Abstract
Salmonella Typhimurium is a common cause of gastroenteritis in humans and also localizes to neoplastic tumors in animals. Invasion of specific eukaryotic cells is a key mechanism of Salmonella interactions with host tissues. Early stages of gastrointestinal cell invasion are mediated by a Salmonella type III secretion system, powered by the adenosine triphosphatase invC. The aim of this work was to characterize the invC dependence of invasion kinetics into disparate eukaryotic cells traditionally used as models of gut epithelium or neoplasms. Thus, a nondestructive real-time assay was developed to report eukaryotic cell invasion kinetics using lux+ Salmonella that contain chromosomally integrated luxCDABE genes. Bioluminescence-based invasion assays using lux+ Salmonella exhibited inoculum dose-response correlation, distinguished invasion-competent from invasion-incompetent Salmonella, and discriminated relative Salmonella invasiveness in accordance with environmental conditions that induce invasion gene expression. In standard gentamicin protection assays, bioluminescence from lux+ Salmonella correlated with recovery of colony-forming units of internalized bacteria and could be visualized by bioluminescence microscopy. Furthermore, this assay distinguished invasion-competent from invasion-incompetent bacteria independent of gentamicin treatment in real time. Bioluminescence reported Salmonella invasion of disparate eukaryotic cell lines, including neoplastic melanoma, colon adenocarcinoma, and glioma cell lines used in animal models of malignancy. In each case, Salmonella invasion of eukaryotic cells was invC dependent.
Eukaryotic cell invasion is used by Salmonella during the initial steps of pathogenesis1 and leads to enteric symptoms and disseminated infection. Salmonella also localize to, and sometimes invade, cancerous tumors in mice.2 One basic tool for dissecting the mechanisms of these bacterial–eukaryotic cell interactions is the in vitro cell invasion assay.
The standard technique to assess Salmonella invasion into cultured cells is the gentamicin protection assay,3 which exploits the poor penetration of this antibiotic into eukaryotic cells.4 Specifically, gentamicin is postulated to kill susceptible extracellular bacteria but not “protected” bacteria that have invaded. Presumably, such selective killing permits the preferential recovery of intracellular bacteria on subsequent culture of lysed cells. Gentamicin protection assays have been used to illuminate genetic and cellular mechanisms of cell invasion by Salmonella.5 For example, the invC gene in Salmonella encodes an adenosine triphosphatase (ATPase) that powers a type III secretion system, triggering eukaryotic actin reorganization and Salmonella invasion of some eukaryotic cell lines.6,7
Despite their widespread use, standard gentamicin protection assays are technically and conceptually limited because they attempt to quantify the invasiveness of individual bacterial strains via direct enumeration of bacterial colony-forming units (CFUs) recovered from lysed eukaryotic cells. The lysis and CFU determination steps consume time, materials, and labor. Colonies are not necessarily correlated with bacterial numbers, so agglomerated organisms might be underenumerated. Additionally, because eukaryotic cells must be destroyed to release invaded bacteria, serial evaluations of bacterial invasion in a single temporal assay are precluded. Furthermore, by definition, current gentamicin protection, CFU-based assays of invasion require that extracellular bacteria of interest are killed by gentamicin, which is a condition not always met.
To attempt to address such limitations, we have modified current gentamicin protection assays of bacterial invasion into eukaryotic cells, including neoplastic lines, by using bioluminescence to report bacterial invasion. Contag and colleagues originally pioneered the use of bacteria expressing luciferase to monitor in vitro and in vivo pathogenesis with organisms containing plasmid-encoded luciferase.8 We employed constitutively bioluminescent Salmonella, which contain chromosomally integrated luxCDABE genes from Photorhabdus luminescens,9 and imaging systems that sensitively and specifically detect bioluminescent Salmonella.10 This nondestructive assay requires neither eukaryotic cell lysis nor gentamicin. Rather, we use bioluminescence to track the invasion of lux+ Salmonella into various eukaryotic cells in tissue culture. These eukaryotic cells include those traditionally used for models of gastroenteritis, as well as cells previously used in whole mouse models of metastatic cancers. To determine the invC dependence of invasion kinetics in these different systems, we compared the invasiveness of lux+ Salmonella that are isogenic except for invC.
Methods
All experiments were performed in accordance with protocols approved by the Washington University animal studies committee.
Bacterial Strains and Eukaryotic Cell Lines
The bacterial strains and eukaryotic cell lines used in this study are listed in Table 1.
Table 1.
Strains and Eukaryotic Cell Lines Used in the Study
| Strain or Cell Line | Description | Source or Reference |
|---|---|---|
|
Salmonella SB300A1 |
Parent strain, not bioluminescent; contains araC-PBAD-regulated T7 RNA polymerase | 11 |
|
Salmonella SB300A1FL6 |
SB300A1, modified by chromosomal integration of luxCDABE to be constitutively bioluminescent | This study |
|
Salmonella SB300A1FL6AM1 |
SB300A1FL6, modified by in-frame excision of invC nucleotides between 506 and 590 | This study |
|
Escherichia coli S17-1 |
Donor strain in conjugation with SB300A1, for delivery of pUT mini-Tn5 lux Km2 | 12 |
| E. coli SM10(λpir) | Donor strain in conjugation with SB300A1FL6, for delivery of plasmid pCVD442(invCΔ506–590) | 17 |
| Henle 407 | Human epithelial cell line | ATCC: CCL-6 |
| B16F10 | Murine melanoma cell line | ATCC: CRL-6475 |
| HT29 | Human colon carcinoma cell line | ATCC: HTB-38 |
| C6 | Rat glioma cell line | ATCC: CCL-107 |
| MC38 | Murine colon adenocarcinoma cell line | Gift: N.O. Davidson |
ATCC = American Type Culture Collection.
Construction of Salmonella Strains with Stably Integrated luxCDABE
Conjugative mating was performed between donor strain Escherichia coli S17-1 (containing the transfer plasmid pUT mini-Tn5 lux Km2; a gift of Michael Winson) and recipient Salmonella enterica serovar typhimurium strain SB300A1.11 Mating was performed as described12 in Luria-Bertani (LB) broth, plated onto LB agar, and incubated at 37°C overnight. Mated colonies were scraped from the LB agar and onto kanamycin (50 μg/mL) MacConkey agar to discriminate Salmonella from E. coli. The isolated candidate Salmonella were grown at dilutions of 10−5, 10−6, and 10−7 on these agar plates for 48 hours. Replating on LB/kanamycin plates documented the kanamycin resistance of the recipients of pUT mini-Tn5 lux Km2. PCR confirmed the gross presence of each gene of luxCDABE in the new strains but not in the parent Salmonella SB300A1.
Identification of Site of luxCDABE Integration into the SalmonellaGenome
First, the general location of luxCDABE integration was determined from sequences of amplicons produced using touchdown PCR13 of the genomic DNA of our new lux+ Salmonella strain. Touchdown PCR used high-fidelity Taq DNA polymerase (Invitrogen, Carlsbad, CA) and thermal cycling conditions (95°C for 5 minutes, then 25 cycles of 95°C for 45 seconds, annealing at variable temperature for 45 seconds [60°C in the first cycle and, at each of the 24 cycles, decreased by 0.5°C per cycle down to 47.5°C], and extension at 72°C for 2 minutes). This was followed by 25 cycles of 95°C for 45 seconds, 50°C for 45 seconds, and 72°C for 2 minutes. Primer pairs included a degenerate primer CCGAATTCCGGATNGAYKSNGGNTC (where N = A, C, G, or T; Y = C or T; K = G or T; and S = C or G), in combination with either an outward facing luxC or luxE primer (outward luxC: CCATCTTTGCCCTACCGTATAGAG and outward luxE: TGAGGATGAAATGCAGCGTA). Sequence data from the resulting amplicons suggested luxCDABE integration between Salmonella chromosomal genes acrB and hha.
The precise integration site of luxCDABE was then identified. PCR amplification from the genomic DNA of our new lux+ Salmonella strain was performed using two sets of primers. One reaction, which produced an amplicon of approximately 2.5 kb, used luxE (TGAGGATGAAATGCAGCGTA) and hha (GCCAGAACGAGGAGGCAGATAACA) primers and PCR conditions of 94°C for 3 minutes; 30 cycles of 94°C for 30 seconds, 51°C for 30 seconds, and 72°C for 3 minutes; and 72°C for 7 minutes. The second reaction produced an amplicon of approximately 3 kb, with luxC (ATCCAATTGGCCTCTAGCTTAGCC) and acrB (ACCTCAACGGATGAGTTTGG) primers, and the PCR conditions directly above. These amplicons above were sequenced by the Protein and Nucleic Acid Chemistry Laboratory at Washington University in St. Louis. Sequences were aligned with the Salmonella enterica serovar Typhimurium strain LT2 complete genome sequence.14
Growth Curves in Liquid Culture
Otherwise isogenic Salmonella with and without chromosomal luxCDABE were grown in overnight liquid cultures and then diluted 1:10 into fresh liquid medium for growth curve analysis. Growth was assessed via serial optical transmission measurements. For growth curve analyses, the growth medium was LB broth, and incubation was at 37°C in a shaker incubator at 200 to 250 rpm.
Construction of an In-Frame invC Deletion Mutant of luxCDABE+ Salmonella
An in-frame excision of invC nucleotides between 506 and 590 was performed using the pCVD442 suicide vector, engineered as previously described,15 for gene allele exchange.16 Here 5′ and 3′ segments of invC were amplified from wild-type invC+ Salmonella SB300A111 by PCR using the respective primer pairs 5′GGAGCGAGCTCACTGCAATATCTGGCCTACCCACA3′ with 5′GGAGCAAGCTTATCAGCATGGTCTTACCGCATCCT3′ and 5′GGAGCAAGCTTGGATATGTTGCGCGCTTCGCATAA3′ with 5′GCTATCTCGAGTTTCGCCAGGACGATATTCTCCCA3′. These four primers contain SacI, HindIII, HindIII, and XhoI sites, respectively, and nucleotides (underlined, above) of the published Salmonella LT2 genomic sequence for invC.14 The resulting PCR products were digested with SacI and HindIII or with HindIII and XhoI, respectively, and then individually cloned into pBSIISK+. Following digestion of these two plasmids with SacI and HindIII or with HindIII and XhoI, respectively, the small fragments were cloned in tandem into SacI and XhoI digested pBSIISK+, producing pBSIISK+ (invCΔ506–590). Finally, the SacI delimited insert of pBSIISK+(invCΔ506–590) was ligated into SacI linearized suicide plasmid pCVD442.16 The resulting pCVD442(invCΔ506–590) was transformed into E. coli SM10(λpir).17 Mating was performed between the donor SM10(λpir) strain and the bioluminescent chromosomal luxCDABE+ Salmonella strain SB300A1FL6 on LB agar. The cells were then scraped from the LB plates, and serial dilutions (to 10−7) were made in LB. One hundred microliters of the dilutions was spread on MacConkey agar containing ampicillin (100 μg/mL) and kanamycin (50 μg/mL) to select merodiploids. Of 30 merodiploid candidates, 5 were picked and grown overnight in LB medium without salt. These cultures were then plated on 5% sucrose plates and incubated overnight at 30°C to select for sacB removal. Presumptive sacB-deficient colonies on sucrose plates were further screened on LB ampicillin (100 μg/mL) plates for a phenotype consistent with concomitant excision of the bla gene. One such ampicillin-susceptible clone was analyzed by PCR amplification using invC flanking primers. The resulting amplicon was sequenced to confirm the anticipated 84-nucleotide deletion from the 1,296 nucleotide long invC, between invC nucleotides 506 and 590. The invC mutant also includes a six nucleotide HindIII site introduced as a by-product of the subcloning steps above (ie, TGCTGATAAGCTTGGATAT, with invC nucleotides 506T and 590G underlined). Accordingly, the predicted InvC protein encoded by our invC mutation is missing intact InvC amino acids 169 to 197 and has an isoleucine-serine-leucine insert encoded by the TAA/GCT/TGG sequence created by the HindIII site insert. This invC mutant does not create a frame shift, so it should not have polar effects on adjacent genes.
Invasion Assays
Standard gentamicin protection assays were performed as described.18 Salmonella grown overnight in LB broth (37°C) were diluted 1:100 or 1:10 and grown to an OD600 of between 0.45 and 0.7, with OD600 matched across samples for a given experiment. Incubations were not shaken, except where noted and as previously described.19 These bacteria were diluted 1:10 in Dulbecco's Modified Eagle's Medium (DMEM) or to a multiplicity of infection of 100 where noted.
Diluted bacterial suspensions were added to tissue culture plates, at 500 μL to each well in 24-well plates or 100 μL to each well in 96-well plates. For 60 minutes, the bacteria were coincubated with adherent tissue culture monolayers at 60 to 100% confluence. Wells were then washed with DMEM and treated with a medium containing gentamicin at a final concentration of 100 μg/mL. The antibiotic-containing medium was replaced with phenol red-free medium after 90 minutes of treatment, and bioluminescence was measured 3.5 hours later (5 hours after the initiation of gentamicin treatment), unless otherwise noted. Gentamicin-free conditions represented use of phenol red-free DMEM lacking gentamicin after the wash step; imaging occurred 3 hours after washing.
In the CFU recovery assay, following bioluminescence imaging, bacteria were quantified by CFU recovery after immediate lysis of tissue culture cells with detergent lysis as described.18
Measurement of Bioluminescence
Bioluminescence measurements were performed as published.20-22 Images were captured with a cooled charge-coupled device (CCD) camera (IVIS 100, Caliper, Hopkinton, MA). Acquisition parameters were exposure time, 30 seconds; binning, 8; no filter; f/stop, 1; field of view, 15 cm. Signals were measured as the radiance (photons/second/cm2/sr). To calculate the bioluminescence from a given well, total photon flux (photons/second) was determined from a region of interest (ROI) positioned over the given well and an empty well, which was subtracted to correct for background machine noise using Living Image (Xenogen) and Igor Pro (WaveMetrics) software. Bioluminescence was presented as mean ± standard deviation of the mean of the total photon flux for replicate well assays.
Bioluminescence Microscopy
Henle cell monolayers cultured on glass-bottomed 35 mm dishes, with or without luxCDABE+ Salmonella, were treated as described for gentamicin protection invasion assays. Two to 3 hours after Salmonella inoculation, these plates were examined on an inverted microscope (Nikon TE 2000-S) housed in a light-tight microscope incubator (In Vivo Scientific) with the temperature maintained at 36°C. Bioluminescence was recorded with a cooled intensified CCD camera (XR/MEGA10-AW, Stanford Photonics) controlled by Piper Imaging software, version 1.3.6 (Agile Automation). Owing to high amplification of the signal (gain set at 400,000 lm/m^2/lx), camera noise was reduced during image acquisition by setting a minimum threshold for the signal that was kept constant for all cultures. Fifteen image sequence frames were obtained per second and integrated later in 40-minute stacks to obtain a single image. Integration, pseudo–color processing, and color merge were performed with ImageJ (National Institutes of Health, Bethesda, MD) and Photoshop CS2 (Adobe) software.
Results
Chromosomal Integration of luxCDABE
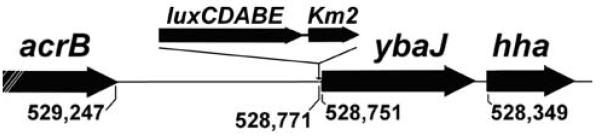
To create a Salmonella strain that constitutively produced bioluminescence, a Tn5 transfer plasmid was used to engineer a strain with chromosomal integration of the luxCDABE operon without disrupting essential genes in this process. Independent PCR assays, followed by amplicon DNA sequencing, defined the luxCDABE integration site in the Salmonella chromosome (Figure 1). According to Salmonella strain LT2 complete genomic sequence annotation convention,14 luxCDABE in our Salmonella integrated at nucleotide 528,771, 20 nucleotides 5′ to the start codon of ybaJ.
Figure 1.
Foreign DNA integration site into the Salmonella genome, at nucleotide 528,771. Nearby loci are acrB, ybaJ, and hha, flanking the insert site of luxCDABE and Km2 as shown. The following DNA sequence from the chromosome of our bioluminescent Salmonella identifies the junction between Salmonella genomic DNA (nonitalics, corresponding to Salmonella LT2 genome nucleotides 529,230 to 528,771) and foreign DNA (italics, with luxC coding nucleotides 1–163 in underlined italics): AAGGCCGCGCAAGCGGCCTTTTTTACGCAAAAATCATAAAATACGCTTATTGTTAGATTGATTATTTTTTGCCATATTAATAAAAGGTATAATCCTTACTGCGTTAAAGGCTTTTCTTAGGAAAGTTGGCCATTTCTTAATTCAGCCATTAATTAAGAAATATTAAGAATATTCCTGGCTATTTTCTCCTGTCAGAGTCTATTGTTTTAGCCTGAAAAGCTAAAAAACGTTAACCCAATGATTACACAAACAATAAAACTGGTTCCTTTTTAGGCGACCGACGATCACTGTTAAAATTCGAAAAAGTATGGCAACACGCGGCTTTCACGCAATTGTAATTTTTAGTAATATGACGATGAAAAGTTTTTTAGAGTAGATTATAGTTAAATCATAAGGTGACGTGGGAAGTACCAGGTTAGTTAGTTGTATCCATCCCGAAGGTGTTCGGTTAGTTTAAGCCCTGACTCTTATACACAAGTGCGGCCGCGTTTAAACCCATGGACGTGTTGACAATTAATCATCGGCATAGTATATCGGCATAGTATAATACGACTCAGGGCCCACTAGTGGTACCCGGGGATCCTCTAGAGTCGACCTGCAGGTCGACGGATCCGGGGAATTCAGGCTTGGAGGATACGTATGACT A A A A A A A T T T C A T T C A T T A T T A A C G GCCAGGTTGAAATCTTTCCCGAAGGTGATGATTTAGTGCAATCCATTAATTTTGGTGATAATAGTGTTTACCTGCCAATATTGAATGACTCTCATGTAAAAAACCATTATTGATTGTAATGGAAATAACGAA
Despite some preference for insertion at G/C pairs,23 Tn5 is considered to mediate near-random integration into bacterial genomes.24 Interestingly, in bioluminescent Salmonella produced by another laboratory using the same suicide vector system, the transposon is reported to have integrated at hha.25 Given that hha is immediately 3′ to ybaJ in the Salmonella genome, perhaps there is preferential integration of the Tn5 luxCDABE element into the Salmonella genome near ybaJ / hha.
Alterations in hha gene expression, secondary to luxCDABE integration, in principle could alter pathogenesis because Hha negatively regulates hilA,26 and hilA regulates the invasive phenotype of Salmonella.27 However, our new luxCDABE+ Salmonella does not exhibit decreased hha mRNA levels, assessed by reverese transcription PCR, compared with its parent (data not shown).
Fitness and Bioluminescence of luxCDABE+ Salmonella
The growth curves of otherwise isogenic Salmonella with and without luxCDABE were identical in LB broth (data not shown). As predicted, the Km2 kanamycin selection marker integrated with luxCDABE did not bestow resistance to gentamicin at concentrations of 100 μg/mL (data not shown). Furthermore, the kanamycin resistance and bioluminescence phenotypes of our luxCDABE+ Salmonella were stably maintained without kanamycin selection, both in long-term in vitro cultures and in mouse infections (data not shown).
Invasion Competence of luxCDABE+ Salmonella
To determine the impact of integrated luxCDABE on Salmonella invasiveness, we performed parallel standard gentamicin protection assays with equal inoculations of otherwise isogenic Salmonella, differing only in the presence or absence of chromosomally integrated luxCDABE. Based on numbers of bacterial CFUs from lysed eukaryotic cells, there was no defect in Salmonella invasion because of luxCDABE integration (data not shown).
Bioluminescence as a Reporter of Invasion by luxCDABE+ Salmonella
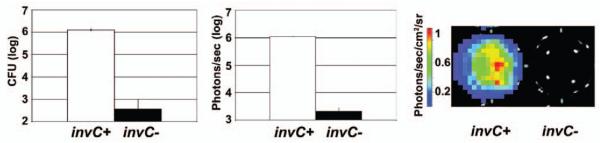
As determinates of host cell invasion, we compared Salmonella bioluminescence assays and CFUs from standard gentamicin invasion assays in tissue culture wells. Following measurement of bioluminescence signals from invasion assay tissue culture wells, we processed the tissue culture wells to obtain CFU data from the same assay wells that had been imaged for bioluminescence. We lysed the eukaryotic cells and used plate counts to recover and enumerate Salmonella CFU. There was concordance between bioluminescence output and CFU recovery in gentamicin protection assays (Figure 2). Furthermore, bioluminescence readily discriminated between invasion-competent and invasion-incompetent luxCDABE+ Salmonella strains. The protein encoded by invC is an ATPase that powers a type III secretion system, triggering eukaryotic actin reorganization and Salmonella invasion.6,7 Bioluminescence distinguished between otherwise isogenic chromosomal luxCDABE+ Salmonella strains that had an invC gene that was either intact (invC+) or ablated by an in-frame deletion (invC−) (see Figure 2). Our invC− strain is more than 200-fold less invasive compared with isogenic invC+ Salmonella concordantly assessed by standard gentamicin protection assay.6,7,18
Figure 2.
Comparison of colony-forming unit (CFU) recovery and bioluminescence (photon flux, in photons/second) from gentamicin protection assays using bioluminescent Salmonella that vary only by invC gene status. Data from a representative gentamicin protection assay performed in triplicate wells are shown. Here invasion of Henle epithelial eukaryotic cells was assessed by bioluminescent Salmonella either with invC (wild type, invC+) or without invC (invC−). In each case, the multiplicity of infection was 100. CFUs report Salmonella grown from lysates, per single wells in a 24-well plate. Photon flux is in units of photons/second, also per single wells in a 24-well plate. CFU and photon flux results are shown as means (± SD). A representative pair of wells from the bioluminescence-based assay is shown with adjacent wells containing either invC+ (left well) or invC− (right well) luxCDABE+ Salmonella; the pseudo-color scale denotes photon intensity radiance. Similar results were obtained in experiments using HT29 rather than Henle eukaryotic cells (see Figure 5 and data not shown).
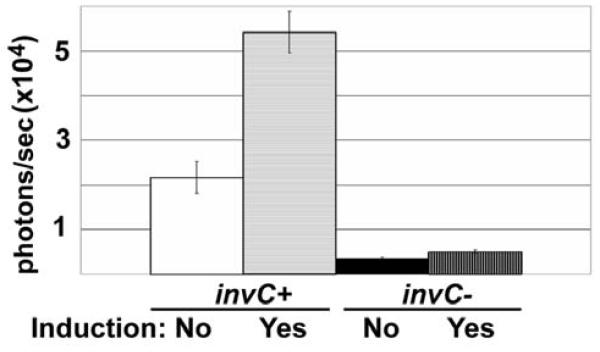
Salmonella invasiveness can also be modified by varying environmental conditions. For example, entry into eukaryotic cells can be significantly enhanced when invasion-competent Salmonella are prepared using standing rather than shaken cultures.19 Using a bioluminescence-based gentamicin protection assay, we could discriminate these invasion phenotype differences for invC+ Salmonella that differ only in culture conditions prior to exposure to eukaryotic cells (Figure 3). By contrast, the otherwise isogenic invC− Salmonella remained minimally invasive when prepared in either standing or shaken cultures (see Figure 3).
Figure 3.
Bioluminescence and gentamicin protection following differing Salmonella growth conditions known to induce enhanced Salmonella invasiveness. The bar graph compares bioluminescence data obtained from a bacterial invasion assay of HT29 eukaryotic cells by bacteria previously grown in cultures that were either shaken or standing. Standing cultures are known to induce enhanced Salmonella invasiveness in cell culture compared with shaken culture conditions.19 Induction state is as indicated. Photon flux data are shown as means (± SD), in units of photons/second, and were obtained 90 minutes after adding gentamicin.
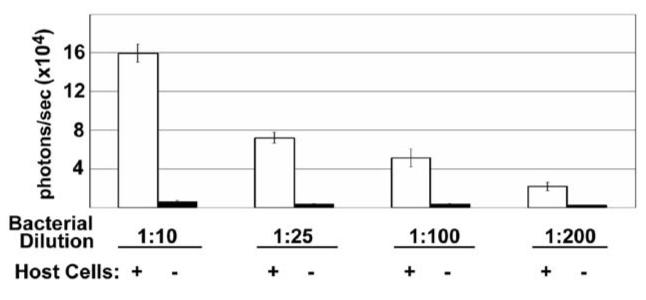
We reproducibly observed bioluminescence enhancement with increasing Salmonella inoculum (Figure 4). In standard gentamicin protection assays, the multiplicity of infection often requires optimization.3 In the bioluminescence assay, we detected wild-type Salmonella invasion into eukaryotic cells over a 20-fold range in multiplicity of infection (see Figure 4).
Figure 4.
Bioluminescence signal intensity correlated with Salmonella inoculum dose. Photon flux signals from invasion assays of HT29 cells across a 20-fold dilution range of invasion-competent (invC+) bioluminescent Salmonella. Because eukaryotic cell numbers per well were constant, this also corresponds to 20-fold range of multiplicity of infection. In wells lacking eukaryotic cells, photon flux signals approach ambient background. The results are from quadruplicate samples, with photon flux data (in units of photons/second) shown as means (± SD).
Salmonella Invasion into Diverse Eukaryotic Cells: Dependence on invC
Salmonella often invade eukaryotic cells postulated to be relevant to cell–cell interactions during pathologic intestinal infections (eg, Henle intestinal epithelial cells,3 HT29 colon carcinoma cells28). Salmonella also localize to cancerous tumors in animals,2,29,30 including nonintestinal cells, such as melanoma, glioma, and breast and prostate neoplastic cells. Salmonella can be recovered from these tumors and in some cases appear by electron microscopy to have invaded the neoplastic cells.2 Here we examined the ability of Salmonella to invade various cells used for models of malignancy in mice. Our wild-type bioluminescent Salmonella invaded not only Henle (see Figure 2) and HT29 colon cells but also eukaryotic cells of diverse origins, including colon adenocarcinoma MC38, melanoma B16F10, and even, albeit to a lesser extent, glioma C6 cells. In each case, this invasion depended on invC (Figure 5).
Figure 5.
Invasion of bioluminescent Salmonella into eukaryotic cell lines of diverse origins. Photon flux data represent invasion of wild-type invC+ and invC− Salmonella into human intestinal HT29, mouse colon adenocarcinoma MC38, mouse melanoma B16F10, or rat glioma C6 cell lines. For invC+ bioluminescent Salmonella, the photon signal following exposure to glioma eukaryotic cells was approximately one-tenth that seen with adenocarcinoma cells (note the different y-axis scale for glioma cells). Data are from triplicate wells, shown as means (± SD), in photons/second.
Real-Time Kinetic Measurements of Bioluminescence during Invasion Assays
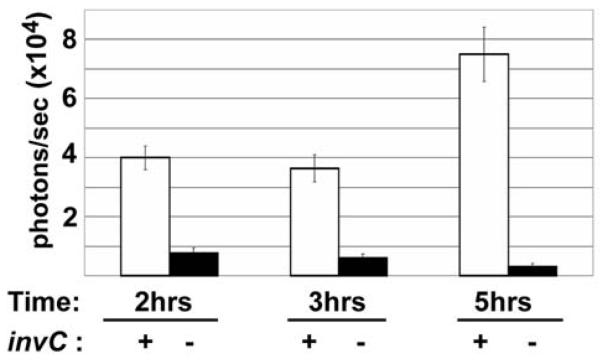
The nondestructive nature of bioluminescence now permits serial assessments of the same invaded cells over time. We assessed the kinetics of Salmonella bioluminescence from single wells of C6 glioma cells during invasion assays. The bioluminescence from a given well reflects several factors, including Salmonella cell numbers and viability. At a given time, Salmonella viability is influenced by the effects of gentamicin and the protection from gentamicin killing afforded by Salmonella invasion into eukaryotic cells. Bioluminescence versus time is shown in Figure 6. In this experiment, the distinction between bioluminescence of invC+ versus invC− Salmonella was most pronounced 5 hours after the initial inoculation.
Figure 6.
Kinetics of bioluminescence during Salmonella infection. Photon flux data of bacteria serially measured from the same invasion assay wells. The times indicated hours after initiation of a 90-minute gentamicin treatment followed by replacement with medium. Salmonella invC+ and invC− strains and C6 glioma eukaryotic cells are as described above. Data are from triplicate wells, shown as means (± SD), in units of photons/second.
Salmonella Invasion Assay, With and Without Gentamicin Protection
Our ability to distinguish invasion-competent from invasion-incompetent Salmonella at time points soon after adding gentamicin (see Figure 6) raised the possibility that bioluminescence could assess invasion independent of gentamicin protection per se. Accordingly, rather than gentamicin treatment, we employed a washing step with three rounds of gentamicin-free medium to physically deplete noninvaded Salmonella from the assay wells. In assays in which gentamicin was not used, invC-dependent invasion competence of Salmonella could still be readily resolved (Figure 7). Although the overall assay time was shorter, the background activity was higher.
Figure 7.
Bioluminescence monitoring of Salmonella interactions with eukaryotic cells, using gentamicin-containing and gentamicin-free media. Photon output from invC+ and invC− Salmonella invasion of Henle cells, from quadruplicate samples, with photon flux data shown in units of photons/second, expressed as means (± SD). Gentamicin-treatment conditions represented 90 minutes of gentamicin incubation, following replacement with phenol red free DMEM and imaging at 5 hours after initiating gentamicin addition. Gentamicin-free conditions represented use of phenol red-free DMEM lacking gentamicin throughout, followed by three washes; imaging occurred 3 hours after washing.
Bioluminescent Salmonella Visualized by Cooled CCD Microscopy
Traditionally, bioluminescent bacteria have been most extensively exploited for imaging studies of bacterial spread within whole animals, such as for Salmonella infections manifesting as gastroenteritis or disseminated infections8,25 or for the targeted localization of Salmonella to malignant tumors.29 By contrast, for microscopic level studies of bacterial localization30 or gene expression,31 fluorescent rather than bioluminescent bacteria have been most widely used. Given recent advances in cooled CCD cameras and our interest in tracking luxCDABE Salmonella microscopically, we attempted to visualize our luxCDABE+ Salmonella using a cooled CCD bioluminescence microscope. Compared to uninfected eukaryotic cell controls, tissue cultures inoculated with luxCDABE+ Salmonella with intact invC exhibited foci of bioluminescence, with maximal intensity foci clustered near or within eukaryotic cells (Figure 8). For luxCDABE+ Salmonella lacking invC, the number and intensity of these foci at the single cell level were much reduced (data not shown). The ability to use microscopy to visualize invasion by our bioluminescent Salmonella provides an additional advantage as bioluminescent microscopy does not require potentially cytotoxic excitation light and typically has low background signal.
Figure 8.
Microscopic detection of bioluminescence from luxCDABE+ Salmonella in eukaryotic cell cultures. Henle cell monolayers are shown either alone (left panels) or following inoculation with luxCDABE invC+ Salmonella (right panels). For each sample, images show the bioluminescence signal alone (top), the phase contrast (bottom), and a merged image of bioluminescence and phase contrast (middle). Within the merged image of the Henle cells inoculated with luxCDABE Salmonella, a box demarcates the image area enlarged in the upper right corner (inset). Yellow arrows outline a Henle cell; maximal intensity foci occur near or within eukaryotic cells. Spectral scales denote pseudo–color representation of bioluminescence signal intensity in relative light units (RLU), ranging from 1 to 50 RLU for samples without Salmonella and 1 to 100 RLU for samples with Salmonella. Processing of these samples followed the same methods as gentamicin protection assays described in the text; images were obtained 2 to 3 hours after inoculation of Salmonella. Bar = 50 μM.
Discussion
We report a new constitutively bioluminescent Salmonella strain (SB300A1FL6). We have subsequently deleted (in frame) specific segments of invasion competence genes in this primary luxCDABE+ Salmonella strain, creating a set of reagents to study the functions of specific Salmonella genes during bacteria–host interactions. In our Salmonella clones, luxCDABE was apparently maintained at low fitness cost. This contrasts with other Salmonella strains that have been engineered to be constitutively fluorescent via the presence of green fluorescent protein. For example, green fluorescent proteins can significantly inhibit Salmonella growth in epithelial cells and macrophages32 and increase Salmonella doubling time.33
Plasmid-based lux constructs have been exploited as reporter systems for bacterial location in vitro and in vivo.8,29 However, plasmid-based lux systems can suffer from instability. Using the pLITE lux expression plasmid in Salmonella infections of mice, loss rates of plasmid (and bioluminescence) exceeding 95% of bacterial colonies have been reported.29 We observed no loss of bioluminescence of our chromosomal luxCDABE+ Salmonella strains after serial passages in bacterial cultures or after prolonged infections in mice, even in the absence of kanamycin selection to maintain the luxCDABE / kanamycin resistance gene insert.
One motivation for constructing and characterizing these luxCDABE+ Salmonella strains was to use bioluminescence to assess Salmonella invasion kinetics into eukaryotic cells. Herein we described a robust and versatile eukaryotic cell invasion assay using chromosomal luxCDABE+ Salmonella. The new assay correlated well with the standard detection methodology over a broad inoculum range. The bioluminescence assay readily discriminated the invasion competencies of invC+ and invC− Salmonella. Indeed, the resolution between invC+ and invC− organisms was reliable across a 20-fold range of bacterial inoculated dose and multiplicities of infection. By contrast, CFU-based assays are notably nonlinear with respect to the number of inoculated bacteria.3 Bioluminescence-based invasion assays also resolved invasion differences among lux+ Salmonella as regulated by environmental stimuli.19
Bioluminescence-based tracking of lux+ Salmonella during eukaryotic cell invasion permits invasion assays to be performed independent of the stringent requirements of gentamicin protection assays. For example, bioluminescence assays need not depend on the use of gentamicin to kill extracellular bacteria or eukaryotic cell lysis to report intracellular “gentamicin-protected” bacteria. This allows studies on bacteria intrinsically resistant to gentamicin or enables analysis in growth conditions that compromise gentamicin activity (eg, acidic pH or divalent cation concentrations34) while also allowing assays in which gentamicin-mediated effects on eukaryotic phenotypes are a concern.35,36
Our assay had similarities to other techniques for assessing bacterial invasion, such as direct observation of internalized bacteria following Giemsa staining37 or direct observation of gfp-labeled bacteria within eukaryotic cells via fluorescent microscopy30 or fluorescence-activated cell sorter (FACS) analysis.31 However, luxCDABE-encoded bioluminescence provided potential advantages to detect intracellular bacteria. For example, in contrast to Giemsa staining of inanimate features of bacterial cell walls or to gfp-based fluorescence, lux bioluminescence only reported bacteria that were alive and biochemically active.38 This direct detection of living bacteria removed the lag time and intermediate maneuvers imposed by experiments that rely on bacterial staining or recovery of bacterial CFUs for data. The real-time and nondestructive nature of lux-based tracking of Salmonella in eukaryotic cells also allowed serial measurements from the same well over time. Hence, it was well suited to kinetic studies of bacterial invasion and intracellular survival. From a technical perspective, bioluminescence-based detection of lux+ Salmonella should readily allow high-throughput experimental scaling in multiwell plate assays, with readout times within minutes.
Furthermore, the components of the experimental system described here for studying bacterial invasion into eukaryotic cells in tissue cultures can also be used to noninvasively detect and localize luxCDABE+ Salmonella during infections in living mice (data not shown). Thus, bioluminescence-based detection of lux+ Salmonella presents opportunities to more directly correlate in vitro and in vivo models of bacteria–host interactions. This can be used to detect Salmonella in experimental mouse models of infection and malignancy. Intriguingly, our bioluminescent Salmonella invade a disparate range of malignant eukaryotic cells in vitro, each in an invC-dependent manner. This suggests that Salmonella interactions with eukaryotic neoplastic cells may recapitulate features of Salmonella interactions with eukaryotic epithelial cells in the host intestinal tract.
Acknowledgments
Gregory Storch, Phillip I. Tarr, Virginia Miller, Kim Walker, and members of the McKinney laboratory provided helpful discussions and review of the manuscript. J.S.M. thanks Dr. Michael Winson for the kind gift of transfer plasmid pUT mini-Tn5 lux Km2, Dr. Nicholas Davidson for the cell line MC38, and Joshua Aranda for technical assistance.
This work was supported by research grants from the National Institutes of Health (NIH) (Washington University Digestive Diseases Research Core Center Grant P30 DK52574, Washington University Molecular Imaging Center Grant P50 CA94056, Washington University Children's Health Research Center Grant K12 HD01487, Neuroscience Blueprint Center Core Grant P30 NS057105), the American Cancer Society, and a March of Dimes Birth Defects Foundation Basil O'Connor Scholar Research Award to J.S.M. We acknowledge student stipend support to K.N.F. from NIH Training Grant T32 GM007067 and to M.Q., F.L., and A.M. from Washington University and the Howard Hughes Medical Institute.
References
- 1.Ohl ME, Miller SI. Salmonella: a model for bacterial pathogenesis. Annu Rev Med. 2001;52:259–74. doi: 10.1146/annurev.med.52.1.259. [DOI] [PubMed] [Google Scholar]
- 2.Pawelek JM, Low KB, Bermudes D. Bacteria as tumour-targeting vectors. Lancet Oncol. 2003;4:548–56. doi: 10.1016/s1470-2045(03)01194-x. [DOI] [PubMed] [Google Scholar]
- 3.Elsinghorst EA. Measurement of invasion by gentamicin resistance. Methods Enzymol. 1994;236:405–20. doi: 10.1016/0076-6879(94)36030-8. [DOI] [PubMed] [Google Scholar]
- 4.Vaudaux P, Waldvogel FA. Gentamicin antibacterial activity in the presence of human polymorphonuclear leukocytes. Antimicrob Agents Chemother. 1979;16:743–9. doi: 10.1128/aac.16.6.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Galan JE, Curtiss R., 3rd Cloning and molecular characterization of genes whose products allow Salmonella typhimurium to penetrate tissue culture cells. Proc Natl Acad Sci U S A. 1989;86:6383–7. doi: 10.1073/pnas.86.16.6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eichelberg K, Ginocchio CC, Galan JE. Molecular and functional characterization of the Salmonella typhimurium invasion genes invB and invC: homology of InvC to the F0F1 ATPase family of proteins. J Bacteriol. 1994;176:4501–10. doi: 10.1128/jb.176.15.4501-4510.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Akeda Y, Galan JE. Genetic analysis of the Salmonella enterica type III secretion-associated ATPase InvC defines discrete functional domains. J Bacteriol. 2004;186:2402–12. doi: 10.1128/JB.186.8.2402-2412.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Contag CH, Contag PR, Mullins JI, et al. Photonic detection of bacterial pathogens in living hosts. Mol Microbiol. 1995;18:593–603. doi: 10.1111/j.1365-2958.1995.mmi_18040593.x. [DOI] [PubMed] [Google Scholar]
- 9.Duchaud E, Rusniok C, Frangeul L, et al. The genome sequence of the entomopathogenic bacterium Photorhabdus luminescens. Nat Biotechnol. 2003;21:1307–13. doi: 10.1038/nbt886. [DOI] [PubMed] [Google Scholar]
- 10.Doyle TC, Burns SM, Contag CH. In vivo bioluminescence imaging for integrated studies of infection. Cell Microbiol. 2004;6:303–17. doi: 10.1111/j.1462-5822.2004.00378.x. [DOI] [PubMed] [Google Scholar]
- 11.McKinney J, Guerrier-Takada C, Galan J, Altman S. Tightly regulated gene expression system in Salmonella enterica serovar Typhimurium. J Bacteriol. 2002;184:6056–9. doi: 10.1128/JB.184.21.6056-6059.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Winson MK, Swift S, Hill PJ, et al. Engineering the luxCDABE genes from Photorhabdus luminescens to provide a bioluminescent reporter for constitutive and promoter probe plasmids and mini-Tn5 constructs. FEMS Microbiol Lett. 1998;163:193–202. doi: 10.1111/j.1574-6968.1998.tb13045.x. [DOI] [PubMed] [Google Scholar]
- 13.Levano-Garcia J, Verjovski-Almeida S, da Silva AC. Mapping transposon insertion sites by touchdown PCR and hybrid degenerate primers. Biotechniques. 2005;38:225–9. doi: 10.2144/05382ST03. [DOI] [PubMed] [Google Scholar]
- 14.McClelland M, Sanderson KE, Spieth J, et al. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature. 2001;413:852–6. doi: 10.1038/35101614. [DOI] [PubMed] [Google Scholar]
- 15.Tarr PI, Bilge SS, Vary JC, Jr, et al. Iha: a novel Escherichia coli O157:H7 adherence-conferring molecule encoded on a recently acquired chromosomal island of conserved structure. Infect Immun. 2000;68:1400–7. doi: 10.1128/iai.68.3.1400-1407.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Donnenberg MS, Kaper JB. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect Immun. 1991;59:4310–7. doi: 10.1128/iai.59.12.4310-4317.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Simon R, Priefer U, Puhler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology. 1983;1:784–91. [Google Scholar]
- 18.McKinney JS, Zhang H, Kubori T, et al. Disruption of type III secretion in Salmonella enterica serovar Typhimurium by external guide sequences. Nucleic Acids Res. 2004;32:848–54. doi: 10.1093/nar/gkh219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee CA, Falkow S. The ability of Salmonella to enter mammalian cells is affected by bacterial growth state. Proc Natl Acad Sci U S A. 1990;87:4304–8. doi: 10.1073/pnas.87.11.4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gross S, Piwnica-Worms D. Monitoring proteasome activity in cellulo and in living animals by bioluminescent imaging: technical considerations for design and use of genetically encoded reporters. Methods Enzymol. 2005;399:512–30. doi: 10.1016/S0076-6879(05)99035-6. [DOI] [PubMed] [Google Scholar]
- 21.Leevy WM, Gammon ST, Levchenko T, et al. Structure-activity relationships, kinetics, selectivity, and mechanistic studies of synthetic hydraphile channels in bacterial and mammalian cells. Org Biomol Chem. 2005;3:3544–50. doi: 10.1039/b508157b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beyer W, Bohm R. Labeling Salmonella live vaccine strains with the lux operon from Vibrio fischeri improves their detection and discrimination from wild type. Microbiol Res. 1996;151:407–19. doi: 10.1016/S0944-5013(96)80011-5. [DOI] [PubMed] [Google Scholar]
- 23.Lodge JK, Weston-Hafer K, Berg DE. Transposon Tn5 target specificity: preference for insertion at G/C pairs. Genetics. 1988;120:645–50. doi: 10.1093/genetics/120.3.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hayes F. Transposon-based strategies for microbial functional genomics and proteomics. Annu Rev Genet. 2003;37:3–29. doi: 10.1146/annurev.genet.37.110801.142807. [DOI] [PubMed] [Google Scholar]
- 25.Burns-Guydish SM, Olomu IN, Zhao H, et al. Monitoring age-related susceptibility of young mice to oral Salmonella enterica serovar Typhimurium infection using an in vivo murine model. Pediatr Res. 2005;58:153–8. doi: 10.1203/01.PDR.0000157725.44213.C4. [DOI] [PubMed] [Google Scholar]
- 26.Fahlen TF, Wilson RL, Boddicker JD, Jones BD. Hha is a negative modulator of transcription of hilA, the Salmonella enterica serovar Typhimurium invasion gene transcriptional activator. J Bacteriol. 2001;183:6620–9. doi: 10.1128/JB.183.22.6620-6629.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bajaj V, Lucas RL, Hwang C, Lee CA. Co-ordinate regulation of Salmonella typhimurium invasion genes by environmental and regulatory factors is mediated by control of hilA expression. Mol Microbiol. 1996;22:703–14. doi: 10.1046/j.1365-2958.1996.d01-1718.x. [DOI] [PubMed] [Google Scholar]
- 28.Raffatellu M, Wilson RP, Chessa D, et al. SipA, SopA, SopB, SopD, and SopE2 contribute to Salmonella enterica serotype typhimurium invasion of epithelial cells. Infect Immun. 2005;73:146–54. doi: 10.1128/IAI.73.1.146-154.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu YA, Shabahang S, Timiryasova TM, et al. Visualization of tumors and metastases in live animals with bacteria and vaccinia virus encoding light-emitting proteins. Nat Biotechnol. 2004;22:313–20. doi: 10.1038/nbt937. [DOI] [PubMed] [Google Scholar]
- 30.Zhao M, Yang M, Li XM, et al. Tumor-targeting bacterial therapy with amino acid auxotrophs of GFP-expressing Salmonella typhimurium. Proc Natl Acad Sci U S A. 2005;102:755–60. doi: 10.1073/pnas.0408422102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bumann D. Examination of Salmonella gene expression in an infected mammalian host using the green fluorescent protein and two-colour flow cytometry. Mol Microbiol. 2002;43:1269–83. doi: 10.1046/j.1365-2958.2002.02821.x. [DOI] [PubMed] [Google Scholar]
- 32.Knodler LA, Bestor A, Ma C, et al. Cloning vectors and fluorescent proteins can significantly inhibit Salmonella enterica virulence in both epithelial cells and macrophages: implications for bacterial pathogenesis studies. Infect Immun. 2005;73:7027–31. doi: 10.1128/IAI.73.10.7027-7031.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rang C, Galen JE, Kaper JB, Chao L. Fitness cost of the green fluorescent protein in gastrointestinal bacteria. Can J Microbiol. 2003;49:531–7. doi: 10.1139/w03-072. [DOI] [PubMed] [Google Scholar]
- 34.Fass RJ, Barnishan J. Effect of divalent cation concentrations on the antibiotic susceptibilities of nonfermenters other than Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1979;16:434–8. doi: 10.1128/aac.16.4.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Keeling KM, Bedwell DM. Clinically relevant aminoglycosides can suppress disease-associated premature stop mutations in the IDUA and P53 cDNAs in a mammalian translation system. J Mol Med. 2002;80:367–6. doi: 10.1007/s00109-001-0317-z. [DOI] [PubMed] [Google Scholar]
- 36.Lai CH, Chun HH, Nahas SA, et al. Correction of ATM gene function by aminoglycoside-induced read-through of premature termination codons. Proc Natl Acad Sci U S A. 2004;101:15676–81. doi: 10.1073/pnas.0405155101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yabuuchi E, Ikedo M, Ezaki T. Invasiveness of Salmonella typhi strains in HeLa S3 monolayer cells. Microbiol Immunol. 1986;30:1213–24. doi: 10.1111/j.1348-0421.1986.tb03050.x. [DOI] [PubMed] [Google Scholar]
- 38.Meighen EA. Molecular biology of bacterial bioluminescence. Microbiol Rev. 1991;55:123–42. doi: 10.1128/mr.55.1.123-142.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]