Abstract
Cell biology research encompasses everything from single cells to whole animals. Recent discoveries concerning particular gene functions can be applied to the whole animal for understanding genotype-phenotype relationships underlying disease mechanisms. For this reason, genetically manipulated mouse models are now considered essential to correctly understand disease processes in whole animals. This unit provides the basic mouse technologies used to generate conventional transgenic mice, which represents gain-of-function approach. First, an overview of the transgenic construct design is presented. This unit then explains basic strategies for the identification and establishment of independent transgenic mouse lines, followed by comments on historical and emerging techniques, and then on typical problems that are encountered when researchers start to generate transgenic mice.
Keywords: transgenic mice, plasmid vector, constructs, gene expression, reporter gene
Introduction
Genetically manipulated mouse models are now considered essential for studying gene functions in whole animals. Gene knockout mice represent a loss-of-function strategy, but the transgenic mice discussed in this unit represent the gain-of-function approach to defining molecular and cellular functions of a gene of interest. This approach can be also used to analyze tissue-specific or developmental stage-specific gene expression by introducing reporter genes, such as β-galactosidase (lacZ) or green fluorescent protein (GFP), under the control of a specific gene promoter. If the functional domain of a gene of interest is well characterized, it is possible to create transgenic mice with gene ablation by introducing the dominant negative forms of the gene into the mouse genome. Typically, transgenic mice are generated by microinjecting the transgenic construct into a fertilized egg (oocyte or zygote). An alternative way to effectively introduce a transgene into an egg is the use of a retrovirus vector. However, the retroviral method has limitations: high titers must be used to obtain the vector, mosaic mice can be generated that do not carry the transgene in each and every cell. Another method to generate transgenic mice is to transfect a transgenic construct into mouse embryonic stem (ES) cells and then inject these cells into mouse blastocysts. This method is particularly useful to obtain a low copy number of the transgene in the mice and also when embryonic lethality is expected in the resultant transgenic mice. Chimeric mice generated by the ES cell transfer method may be able to transmit the transgene to embryos, allowing analysis of transgenic embryos in the dams that have mated with the chimera.
This unit presents the most widely used strategies for generating plasmid-based transgenic constructs to be injected into fertilized mouse eggs, identifying of potential founders by genotyping, and establishing of separate founder lines. Frequent problems, which will be encountered when researchers start to generate transgenic mice, will be discussed in the Commentary sections.
Design of the Transgenic Construct
Like other plasmid constructs used to express proteins in a cell culture system, transgenic constructs must have all the critical elements for gene expression to occur, such as promoters, introns, the protein coding sequence of interest (cDNA or genome) and a poly (A) site (Figure 1A). A cloning strategy should be clearly planned with a good understanding of each functional element so as to obtain the expected expression of a transgene. Well-defined expression in a cell line is certainly a good indicator that a transgene was properly constructed, although this will not guarantee transgene expression in vivo. A transgene must be excised out of the vector backbone prior to injection into mouse eggs (see following unit). Unnecessary sequences from a plasmid vector (derived from prokaryotic sequences) should be removed as much as possible to increase the probability of transgene expression in mice, although the expression seems unaffected by up to 100 extra bases. Inclusion of extra sequences between each element of a transgene does not usually affect the transgene's function as long as some regulatory elements are not inadvertently created. To release a transgenic cassette from a vector, the construct must be designed with unique restriction enzyme sites flanking the transgenic cassette (Figure 1A). The size of a transgene does not affect efficiency of generating transgenic mice.
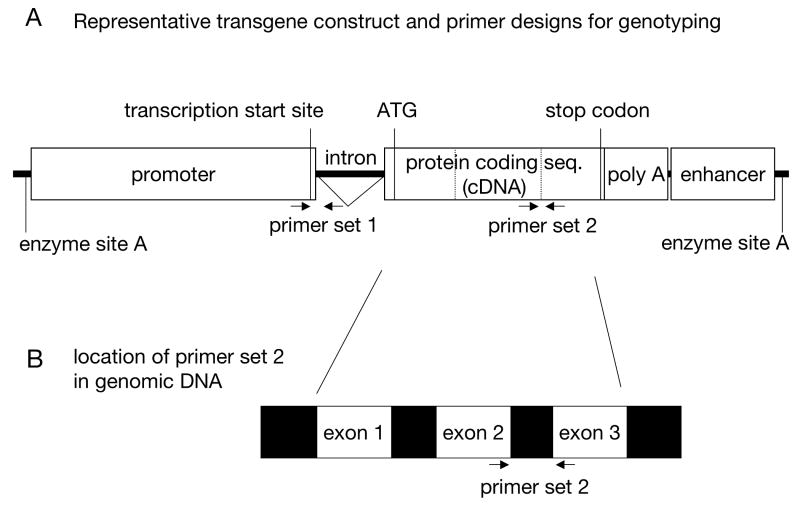
Figure 1.
Representative transgene construct and primer designs for genotyping. (A) Typical transgene construct is depicted, consisting of promoter, intron, protein coding sequence (cDNA) from a gene of interest, poly (A) seqence, and enhancer sequences. The promoter sequence normally contains the transcriptional start site, although there are variations. The inclusion of an intron leads to a greater percentage of active transgene expression in mouse lines. The protein coding sequence (cDNA) needs to have a translation start site, typically consisting of a Kozak consensus sequence in addition to an ATG start codon. The entire fragment can be cut out from the vector backbone by a single restriction enzyme (site A) prior to zygote injection. A restriction enzyme site that can be cut only once inside of the fragment should be considered at the construct designing stage for future genotyping by Southern blotting. Primer set 1, designed at the junction of 2 different elements, can amplify a unique sequence. (B) Alternative strategy of the primer design. The primers for genotyping can also be designed in cDNA to bridge between 2 or more different exons so that the endogenous gene would be amplified with a larger size at low efficiency. ATG, translation start codon; stop, translation stop codon; seq., sequence.
Promoter
A promoter is a critical sequence required for regulating the spatial and temporal expression pattern of a transgene. Promoter sequences are isolated from upstream regions of endogenous mammalian genes. A promoter sequence normally includes a transcriptional start site as well as transcription regulatory sequences. Many promoters have been reported that can successfully achieve tissue- specific and developmental stage-specific expression of transgenes. Database search such as PubMed or International Mouse Strain Resource (See Internet resources with annotations in this unit) is preferable way to find out the promoters of tissue or cell type of interest. Promoters should be tested to determine if they contain the appropriate transcriptional regulatory elements. For example, transgenic mice harboring 0.9 kb of the type I collagen (Col1a1) promoter expressed the transgene at relatively low levels almost exclusively in the skin. On the other hand, mice with a sequence containing 2.3 kb of this proximal promoter expressed the transgene at high levels in mineralized tissues, but not in other type I collagen-producing cells. Transgenic mice harboring 3.2 kb of the proximal promoter showed an additional high-level expression of the transgene in tendon and fascia fibroblasts (Rossert et al. 1995).
The best way to choose a promoter to generate transgenic mice is to review original papers that examined endogenous expression patterns. To prevent any significant difference from the original work, investigators should ask the group that published the work to provide the vector with detailed information about the promoter sequence. Inducible expression of a transgene in mice is possible by utilizing inducible promoters. For example, the metallothionein-1 promoter is induced by heavy metals like zinc (Mayo et al. 1982; Sumarsono et al. 1996), and interferon-inducible gene expression has been reported for Cre (P1 bacteriophage cyclization recombinase) in transgenic mice (Kühn et al. 1995). Tetracycline, used for tightly regulated expression of a transgene, is the most frequently used promoter and activator of binary systems (Gossen and Bujard 1992). In this system, the expression of a fusion protein consisting the viral transactivation domain and the prokaryotic tetracycline repressor (tetR) domain, is controlled by a tissue-specific promoter. Because the fusion protein binds to both tetracycline and the operator sequences (tetO) of the tet operon in the target transgene, the target transgene can be controlled by a minimal promoter that contains only a TATA box, depending on the tetracycline concentrations in tissues. There are 2 options in this system: in the first one, the tetracycline-controlled transactivator (tTA) cannot bind to DNA when the tetracycline is present (tet-off), whereas in the second one, the reverse tTA (rtTA) binds to DNA only when tetracycline is present (tet-on).
Protein coding sequence and transcription start site
A protein coding sequence is usually a full-length cDNA derived from the RNA of a gene of interest. This sequence normally contains a translational start codon (ATG), a translational stop codon, and a Kozak sequence upstream of the start codon so that the ribosome can scan and recognize the proper translation start and stop sites on mRNA. An authentic 10-15 nucleotides sequence found at the 5′ end of the start codon in a gene of interest can be assumed to have appropriate translational start signals. Alternatively, a Kozak consensus sequence such as GCCGCC (G/A) NN ATG G could be incorporated as a translational start site (Kozak 1987). The sequence between the transcriptional start site and the translational start site should be checked for inadvertent ATG start codons, stop codons, or other potential regulatory elements created during construction of a transgene.
Adding protein tag sequences such as VSV, 6xHis, HA, V5, or Myc to the transgene may be useful to detect transgene expression because reliable antibodies to these epitopes are available. Additionally, a tag sequence provides a way to differentiate between endogenous gene expression and transgene expression. It is important to avoid disruption of the transgene function while adding a tag sequence. Introducing an internal ribosomal entry site (IRES) followed by a reporter gene such as lacZ or GFP is also useful for checking the expression pattern and for genotyping, although the efficiency of IRES-mediated bicistronic expression may require additional studies.
If cDNA cloning is difficult because of highly repetitive sequences, and if a genomic clone containing all of the required exons and introns is available, the promoter can be ligated with the genomic sequence (Tanaka et al. 2001).
Roles of intron, stop, poly (A) addition and enhancer sequences
Variation in transgene expression levels is influenced by a number of factors. However, inclusion of an intron in a transgene construct leads to significantly greater transgene expression (Brinster et al. 1988) (Figure 1). The intron seems to have important effects on mRNA stabilization and the efficient translocation from nucleus to cytoplasm (Huang and Gorman 1990). Examples of introns commonly used are the rabbit beta-globin intron or the simian virus 40 (SV40) intron. An authentic intron 1 located upstream of exon 2 which often contains a translational start site can also be used in experiments where tissue- specific expression is important. Each transgene must also contain a transcriptional stop signal, typically included in the promoter, to match the start signal. Eukaryotic transcriptional stop signals include a poly (A) addition sequence (AAAUAA) as well as hundreds of downstream nucleotides where function is important but not clearly understood. The transcriptional stop signal frequently designated as poly (A) (Figure 1) is included at the end of the protein coding sequence. Examples of transcriptional stop sequences are those from SV40, bovine growth hormone (BGH), and human growth hormone (hGH) (Goodwin and Rottman 1992; Sheets et al. 1987). If an enhancer sequence for the gene of interest is identified, it can be included at either end of all the fragments mentioned above.
Identification of Potential Founders and Genotyping Strategy
About 3 weeks after the zygote injection, potential founders (F0) should be delivered. Genomic DNA for genotyping should be isolated from either ear-punch or snipped tail according to the approved animal study protocol. All the founders should be screened for the presence of the transgene to establish separate transgenic lines. The founders will have variable copy numbers and unique transgene insertion sites because of the random integration.
Polymerase chain reaction (PCR) and Southern blotting are the most commonly used methods to detect the presence of the transgene. Southern blotting is highly recommended to identify founder animals because of the low false positive rate and because more information can be obtained from the blot. If investigators receive too many potential founders to be genotyped, a PCR assay can be used to screen the positive mice first, and Southern blotting can be used with the reduced sample size. PCR is probably the best method for the offspring of the founders, but the result of the PCR assay should be validated with Southern blotting at least once. In either case, the validation protocol must ensure that the methods have enough sensitivity to detect one copy of the transgene and that established lines can be maintained without loss in the future.
PCR method to identify the transgene
Because of the high sensitivity of PCR, cross-contamination between samples and low primer sensitivity should be eliminated prior to genotyping. It is therefore, always preferable to use wild-type mouse genomic DNA as a negative control and a positive control of wild-type mouse genomic DNA spiked with a transgene at 1 and 10 copies per diploid genome. Neither the pure transgene fragment nor the plasmid should be used for the positive control.
The input DNA for PCR should be 20 to 200 ng of genomic DNA for a 25-microlitter reaction. If the PCR products smear in the electrophoresed gel, the amount of input DNA should be decreased. PCR primers should consist of around 30 nucleotides and produce a short amplicon of between 100 and 500 nucleotides for robust and specific amplification.
The best primer pair can be designed in flanking sequences of the junction between 2 elements uniquely combined in the transgene (primer set 1 in Figure 1A). This primer pair distinguishes the transgene from the endogenous gene. Alternatively, if a transgene construct has a foreign sequence from other species such as a GFP, the primers designed in the sequence can amplify the unique product. If a cDNA is used as a coding sequence, the primers can be designed to bridge between two or more different exons so that the endogenous gene will be amplified as a larger product (primer set 2 in Figure 1A and B).
Southern blotting method to identify the founders
A restriction enzyme that can cut only once in the transgene fragment is frequently used to digest genomic DNA for the genotyping of transgenic mice by Southern blotting. Since the majority of cytosines in the context of the CpG dinucleotide are methylated in normal adult somatic tissues, a restriction enzyme that is CpG methylation-sensitive may not be used. Because the transgenic construct integrates as multiple head-to-tail tandem copies in the mouse genome in most cases, digestion with the restriction enzyme for one site in the transgene will result in a fragment that is the same size as the original transgene (enzyme site A in Figure 2A). These fragments will appear as the most intense band by using either a transgene-specific probe or a probe made from the entire injected fragment. The transgene flanking fragments, which consist of the genomic sequence and a part of the transgene, can also be detected as 2 obvious bands with different sizes depending on the integration sites. These bands can be used as single copy standards to calculate the absolute copy numbers of the transgene. If more than 3 different flanking sequences appear, the founder mouse may have multichromosomal integration (Figure 2B, Lane E).
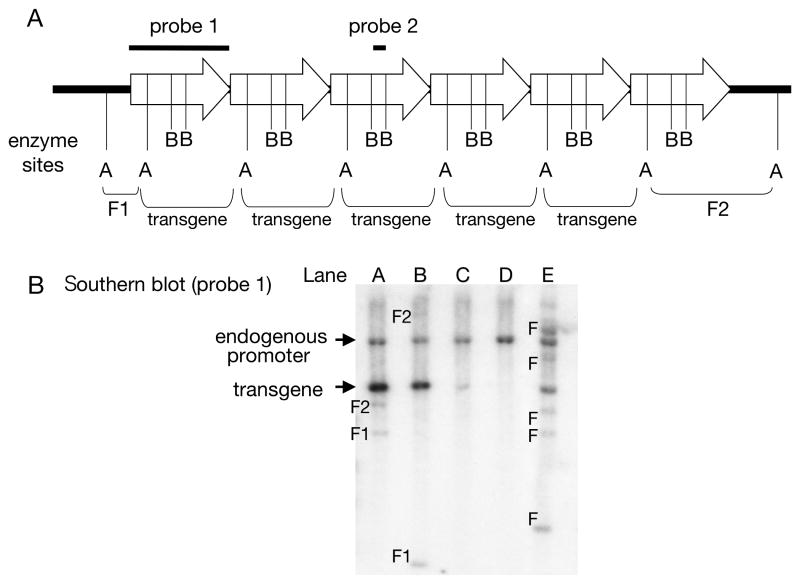
Figure 2.
Diagrams of transgenes integrated in the mouse genome and the strategy for genotyping by Southern blotting. (A) The scheme assumes there are 5 copies of transgenes integrated head-to-tail. (B) Example for genotyping of transgenic mouse by Southern blotting. Enzyme sites A and B in panel (A) represent hypothetical locations of the restriction enzyme sites in integrated locus. If the genomic DNA containing the transgenes is digested with enzyme A, 5 copies of transgenes with the size of the injection fragment (transgene) and 2 flanking sequences (F1 and F2) will be released from the genomic DNA. The 5 copies of transgene fragments will be detected as the most intense band by using a probe made from the entire injected fragment (probe 1) (transgene in panel (B) lane A). The flanking fragments of the transgene, which consist of the genomic sequence and a part of the transgene, can also be detected as 2 obvious bands with unique sizes depending on the integration sites (bands F1 and F2 in panel (B) lane A). These bands can be used as single copy standards to calculate the absolute copy numbers of the transgene. Because the transgene is randomly integrated in the mouse genome, different lines show different sizes of flanking sequences (bands F1 and F2 in panel (B) lane B). The band from an endogenous promoter may also be detected in all the samples (endogenous promoter in panel B). Lane C in panel B shows a transgene band with less intensity (∼1 copy), suggesting that this line may be a mosaic mouse. If more than 3 different flanking sequences appear, the founder mouse may have multichromosomal integration (bands F in panel (B) lane E). Lane D represents the wild-type mouse control. If the transgene fragment is too long to resolve using standard Southern blotting (more than 15-20 kb), a restriction enzyme that cuts 2 or more times in the injected fragment can be used (enzyme site B in Figure 2A). In this case, probe 2 detects 6 copies of transgene fragments generated by enzyme B with no flanking sequences.
We recommend using the entire injected fragment as a probe (Figure 2A). Because the promoter sequence in the probe fragment binds to the endogenous promoter region (the size varies depending upon the restriction enzyme used for genomic DNA digestion), the band derived from the endogenous promoter region can be used as a control to check whether equal amounts of DNA were used between samples. In addition, the band from the endogenous promoter could be used as a single copy standard in case only one endogenous promoter with a unique sequence exists in the mouse genome. For example, if the transgene band has an apparent low intensity compared to the promoter band, the founder may be mosaic (Figure 2B, Lane C).
In some cases, it may not be possible to use a restriction enzyme that can cut only once in the transgene. Especially if the transgene is longer than 15-20 kb, the fragment will be too long to resolve using standard Southern blotting (pulse field gel electrophoresis might be used). If that is the case, a restriction enzyme that cuts 2 or more times in the injected fragment can be used (enzyme site B in Figure 2A). A probe designed in one of the internal fragments can hybridize and detect the intense band provided by the transgene. A series of wild-type genomic DNA samples spiked with 1 to 100 copies of the transgene can be used to generate a standard curve for calculating the copy numbers.
Calculation of transgene mass to spike one copy per diploid genome
Assume the mouse genome has around 6.0 × 109 bp/cell and the injected transgenic fragment has 6.0 kb (6.0 × 103 bp).
The ratio (the size difference) between the foreign DNA fragment and the mouse genome is
To obtain a single copy of the fragment, the size difference or the ratio between the fragment and the total genome should be translated into the concentration of the DNA. For example, if 10 micrograms of genomic DNA is to be analyzed by Southern blotting, multiply the difference to obtain a single copy amount of transgene, i.e., 10 micrograms of genomic DNA × 1.0× 10-6 = 1.0 × 10-5 = 10 picograms.
Therefore, 10 picograms of the 6.0 kb transgene fragment are equal to 1 copy.
If 200 nanograms of genomic DNA are used for the PCR reaction, the single copy amount for spiking is 0.20 picograms. Note: The copy number of transgene usually has a strong correlation with the transgene expression level, although it will not guarantee the expression level of protein (see Transgene silencing and leaky expression in this unit).
Establishment of Independent Mouse Lines for Experimental Use
Once the founders reach sexual maturity, they can be crossed with wild-type mice to establish separate transgenic lines. However, the wild-type mouse strain should be considered before the founders are mated. Because the phenotype may vary depending upon the strain background, generating a littermate with a variety of background ratio should be prevented. If the founders are generated from inbred strains such as C57BL6/J or FVB, the same background should be maintained by crossing with the same strain so that the no mixed backgrounds will result. After verifying transmission from founder mice (F0) to offspring (F1), transgene expression within each line of the F1 or F2 generation has to be examined in most of the organs to prove that the line has proper temporal and spatial gene expression patterns and no leaky expression. Undesirable leaky (ectopic) expression in organs or tissues may result in unexpected phenotypes. In case the transgene is integrated into multiple chromosomes, further crossing with wild-type mice is necessary for the transgene to be segregated into different chromosomes to establish separate mouse lines.
Reverse transcriptase-polymerase chain reaction (RT-PCR) and/or northern blots can be used for the screening of expression patterns. It is advisable to perform the northern blots to minimize the effect of trace amounts of genomic DNA contaminations in RNA, especially when cDNA is used as a transgene. Western blots should also be performed to verify that the desired proteins were translated from transgenes.
At least 2 or 3 different lines generated from the same construct should be established and analyzed. Because each line should have different transgene insertion sites, copy numbers and protein expression levels, the analysis of all the transgenic lines may prove dose-dependent phenotypes. The multiple-line analysis also rules out the possibility of analyzing phenotypes from an unknown gene mutation and ectopic expression of the transgene.
Commentary
Background information
The generation of the first transgenic mouse was reported by Gordon et al. (1980). They produced transgenic mice by pronuclear injection of a pBR322-based plasmid containing HSV (herpes simplex virus) and the SV40 DNA sequence. After this report, the technique spread immediately and was well pursued. It is now an indispensable tool when gene functions needs to be analyzed at the whole-body level, such as in developmental biology, immunology, and neuroscience. A large number of promoters have been characterized that direct a wide variety of transgene expression patterns. Varieties of transgenic mice that express Cre (P1 bacteriophage cyclization recombinase) in a tissue-specific manner are available for creating conditional knockout mice utilizing the Cre-loxP system (Gu et al. 1994; Sauer and Henderson 1988).
Historically, the majority of transgenic mice were generated by injecting plasmid-based recombinant DNA, which limited the insert size to less than 20 kb because of the cloning capability of bacteria, but investigators could still expect faithful amplification, easy manipulation, and a good yield of recombinants. Relatively shorter inserts excised out from plasmids can integrate as tandem repeat concatemers resulting in high copy number in most cases. To a certain extent, transgene silencing is inevitable from the position effects caused by the use of shorter transgenes (see Transgene silencing and leaky expression in this unit).
To overcome the problems related to size-restricted plasmid vectors, larger constructs can be made in YAC (yeast artificial chromosome), BAC (bacterial artificial chromosome), or PAC (P1-derived artificial chromosome). A premade BAC library can especially facilitate the cloning and identification of the gene of interest. The advantages of using a large clone such as YAC are reported in a number of papers (i.e., Giraldo and Montoliu 2001). Transgenic mice created from large genomic clones showed position-independent, copy number-dependent expression of the transgene, suggesting that most of the regulatory sequences are needed to stably maintain the expression. Recently, Lois et al. (2002) reported that an alternative method of introducing a transgene by lentiviral vectors was successfully used to generate transgenic mice. This virus can be introduced either by microinjection in the perivitelline space or by coincubation with a denuded zygote. Furthermore, this method could be adapted with tightly regulated tet-on/off RNA interference (RNAi) technology to achieve conditional gene expression and knock down in mice (Szulc et al. 2006). These emerging technologies will provide further insights for analyzing gene functions and developing therapeutics.
Critical parameters and troubleshooting
No founders born
If no founder mice are obtained, it may be the result of technical problems with microinjection. Another possibility is that the transgene functions may be critical for the development of mouse embryos or for survival before the weaning stage. Investigators may need to reconsider these issues prior to repeating microinjections. There is also a possibility that the potential founder may be a false positive because of poor specificity for PCR primer sets or probes (see IDENTIFICATION OF POTENTIAL FOUNDERS AND GENOTYPING STRATEGY). Lethal phenotypes can be analyzed at the embryonic stage of founder mice.
Germline transmission
The offspring (F1) from founder mice (F0) should inherit the transgene at almost 50% according to the Mendelian ratio, if the transgene is integrated into a single chromosome. If investigators encounter no germline transmission, the genotype of the founder should be verified again. If F1 inherit the transgene a lot less than 50%, the founder is most likely a mosaic mouse, which means that not all the cells carry the transgene because of late integration. The mosaic founder may never transmit the transgene to offspring, or it may transmits it at very low efficiency, depending upon how many germ cells have the transgene. If the transgene is integrated into multiple chromosomes, more than 50% of the offspring will inherit the transgene. Separate lines may be obtained if the transgenes could be successfully segregated to different chromosomes in the F1 generation. If the transgene integrates into the X chromosome of male founders, the female offspring will inherit the transgene at 100%, but the male offspring will inherit none.
Transgene silencing and leaky expression
Transgene silencing occurs when a transgene is inserted into a transcriptionally inactive region in the mouse genome. Even though the transgene is present in the genome, the transcript (mRNA) cannot be made because of the positional effect. Thus, the copy numbers of integrated transgene will not necessarily correlate with the transgene expression level. Transcriptionally inactive regions may be due to repressor sites near the inserted promoter, insertion of the transgene into a gene with a repressed promoter, DNA methylation, X-inactivation, or genome imprinting. The use of an insulator sequence might decrease the probability of transgene silencing caused by the positional effects (Rivella et al., 2000). On the contrary, leaky (ectopic) expression of a transgene in unexpected tissues or unrestricted temporal expression manner is the result of insertion into the vicinity of an endogenous enhancer or promoter. The transgene expression level may decrease after several generations and may eventually stop in some cases. This epigenetic silencing happens when the animals with high copy number become older. To reduce this risk, creating 2 or more lines of transgenic mice is preferable.
Unknown gene mutations as a result of random integration
Because the transgene integrates randomly into mouse genomes, there is a possibility of disrupting a normal gene sequence. Such insertion may lead to an unexpected phenotype even in heterozygous mice because of haploinsufficiency. Since the phenotype caused by most of the mutations should be recessive, the phenotype may not appear until homozygotes are generated by crossing heterozygotes. Homozygotes may also have higher transgene expression compared to hemizogotes. To rule out the possibility of an unknown gene knock down phenotypes, more than 2 independent lines should be examined because the independent lines should not have the same insertion site.
Mixing different lines
The transgenic mouse created from the same construct may have similar but different phenotypes. It is always important to maintain the independent lines without mixing. The result from the PCR genotyping will produce the same amplicon from different lines with same transgene, so regular PCR genotyping cannot distinguish the lines accidentally mixed. In this case, the southern blotting can distinguish by comparing the different size from flanking fragments.
Time considerations
Potential founders are born 3 weeks after zygote injection. Pups are typically genotyped before weaning. After founders are identified, mating can be set up once the founders reach sexual maturity around 9 weeks of age. The founders' offspring (F1) can be used for the first experiment. The germline transmission and transgene copy numbers should also be verified in this generation. If all F1 pups from a founder have the same copy numbers and flanking sequences, any positive mouse in the F1 littermates could be used as a single established line.
Table (unit number) Time line for identifying founders and establishing lines
| Week | Action |
|---|---|
| 0 | Microinjection |
| 3 | birth of potential founders (F0) |
| 4∼6 | tag (tattooing), tail biopsy, genotyping for founder identification |
| 6 | Wean |
| 9 | set up founder mating |
| 12 | birth of first founder offspring (F1) |
| 13∼15 | tag (tattooing), tail biopsy, genotyping for transgenic mouse identification |
Acknowledgments
This work was supported by the Division of Intramural Research of the National Institute of Dental and Craniofacial Research, National Institutes of Health, Bethesda, MD.
Literature cited
- Brinster RL, Allen JM, Behringer RR, Gelinas RE, Palmiter RD. Introns increase transcriptional efficiency in transgenic mice. Proc Natl Acad Sci U S A. 1988;85:836–840. doi: 10.1073/pnas.85.3.836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldo P, Montoliu L. Size matters: use of YACs, BACs and PACs in transgenic animals. Transgenic Res. 2001;10:83–103. doi: 10.1023/a:1008918913249. [DOI] [PubMed] [Google Scholar]
- Goodwin EC, Rottman FM. The 3′-flanking sequence of the bovine growth hormone gene contains novel elements required for efficient and accurate polyadenylation. J Biol Chem. 1992;267:16330–16334. [PubMed] [Google Scholar]
- Gordon JW, Scangos GA, Plotkin DJ, Barbosa JA, Ruddle FH. Genetic transformation of mouse embryos by microinjection of purified DNA. Proc Natl Acad Sci U S A. 1980;77:7380–7384. doi: 10.1073/pnas.77.12.7380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci U S A. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu H, Marth JD, Orban PC, Mossmann H, Rajewsky K. Deletion of a DNA polymerase beta gene segment in T cells using cell type-specific gene targeting. Science. 1994;265:103–106. doi: 10.1126/science.8016642. [DOI] [PubMed] [Google Scholar]
- Huang MT, Gorman CM. Intervening sequences increase efficiency of RNA 3′ processing and accumulation of cytoplasmic RNA. Nucleic Acids Res. 1990;18:937–947. doi: 10.1093/nar/18.4.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühn R, Schwenk F, Aguet M, Rajewsky K. Inducible gene targeting in mice. Science. 1995;269:1427–1429. doi: 10.1126/science.7660125. [DOI] [PubMed] [Google Scholar]
- Lois C, Hong EJ, Pease S, Brown EJ, Baltimore D. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science. 2002;295:868–872. doi: 10.1126/science.1067081. [DOI] [PubMed] [Google Scholar]
- Mayo KE, Warren R, Palmiter RD. The mouse metallothionein-I gene is transcriptionally regulated by cadmium following transfection into human or mouse cells. Cell. 1982;29:99–108. doi: 10.1016/0092-8674(82)90094-0. [DOI] [PubMed] [Google Scholar]
- Rivella S, Callegari JA, May C, Tan CW, Sadelain M. The cHS4 insulator increases the probability of retroviral expression at random chromosomal integration sites. J Virol. 2000;74:4679–4687. doi: 10.1128/jvi.74.10.4679-4687.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossert J, Eberspaecher H, de Crombrugghe B. Separate cis-acting DNA elements of the mouse pro-alpha 1(I) collagen promoter direct expression of reporter genes to different type I collagen-producing cells in transgenic mice. J Cell Biol. 1995;129:1421–1432. doi: 10.1083/jcb.129.5.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer B, Henderson N. Site-specific DNA recombination in mammalian cells by the Cre recombinase of bacteriophage P1. Proc Natl Acad Sci U S A. 1988;85:5166–5170. doi: 10.1073/pnas.85.14.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheets MD, Stephenson P, Wickens MP. Products of in vitro cleavage and polyadenylation of simian virus 40 late pre-mRNAs. Mol Cell Biol. 1987;7:1518–1529. doi: 10.1128/mcb.7.4.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumarsono SH, Wilson TJ, Tymms MJ, Venter DJ, Corrick CM, Kola R, Lahoud MH, Papas TS, Seth A, Kola I. Down's syndrome-like skeletal abnormalities in Ets2 transgenic mice. Nature. 1996;379:534–537. doi: 10.1038/379534a0. [DOI] [PubMed] [Google Scholar]
- Szulc J, Wiznerowicz M, Sauvain MO, Trono D, Aebischer P. A versatile tool for conditional gene expression and knockdown. Nat Methods. 2006;3:109–116. doi: 10.1038/nmeth846. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Veeranna O, Ohshima T, Rajan P, Amin ND, Cho A, Sreenath T, Pant HC, Brady RO, Kulkarni AB. Neuronal cyclin-dependent kinase 5 activity is critical for survival. J Neurosci. 2001;21:550–558. doi: 10.1523/JNEUROSCI.21-02-00550.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Key references with annotations
- Nagy A, Gertsenstein M, Vintersten K, Behringer R. Manupulating the Mouse Embryo: A laboratory manual. 3rd. Cold Spring Harbor Laboratory Press; New York: 2003. Production of transgenic mouse; pp. 289–358. [Google Scholar]; This book is one of the most read lab manuals for the production of transgenic mice.
- Voncken JW. In: Transgenic Mouse: Methods and Protocols. Hofker MH, van Deursen J, editors. Humana Press; Totawa, New Jersey: 2003. pp. 9–34. [Google Scholar]; General mouse technologies including basic husbandry and microinjections are very well described.
Internet resources with annotations
- PubMed: http://www.ncbi.nlm.nih.gov/sites/entrez?db=PubMedThe world largest literature database search. Use key words ‘transgenic’, ‘mouse’, and tissue or cell type of interest to find out the preferable promoters.
- Cre and Flox mouse database: http://www.mshri.on.ca/nagy/International mouse strain resource: http://www.informatics.jax.org/imsr/index.jspResearch resources: http://www.jax.org/resources/index.htmlMouse Genome Informatics: http://www.informatics.jax.org/These Jackson Laboratory website provide links to a variety of mouse-related information, such as mutant resources and literature pertaining to mouse genetics.