Abstract
15-Lipoxygenase-1 (15-LOX-1) is transcriptionally silenced in cancer cells, and its transcription reactivation (e.g., via histone deacetylase inhibitors [HDACIs]) restores apoptosis to cancer cells. However, the exact mechanism underlying 15-LOX-1 transcription reactivation in cancer cells is still undefined. Therefore, we evaluated the critical mechanisms required for 15-LOX-1 transcription reactivation in colon cancer cells. Specific HDAC1 and HDAC2 inhibition activated 15-LOX-1 transcription. 15-LOX-1 transcription was repressed through transcription repressor complex recruitment in the region of −120 to −391 of the 15-LOX-1 promoter. The nucleosome remodeling and histone deacetylase (NuRD) repression complex was recruited to this region. Depsipeptide significantly reduced the recruitment of NuRD key components (e.g., MTA1 and HDAC1) to the 15-LOX-1 promoter prior to 15-LOX-1 transcriptional activation. Knock-down of NuRD key components (e.g., MTA1 and HDAC1) by small interfering RNA activated 15-LOX-1 transcription, as measured by luciferase reporter assays in stably transfected SW480 cells with the 15-LOX-1 promoter construct of the −391 but not the −120 region. Relative to expression in normal tissue, MTA1 expression in colorectal cancer mucosa from colorectal cancer patients was negatively related to 15-LOX-1 expression. Thus, our results demonstrate that NuRD contributes to 15-LOX-1 transcription suppression in colon cancer cells and that HDACIs can inhibit NuRD recruitment to a promoter to activate gene transcription, as in the case of 15-LOX-1.
INTRODUCTION
The marked improvement in our understanding of the molecular mechanisms underlying tumorigenesis has led to the emergence of molecular therapeutic targeting, which holds promise for the development of more effective and better-tolerated anticancer therapies (Gibbs, 2000). 15-Lipoxygenase-1 (15-LOX-1) is a critical enzyme for the production of various inflammation-regulatory lipid signaling mediators, including 13-S-hydroxyoctadecadienoic acid from linoleic acid (Baer et al., 1991; Brash et al., 1997), 15-hydroxyeicosatetraenoic acid (15-HETE) and lipoxins from arachidonic acid (Serhan et al., 2003; Takata et al., 1994), and resolvins and protectins from docosahexaenoic acid (Ariel & Serhan, 2007). Apoptosis is restored in cancer cells and tumor cell growth is inhibited by 13-S-HODE (Shureiqi et al., 1999) (Shureiqi et al., 2000b; Wu et al., 2003) and 15-HETE(Chen et al., 2003; Hennig et al., 2007). 15-LOX-1 is a promising molecular target because 15-LOX-1 is downregulated in various human cancers [colon (Heslin et al., 2005; Nixon et al., 2004; Shureiqi et al., 1999), esophageal (Shureiqi et al., 2001), breast (Jiang et al., 2006), and pancreatic (Hennig et al., 2007) cancers] and because 15-LOX-1 re-expression via pharmaceutical agents or plasmid or adenoviral vectors induces growth inhibition and reestablishes terminal differentiation and apoptosis in cancer cells (Deguchi et al., 2005; Heslin et al., 2005; Hsi et al., 2004; Nixon et al., 2004; Shureiqi et al., 2000a; Shureiqi et al., 2003; Shureiqi et al., 2005; Shureiqi et al., 2001; Wu et al., 2003; Wu et al., 2008; Zuo et al., 2006). Identifying the crucial cellular events for maintaining 15-LOX-1 transcription suppression in cancer cells will allow us to identify targets for pharmaceutical inhibition to reconstitute 15-LOX-1 expression and thereby induce apoptosis in cancer cells.
Several mechanisms for the activation of 15-LOX-1 transcription by therapeutic agents have been proposed: global histone 4 acetylation (Kamitani et al., 2001), STAT-6 acetylation and phosphorylation (Conrad & Lu, 2000; Shankaranarayanan et al., 2001), inhibition of GATA-6 transcription repression of the 15-LOX-1 promoter (Kamitani et al., 2000; Shureiqi et al., 2002), and 15-LOX-1 promoter DNA demethylation (Hsi et al., 2005; Liu et al., 2004). Nonetheless, the exact mechanisms underlying the transcription reactivation of 15-LOX-1 remain unknown. For example, it is unclear whether global histone 4 acetylation activates 15-LOX-1 transcription through direct 15-LOX-1 promoter chromatin remodeling or through the expression of transcription factors that subsequently influence the 15-LOX-1 promoter. The requirement for STAT-6 to activate 15-LOX-1 transcription has been questioned on the basis of the observation that suberoylanilide hydroxamic acid (SAHA) downregulates STAT-6 (Zhang et al., 2005a) but still activates 15-LOX-1 transcription (Hsi et al., 2004). GATA-6 knockdown inhibits GATA-6 binding to the 15-LOX-1 promoter but fails to activate 15-LOX-1 transcription (Shureiqi et al., 2007). 15-LOX-1 promoter DNA demethylation is insufficient to reactivate 15-LOX-1 expression (Zuo et al., 2008a).
To identify the crucial molecular mechanisms underlying 15-LOX-1 transcription reactivation in cancer cells, we evaluated the mechanisms by which histone deacetylase inhibitors (HDACIs) activate 15-LOX-1 transcription. HDACIs, a promising class of antitumorigenic agents (Bolden et al., 2006), are the most potent known inducers of 15-LOX-1 transcription activation (Hsi et al., 2004; Kamitani et al., 1998; Kamitani et al., 2000; Kamitani et al., 2001; Shankaranarayanan et al., 2001; Shureiqi et al., 2005; Shureiqi et al., 2007; Zuo et al., 2008a). Both specific and nonspecific inhibitors of a large number of known HDACs have been identified (Bolden et al., 2006). Prior to the current study, reports of 15-LOX-1 activation by HDACIs had been limited to nonspecific HDACIs (Hsi et al., 2004; Kamitani et al., 1998; Kamitani et al., 2000; Kamitani et al., 2001; Shankaranarayanan et al., 2001; Shureiqi et al., 2005; Shureiqi et al., 2007; Zuo et al., 2008a). Therefore, we first determined whether depsipeptide, a selective HDAC1 and HDAC2 inhibitor (Bolden et al., 2006; Furumai et al., 2002), activated 15-LOX-1 transcription. In a subsequent series of experiments, we identified a region in the 15-LOX-1 promoter that is targeted by the nucleosome remodeling and histone deacetylase (NuRD) complex to mediate 15-LOX-1 transcription suppression.
RESULTS
Specific HDAC1 and HDAC2 inhibition and 15-LOX-1 transcription in colon cancer cells
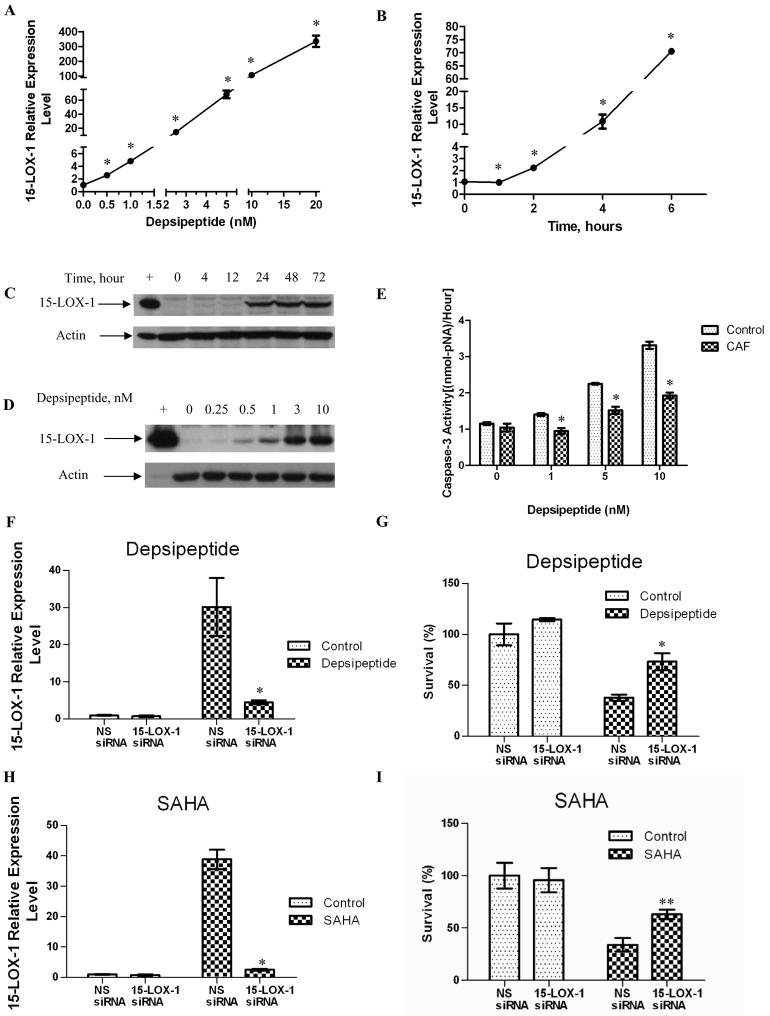
Treatment of Caco-2 and SW480 colon cancer cells with depsipeptide induced 15-LOX-1 mRNA expression in a concentration- and time-dependent manner starting at 0.5 nM (higher than for control cells by 2.83-fold for Caco-2 and 14-fold for SW480 cells; P < 0.0001 for all comparisons) and within 2 h in Caco-2 cells and 4 h in SW480 cells (higher than for control cells by 2.14-fold for Caco-2 and 37-fold for SW480 cells; P < 0.0001 for all comparisons) (Figs. 1A and B for Caco-2; Supplementary Figs. 1A and B for SW480). Depsipeptide also increased 15-LOX-1 protein expression in Caco-2 and SW480 cells in a concentration- and time-dependent manner (Fig. 1C and D for Caco-2; Supplementary Figs. 1C and D for SW480). Protein expression became clearly evident at 24 h in Caco-2 cells (Fig. 1C) and at 12 h in SW480 cells (Supplementary Fig. 1C). Depsipeptide induced apoptosis in Caco-2 and SW480 colon cancer cell lines (Fig. 1E and Supplementary Fig. 1E). Caffeic acid, a direct enzymatic inhibitor of 15-LOX-1 (Gleason et al., 1995), specifically inhibits 15-LOX-1 enzymatic activity at a concentration of 2.2 μM (Shureiqi et al., 2000a), but it had no significant effect on 15-LOX-1 transcriptional activation (Supplementary Fig. 2). Caffeic acid inhibited depsipeptide induction of apoptosis, as measured by a caspase 3 enzymatic activity assay, for all tested concentrations of depsipeptide (Fig. 1E for Caco-2; Supplementary Fig. 1E for SW480). Furthermore, small interfering (siRNA) downregulation of 15-LOX-1 expression induced by depsipeptide or SAHA (Figs. 1F and H) significantly reduced cell growth inhibition by depsipeptide (Fig. 1G) and SAHA (Fig. 1I) in Caco-2 cells. Similar results were observed in SW480 cells (Supplementary Figs. 1F-I).
Fig. 1.
HDAC1 and HDAC2 inhibition and 15-LOX-1 transcription activation. (A and B) Depsipeptide effects on 15-LOX-1 mRNA expression in Caco-2 colon cancer cells. Caco-2 cells were treated with depsipeptide at various concentrations (0-20 nM) for 24 hours (A) and for different time periods (B) at a concentration of 5 nM, and 15-LOX-1 mRNA expression was measured with real-time PCR. The relative expression levels were calculated as the values relative to that of the calibrator sample (time point or concentration 0). Values are the means ± SDs of triplicate experiments. (C and D) Depsipeptide induction of 15-LOX-1 protein expression in Caco-2 cancer cells. Caco-2 cells were treated for various times (at 5 nM depsipeptide concentration, Fig. 1C) and with various concentrations (for 24 h. Fig. 1D) as indicated and processed for 15-LOX-1 Western blotting. + indicates positive control (HCT-116 cells transfected with a 15-LOX-1 expression vector). (E) Effects of 15-LOX-1 enzymatic activity on depsipeptide-induced apoptosis in Caco-2 cancer cells. Caco-2 cells treated with various depsipeptide concentrations, as indicated, were cultured with or without caffeic acid (CAF) at the 2.2 μM concentration that specifically inhibits 15-LOX-1 enzymatic activity. Apoptosis was assessed by measuring caspase 3 activity levels. Values are the means ± SDs of triplicate measurements. F-I. Effects of 15-LOX-1 downregulation on depsipeptide- and SAHA- induced survival inhibition in colon cancer cells. Caco-2 colon cancer cells were transfected with a pool of four siRNA duplexes for 15-LOX-1 or nonspecific siRNA sequence (NS siRNA). 24 h later, they were treated with either (F and G) depsipeptide (5 nM) or (H and I) SAHA (1μM). Control indicates vehicle-solvent treated cells. 48 h later, 15-LOX-1 mRNA was measured by real-time PCR (depsipeptide [F], SAHA treatment [H]). G and I. Cell survival was assessed by SRB assays 72 h after treatment. Values are presented as the survival percentages relative to control (solvent)-treated cells transfected with nonspecific siRNA. Values shown are the means ± SDs of triplicate measurements. * P< 0.0001; ** P = 0.0005.
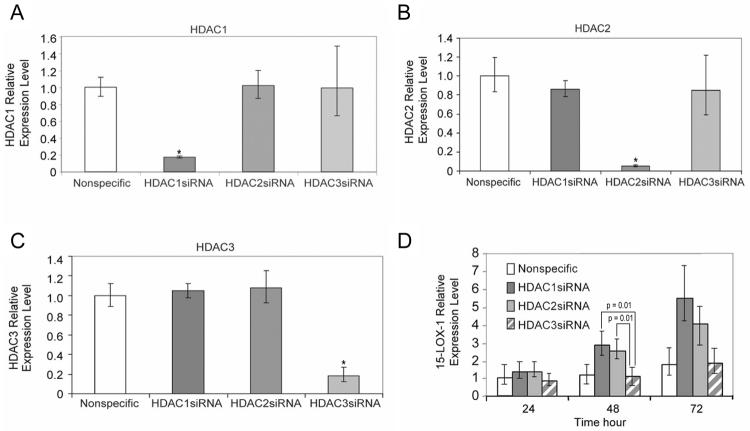
We further tested the specificity of the effects of HDAC1 and HDAC2 on 15-LOX-1 transcription activation relative to that of other HDACs by using a siRNA approach to inhibit HDAC1–11 via siRNA transfection into Caco-2 cells. The expression of 15-LOX-1 mRNA was two or more times higher than that of nonspecific siRNA only in the case of HDAC1 and HDAC2 siRNA transfection (data not shown). HDAC1–3 siRNAs reduced targeted HDAC mRNA expression (ratio to nonspecific siRNA) but not the expression of the other HDACs (83% for HDAC1, 95% for HDAC2, and 82% for HDAC3) (Fig. 2A–C). 15-LOX-1 expression levels were higher for HDAC1 and HDAC2 siRNA than they were for HDAC3 siRNA 48 h after transfection (Fig. 2D).
Fig. 2.
Effects of HDAC1–3 siRNAs on 15-LOX-1 expression. (A–C) Effects of HDAC1–3 siRNA transfection on HDAC expression. Caco-2 cells were transfected with pools of four siRNA duplexes, each designed specifically against HDAC1, HDAC2, or HDAC3 or with a nonspecific siRNA sequence. Cells were harvested 24 h later. HDAC1 (A), HDAC2 (B), and HDAC3 (C) mRNA levels were measured by real-time PCR. The relative expression levels were calculated as the values relative to those of the calibrator samples (nonspecific siRNA). Values shown are the means ± SDs of triplicate experiments. (D) Effects of HDAC1–3 downregulation on 15-LOX-1 expression. Caco-2 cells were transfected as in panel A-C. Cells were harvested 24, 48, and 72 h after transfection and processed for 15-LOX-1 measurements by real-time PCR. Values shown are the means ± SDs of triplicate experiments. * P < 0.0001.
15-LOX-1 transcription is repressed through transcription repressor complex recruitment in the region of −120 to −391 of the 15-LOX-1 promoter.
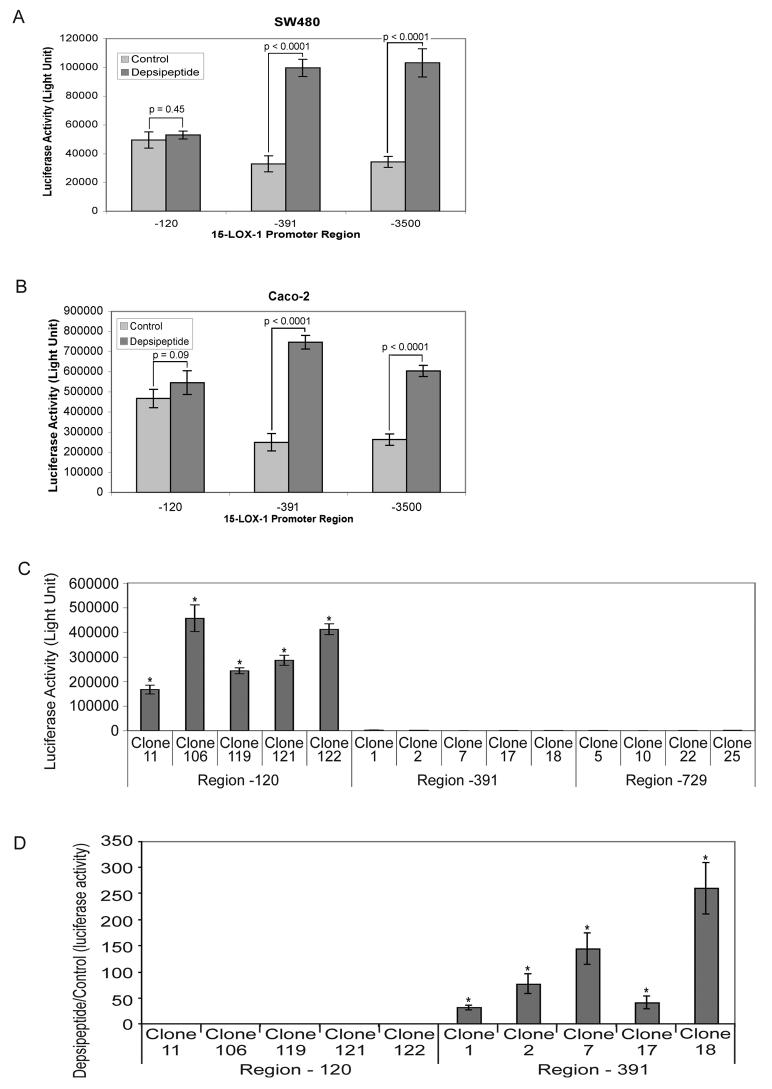
We used promoter-deletion analyses to identify the region in the 15-LOX-1 promoter in which HDAC1 and HDAC2 exert their repressive effects. In transient-transfection assays of luciferase reporter constructs for the −120, −391, and −3500 to +18 bp regions of the 15-LOX-1 promoter, the −120 region had higher basal activity than did the −391 or −3500 regions, and depsipeptide treatment significantly increased transcription activation of the −391 and −3500 regions but not the −120 region in SW480 and Caco-2 cell lines (Fig. 3A and B). (Control −120 vs. −391 and −3500 regions, P < 0.0013 for both Caco-2 and SW480.)
Fig. 3.
Transcription activity of 15-LOX-1 promoter deletion constructs. (A) SW480 cells and (B) Caco-2 cells were transiently transfected with PGL4.16 luciferase reporter vectors containing the −120 to +18 bp (−120), −391 to +18 bp (−391), or −3500 to +18 bp (−3500) regions of the 15-LOX-1 promoter and treated with either depsipeptide or the control vehicle. Transcription activity was measured 24 h after treatment. (C and D) PGL4.16 vectors containing −120, −391, or −729 to +18 bp (−729) regions of the 15-LOX-1 promoter were stably transfected into SW480 cells. (C) Basal transcription levels (measured as luciferase reporter activity) for representative stably transfected clones with −120, −391, and −729 region vectors are shown. Values are the means ± SDs of triplicate experiments. (D) Depsipeptide effects on transcription activation for the −120 and −391 regions of the 15-LOX-1 promoter. SW480 stably transfected clones with −120 or −391 region vectors (as described in panel C) were treated with either depsipeptide or DMSO only (control). Luciferase activity was measured 24 h later. Values are the ratios of depsipeptide- to control-treated cells (means ± SDs of triplicate experiments). * P < 0.0001.
Several representative clones of SW480 cells stably transfected with one of the three 15-LOX-1 promoter deletion constructs (−120, −391, or −729 to +18 bp sequence of the 15-LOX-1 promoter) were used for characterization. Transcription activities were 226 to 363 times higher for the −120 region of the 15-LOX-1 promoter than for the −391 and −729 regions (Fig. 3C). Depsipeptide treatment markedly increased 15-LOX-1 promoter transcription activation for the −391 region (median of depsipeptide treated-to-control ratio, 78; range, 32–260) but only minimally for the −120 region (median of depsipeptide treated-to-control ratio, 1; range, 0.94–1.2; P = 0.8) (Fig. 3D).
NuRD repression complex recruitment to the 15-LOX-1 promoter and 15-LOX-1 transcription repression
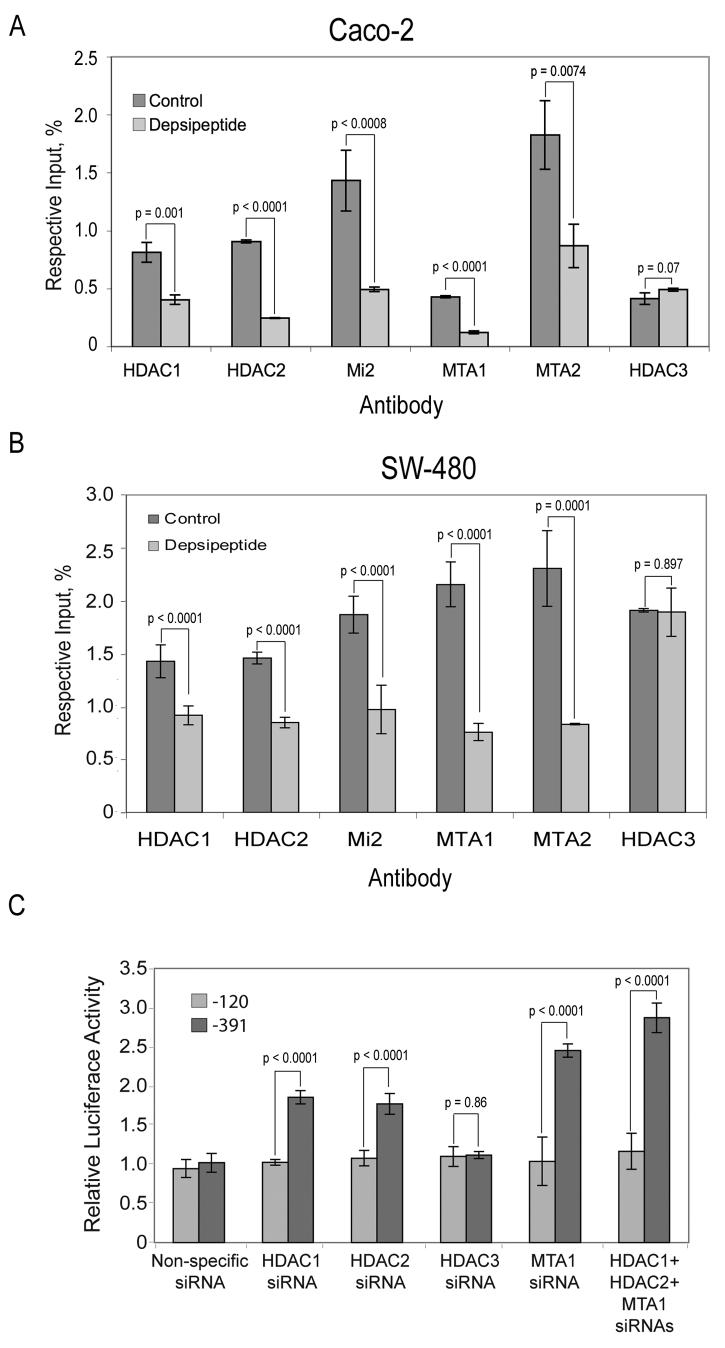
Depsipeptide significantly reduced the recruitment of essential components of the NuRD repression complex from the 15-LOX-1 promoter region between −215 and −283 bp, including HDAC1, HDAC2, Mi2, MTA1, and MTA2 (Fig. 4A for Caco-2; Fig. 4B for SW480). In contrast, depsipeptide treatment did not decrease the recruitment of HDAC3, a component of the nuclear compressor complex, (NCoR) (Li et al., 2000), to the same 15-LOX-1 promoter region (Fig. 4A for Caco-2; Fig. 4B for SW480). Depsipeptide treatment had no effects on HDAC1 or MTA1 protein expression (Supplementary Fig. 3). HDAC1 and HDAC2 siRNA, but not HDAC3 siRNA, activated 15-LOX-1 transcription, as measured by a luciferase reporter assay, in stably transfected SW480 cells with the 15-LOX-1 promoter −391 construct but not the −120 construct (Fig. 4C). MTA1 siRNA activated 15-LOX-1 transcription in the same system (Fig. 4C). HDAC1 + HDAC2 + MTA1 siRNA resulted in a further increase in 15-LOX-1 transcription activation compared with HDAC1 or HDAC2 siRNA alone (P < 0.001; Fig. 4C).
Fig. 4.
Dissociation of the NuRD complex from the 15-LOX-1 promoter and 15-LOX-1 transcription activation. (A and B) Inhibition of NuRD complex recruitment to the 15-LOX-1 promoter during 15-LOX-1 transcription activation. (A) Caco-2 cells were treated with depsipeptide (5 nM) or DMSO for 15 min. Cells were subjected to ChIP/real-time PCR assays using HDAC1, HDAC2, HDAC3, Mi2, MTA1, or MTA2 antibodies. Data are given as percentages of the respective input genomic DNA for the 15-LOX-1 promoter. Values are the means ± SDs of triplicate measurements. (B) SW480 cells were treated with depsipeptide (5 nM) or DMSO for 3.5 h. Cells were subjected to the same assays described in A, and data are presented in the same way. (C) Effects of HDAC1–3 and MTA1 downregulation on 15-LOX-1 expression. SW480 cells stably transfected with PGL4.16 luciferase reporter vector containing the −120 or −391 bp region of the 15-LOX-1 promoter were transfected with pools of four siRNA duplexes. Each pool was designed specifically against HDAC1, HDAC2, HDAC3, or MTA1 or a nonspecific siRNA sequence (control). Luciferase activity was measured 72 h later. Relative luciferase activity represents the ratios to that of the mock experiment (transfection medium alone). Values shown are the means ± SDs of triplicate experiments.
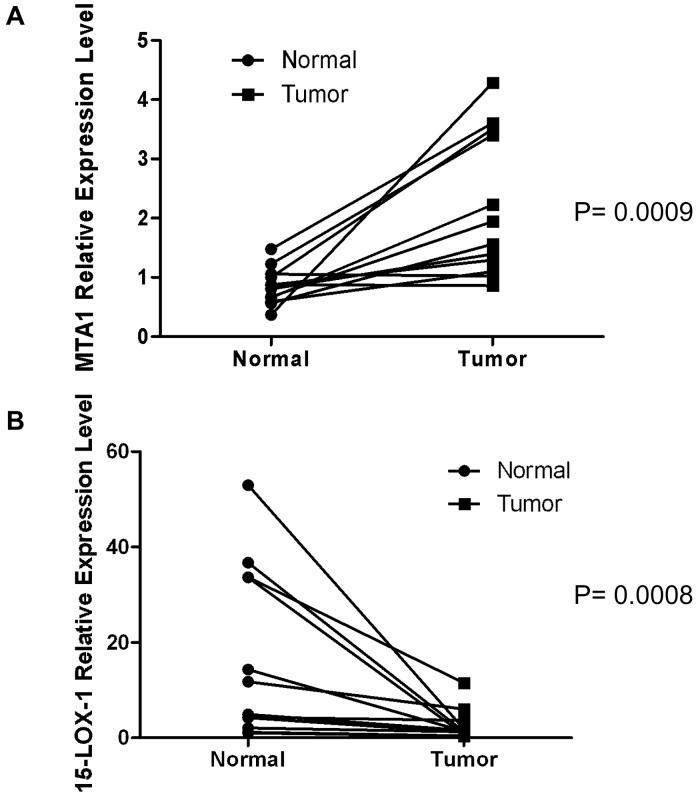
MTA1 and 15-LOX-1 expression in colorectal cancer
MTA1 mRNA was higher in cancerous colorectal mucosa than in paired normal mucosa in samples from 10 of 12 tested patients (Fig. 5A) (P = 0.0009). In contrast, 15-LOX-1 mRNA expression levels were higher in normal than cancerous mucosa in samples from 11 of 12 of the same patients (Fig. 5B) (P = 0.0008). MTA-1 protein expression, measured by immunohistochemistry staining, was higher in all examined cases (10/10, P = 0.001) (Supplementary Fig. 4). The median intensity of staining was 3 in the tumors and 0.5 in the normal tissues.
Fig. 5.
MTA1 and 15-LOX-1 expression in colorectal cancer. (A) MTA1 mRNA expression levels in cancerous and normal colorectal mucosa. MTA1 was measured by real-time PCR in paired colorectal cancer and normal tissue samples from 12 patients. Values shown are the means ± SDs of duplicate measurements. (B) 15-LOX-1 mRNA expression levels in cancerous and normal colorectal mucosa. 15-LOX-1 was measured by real-time PCR from the same paired colorectal cancer and normal tissue samples used for MTA1 measurements in Fig. 5A. Values shown are the means ± SDs of triplicate measurements.
DISCUSSION
Our findings demonstrate that NuRD recruitment to the 15-LOX-1 promoter contributes to 15-LOX-1 transcription suppression and that HDACIs can activate gene transcription via NuRD dissociation from a promoter, as in the case of 15-LOX-1 in colon cancer cells.
In our study, 15-LOX-1 transcription activation occurred through selective HDAC1 and HDAC2 inhibition. Depsipeptide, a selective HDAC1 and HDAC2 inhibitor (Bolden et al., 2006), activated 15-LOX-1 transcription at much lower concentrations (starting at 0.5 nM) than those required for nonspecific HDACIs (e.g., 5 mM for sodium butyrate (Kamitani et al., 1998) and 1–2.5 μM for SAHA (Hsi et al., 2004) and this report]). These data suggest that 15-LOX-1 transcription suppression is more specifically related to HDAC1 and HDAC2. Early transcription activation by depsipeptide (2–4 h) suggests that depsipeptide directly modulates 15-LOX-1 transcription. Similar to nonspecific HDACIs (Hsi et al., 2004), depsipeptide activation of 15-LOX-1 significantly contributed to its anti-tumorigenic effects in vitro. Depsipeptide activation of 15-LOX-1 transcription and apoptosis occurred within the same concentration range, and specific inhibition of 15-LOX-1 enzymatic activity suppressed apoptosis induction by depsipeptide. 15-LOX-1 downregulation by siRNA significantly repressed SAHA and depsipeptide ability to inhibit colon cancer cell growth. The incomplete inhibition of apoptosis is likely related to the ability of HDACIs to trigger apoptosis via mechanisms (Bolden et al., 2006) besides 15-LOX-1 transcription activation, but the current data demonstrate the significance of 15-LOX-1 as a mechanism for HDACIs' antitumorigenic effects. The delay between 15-LOX-1 mRNA (real-time PCR) and protein (Western blotting) expression detection may be partly due to the higher detection sensitivity of real-time PCR than of Western blotting. In addition, 15-LOX-1 is tightly regulated at the translational levels by hnRNP K and hnRNP E1 in normal cells (Ostareck et al., 1997). Whether an element of this translational regulation contributed to the apparent lag time between 15-LOX-1 mRNA and protein synthesis in depsipeptide-treated cancer cells cannot be determined on the basis of our current studies, which were focused on studying transcriptional regulation of 15-LOX-1.
We further confirmed the specific role of HDAC1 and HDAC2 in 15-LOX-1 transcription suppression by siRNA knockdown experiments. We found that HDAC1 and HDAC2 downregulation activated 15-LOX-1, in contrast to other HDACs, including the other members of HDAC class I (e.g., HDAC3). Thus, our data demonstrate, for the first time, the specific role of HDAC1 and HDAC2 in 15-LOX-1 transcription repression. In addition, these results suggest a mechanistic link between HDAC1 and HDAC2 upregulation (Huang et al., 2005; Zhu et al., 2004) and 15-LOX-1 downregulation (Heslin et al., 2005; Nixon et al., 2004; Shureiqi et al., 1999; Shureiqi et al., 2005; Takamitsu et al., 2006) in human colonic tumorigenesis.
HDAC1 and HDAC2 act within the NuRD transcription complex to suppress 15-LOX-1 transcription. HDACs have no direct DNA binding sites and are recruited as part of transcription repression complexes to promoters (Marks et al., 2004). We identified the region between −120 and −391 bp of the 15-LOX-1 promoter as being the target for repression complex recruitment on the basis of our observations that basal transcription activity was profoundly suppressed for the regions upstream of −120 and that depsipeptide activated transcription for the −391 but not the −120 region.
The NuRD repression complex, which contains HDAC1 and HDAC2 as core proteins (Brehm et al., 1999; Manavathi & Kumar, 2007; Xue et al., 1998; Zhang et al., 2005b), is a strong candidate for being the repression complex recruited to this 15-LOX-1 promoter region, according to our results. NuRD components, including the distinctive NuRD proteins MTA1 and MTA2 (Sharma et al., 2006), were recruited to the 15-LOX-1 promoter region that we identified as the site for repressor complex recruitment. Depsipeptide induced dissociation of the NuRD components from this region prior to 15-LOX-1 transcription activation; however, in the control experiment, depsipeptide had no effect on HDAC3, a component of the NCoR complex. The downregulation of essential NuRD complex components such as HDAC1, HDAC2, and MTA1 by siRNA activated transcription in the −391 but not the −120 bp region of the 15-LOX-1 promoter, whereas HDAC3 siRNA had no effect. These findings indicate that NuRD has an important and specific role in 15-LOX-1 repression. Depsipeptide effects on NuRD were not secondary to downregulation of NuRD complex components, as the expression of essential NuRD components (e.g., HDAC1 and MTA1) remained unchanged by depsipeptide treatment. Further studies are needed to determine the mechanisms by which depsipeptide causes dissociation of the NuRD complex from the promoter. Our findings also demonstrate, for the first time to our knowledge, a new mechanism for gene transcription activation by HDACIs through inhibiting NuRD recruitment to a promoter.
MTA1 overexpression in colon cancer was associated with 15-LOX-1 transcriptional suppression in human colorectal cancers. MTA1 is overexpressed in various human cancers, and MTA1 overexpression in mouse models promotes lymphoma and breast tumorigenesis (Manavathi & Kumar, 2007). MTA1 has been reported to be overexpressed in human colon cancer at mRNA levels (Giannini & Cavallini, 2005), but the relevance of this finding to colon cancer biology has remained undefined prior to our current study. Our clinical findings, which also included the newly demonstrated MTA1 upregulation at protein levels in colon cancer cells in vivo, support those of our earlier in vitro studies and suggest that MTA1, acting as part of the NuRD complex, contributes to colonic tumorigenesis by suppressing 15-LOX-1 transcription.
In summary, our findings demonstrate that HDAC1 and HDAC2 act as part of the NuRD repression complex to suppress 15-LOX-1 transcription in cancer cells. Further studies of the mechanisms of NuRD recruitment into the 15-LOX-1 promoter will help to not only improve our understanding of the mechanism of 15-LOX-1 silencing in cancer cells but also identify novel molecular targets for inhibiting colonic tumorigenesis.
MATERIALS AND METHODS
Materials
SAHA was provided by Merck and Company, Inc., and the National Cancer Institute; depsipeptide was provided by Gloucester Pharmaceuticals, Inc., and the National Cancer Institute. Antibodies against HDAC1, HDAC2, and HDAC3 were purchased from Upstate Cell Signaling Solutions (Lake Placid, NY). Anti-MTA1, anti-MTA2, and anti-Mi2 antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Rabbit polyclonal antiserum to recombinant human 15-LOX-1 protein was obtained as described previously (Shureiqi et al., 2003). Caco-2 and SW480 colon cancer cell lines were purchased from the American Type Culture Collection (Manassas, VA). Other reagents, molecular-grade solvents, and chemicals were obtained from commercial manufacturers or as specified.
Acquisition of clinical samples
Colon biopsy specimens were obtained from colorectal cancer patients after approval by the institutional review board of each institution, as described previously (Zuo et al., 2008a). Patients' demographic and clinical characteristics were included in the analyses and are shown in Table 1. No patients were being treated with NSAIDs at the time of the study.
TABLE 1.
Patients' demographic and clinical characteristics
| No. | Age (years) |
Sex | Ethnicity | BMI | Tumor stage |
Tumor grade |
|---|---|---|---|---|---|---|
| 1 | 67 | Female | Hispanic | 22.32 | 4 | Well differentiated adenocarcinoma |
| 2 | 65 | Female | White | 23.20 | 4 | Moderately differentiated adenocarcinoma |
| 3 | 55 | Male | White | 33.38 | 2 | Moderately differentiated adenocarcinoma |
| 4 | 47 | Female | White | 28.49 | 2 | Moderately differentiated adenocarcinoma |
| 5 | 80 | Female | White | 29.95 | 2 | Moderately differentiated adenocarcinoma |
| 6 | 80 | Male | Asian | 23.33 | 3 | Moderately differentiated adenocarcinoma |
| 7 | 51 | Female | White | 20.97 | 3 | Moderately differentiated adenocarcinoma |
| 8 | 73 | Female | White | 23.24 | 2 | Moderate to Poorly differentiated adenocarcinoma |
| 9 | 54 | Female | White | 28.72 | 2 | Moderately differentiated adenocarcinoma |
| 10 | 63 | Female | Hispanic | 20.68 | 2 | Poorly differentiated adenocarcinoma |
| 11 | 56 | Female | White | 35.61 | 2 | Moderately differentiated adenocarcinoma |
| 12 | 63 | Male | White | 24.00 | 3 | Moderately differentiated adenocarcinoma |
Cell cultures and treatments
SW480 cells were grown in RPMI 1640 medium, and Caco-2 cells were grown in Eagle's minimal essential medium (Cambrex, Walkersville, MD) with L-glutamine in a humidified atmosphere containing 5% CO2 at 37°C. Both media contained 10% fetal bovine serum and were supplemented with 1% penicillin-streptomycin, as described previously (Shureiqi et al., 2003). Depsipeptide and SAHA were dissolved in dimethyl sulfoxide (DMSO) and added to the culture media at the indicated concentrations.
Transient transfection of 15-LOX-1 deletion constructs
To identify the region of repressor recruitment in the 15-LOX-1 promoter, SW480 and Caco-2 cells were cotransfected with −120, −391, or −3500 bp to +18 sequence of the 15-LOX-1 promoter subcloned into the pGL4.16 (luc2CP/Hygro) vector, which has a luciferase reporter system (Promega, Madison, WI), and the pSV-β-galactosidase vector (Promega) using Lipofectamine 2000 (Invitrogen). Cells were treated with 5 nM depsipeptide 24 h after transfection, and luciferase activity was measured 24 h after depsipeptide treatment, as described previously (Shureiqi et al., 2003).
Establishment of stably transfected cell lines with 15-LOX-1 promoter deletion constructs
We transfected SW480 cells with 15-LOX-1 promoter deletion constructs that contained the −120, −391, or −729 bp to +18 bp sequence of the 15-LOX-1 promoter subcloned into the pGL4.16 (luc2CP/Hygro) vector (Promega). Stably transfected clones were selected using hygromycin B (Invitrogen, Carlsbad, CA). Representative clones for each of the three deletion constructs were used for characterization. Cells were harvested, and luciferase activity was measured using a luciferase assay kit (Promega).
siRNA transfection
SW480 and Caco-2 cells were cultured to 40% to 50% confluence and transfected with 100 nM of a pooled mixture of four siRNA duplexes (SMARTselected or OnTarget; Dharmacon, Inc., Lafayette, CO) for the targeted gene (e.g., MTA1 or HDACs) or with nonspecific control siRNA (Dharmacon, Inc.) using Lipofectamine 2000 (Invitrogen).
RNA extraction and real-time polymerase chain reaction (PCR)
Total RNA was extracted from cells using TRI reagent (Molecular Research Center, Inc., Cincinnati, OH). RNA from each sample was reverse transcribed and then measured quantitatively by real-time PCR using a comparative Ct method, as described previously (Shureiqi et al., 2007).
Western blot analyses
For Western blotting, protein samples were prepared, subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis under reducing conditions, and probed with rabbit polyclonal antibody to human 15-LOX-1, MTA1, or HDAC1 using methods similar to those described previously (Shureiqi et al., 2005).
Chromatin immunoprecipitation (ChIP) and ChIP/real-time PCR assays
Cross-linking was performed by adding formaldehyde to the cell culture medium to a final concentration of 1% and incubating the medium for 10 min at 37°C. ChIP assays were performed using a commercial kit according to the manufacturer's protocol (Upstate Cell Signaling Solutions). Chromatin was immunoprecipitated using the indicated antibodies. The relative enrichment of transcription factors (e.g., MTA1 and HDACs) was measured using ChIP/real-time PCR with the following primers and FAM dye-labeling probe: forward, GTGTTTTCGGTCCAAATCCTTTTCT; reverse, GAGAGCAGGGAGTGGAAACC; and probe, CCTCCCGTCAAGATAGT to amplify the −283 bp to −215 bp region of the 15-LOX-1 promoter relative to the ATG site. Real-time PCR was performed as described previously (Zuo et al., 2008b).
Immunohistochemical evaluation of MTA1 expression
Paraffin-embedded tissue blocks were cut into 4-mm-thick sections, incubated at 60° C overnight, and deparaffinized in xylene. Sections were stained for MTA-1, as previously described (Balasenthil et al., 2006) with minor modifications that included: (a) Slides were brought to a boiling temperature in 10 mM sodium citrate buffer pH 6.0 then maintained at a sub-boiling temperature for 20 minutes; (b) Sections were incubated with 3% hydrogen peroxide and methanol for 15 minutes; (c) a anti human MTA-1 primary antibody (anti-MTA1 polyclonal sc-17773, Santa Cruz Biotechnology) was used at 1:100 concentration. The specific staining of MTA1 antibody was assessed by an experienced pathologist (R.B)(Balasenthil et al., 2006) semiquantitatively using a 4-point scale: 0 = no staining, 1 = mild staining, 2 = moderate staining, and 3 = intense staining.
Cell survival and apoptosis assays
We measured cell survival using the sulforhodamine B assay and apoptosis by measuring caspase 3 activation with a commercial kit (BD Biosciences Clontech, Palo Alto, CA), as described previously (Shureiqi et al., 2007).
Statistical analyses
We used the t-test for two-group comparisons. For analyses involving single factors and more than two groups, we performed a one-way analysis of variance. If the overall analysis of variance test result was significant, we performed pairwise comparisons, adjusting for multiplicities using the Bonferroni correction. We analyzed data involving the simultaneous consideration of two factors using two-way analysis of variance. We first tested the interaction effect, and if it was statistically significant, we performed specific comparisons to investigate which differences were driving the effect, using the Bonferroni correction to adjust for the multiple testing problem. An individual comparison was considered significant only if the P value was less than 0.05/k, with k representing the number of comparisons performed. If the interaction effect was not significant, we tested the individual main effects. If those were significant, we determined which pairwise comparisons were significant, again adjusting for multiplicities using the Bonferroni correction. We used the sign test for nonparametric data analyses for MTA-1 protein expression (immunohistochemistry). Quantitative data log-transformation was used to account for the normal-distributional assumptions underlying the methods. Data were analyzed using SAS software (SAS Institute, Cary, NC).
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by the National Cancer Institute, National Institutes of Health, Department of Health and Human Services R01 grants CA106577 and CA104278; the Caroline Wiess Law Endowment for Cancer Prevention; The Jerry and Maury Rubenstein Foundation and the National Colorectal Cancer Research Alliance. We thank Merck and Company, Inc., Gloucester Pharmaceuticals, and the National Cancer Institute for providing SAHA and depsipeptide. We also thank Ann M. Sutton of the Department of Scientific Publications at The University of Texas M. D. Anderson Cancer Center for editing the manuscript.
REFERENCES
- Ariel A, Serhan CN. Resolvins and protectins in the termination program of acute inflammation. Trends Immunol. 2007;28:176–83. doi: 10.1016/j.it.2007.02.007. [DOI] [PubMed] [Google Scholar]
- Baer AN, Costello PB, Green FA. In vivo activation of an omega-6 oxygenase in human skin. Biochem Biophys Res Commun. 1991;180:98–104. doi: 10.1016/s0006-291x(05)81260-4. [DOI] [PubMed] [Google Scholar]
- Balasenthil S, Broaddus RR, Kumar R. Expression of metastasis-associated protein 1 (MTA1) in benign endometrium and endometrial adenocarcinomas. Human Pathology. 2006;37:656–661. doi: 10.1016/j.humpath.2006.01.024. [DOI] [PubMed] [Google Scholar]
- Bolden JE, Peart MJ, Johnstone RW. Anticancer activities of histone deacetylase inhibitors. Nat Rev Drug Discov. 2006;5:769–84. doi: 10.1038/nrd2133. [DOI] [PubMed] [Google Scholar]
- Brash AR, Boeglin WE, Chang MS. Discovery of a second 15S-lipoxygenase in humans. Proc Natl Acad Sci U S A. 1997;94:6148–52. doi: 10.1073/pnas.94.12.6148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brehm A, Nielsen SJ, Miska EA, McCance DJ, Reid JL, Bannister AJ, et al. The E7 oncoprotein associates with Mi2 and histone deacetylase activity to promote cell growth. Embo J. 1999;18:2449–58. doi: 10.1093/emboj/18.9.2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen GG, Xu H, Lee JF, Subramaniam M, Leung KL, Wang SH, et al. 15-hydroxy-eicosatetraenoic acid arrests growth of colorectal cancer cells via a peroxisome proliferator-activated receptor gamma-dependent pathway. Int J Cancer. 2003;107:837–43. doi: 10.1002/ijc.11447. [DOI] [PubMed] [Google Scholar]
- Conrad DJ, Lu M. Regulation of Human 12/15-Lipoxygenase by Stat6-Dependent Transcription. Am. J. Respir. Cell Mol. Biol. 2000;22:226–234. doi: 10.1165/ajrcmb.22.2.3786. [DOI] [PubMed] [Google Scholar]
- Deguchi A, Xing SW, Shureiqi I, Yang P, Newman RA, Lippman SM, et al. Activation of protein kinase G up-regulates expression of 15-lipoxygenase-1 in human colon cancer cells. Cancer Res. 2005;65:8442–7. doi: 10.1158/0008-5472.CAN-05-1109. [DOI] [PubMed] [Google Scholar]
- Furumai R, Matsuyama A, Kobashi N, Lee KH, Nishiyama M, Nakajima H, et al. FK228 (depsipeptide) as a natural prodrug that inhibits class I histone deacetylases. Cancer Res. 2002;62:4916–21. [PubMed] [Google Scholar]
- Giannini R, Cavallini A. Expression analysis of a subset of coregulators and three nuclear receptors in human colorectal carcinoma. Anticancer Res. 2005;25:4287–92. [PubMed] [Google Scholar]
- Gibbs JB. Mechanism-based target identification and drug discovery in cancer research. Science. 2000;287:1969–73. doi: 10.1126/science.287.5460.1969. [DOI] [PubMed] [Google Scholar]
- Gleason MM, Rojas CJ, Learn KS, Perrone MH, Bilder GE. Characterization and inhibition of 15-lipoxygenase in human monocytes: comparison with soybean 15-lipoxygenase. Am J Physiol. 1995;268:C1301–7. doi: 10.1152/ajpcell.1995.268.5.C1301. [DOI] [PubMed] [Google Scholar]
- Hennig R, Kehl T, Noor S, Ding XZ, Rao SM, Bergmann F, et al. 15-Lipoxygenase-1 Production is Lost in Pancreatic Cancer and Overexpression of the Gene Inhibits Tumor Cell Growth. Neoplasia. 2007;9:917–26. doi: 10.1593/neo.07565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heslin MJ, Hawkins A, Boedefeld W, Arnoletti JP, Frolov A, Soong R, et al. Tumor-associated down-regulation of 15-lipoxygenase-1 is reversed by celecoxib in colorectal cancer. Ann Surg. 2005;241:941–6. doi: 10.1097/01.sla.0000164177.95620.c1. discussion 946-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsi LC, Xi X, Lotan R, Shureiqi I, Lippman SM. The histone deacetylase inhibitor suberoylanilide hydroxamic acid induces apoptosis via induction of 15-lipoxygenase-1 in colorectal cancer cells. Cancer Res. 2004;64:8778–81. doi: 10.1158/0008-5472.CAN-04-1867. [DOI] [PubMed] [Google Scholar]
- Hsi LC, Xi X, Wu Y, Lippman SM. The methyltransferase inhibitor 5-aza-2-deoxycytidine induces apoptosis via induction of 15-lipoxygenase-1 in colorectal cancer cells. Mol Cancer Ther. 2005;4:1740–6. doi: 10.1158/1535-7163.MCT-05-0218. [DOI] [PubMed] [Google Scholar]
- Huang BH, Laban M, Leung CHW, Lee L, Lee CK, Salto-Tellez M, et al. Inhibition of histone deacetylase 2 increases apoptosis and p21Cip1//WAF1 expression, independent of histone deacetylase 1. Cell Death Differ. 2005;12:395–404. doi: 10.1038/sj.cdd.4401567. [DOI] [PubMed] [Google Scholar]
- Jiang WG, Watkins G, Douglas-Jones A, Mansel RE. Reduction of isoforms of 15-lipoxygenase (15-LOX)-1 and 15-LOX-2 in human breast cancer. Prostaglandins, Leukotrienes and Essential Fatty Acids. 2006;74:235–245. doi: 10.1016/j.plefa.2006.01.009. [DOI] [PubMed] [Google Scholar]
- Kamitani H, Geller M, Eling T. Expression of 15-lipoxygenase by human colorectal carcinoma Caco-2 cells during apoptosis and cell differentiation. J Biol Chem. 1998;273:21569–77. doi: 10.1074/jbc.273.34.21569. [DOI] [PubMed] [Google Scholar]
- Kamitani H, Kameda H, Kelavkar UP, Eling TE. A GATA binding site is involved in the regulation of 15-lipoxygenase-1 expression in human colorectal carcinoma cell line, caco-2. FEBS Lett. 2000;467:341–7. doi: 10.1016/s0014-5793(00)01155-8. [DOI] [PubMed] [Google Scholar]
- Kamitani H, Taniura S, Ikawa H, Watanabe T, Kelavkar UP, Eling TE. Expression of 15-lipoxygenase-1 is regulated by histone acetylation in human colorectal carcinoma. Carcinogenesis. 2001;22:187–91. doi: 10.1093/carcin/22.1.187. [DOI] [PubMed] [Google Scholar]
- Li J, Wang J, Wang J, Nawaz Z, Liu JM, Qin J, et al. Both corepressor proteins SMRT and N-CoR exist in large protein complexes containing HDAC3. Embo J. 2000;19:4342–50. doi: 10.1093/emboj/19.16.4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Xu D, Sjoberg J, Forsell P, Bjorkholm M, Claesson HE. Transcriptional regulation of 15-lipoxygenase expression by promoter methylation. Exp Cell Res. 2004;297:61–7. doi: 10.1016/j.yexcr.2004.02.014. [DOI] [PubMed] [Google Scholar]
- Manavathi B, Kumar R. Metastasis tumor antigens, an emerging family of multifaceted master coregulators. J Biol Chem. 2007;282:1529–33. doi: 10.1074/jbc.R600029200. [DOI] [PubMed] [Google Scholar]
- Marks PA, Richon VM, Miller T, Kelly WK. Histone Deacetylase Inhibitors. Adv Cancer Res. 2004;91:137–168. doi: 10.1016/S0065-230X(04)91004-4. [DOI] [PubMed] [Google Scholar]
- Nixon JB, Kim KS, Lamb PW, Bottone FG, Eling TE. 15-Lipoxygenase-1 has anti-tumorigenic effects in colorectal cancer. Prostaglandins Leukot Essent Fatty Acids. 2004;70:7–15. doi: 10.1016/j.plefa.2003.06.001. [DOI] [PubMed] [Google Scholar]
- Ostareck DH, Ostareck-Lederer A, Wilm M, Thiele BJ, Mann M, Hentze MW. mRNA Silencing in Erythroid Differentiation: hnRNP K and hnRNP E1 Regulate 15-Lipoxygenase Translation from the 3′ End. Cell. 1997;89:597–606. doi: 10.1016/s0092-8674(00)80241-x. [DOI] [PubMed] [Google Scholar]
- Serhan CN, Jain A, Marleau S, Clish C, Kantarci A, Behbehani B, et al. Reduced inflammation and tissue damage in transgenic rabbits overexpressing 15-lipoxygenase and endogenous anti-inflammatory lipid mediators. J Immunol. 2003;171:6856–65. doi: 10.4049/jimmunol.171.12.6856. [DOI] [PubMed] [Google Scholar]
- Shankaranarayanan P, Chaitidis P, Kuhn H, Nigam S. Acetylation by histone acetyltransferase CREB-binding protein/p300 of STAT6 is required for transcriptional activation of the 15-lipoxygenase-1 gene. J Biol Chem. 2001;276:42753–60. doi: 10.1074/jbc.M102626200. [DOI] [PubMed] [Google Scholar]
- Sharma D, Saxena NK, Davidson NE, Vertino PM. Restoration of tamoxifen sensitivity in estrogen receptor-negative breast cancer cells: tamoxifen-bound reactivated ER recruits distinctive corepressor complexes. Cancer Res. 2006;66:6370–8. doi: 10.1158/0008-5472.CAN-06-0402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shureiqi I, Chen D, Lee JJ, Yang P, Newman RA, Brenner DE, et al. 15-LOX-1: a novel molecular target of nonsteroidal anti-inflammatory drug-induced apoptosis in colorectal cancer cells. J Natl Cancer Inst. 2000a;92:1136–42. doi: 10.1093/jnci/92.14.1136. [DOI] [PubMed] [Google Scholar]
- Shureiqi I, Chen D, Lotan R, Yang P, Newman RA, Fischer SM, et al. 15-Lipoxygenase-1 mediates nonsteroidal anti-inflammatory drug-induced apoptosis independently of cyclooxygenase-2 in colon cancer cells. Cancer Res. 2000b;60:6846–50. [PubMed] [Google Scholar]
- Shureiqi I, Jiang W, Fischer SM, Xu X, Chen D, Lee JJ, et al. GATA-6 transcriptional regulation of 15-lipoxygenase-1 during NSAID-induced apoptosis in colorectal cancer cells. Cancer Res. 2002;62:1178–83. [PubMed] [Google Scholar]
- Shureiqi I, Jiang W, Zuo X, Wu Y, Stimmel JB, Leesnitzer LM, et al. The 15-lipoxygenase-1 product 13-S-hydroxyoctadecadienoic acid down-regulates PPAR-delta to induce apoptosis in colorectal cancer cells. Proc Natl Acad Sci U S A. 2003;100:9968–73. doi: 10.1073/pnas.1631086100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shureiqi I, Wojno KJ, Poore JA, Reddy RG, Moussalli MJ, Spindler SA, et al. Decreased 13-S-hydroxyoctadecadienoic acid levels and 15-lipoxygenase-1 expression in human colon cancers. Carcinogenesis. 1999;20:1985–95. doi: 10.1093/carcin/20.10.1985. [DOI] [PubMed] [Google Scholar]
- Shureiqi I, Wu Y, Chen D, Yang XL, Guan B, Morris JS, et al. The Critical Role of 15-Lipoxygenase-1 in Colorectal Epithelial Cell Terminal Differentiation and Tumorigenesis. Cancer Res. 2005;65:11486–11492. doi: 10.1158/0008-5472.CAN-05-2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shureiqi I, Xu X, Chen D, Lotan R, Morris JS, Fischer SM, et al. Nonsteroidal anti-inflammatory drugs induce apoptosis in esophageal cancer cells by restoring 15-lipoxygenase-1 expression. Cancer Res. 2001;61:4879–84. [PubMed] [Google Scholar]
- Shureiqi I, Zuo X, Broaddus R, Wu Y, Guan B, Morris JS, et al. The transcription factor GATA-6 is overexpressed in vivo and contributes to silencing 15-LOX-1 in vitro in human colon cancer. Faseb J. 2007;21:743–53. doi: 10.1096/fj.06-6830com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamitsu S, Kiyomu F, Kazuhiro Y, Hideo S, Tomonori S, Hitoshi O, et al. Peritoneal metastasis inhibition by linoleic acid with activation of PPARγ in human gastrointestinal cancer cells. Virchows Archiv. 2006;448:422–427. doi: 10.1007/s00428-005-0110-4. [DOI] [PubMed] [Google Scholar]
- Takata S, Matsubara M, Allen PG, Janmey PA, Serhan CN, Brady HR. Remodeling of neutrophil phospholipids with 15(S)- hydroxyeicosatetraenoic acid inhibits leukotriene B4-induced neutrophil migration across endothelium. J Clin Invest. 1994;93:499–508. doi: 10.1172/JCI116999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Xia HH, Tu SP, Fan DM, Lin MC, Kung HF, et al. 15-Lipoxygenase-1 mediates cyclooxygenase-2 inhibitor-induced apoptosis in gastric cancer. Carcinogenesis. 2003;24:243–7. doi: 10.1093/carcin/24.2.243. [DOI] [PubMed] [Google Scholar]
- Wu Y, Fang B, Yang XQ, Wang L, Chen D, Krasnykh V, et al. Therapeutic Molecular Targetingof 15-Lipoxygenase-1 in Colon Cancer. Mol Ther. 2008;16:886–892. doi: 10.1038/mt.2008.44. [DOI] [PubMed] [Google Scholar]
- Xue Y, Wong J, Moreno GT, Young MK, Cote J, Wang W. NURD, a Novel Complex with Both ATP-Dependent Chromatin-Remodeling and Histone Deacetylase Activities. Molecular Cell. 1998;2:851–861. doi: 10.1016/s1097-2765(00)80299-3. [DOI] [PubMed] [Google Scholar]
- Zhang C, Richon V, Ni X, Talpur R, Duvic M. Selective Induction of Apoptosis by Histone Deacetylase Inhibitor SAHA in Cutaneous T-Cell Lymphoma Cells: Relevance to Mechanism of Therapeutic Action. J Investig Dermatol. 2005a;125:1045–1052. doi: 10.1111/j.0022-202X.2005.23925.x. [DOI] [PubMed] [Google Scholar]
- Zhang X-y, DeSalle LM, Patel JH, Capobianco AJ, Yu D, Thomas-Tikhonenko A, et al. Metastasis-associated protein 1 (MTA1) is an essential downstream effector of the c-MYC oncoprotein. PNAS. 2005b;102:13968–13973. doi: 10.1073/pnas.0502330102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu P, Martin E, Mengwasser J, Schlag P, Janssen KP, Gottlicher M. Induction of HDAC2 expression upon loss of APC in colorectal tumorigenesis. Cancer Cell. 2004;5:455–63. doi: 10.1016/s1535-6108(04)00114-x. [DOI] [PubMed] [Google Scholar]
- Zuo X, Shen L, Issa J-P, Moy O, Morris JS, Lippman SM, et al. 15-Lipoxygenase-1 transcriptional silencing by DNA methyltransferase-1 independently of DNA methylation. FASEB J. 2008a doi: 10.1096/fj.07-098301. fj.07-098301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo X, Shen L, Issa JP, Moy O, Morris JS, Lippman SM, et al. 15-Lipoxygenase-1 transcriptional silencing by DNA methyltransferase-1 independently of DNA methylation. Faseb J. 2008b doi: 10.1096/fj.07-098301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo X, Wu Y, Morris JS, Stimmel JB, Leesnitzer LM, Fischer SM, et al. Oxidative metabolism of linoleic acid modulates PPAR-beta/delta suppression of PPAR-gamma activity. Oncogene. 2006;25:1225–41. doi: 10.1038/sj.onc.1209160. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.