolve lipid in buffer and add two drops to each plate then heat in oven, Step 3-wipe plate with kimwipe, Step 4-add pure buffer to each plate and heat in oven, Step 5-wipe a second time, Step 6-when a change from translucency to opacity occurs use small vacuum to stack each plate, Step 7-wrap stack in parafilm after adding a drop of water to the top plate and a glass plate to the top, Step 8-put in NMR spectrometer. The diagram after step 8 illustrates the alignment of lipid bilayers in one pore of the nanoporous aluminum oxide. The bilayer normals are perpendicular to the magnetic field B0. Note that more layers of concentric rings of bilayer may be present in a pore and were omitted for clarity.
Acknowledgments
The 500 MHz wide bore NMR spectrometer was obtained from National Science Foundation Grant (No. 10116333). Prof. Gary A. Lorigan acknowledges funding from ACS PRF type AC (45340). E.S.K. acknowledges support from the Arnold Beckman Foundation, the Astronaut Scholars Program, Miami University, and the Barry M. Goldwater Foundation.
Abbreviations
- NMR
nuclear magnetic resonance
- DMPC
1,2-dimyristoyl-sn-glycero-3-phosphatidylcholine
- POPC
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphatidylcholine
- DPPC
1,2-dipalmitoyl-3-sn-phosphatidylcholine
- AAO
anodized aluminum oxide
- DHPC
1,2-dihexanoyl-sn-glycero-3-phosphocholine
- CSA
chemical shift anisotropy
- EPR
electron paramagnetic resonance
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Karp ES, Tiburu EK, Baker SA, Lorigan GA. Determination of secondary structure of the integral membrane protein, phospholamban, using solid-state NMR spectroscopy. BBA Biomembranes. 2006;1758:772–780. doi: 10.1016/j.bbamem.2006.04.016. [DOI] [PubMed] [Google Scholar]
- 2.De Angelis AA, Nevzorov AA, Park SH, Howell SC, Mrse AA, Opella SJ. High-resolution NMR spectroscopy of membrane proteins in aligned bicelles. J Amer Chem Soc. 2004;126:15340–15341. doi: 10.1021/ja045631y. [DOI] [PubMed] [Google Scholar]
- 3.Chekmenev EY, Gor’kov PL, Cross TA, Alaouie AM, Smirnov AI. Flow-through lipid nanotbue arrays for structure-function studies of membrane proteins by solid-state NMR spectroscopy. Biophys J. 2006;91:3076–3084. doi: 10.1529/biophysj.106.085191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seelig J. Deuterium magnetic resonance: theory and applications to lipid membranes. Q Rev Biophys. 1977;10:353–418. doi: 10.1017/s0033583500002948. [DOI] [PubMed] [Google Scholar]
- 5.Seelig J. 31P nuclear magnetic resonance and the head group structure off phospholipids in membranes. Biochim Biophys Acta. 1978;515:105–140. doi: 10.1016/0304-4157(78)90001-1. [DOI] [PubMed] [Google Scholar]
- 6.Tiburu EK, Karp ES, Dave PC, Damodaran K, Lorigan GA. Probing the dynamics and side chain motion of leucine residue in phospholamban using 2H and 15N solid-state NMR spectroscopic techniques. Biochemisty. 2004;43:13899–13909. doi: 10.1021/bi0490993. [DOI] [PubMed] [Google Scholar]
- 7.Sanders CR, Hare BJ, Howard KP, Prestegard JH. Magnetically-oriented phospholipid micelles as a tool for the study of membrane-associated molecules. Prog NMR Spect. 1994;26:421–444. [Google Scholar]
- 8.Nieh M, Raghunathan VA, Glinka CJ, Harroun TA, Pabst G, Katsara J. Magnetically alignable phase of phospholipid “Bicelle” mixtures is a chiral nematic made up of wormlike micelles. Langmuir. 2004;20:7893–7897. doi: 10.1021/la048641l. [DOI] [PubMed] [Google Scholar]
- 9.Smirnov AI, Poluektov OG. Substrate-supported lipid nanotube arrays. J Amer Chem Soc. 2003;125:8434–8435. doi: 10.1021/ja0349406. [DOI] [PubMed] [Google Scholar]
- 10.Karp ES, Inbaraj JJ, Laryukhin M, Lorigan GA. Electron paramagnetic resonance studies of an integral membrane protein inserted into aligned phospholipid bilayer nanotube arrays. J Amer Chem Soc. 2006;128:12070–12071. doi: 10.1021/ja064077k. [DOI] [PubMed] [Google Scholar]
- 11.Lorigan GA, Dave PC, Tiburu EK, Damodaran K, Abu-Baker S, Karp ES, Gibbons WJ, Minto RE. Solid-state NMR spectroscopic studies of an integral membrane protein inserted into aligned phospholipid bilayer nanotube arrays. J Amer Chem Soc. 2004;126:9504–9505. doi: 10.1021/ja047317+. [DOI] [PubMed] [Google Scholar]
- 12.Gaede HC, Luckett KM, Polozov IV, Gawrisch K. Multinuclear NMR studies of single lipid bilayers supported in cylindrical aluminum oxide nanopores. Langmuir. 2004;20:7711–7719. doi: 10.1021/la0493114. [DOI] [PubMed] [Google Scholar]
- 13.Chekmenev EY, Hu J, Gor’kov PL, Brey WW, Cross TA, Ruuge A, Smirnov AI. 15N and 31P solid-state NMR study of transmembrane domain alignment of M2 protein of influenza A virus in hydrated cylindrical lipid bilayers confined to anodic aluminum oxide nanopores. J Magn Reson. 2005;173:322–327. doi: 10.1016/j.jmr.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 14.Takmakov P, Vlassiouk I, Smirnov AI. Application of anodized aluminum in fluorescence detection of biological species. Anal Bioanal Chem. 2006;385:954–958. doi: 10.1007/s00216-006-0504-4. [DOI] [PubMed] [Google Scholar]
- 15.Predki PF. Functional protein microarrays: ripe for discovery. Curr Opin Chem Biol. 2004;8:8–13. doi: 10.1016/j.cbpa.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 16.Brown DA, London E. Structure and origin of ordered lipid domains in biological membranes. J Membrane Biol. 1998;164:1103–1114. doi: 10.1007/s002329900397. [DOI] [PubMed] [Google Scholar]
- 17.Keller CA, Glasmastar K, Zhdanov VP, Kasemo B. Formation of supported membranes from vesicles. Phys Rev Lett. 2000;84:5443–5446. doi: 10.1103/PhysRevLett.84.5443. [DOI] [PubMed] [Google Scholar]
- 18.Liew KY, Seng CE, Ng BH. Viscosities of long chain n-alcohols from 15 to 80 degrees C. J Soln Chem. 1993;22:1033–1040. [Google Scholar]
- 19.Takahash H, Fujimoto K, Konno H, Nogayama M. Distribution of anions and protons in oxide films formed anodically on aluminum in a phosphate solution. J Electochemical Society. 1983;131:1856–1861. [Google Scholar]
- 20.Macdonald PM, Seelig J. Anion Binding to Neutral and Positively Charged Lipid Membranes. Biochemistry. 1988;27:6769–6775. doi: 10.1021/bi00418a019. [DOI] [PubMed] [Google Scholar]
- 21.Scherer PG, Seelig J. Electric charge effects on phospholipid headgroups. Phosphatidylcholine in mixtures with cationic and anionic amphiphiles. Biochemistry. 1989;28:7720–7728. doi: 10.1021/bi00445a030. [DOI] [PubMed] [Google Scholar]
- 22.Karlsson M, Johansson A, Tang L, Boman M. Nanoporous aluminum oxide affects neutrophil behaviour. Microscopy research and technique. 2004;63:259–265. doi: 10.1002/jemt.20040. [DOI] [PubMed] [Google Scholar]
- 23.Alaouie AM, Smirnov AI. Cooperativity and kinetics of phase transitions in nanopore-confined bilayers studied by differential scanning calorimetry. Biophys J. 2004 doi: 10.1529/biophysj.104.056523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hamai C, Yang T, Kataoka S, Cremer PS, Siegfried MM. Effect of average phospholipid cuvature on supported bilayer formation on glass by vesicle fusion. Biophys J. 2006;90:1241–1248. doi: 10.1529/biophysj.105.069435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gruner SM. Membrane fusion: Caught in the act. Science. 2002;13:1817–1818. doi: 10.1126/science.1076394. [DOI] [PubMed] [Google Scholar]
- 26.Cremer PS, Boxer SG. Formation and spreading of lipid bilayers on planar glass supports. J Phys Chem B. 1999;103:2554–2559. [Google Scholar]
- 27.Cevc G. How membrane chain-melting phase-transition temperature is affected by the lipid chain asymmetry and degree of unsaturation: An effective chain-length model. Biochemistry. 1991;20:7186–7193. doi: 10.1021/bi00243a021. [DOI] [PubMed] [Google Scholar]



