Abstract
A multi-species based taxonomic microarray targeting coding sequences of diverged orthologous genes in Saccharomyces cerevisiae, S. paradoxus, S. mikatae, S. bayanus, S. kudriavzevii, Naumovia castellii, Lachancea kluyveri and Candida glabrata was designed to allow identification of isolates of these species and their interspecies hybrids. Analysis of isolates of several Saccharomyces species and interspecies hybrids demonstrated the ability of the microarray to differentiate these yeasts on the basis of their specific hybridization patterns. Subsequent analysis of 183 supposed S. cerevisiae isolates of various ecological and geographical backgrounds revealed one misclassified S. bayanus or S. uvarum isolate and four aneuploid interspecies hybrids, one between S. cerevisiae and S. bayanus and three between S. cerevisiae and S. kudriavzevii. Furthermore, this microarray design allowed the detection of multiple introgressed S. paradoxus DNA fragments in the genomes of three different S. cerevisiae isolates. These results show the power of multi-species based microarrays as taxonomic tools for the identification of species and interspecies hybrids, and their ability to provide a more detailed characterization of interspecies hybrids and recombinants.
Keywords: Saccharomyces cerevisiae, taxonomy, multi-species microarray, interspecies hybridization, introgression
Introduction
Formerly, the Saccharomyces genus was divided into two subgroups according to complex criteria, a sensu stricto group comprised of yeast species closely related to S. cerevisiae (S. cerevisiae, S. paradoxus, S. bayanus, S. kudriavzevii and S. mikatae) and a sensu lato group which involved heterogeneous yeast species that are more diverged from S. cerevisiae (Naumov, 1996; Naumov et al., 2000; Kurtzman & Robnett, 2003). However, recent changes in the taxonomic status of the yeast species in the sensu lato group abandoned this subdivision and assigned these species to other genera. According to this new classification, the former S. castellii and S. kluyveri, isolates of which are used in this study, respectively belong to the genera Naumovia and Lachancea and were renamed N. castellii and L. kluyveri (Kurtzman & Robnett, 1998; Kurtzman, 2003; Suh et al., 2006).
Yeast species within the Saccharomyces genus have been shown to be interfertile, in contrast to the former sensu lato species which give rise to unstable hybrids in interspecies crosses (Marinoni et al., 1999; Barnett, 1992), and interspecies hybridization has been proposed to be the result of evolutionary adaptation to different industrial environments (Matzke et al., 1999). An example of a well-known industrial interspecies hybrid is the lager yeast S. pastorianus, formed by the union of S. cerevisiae and S. bayanus-related yeasts in response to selective pressures from brewing at low temperatures (Kodama et al., 2005). Although interspecies hybrids are quite common among beer and wine-making Saccharomyces strains (Vaughan Martini & Martini, 1987; Groth et al., 1999; Masneuf et al., 1998; Kielland-Brandt et al., 1995; González et al., 2007), it remains unclear to what extent hybridization occurs in natural Saccharomyces populations (Landry et al., 2006).
Traditionally, identification of yeast isolates relied on morphological and physiological observations (Barnett et al., 2000). However, because of ambiguity due to intraspecies variability and interspecies hybridization, these methods are now considered unsatisfactory and more importance is given to genotypic methods for the identification of yeast isolates to the species level. With the recent release of the genome sequences of various ascomycete yeast species, species-specific DNA markers can be readily developed and high-throughput techniques, such as DNA microarrays, allow for rapid and accurate taxonomic identification of yeast isolates.
Here, we report the development of a multi-species based taxonomic microarray consisting of features targeted to multiple orthologous genes from S. cerevisiae, S. paradoxus, S. mikatae, S. bayanus, S. kudriavzevii, N. castellii, L. kluyveri and the closely related Candida glabrata. Using a set of reference isolates of various Saccharomyces species and a sample of 183 supposed S. cerevisiae isolates of diverse ecological and geographical origins, we show that this multi-species based microarray design is well suited for the routine identification and characterization of yeast species and their interspecies hybrids.
Material and methods
Microarray probe design
A multi-species based microarray was designed that, apart from the control features, consisted of DNA oligonucleotide probes targeted to the coding sequences of orthologs of 131 genes in S. cerevisiae, S. paradoxus, S. mikatae, S. bayanus, S. kudriavzevii, N. castellii, L. kluyveri and C. glabrata. Based on data from the Saccharomyces Genome Database (SGD; http://www.yeastgenome.org/) and the Yeast Gene Order Browser (Byrne & Wolfe, 2005), these genes were selected to be more-or-less evenly spread throughout the genome and to have orthologous DNA sequences diverged enough as to allow species-specific hybridization. The genomic distribution of the selected genes in S. cerevisiae S288c is illustrated in Figure S1 (Supplementary material).
DNA sequences of the orthologs in S. cerevisiae S288c, S. paradoxus NRRL Y-17217T, S. mikatae IFO 1815T, S. bayanus MCYC 623, S. kudriavzevii IFO 1802T, N. castellii NRRL Y-12630T, L. kluyveri NRRL Y-12651T and C. glabrata CBS 138T were retrieved from the NCBI database (National Center for Biotechnology Service; http://www.ncbi.nlm.nih.gov/) and species-specific oligonucleotide probes were designed using OligoArray v2.1 (Rouillard et al., 2003) with the following oligonucleotide primer design parameters: length of 30-35 nucleotides, melting temperature (Tm) of 80 ± 5°C, G+C content of 35-50% and maximum Tm of secondary structures and cross-hybridizations of 65°C. An overview of the selected probe sequences is given in Table S1 (Supplementary material).
Study and control samples
The study sample consisted of 183 S. cerevisiae isolates of various geographical and ecological origins, obtained from our own collection or from other laboratories and public collections (please direct strain requests to the appropriate culture collections; see Table S2, Supplementary material). Reference isolates of S. cerevisiae, S. paradoxus, S. uvarum (formerly a subgroup of the S. bayanus taxon, but now considered a separate species; Nguyen & Gaillardin, 2005), S. cariocanus (now considered to be a member of the S. paradoxus species; Liti et al., 2006), S. mikatae and S. kudriavzevii were used as control genotypes, as well as isolates of the natural interspecies hybrid S. pastorianus and laboratory-generated hybrids between various sibling species and S. cerevisiae (see Table 1). Interspecies hybrids were obtained by crossing haploid spores of HO lys2 or lys5 mutants of the non-S. cerevisiae parental species with haploid lys2 or lys5 strains of S. cerevisiae S288c and selecting for Lys+ diploids on synthetic dextrose (SD) medium. Lysine auxotrophs were selected by plating spores of the non-S. cerevisiae parental species on α-aminoadipate medium (Chattoo & Sherman, 1979). All isolates were kept in glycerol (15% v/v) at -80°C, or for short-term storage on yeast extract, peptone, and 2% dextrose (YPD) agar medium.
Table 1. Overview of Saccharomyces spp. reference samples.
Isolates of different Saccharomyces species and natural or laboratory-generated interspecies hybrids used to assess the performance of a multi-species based microarray as a taxonomic tool; interspecies hybrids were generated by crossing lys2 and lys5 mutants of both parental isolates and selecting for the Lys+ phenotype on synthetic dextrose medium; original names of isolates obtained from other culture collections are given in parenthesis.
| Isolate # | Species name and genetic background |
|---|---|
| S1 | S. cerevisiae S288c |
| YJM493 (CBS 7400) | S. paradoxus |
| YJM494 (CBS 2980) | S. paradoxus |
| YJM518 (CBS 395T) | S. uvarum |
| YJM789 | S. cerevisiae YJM145 |
| YJM793 (CBS 1503) | S. pastorianus |
| YJM794 (CBS 1538T) | S. pastorianus |
| YJM1148 (UFRJ 50791) | S. cariocanus |
| YJM1150 (IFO 1802T) | S. kudriavzevii |
| YJM1151 (IFO 1803) | S. kudriavzevii |
| YJM1152 (IFO 1815T) | S. mikatae |
| YJM1153 (IFO 1816) | S. mikatae |
| YJM1292 | S. cerevisiae RM11 |
| Hpar1 | S. paradoxus YJM508 (HO lys2) x S. cerevisiae S31 (α lys5) |
| Hpar2 | S. paradoxus YJM508 (HO lys2) x S. cerevisiae S31 (α lys5) |
| Hpar3 | S. paradoxus YJM509 (HO lys5) x S. cerevisiae S95 (a lys2) |
| Hkud1 | S. kudriavzevii YJM1176 (HO lys2) x S. cerevisiae S31 (α lys5) |
| Hkud2 | S. kudriavzevii YJM1177 (HO lys5) x S. cerevisiae S95 (a lys2) |
| Hkud3 | S. kudriavzevii YJM1177 (HO lys5) x S. cerevisiae S95 (a lys2) |
| Hbay1 | S. bayanus YJM295 (HO lys2) x S. cerevisiae S31 (α lys5) |
| Hbay2 | S. bayanus YJM295 (HO lys2) x S. cerevisiae S31 (α lys5) |
| Hmik1 | S. mikatae YJM1172 (HO lys2) x S. cerevisiae S31 (α lys5) |
| Hmik2 | S. mikatae YJM1172 (HO lys2) x S. cerevisiae S31 (α lys5) |
DNA extraction and microarray hybridization
DNA was extracted from overnight cultures grown in 50 ml YPD medium using the QIAGEN Genomic-tip 100/G (QIAGEN) following the manufacturer’s instructions. Ten micrograms of purified DNA were digested with 1 U of DNase I (New England Biolabs) for 1.5 minutes at 37°C in 1x DNase I Reaction buffer (New England Biolabs) to obtain fragments of 50 – 100 bp. DNase I was heat inactivated at 75°C for 25 minutes, and the digestion products were analyzed on a 2% agarose gel. Fragmented DNA was end-labeled by incubation at 37°C for 1 hour with 20 U terminal deoxynucleotidyl transferase (New England Biolabs) and 1 nmol of biotin-11-ddATP (Perkin Elmer) in 1x NEBuffer 4 (New England Biolabs), and the labeling reaction was terminated by heat inactivation of the terminal transferase at 75°C for 25 minutes.
The digested and labeled DNA was hybridized to custom-made CustomArray™ 4x2K microarrays (Combimatrix) following standard protocols from Combimatrix for hybridization, washing, and staining. In short, approximately 2 μg of DNA in 35 μl of hybridization buffer (6x SSPE, 0.05% Tween-20, 20 mM EDTA, 0.05% SDS) was denatured at 95°C for 5 minutes, quickly cooled on ice, and applied to the CustomArray™ 4x2K array which had been pre-incubated with pre-hybridization buffer (6x SSPE, 0.05% Tween-20, 20 mM EDTA, 5x Denhardt’s solution, 100 ng/μl salmon sperm DNA, 0.05% SDS) at 50°C for 30 minutes. Hybridization occurred at 50°C for 15 – 17 hours, after which the reaction solution was removed and several wash steps were performed: a first wash step at 50°C for 15 minutes using SSPE wash solution 1 (6x SSPE, 0.05% Tween-20), followed by three wash steps at room temperature for 1 minute using SSPE wash solution 2 (3x SSPE, 0.05% Tween-20), SSPE wash solution 3 (0.5x SSPE, 0.05% Tween-20) and PBST wash solution (2x PBS, 0.1% Tween-20) consecutively. The arrays were stained using 2 μg/ml Fluorolink Cy5-labeled streptavidin (GE Healthcare) in biotin blocking solution (2x PBS, 0.1% Tween-20, 1% BSA).
Image acquisition and data analysis
The microarrays were scanned at 5 μm resolution using a GenePix 4000B scanner (Axon) in combination with GenePix Pro v6.0 imaging software and data extraction was performed with Microarray Imager v5.8.1 software (Combimatrix). Hybridization intensity data were binary recoded into genotypes using a threshold value calculated as t = m + 5 * sd, with m and sd respectively equal to the mean and the standard deviation of the negative control data. Orthologous genes corresponding to microarray features with hybridization intensities above the threshold value were designated as being present (“1”), while genes corresponding to features with intensity values below the threshold were regarded as absent or highly divergent (“0”). Data reproducibility was assessed by calculating the percentage of loci with identical genotypes in two replicated experiments for each of three different isolates (YJM128, YJM180, and YJM1152; see Table 1 and Table S2).
Correspondence analysis was performed using the dudi.coa routine in R (R Development Core Team, 2007) to graphically represent the genetic relationships between the different isolates and control samples. In addition, the percentages of orthologs designated to be present in each isolate were calculated separately for the different species represented on the microarray.
Confirmation of interspecies hybrids
The hybrid nature of isolates YJM187, YJM264, YJM337 and YJM338 (see Results) was confirmed by DNA sequencing of fragments of selected genes: CAT2 and PRD1 for YJM187; XPT1, CTR9, and MED4 for YJM264, YJM337 and YJM338. Based on the sequences of these genes in the non-S. cerevisiae parental background (S. bayanus for YJM187; S. kudriavzevii for YJM264, YJM337 and YJM338), oligonucleotide primers were designed using Primer3 software (Rozen & Skaletsky, 2000). Polymerase chain reactions (PCRs) were set up in total volumes of 20 μl containing 0.5 μM of each primer (see Table S3), 1 U Taq DNA polymerase, 10 mM Tris-HCl pH 9, 15 mM MgCl2, 50 mM KCl, 200 μM of all four dNTPs and approximately 2 ng of genomic DNA template. The following PCR temperature profile was used: initial denaturation at 95 °C for 2 min; 30 cycles of 95 °C for 20 s, 55 °C for 20 s, 72 °C for 1 min; final extension at 72 °C for 10 min. The PCR products were subcloned using the TOPO TA Cloning® Kit with pCR®2.1-TOPO® vector and TOP 10 cells (Invitrogen Corporation), and cloned DNA fragments were reamplified by direct PCR of the colony with M13f and M13r primers, and sequenced using ABI BigDye® Terminator reaction mixture and an ABI PRISM® 3730 DNA Analyzer (Applied Biosystems). Obtained nucleotide sequences were compared to fungal genome sequences using the BLAST Network Service (NCBI, National Center for Biotechnology Service; http://www.ncbi.nlm.nih.gov/BLAST).
Confirmation of introgression events
Introgression from S. paradoxus into S. cerevisiae isolates YJM218, YJM220 and YJM1075 (see Results) was confirmed by DNA sequencing of selected genes in the introgressed fragments. In addition, locations and sizes of the introgressed fragments were estimated by DNA hybridization onto GeneChip® S. cerevisiae Tiling 1.0R Arrays (Affymetrix), and flanking sites of the introgressed fragments were sequenced. Involved open reading frames (ORFs) were retrieved using the Synteny Viewer of the Saccharomyces Genome Database (SGD; http://www.yeastgenome.org/). Oligonucleotide primers were designed from the DNA sequences of CDC36, CCA1, DST1, UIP5 and RRP15 in S. paradoxus using Primer3 software and fragments of these genes were amplified by PCR in total volumes of 20 μl containing 0.5 μM of each primer (see Table S4), 1 U Taq DNA polymerase, 10 mM Tris-HCl pH 9, 15 mM MgCl2, 50 mM KCl, 200 μM of all four dNTPs and approximately 2 ng of genomic DNA template. The PCR temperature profile was: initial denaturation at 95 °C for 2 min; 30 cycles of 95 °C for 20 s, 55 °C for 20 s, 72 °C for 1 min; final extension at 72 °C for 10 min. The PCR products were subcloned using the TOPO TA Cloning® Kit with pCR®2.1-TOPO® vector and TOP 10 cells (Invitrogen Corporation), and cloned DNA fragments were reamplified by direct PCR of the colony with M13f and M13r primers, and sequenced using ABI BigDye® Terminator reaction mixture and an ABI PRISM® 3730 DNA Analyzer (Applied Biosystems).
Genomic DNA of the homothallic segregants YJM247, YJM248 and YJM1078 (respectively obtained from spores of YJM220, YJM218 and YJM1075) was extracted from overnight cultures grown in 50 mL of YPD-medium using the QIAGEN Genomic-tip 100/G (QIAGEN) according to the manufacturer’s instructions. Ten micrograms of purified DNA were digested with 1 U of DNase I (New England Biolabs) for 2 minutes at 37°C in 1x DNase I Reaction buffer (New England Biolabs) to obtain fragments of 25 –50 bp. DNase I was heat inactivated at 95°C for 20 minutes, and the digestion products were analyzed on a 2% agarose gel. Fragmented DNA was end-labeled by incubation at 37°C for 1 hour with 20 U terminal deoxynucleotidyl transferase (New England Biolabs) and 1 nmol of biotin-11-ddATP (Perkin Elmer) in 1x NEBuffer 4 (New England Biolabs), and the labeling reaction was terminated by heat inactivation of the terminal transferase at 75°C for 25 minutes.
Labeled DNA was hybridized onto GeneChip® S. cerevisiae Tiling 1.0R Arrays (Affymetrix) following Gresham et al. (2006), and the arrays were scanned using the Affymetrix scanner at 0.7 μm resolution. The hybridization intensity of the 9 central pixels was determined, and an average intensity at each feature was computed using the GeneChip® Operating Software (Affymetrix). Intensity values were analyzed and compared between the three strains using the Integrated Genome Browser software (Affymetrix).
Three introgressed regions located on chromosomes IV, V and XI, and containing respectively CDC36, CCA1 and UIP5, were PCR amplified with the Expand Long Template PCR System (Roche Applied Science) following the manufacturer’s protocol. Fifty microliter reactions were set up with 500 μM of all four dNTPs, 300 nM of each primer (see Table S5), 0.75 μl Expand Long Template enzyme mix and 300 ng of template DNA in 1x PCR buffer 3, and the following PCR temperature profile was used: initial denaturation at 94 °C for 2 min; 10 cycles of 94 °C for 10 s, 55 °C for 30 s, 68 °C for 17 min; 25 cycles of 94 °C for 15 s, 55 °C for 30 s, 68 °C for 17 min + 20 s for each successive cycle; final extension at 68 °C for 7 min. The PCR products were diluted ten-fold and sequenced partially using the forementioned primers and ABI BigDye® Terminator reaction mixture and an ABI PRISM® 3730 DNA Analyzer (Applied Biosystems). Obtained nucleotide sequences were compared to fungal genome sequences using the BLAST Network Service (NCBI, National Center for Biotechnology Service; http://www.ncbi.nlm.nih.gov/BLAST) and the CLUSTAL W algorithm (Thompson et al., 1994) as implemented in BioEdit v7.0.9.0 (Hall, 1999).
Results
Microarray performance
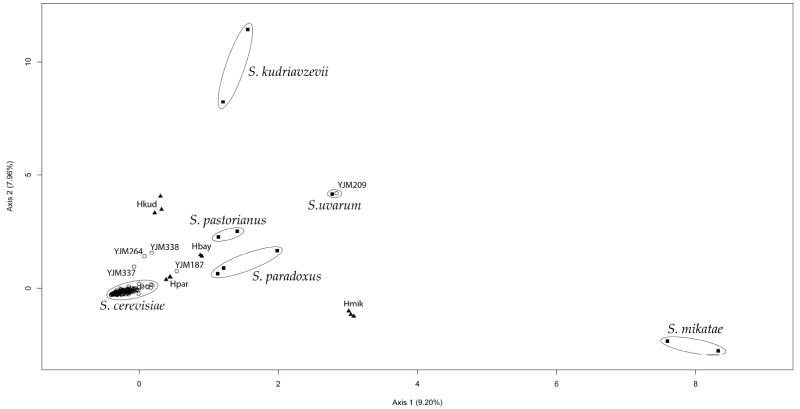
A multi-species based microarray aimed at taxonomic identification of several yeast species and interspecies hybrids of the Saccharomycetaceae was designed and tested using a set of 23 reference isolates (see Table 1). Reproducibility of the genotypes was high, with identical presence/absence scores obtained for approximately 94% of the loci in replicated experiments. Correspondence analysis was able to discriminate between all the different species and interspecies hybrids represented in the reference sample set, and all samples clustered according to their taxonomic identities (including the S. cariocanus isolate which clustered together with the S. paradoxus isolates, consistent with the recent change in its taxonomic status; see Figure 1).
Figure 1.
Results of a correspondence analysis performed using the dudi.coa routine in R (R Development Core Team, 2007) with hybridization data of microarray features corrresponding to a set of 131 orthologous genes in 5 Saccharomyces species, Naumovia castellii, Lachancea kluyveri and Candida glabrata; data for 23 reference isolates, including interspecies hybrids, and 183 test strains were included; “■”, reference isolate; “▲”, laboratory-generated interspecies hybrid; “○”, test isolate.
Although some cross-hybridization occurred, calculated percentages of orthologs designated to be present in the reference isolates were in concordance with their taxonomy. For the laboratory-generated interspecies hybrids, approximately equal values were obtained for the ortholog sets corresponding to each of the parental species. The relatively low percentage values thereby observed were entirely due to a concentration effect: doubling the amount of DNA hybridized to the microarray led to a two-to-three fold increase of the percentage values for the respective parental species (data not shown). In contrast to the laboratory-generated hybrids, the percentage values obtained for the S. pastorianus isolates, known hybrids between S. cerevisiae and S. bayanus used in brewing (Marinoni et al., 1999; Rainieri et al., 2006), were not equal for the ortholog sets corresponding to the parental species, and indicated a higher content of S. bayanus alleles (see Table 2).
Table 2. Summary of orthologous genes presence.
Percentages of orthologous genes designated to be present in different yeast strains based on microarray hybridization data; percentages were calculated separately for each species represented on a multi-species based taxonomic microarray; single highest and the two highest values are depicted in bold for species and interspecies hybrids respectively.
| S. cerevisiae specific | S. bayanus specific | S. mikatae specific | S. paradoxus specific | N. castellii specific | L. kluyveri specific | S. kudriavzevii specific | C. glabrata specific | |
|---|---|---|---|---|---|---|---|---|
| S1 | 100.0 | 4.6 | 7.6 | 19.1 | 6.1 | 9.2 | 7.6 | 3.8 |
| YJM493 | 13.7 | 10.7 | 15.3 | 58.8 | 17.6 | 17.6 | 9.9 | 10.7 |
| YJM494 | 11.6 | 8.6 | 12.8 | 49.5 | 13.3 | 14.7 | 6.8 | 7.3 |
| YJM518 | 0.8 | 44.3 | 3.8 | 3.1 | 4.6 | 6.1 | 1.5 | 1.5 |
| YJM793 | 37.4 | 63.2 | 10.5 | 8.6 | 5.4 | 7.4 | 4.1 | 1.8 |
| YJM794 | 26.8 | 68.5 | 12.1 | 9.4 | 6.8 | 6.2 | 3.8 | 1.5 |
| YJM1148 | 11.5 | 3.8 | 6.9 | 61.8 | 1.5 | 3.1 | 2.3 | 0.8 |
| YJM1150 | 0.8 | 0.8 | 0.8 | 3.1 | 3.8 | 2.3 | 61.1 | 0.8 |
| YJM1151 | 2.3 | 3.1 | 2.3 | 0.8 | 4.6 | 1.5 | 35.9 | 1.5 |
| YJM1152 | 3.1 | 7.6 | 95.4 | 6.1 | 2.3 | 3.1 | 2.3 | 0.0 |
| YJM1153 | 5.3 | 6.9 | 98.5 | 6.9 | 4.6 | 5.3 | 4.6 | 0.8 |
| Hpar1 | 33.6 | 1.5 | 3.8 | 30.5 | 2.3 | 0.8 | 2.3 | 0.0 |
| Hpar2 | 42.0 | 1.5 | 6.1 | 38.9 | 1.5 | 0.8 | 0.8 | 0.0 |
| Hpar3 | 27.5 | 0.8 | 4.6 | 29.8 | 1.5 | 0.0 | 0.8 | 0.0 |
| Hkud1 | 32.8 | 2.3 | 1.5 | 2.3 | 0.8 | 0.0 | 27.5 | 0.0 |
| Hkud2 | 30.5 | 0.8 | 1.5 | 1.5 | 0.8 | 0.8 | 31.3 | 0.0 |
| Hkud3 | 26.0 | 0.8 | 1.5 | 1.5 | 0.8 | 0.0 | 22.9 | 0.0 |
| Hbay1 | 26.0 | 23.7 | 0.8 | 2.3 | 1.5 | 0.0 | 0.8 | 0.0 |
| Hbay2 | 32.8 | 35.1 | 1.5 | 2.3 | 0.8 | 0.0 | 0.0 | 0.0 |
| Hmik1 | 27.5 | 0.8 | 22.9 | 2.3 | 0.8 | 0.0 | 0.8 | 0.0 |
| Hmik2 | 16.8 | 0.0 | 15.3 | 1.5 | 0.8 | 0.0 | 0.8 | 0.0 |
| YJM187 | 67.2 | 32.8 | 10.7 | 12.2 | 6.9 | 7.6 | 6.1 | 4.6 |
| YJM264 | 64.9 | 7.6 | 8.4 | 12.9 | 9.2 | 6.8 | 32.8 | 3.8 |
| YJM337 | 70.2 | 6.8 | 6.1 | 9.1 | 9.1 | 6.8 | 27.5 | 3.8 |
| YJM338 | 56.5 | 6.1 | 6.8 | 6.1 | 6.9 | 5.3 | 24.4 | 3.8 |
Interspecies hybridization
Having validated the microarray design, genotype data were then collected for 183 test isolates of various geographical and ecological origins, which were previously identified as S. cerevisiae using conventional techniques. Most of these isolates formed a continuous S. cerevisiae cluster with S1, which is isogenic with S288c, except for YJM187, YJM209, YJM264, YJM337 and YJM338. YJM209, originally classified as S. cerevisiae by the National Collection of Yeast Cultures, clustered together with YJM518 (S. uvarum type strain) and was therefore considered to have been misclassified by the culture collection.
The clustering positions of YJM187, YJM264, YJM337 and YJM338, originally classified as S. cerevisiae, were indicative of a hybrid nature (See Figure 1). In contrast to the results obtained with the laboratory-generated hybrids but similar to the results obtained with the S. pastorianus isolates, the percentage values of the orthologous genes designated to be present in these samples were not equal for the microarray feature sets corresponding to the suspected parental species. YJM187 was found to be an interspecies hybrid between S. cerevisiae and S. bayanus, while YJM264, YJM337 and YJM338 were found to be hybrids between S. cerevisiae and S. kudriavzevii (see Table 2). A comparison of the nucleotide sequences of selected genes in these isolates (CAT2 and PRD1 for YJM187; XPT1, CTR9 and MED4 for YJM264, YJM337 and YJM338) with all available genome sequences of Saccharomyces species confirmed the presence of non-S. cerevisiae DNA with highest homology to the corrresponding sequences in the non-S. cerevisiae parental species suggested by the microarray data (data not shown). A graphical overview of the hybridization data for isolates YJM187, YJM264, YJM337 and YJM338 is presented in Figure S2 (Supplementary material).
Introgression
Evidence for introgression of S. paradoxus DNA fragments into the genome of S. cerevisiae was found in the hybridization data of isolates YJM218, YJM220 and YJM1075. S. paradoxus alleles of CCA1, DST1 and UIP5 (respectively located on chromosomes V, VII and XI) were designated to be present in isolates YJM218, YJM220 and YJM1075, while an S. paradoxus allele of CDC36 (chromosome IV) was found in YJM220 and YJM1075, and an S. paradoxus allele of RRP15 (chromosome XVI) in YJM218 and YJM1075. Confirmation of the presence of these alleles was obtained by DNA sequencing and comparison of the obtained sequences with all available genome sequences of Saccharomyces species (data not shown).
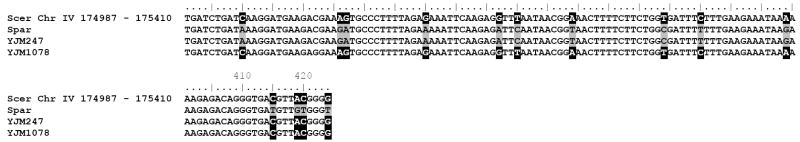
The divergent nature of these loci was also apparent in the hybridization profiles of the homothallic segregants of these isolates on GeneChip® S. cerevisiae Tiling 1.0R Arrays, and these profiles allowed estimation of the genomic coordinates of the introgressed fragments (see Figure 2 and Table 3). Based on the hybridization data of the GeneChip® S. cerevisiae Tiling 1.0R Arrays, primers were designed to PCR-amplify the complete introgressed fragments containing loci CDC36, CCA1 and UIP5, and comparison of the sequences of the flanking sites of these fragments with the genome sequences of S. cerevisiae and S. paradoxus confirmed the estimated extent of the respective introgression events (see Figure 3). As can be seen in Figures 2 and 3, introgressed fragments were not always of identical size: although the 5’ ends were identical in both isolates, the introgressed fragment containing CDC36 on chromosome IV extended approximately 300 basepairs further in YJM1075 compared to YJM218 (see Figure 3b).
Figure 2.
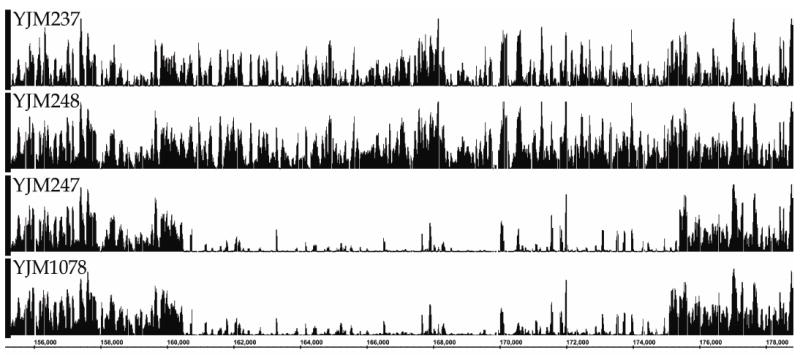
Graphical representation of hybridization intensity values for S. cerevisiae strain S288c (YJM237) and three segregants of S. cerevisiae isolates YJM218, YJM220 and YJM1075 of which the DNA was hybridized onto GeneChip® S. cerevisiae Tiling 1.0R Arrays (Affymetrix); the graphs cover the genome region between nucleotide positions 160475 and 175080 on chromosome IV, which contains introgressed S. paradoxus DNA fragments in YJM247 (segregant of YJM220) and YJM1078 (segregant of YJM1075).
Table 3. Characteristics of introgressed Saccharomyces paradoxus DNA segments in S. cerevisiae.
Characteristics of 5 introgressed Saccharomyces paradoxus DNA segments found in the genomes of three S. cerevisiae strains; (1), S. cerevisiae open reading frame (ORF) absent in S. paradoxus; (2), S. paradoxus ORF absent in S. cerevisiae.
| Present in | Coordinates | ||||||
|---|---|---|---|---|---|---|---|
| Chromosome | YJM218 | YJM220 | YJM1075 | Start | Stop | Involved ORFs | Note |
| IV | - | + | + | 160475 | 175080 | SFA1, PORF3155(2), NRP1, FAP7, CDC36, CDC9, YDL163W, YDL162C(1), ENT1, YDL160C-A(1), DHH1, YDL159W-A(1), STE7, YDL158C(1), YDL157C, YDL156W | Ends at position 175400 in YJM1075 |
| V | + | + | + | 519670 | 532980 | BCK2, CCA1, RPH1, ADK2, RAD3, BRR2 | Ends at position 522360 in YJM1075 |
| VII | + | + | + | 418380 | 430930 | DST1, PORF8516(2), YGL042C(1), YGL041C-B(1), YGL041C, YGL041W-A(1), HEM2, PORF8510(2), YGL039W, OCH1, PNC1, YGL036W | |
| XI | + | + | + | 512700 | 524680 | KAE1, GAP1, YKR040C, YKR041W, UTH1, PORF13342(2), YKR043C, UIP5, YKR045C, PET10 | Starts at position 511750 in YJM218 |
| XVI | + | - | + | 817610 | 837960 | KAR3, YPR142C, RRP15, NOC4, PORF22909(2), SNR45(1), ASN1, YPR145C-A(1), YPR146C(1), YPR147C, YPR148C, PORF22893(2), PORF22891(2), NCE102, YPR150W, SUE1, URN1, YPR153W, PORF22876(2), PIN3, NCA2, TPO3 | Starts at position 835600 in YJM220 |
Figure 3.
Clustal W alignment of (a) left and (b) right flanking sequences of introgressed S. paradoxus DNA segments in S. cerevisiae strains YJM247 and YJM1078; single nucleotide polymorphisms (SNPs) corresponding to the S. cerevisiae allele are indicated by a black background, and SNPs corresponding to the S. paradoxus allele are indicated by a grey background.
Discussion
Although the microarray technology has been mainly used for gene expression profiling, its value as a taxonomic tool for the identification and characterization of microorganisms, especially bacteria, has been shown in various studies. In these mostly clinical studies, targets used included virulence genes, intergenic regions and ribosomal genes (e.g. Volokhov et al., 2003; Giammarinaro et al., 2005; Lehner et al., 2005). Here, we report the development of a taxonomic microarray consisting of oligonucleotide probes targeted to the coding sequences of multiple orthologous genes, approximately evenly distributed throughout the genomes of S. cerevisiae, S. paradoxus, S. bayanus, S. kudriavzevii, S. mikatae, L. kluyveri, N. castellii and C. glabrata. Because interspecies nucleotide variability of the targeted sequences is usually larger than their intraspecies variability, this design allowed species-specific hybridization and correct taxonomic identification of a reference set of isolates of the represented species and their interspecies hybrids. In addition, the multi-gene based design allowed a more detailed characterization of interspecies hybrids and recombinants, including the detection of isolated introgressed DNA segments.
Among a study sample of 183 supposed S. cerevisiae isolates, of which one had been misclassified, one S. cerevisiae x S. bayanus and three S. cerevisiae x S. kudriavzevii interspecies hybrids were identified. One of these hybrids is a commercial strain used in baking (YJM264), while the others are used in the production of beer (YJM187) and wine (YJM337 and YJM338). Marinoni et al. (1999) showed that laboratory-generated interspecies hybrids among Saccharomyces yeasts are stable and display both sets of parental chromosomes, which is consistent with our results. However, microarray hybridization patterns of all non-laboratory generated hybrids identified in this study clearly differed from the hybridization patterns obtained with the laboratory-generated hybrids and were indicative of an aneuploid nature. It appears that these hybrids contain a complete set of S. cerevisiae chromosomes, as well as extra chromosomes from either S. bayanus or S. kudriavzevii. This is in contrast to the S. pastorianus isolates included in this study, which apparently contain a higher content of alleles derived from the non-S. cerevisiae parental species, S. bayanus (see also Rainieri et al., 2006). Nevertheless, these observations are consistent with earlier studies that reported differences in genome size and genomic constitution of industrial yeast strains, the majority of which are believed to be aneuploid (Guijo et al., 1997; Casaregola et al., 2001; Naumova et al., 2005). S. cerevisiae strains and their hybrids are known to have a high tolerance to spontaneous loss or gain of chromosomes (Bakalinsky & Snow, 1990), but aneuploidy renders meiotic segregation ineffective and can cause sterility (Greig et al., 2002). Despite the low fertility of hybrids between Saccharomyces species, introgression following hybridization has been described (Naumova et al., 2005; Liti et al., 2006; Wei et al., 2007). In this study, we found evidence for introgression from S. paradoxus into the genome of S. cerevisiae. While average genomic sequence identity between S. paradoxus and S. cerevisiae is about 85% across chromosomes (Kellis et al., 2003), we identified five regions with an average identity to S. paradoxus of 99% or more in three different S. cerevisiae isolates. These regions varied in length between 2 kbp and 20 kbp and were located on five different chromosomes. Since horizontal gene transfer is quite rare in yeast and large DNA segments are unlikely to be transferred directly (Dujon, 2005), these regions are likely to result from introgression through homologous recombination after a hybridization event between S. cerevisiae and S. paradoxus. Although the involved strains were isolated from human faeces and pig rectum, niches where S. paradoxus is not commonly found, S. cerevisiae and S. paradoxus can be found in overlapping habitats (Barnett et al., 2000; Sniegowski et al., 2002), and introgression between these species has been identified (Liti et al., 2006; Wei et al., 2007). Because the flanking sequences of the introgressed segments were often identical in at least two of the three isolates, it is believed that they are the result of a single hybridization and introgression event that took place in their common ancestor. This common ancestor would have been produced by a rare viable gamete from an S. cerevisiae x S. paradoxus hybrid, possibly containing a majority of S. cerevisiae sequences, that underwent repeated backcrosses with S. cerevisiae, leaving isolated S. paradoxus DNA segments.
In conclusion, this study showed that multi-species based taxonomic microarrays consisting of DNA oligonucleotide probes targeted to the coding sequences of diverged orthologs, provide a reliable alternative to already established molecular typing methods such as chromosome karyotyping, DNA fingerprinting and DNA sequencing. Besides taxonomic identification, this method also allows a more detailed characterization of interspecies hybrids and recombinants, and is in this regard easily extendable to higher coverages of the represented genomes, thus providing a powerful method for the routine screening for introgression events.
Supplementary Material
Acknowledgments
The authors wish to thank the many colleagues and culture collection curators who provided yeast isolates. The authors also wish to thank F. Dietrich and P. Magwene for helpful comments and suggestions. The work was supported by the National Institutes of Health [GM070541].
References
- Bakalinsky AT, Snow R. The chromosomal constitution of wine strains of Saccharomyces cerevisiae. Yeast. 1990;6:367–382. doi: 10.1002/yea.320060503. [DOI] [PubMed] [Google Scholar]
- Barnett JA. The taxonomy of the genus Saccharomyces Meyen ex Reess: a short review for non-taxonomists. Yeast. 1992;8:1–23. [Google Scholar]
- Barnett JA, Payne RW, Yarrow D. Yeasts: Characteristics and Identification. 3. Cambridge University Press; Cambridge: 2000. [Google Scholar]
- Byrne KP, Wolfe KH. The Yeast Gene Order Browser: combining curated homology and syntenic context reveals gene fate in polyploid species. Genome Res. 2005;15:1456–1461. doi: 10.1101/gr.3672305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casaregola S, Nguyen H-V, Lapathitis G, Kotyk A, Gaillardin C. Analysis of the constitution of the beer yeast genome by PCR, sequencing and subtelomeric sequence hybridization. Int J Syst Evol Microbiol. 2001;51:1607–1618. doi: 10.1099/00207713-51-4-1607. [DOI] [PubMed] [Google Scholar]
- Chattoo BB, Sherman F. Selection of lys2 mutants of the yeast Saccharomyces cerevisiae by the utilization of α-aminoadipate. Genetics. 1979;93:51–65. doi: 10.1093/genetics/93.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dujon B. Hemiascomycetous yeasts at the forefront of comparative genomics. Curr Opin Genetics Dev. 2005;15:614–620. doi: 10.1016/j.gde.2005.09.005. [DOI] [PubMed] [Google Scholar]
- Giammarinaro P, Leroy S, Chacornac JP, Delmas J, Talon R. Development of a new oligonucleotide array to identify staphylococcal strains at species level. J Clin Microbiol. 2005;43:3673–3680. doi: 10.1128/JCM.43.8.3673-3680.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González SS, Gallo L, Climent MA, Barrio E, Querol A. Enological characterization of natural hybrids from Saccharomyces cerevisiae and S. kudriavzevii. Int J Food Microbiol. 2007;116:11–18. doi: 10.1016/j.ijfoodmicro.2006.10.047. [DOI] [PubMed] [Google Scholar]
- Greig D, Borts RH, Louis EJ, Travisano M. Hybrid speciation in experimental populations of yeast. Science. 2002;298:1773–1775. doi: 10.1126/science.1076374. [DOI] [PubMed] [Google Scholar]
- Gresham D, Ruderfer DM, Pratt SC, Schacherer J, Dunham MJ, Botstein D, Kruglyak L. Genome-wide detection of polymorphisms at nucleotide resolution with a single DNA microarray. Science. 2006;311:1932–1936. doi: 10.1126/science.1123726. [DOI] [PubMed] [Google Scholar]
- Groth C, Hansen J, Piskur J. A natural chimeric yeast containing genetic material from three species. International Journal of Systematic Bacteriology. 1999;49:1933–1938. doi: 10.1099/00207713-49-4-1933. [DOI] [PubMed] [Google Scholar]
- Guijo S, Mauricio JC, Salmon JM, Ortega JM. Determination of the relative ploidy in different Saccharomyces cerevisiae strains used for fermentation and ‘flor’ film ageing of dry sherry-type wines. Yeast. 1997;13:101–117. doi: 10.1002/(SICI)1097-0061(199702)13:2<101::AID-YEA66>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Kellis M, Patterson N, Endrizzi M, Birren B, Lander ES. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature. 2003;423:241–254. doi: 10.1038/nature01644. [DOI] [PubMed] [Google Scholar]
- Kielland-Brandt M, Nilsson-Tillgren T, Gjermansen C, Holmberg S, Pedersen MB. Genetics of brewing yeasts. In: Rose AH, Wheals E, Harrison JS, editors. The yeasts. Vol. 6. Academic Press; London: 1995. pp. 223–254. [Google Scholar]
- Kodama Y, Kielland-Brandt MC, Hansen J. Lager brewing yeast. In: Sunnerhagen P, Piškur J, editors. Comparative genomics. Springer-Verlag; Berlin: 2005. pp. 145–164. [Google Scholar]
- Kurtzman CP. Phylogenetic circumscription of Saccharomyces, Kluyveromyces and other members of the Saccharomycetaceae, and the proposal of the new genera Lachancea, Nakaseomyces, Naumovia, Vanderwaltozyma and Zygotorulaspora. FEMS Yeast Research. 2003;4:233–245. doi: 10.1016/S1567-1356(03)00175-2. [DOI] [PubMed] [Google Scholar]
- Kurtzman CP, Robnett CJ. Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie van Leeuwenhoek. 1998;73:331–371. doi: 10.1023/a:1001761008817. [DOI] [PubMed] [Google Scholar]
- Kurtzman CP, Robnett CJ. Phylogenetic relationships among yeasts of the ‘Saccharomyces complex’ determined from multigene sequence analyses. FEMS Yeast Res. 2003;3:417–432. doi: 10.1016/S1567-1356(03)00012-6. [DOI] [PubMed] [Google Scholar]
- Landry CR, Towsend JP, Hartl DL, Cavalieri D. Ecological and evolutionary genomics of Saccharomyces cerevisiae. Mol Ecol. 2006;15:575–591. doi: 10.1111/j.1365-294X.2006.02778.x. [DOI] [PubMed] [Google Scholar]
- Lehner A, Loy A, Behr T, Gaenge H, Ludwig W, Wagner M, Schleifer KH. Oligonucleotide microarray for identification of Enterococcus species. FEMS Microbiol Lett. 2005;246:133–142. doi: 10.1016/j.femsle.2005.04.002. [DOI] [PubMed] [Google Scholar]
- Liti G, Barton DBH, Louis EJ. Sequence diversity, reproductive isolation and species concepts in Saccharomyces. Genetics. 2006;174:839–850. doi: 10.1534/genetics.106.062166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinoni G, Manuel M, Petersen RF, Hvidtfeldt J, Sulo P, Piškur J. Horizontal transfer of genetic material among Saccharomyces yeasts. J Bacteriol. 1999;181:6488–6496. doi: 10.1128/jb.181.20.6488-6496.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masneuf I, Hansen J, Groth C, Piskur J, Dubourdieu D. New hybrids between Saccharomyces sensu stricto yeast species found among wine and cider production strains. Applied and Environmental Microbiology. 1998;64:3887–3892. doi: 10.1128/aem.64.10.3887-3892.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke MA, Scheid MO, Matzke AJ. Rapid structural and epigenetic changes in polyploid and aneuploid genomes. Bioessays. 1999;21:761–767. doi: 10.1002/(SICI)1521-1878(199909)21:9<761::AID-BIES7>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Naumov GI. Genetic identification of biological species in the Saccharomyces sensu stricto complex. J Ind Microbiol Biotechnol. 1996;17:295–302. [Google Scholar]
- Naumov GI, James SA, Naumova ES, Louis EJ, Roberts IN. Three new species in the Saccharomyces sensu stricto complex: Saccharomyces cariocanus, Saccharomyces kudriavzevii and Saccharomyces mikatae. Int J Syst Evol Microbiol. 2000;50:1931–1942. doi: 10.1099/00207713-50-5-1931. [DOI] [PubMed] [Google Scholar]
- Naumova ES, Naumov GI, Masneuf-Pomarède I, Aigle M, Dubourdieu D. Molecular genetic study of introgression between Saccharomyces bayanus and S. cerevisiae. Yeast. 2005;22:1099–1115. doi: 10.1002/yea.1298. [DOI] [PubMed] [Google Scholar]
- Nguyen H-V, Gaillardin C. Evolutionary relationships between the former species Saccharomyces uvarum and the hybrids Saccharomyces bayanus and Saccharomyces pastorianus; reinstatement of Saccharomyces uvarum (Beijerinck) as a distinct species. FEMS Yeast Research. 2005;5:471–483. doi: 10.1016/j.femsyr.2004.12.004. [DOI] [PubMed] [Google Scholar]
- R Development Core Team. R: a language and environment for statistical computing. R Foundation for Statistical Computing; Vienna: 2007. [Google Scholar]
- Rouillard JM, Zuker M, Gulari E. OligoArray 2.0: design of oligonucleotide probes for DNA microarrays using a thermodynamic approach. Nucleic Acids Res. 2003;31:3057–3062. doi: 10.1093/nar/gkg426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rainieri S, Kodama Y, Kaneko Y, Mikata K, Nakao Y, Ashikari T. Pure and mixed lines of Saccharomyces bayanus and Saccharomyces pastorianus and their contribution to the lager brewing strain genome. Appl Environ Microbiol. 2006;72:3968–3974. doi: 10.1128/AEM.02769-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S, editors. Bioinformatics methods and protocols: methods in molecular biology. Humana Press; Totowa, NJ: 2000. pp. 365–386. [DOI] [PubMed] [Google Scholar]
- Sniegowski PD, Dombrowski PG, Fingerman E. Saccharomyces cerevisiae and Saccharomyces paradoxus coexist in a natural woodland site in North America and display different levels of reproductive isolation from European conspecifics. FEMS Yeast Res. 2002;1:299–306. doi: 10.1111/j.1567-1364.2002.tb00048.x. [DOI] [PubMed] [Google Scholar]
- Suh S-O, Blackwell M, Kurtzman CP, Lachance M-A. Phylogenetics of Saccharomycetales, the ascomycete yeasts. Mycologia. 2006;98:1006–1017. doi: 10.3852/mycologia.98.6.1006. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan Martini A, Martini A. Three newly delimited species of Saccharomyces sensu stricto. Antonie van Leeuwenhoek. 1987;53:77–84. doi: 10.1007/BF00419503. [DOI] [PubMed] [Google Scholar]
- Volokhov D, Chizhikov V, Chumakov K, Rasooly A. Microarray-based identification of thermophilic Campylobacter jejuni, C. coli, C. lari, and C. upsaliensis. J Clin Microbiol. 2003;41:4071–4080. doi: 10.1128/JCM.41.9.4071-4080.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W, McCusker JH, Hyman RW, Jones T, Ning Y, Cao Z, Gu Z, Bruno D, Miranda M, Nguyen M, et al. Genome sequencing and comparative analysis of Saccharomyces cerevisiae strain YJM789. Proc Natl Acad Sci USA. 2007;104:12825–12830. doi: 10.1073/pnas.0701291104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.