Abstract
Nucleotide binding domains (NBDs) secure ATP-binding cassette (ABC) transporter function. Distinct from traditional ABC transporters, ABCC9-encoded sulfonylurea receptors (SUR2A) form, with Kir6.2 potassium channels, ATP-sensitive K+ (KATP) channel complexes. SUR2A contains ATPase activity harbored within NBD2 and, to a lesser degree, NBD1, with catalytically driven conformations exerting determinate linkage on the Kir6.2 channel pore. While homodomain interactions typify NBDs of conventional ABC transporters, heterodomain NBD interactions and their functional consequence have not been resolved for the atypical SUR2A protein. Here, nanoscale protein topography mapped assembly of monodisperse purified recombinant SUR2A NBD1/NBD2 domains, precharacterized by dynamic light scattering. Heterodomain interaction produced conformational rearrangements inferred by secondary structural change in circular dichroism, and validated by atomic force and transmission electron microscopy. Physical engagement of NBD1 with NBD2 translated into enhanced intrinsic ATPase activity. Molecular modeling delineated a complemental asymmetry of NBD1/NBD2 ATP-binding sites. Mutation in the predicted catalytic base residue, D834E of NBD1, altered NBD1 ATPase activity disrupting potentiation of catalytic behavior in the NBD1/NBD2 interactome. Thus, NBD1/NBD2 assembly, resolved by a panel of proteomic approaches, provides a molecular substrate that determines the optimal catalytic activity in SUR2A, establishing a paradigm for the structure–function relationship within the KATP channel complex.
Keywords: ABC proteins, atomic force microscopy, ATP-sensitive K+ channel, circular dichroism, dynamic light scattering
Introduction
ABC genes encode one of the largest transmembrane proteome, the ATP-binding cassette (ABC) protein superfamily.1,2 ABC genes occupy ~5% of the bacterial genome, and 48 human orthologs serve vital roles in cell physiology.3–6 ABC proteins are typically described as transporters requiring the energy of ATP hydrolysis to translocate substrates across cell membranes.3 Recently, homologues have been identified that couple conformational changes induced by ATP binding and hydrolysis to processes distinct from active transport, such as ion gating.7,8
Critical for ABC function are two hydrophilic nucleotide binding domains (NBDs), contiguous to hydrophobic transmembrane domains (TMDs)2 where TMDs share the architecture of membrane-spanning α-helices, but little sequence similarity. Evolutionarily conserved NBDs display extensive amino acid sequence identity and, in typical ABC transporters, form homodimers.3,4 Each NBD comprises conserved Walker A (GX4GKS/T), Walker B (φ4DD/E; φ represents hydrophobic residues), and signature (LSGGQ) motifs, regrouped into a functional unit.3,9 Nucleotide binding and ATP hydrolysis occur in the Walker motifs-delineated active pocket, with the NBDs ATPase essential for the execution of protein functions.2,4,8,10–12 While homodimer interaction of NBDs in conventional ABC transporters is increasingly understood, less is known for atypical ABC proteins where intrinsic ATPase activity regulates processes distinct from active transport.
A case in point is the catalytic activity harbored in the second nucleotide binding domain, NBD2, of the sulfonylurea receptor 2A (SUR2A), encoded by the ABCC9 gene.8,13–16 Although an assigned member of the ABC proteome, SUR2A uniquely assembles with the pore-forming Kir6.2 channel, and serves as the regulatory subunit of the cardiac ATP-sensitive potassium (KATP) channel complex.11,13,17–21 Cardiac KATP channels work as molecular rheostats adjusting membrane potential-dependent functions to match cellular energetic demands.22–27 KATP channel knockout translates into stress intolerance and susceptibility to organ failure,28,29 while overexpression of the regulatory SUR2A subunit endows enhanced protection against cell injury.30 SUR2A recognizes and processes intracellular energetic signals, endowing KATP channels with a metabolic decoding capacity.7,8,31,32 Potassium movement through Kir6.2 does not require energy expenditure, yet ATP hydrolysis at SUR2A is integral in the transduction of metabolic signals to the channel pore.7,16,33 Suppression of SUR2A ATPase activity, induced by mutations in Walker NBD2 motifs, alters KATP channel nucleotide sensitivity.14 SUR2A mutations that disrupt proper catalytic activity have been recently linked in humans to a cardiomyopathic syndrome underscoring the significance of ATPase-dependent KATP channel regulation.34
Compared to NBD2, NBD1 in SUR2A displays a more limited catalytic activity.14,35,36 Such asymmetry further distinguishes SUR2A from traditional ABC transporters.7,8,16 Site-directed mutagenesis indicates that NBD1 is mandatory for NBD2-dependent KATP channel gating, with ATP binding to NBD1, cooperatively supported by catalytic hydrolysis of ATP at NBD2, a necessary step in channel gating.16 However, direct demonstration of physical interaction of SUR2A NBD1 and NBD2 has not been documented, and the implications on ATPase activity remain untested. In fact, architectural mapping of ABC proteins has been so far limited to bacterial homodimer interactions in typical family members.
Here, we applied a panel of proteomic methods to address the interaction of mammalian ABCC9-encoded SUR2A NBD1 and NBD2 heterodomains. Visualized physical assembly produced conformational changes in the secondary structure enabling enhanced catalytic ATPase activity, a previously unrecognized consequence of the NBD1/NBD2 interaction. Point mutation in the catalytic base altered ATPase activity, disrupting regulation of catalytic behavior induced by interdomain interaction. The resolved NBD1/NBD2 interactome provides thereby a molecular substrate for the structure–function relationship within the KATP channel complex.
Experimental Section
Expression and Purification of Recombinant SUR2A NBDs
The constructs coding for SUR2A NBD1 (D666-E890) and NBD2 (G1306-K1546) were selected based on the Predictor of Natural Disordered Regions (PONDR) algorithm (http://www.pondr.com) to increase stability of the protein by excluding disordered regions. The cDNA fragments of NBD1 and NBD2 were amplified by PCR reaction from mouse SUR2A (GenBank accession no. D86037; kindly provided by Dr. S. Seino, Kobe University), and incorporated into a pET-43.1(a) and pET-44(a) vector (Novagen), resulting in proteins that have NusA-(His)6-tag and (His)6-NusA-(His)6-tag at the N-terminus, respectively. The identity of fusion constructs was confirmed by DNA sequencing. Plasmids containing NBD1 or NBD2 were transformed in the Escherichia coli C41(DE3) strain (OverExpress).37 Cells were grown in Luria–Bertani (LB) broth at 37 °C to an A600 of ~0.6, transferred to an 18 °C incubator, and then, after 1 h, induced by addition of 1 mM isopropyl-β-D-thioga-lactoside (IPTG).14 Induced cells were harvested 17 h later by centrifugation at 4000g for 20 min. Pelleted cells were resuspended in 50 mM sodium phosphate buffer containing 300 mM NaCl (pH 7.5), lyzed with a high-pressure microfluidizer Emulsiflex C-5 (Avestin), and centrifuged at 20000g for 30 min. The supernatant was then passed through a Ni-loaded NTA column (Qiagen), washed, and eluted with 50 mM sodium phosphate and 300 mM NaCl (pH 7.5), containing 20 mM and 500 mM imidazole, respectively. For enhanced solubility and to avoid aggregation, the expressed protein was used without removal of NusA. The eluted NBDs were further purified through a preparative Superdex 200 column (Amersham Biosciences) with 25 mM HEPES, 300 mM NaCl, and 1 mM CHAPS (pH 7.4). Protein concentration was quantified by the Bradford Protein Assay (Bio-Rad), and purity was determined by densitometry. The homogeneity of NBD fusion proteins was tested by dynamic light scattering, and purified proteins were used to characterize physical interaction and enzymatic properties prior and upon heterodomain interaction. In a separate set of experiments prior to expression and purification, point mutations in SUR2A Walker motifs of NBD1 and NBD2 were introduced in pET plasmid by PCR amplification of both DNA strands with complementary primers containing desired amino acid changes (QuickChange, Stratagene). Constructs were sequenced to confirm site-directed mutagenesis.
ATPase Activity Assays
The ATPase activity of recombinant purified NBDs was measured based on organic phosphate production detected by colorimetry.38 Assays were performed in 100 μL of 25 mM HEPES, 300 mM NaCl, 10 mM MgCl2, and 10% glycerol (pH 7.4) with 10 μg of purified NBDs. Reactions were initiated by addition of 4 mM ATP, continued for 50 min at 37 °C, and terminated by addition of 100 μL of 12% SDS. Color development was initiated with 200 μL of a solution (6% ascorbic acid in 1 N HCl and 1% ammonium molybdate), incubated for 10 min at room temperature, and followed by addition of 300 μL of 2% sodium citrate, 2% sodium metaarsenite, and 2% acetic acid mixture solution. Absorbance was measured at 850 nm after 10 min-long incubation at 37 °C. Because of contaminating phosphate in ATP preparations, samples containing ATP in the absence of NBDs were used as negative controls. A nonsignificant amount of ATP hydrolysis was detected in the absence of MgCl2.39 In a separate set of experiments, ATP hydrolysis was measured in the presence of BeFx, prepared as a 33% stock beryllium fluoride solution and dissolved in buffer containing 50 mM KF.7
Dynamic Light Scattering
Dynamic light scattering (DLS) measurements were performed at 20 °C using a PDDLS/CoolBatch and PD2000/DLS system (Precision Detectors). This DLS system is equipped with an 800 nm laser diode, and a coherent detector of a scattered light at an angle of 90° to the incident laser beam. Purified NBD1 and NBD2 proteins (0.1–0.2 mg/mL in 25 mM HEPES and 300 mM NaCl; pH 7.4) were centrifuged at 20000g for 10 min at 4 °C to remove particulate matter. Samples (150 μL) were placed in a quartz cuvette, and 50 measurements were averaged per sample. The diffusion constant was extracted from the autocorrelation function, and the dimension of hydrodynamic diameters (dH, nm) of NBD fusion proteins were calculated from the diffusion coefficient (D) using the Stokes–Einstein equation, that is, dH = (kBT)/(3πηD) where kB is the Boltzmann constant; T, the absolute temperature (K); and η, the buffer viscosity in the Precision Deconview program (Precision Detectors).40
Circular Dichroism
Far-UV circular dichroism (CD) spectra of NBDs were recorded within the 250–200 nm range on a JASCO J715 CD spectropolarimeter (Jasco). The parameters used for each measurement were bandwidth, 1 nm; step resolution, 1 nm; scan speed, 50 nm/min; response time, 1 s; and number of scans, 8–16. NBD1, NBD2, or a 1:1 mixture of NBD1/NBD2 (0.4–0.7 mg/mL protein in 25 mM HEPES, 300 mM NaCl, and 1 mM CHAPS; pH 7.4) was incubated for 10 min at 37 °C. Spectra derived from recombinant proteins were measured in a 1 mm thermostatted quartz cuvette at 4 °C containing 0.4–0.7 mg/mL of NBD1, NBD2, or NBD1 + NBD2 in 25 mM HEPES, 300 mM NaCl, and 1 mM CHAPS (pH 7.4). The background CD spectrum was recorded in buffer and used for correction of protein CD spectra as described.39 The mean residue ellipticity ([θ], expressed as deg cm2 dmol−1) was calculated as (mdeg × MRW)/(10lc), where mdeg is the observed ellipticity in millidegrees; MRW, the mean residue weight set at 115;41 l, the optical path in cm; and c, concentration in mg/mL. On the basis of UV circular dichroism data, the secondary structure composition of NBDs was analyzed using the neural network algorithm SOMCD method (http://geneura.ugr.es/cgi-bin/somcd/index.cgi).
Atomic Force Microscopy
To investigate the dynamic forms of protein interactions,42–45 atomic force microscopy in liquid tapping mode was performed using a Nanoscope IV PicoForce Multimode AFM equipped with an E-scanner and V shaped silicon cantilevers with a 0.06 N/m spring constant (Veeco Instruments). NBD1 (4–7 μg/mL) and/or NBD2 (4–7 μg/mL) were injected into the fluid cell of the atomic force microscope and allowed to adsorb onto freshly cleaved mica discs. Images (512 × 512 pixels) were taken 5 min after each injection. Separately, NBD1 and NBD2 (0.4–0.7 mg/mL) were coincubated at 37 °C for 10 min and injected into the fluid cell after a 100× dilution prior to imaging. Data were collected in both height and phase modes. Analysis was performed using the Nano-scope Version 6.13 software.46 Height images were used for measurement of lateral dimensions of the NBD1 and NBD2 proteins.
Electron Microscopy
Five microliters of NBDs (0.1 mg/mL protein in 25 mM HEPES, 300 mM NaCl, and 1 mM CHAPS; pH 7.4) was placed onto carbon-coated grids and allowed to dry. Each grid was washed three times with distilled water and negatively stained with 1% phosphotungstic acid (pH 7.2). Samples were viewed on an FEI Tecnai 12 transmission electron microscope at 80 kV.47
Molecular Modeling of NBDs
SUR2A NBDs sequences were aligned with hemolysin B (HlyB)48 using ClustalX (http://www.ebi.ac.uk/clustalw). A three-dimensional model of the SUR2A NBD1 and NBD2 dimer was generated using the homology modeling program MODELLER 649 and a crystal structure of the HlyB-NBD dimer, a prototypic ABC transporter, used as a template (PDB code:1XEF) identified by the FASTA search.
Statistics
Data are expressed as mean ± SE; P-values were calculated using the unpaired Student’s t-test. P < 0.05 was predetermined.
Results
Nanoscale Resolution of SUR2A NBD1 and NBD2 Structures
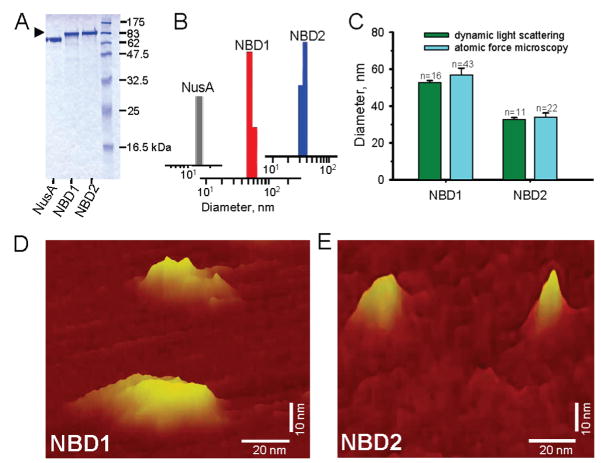
Recombinant nucleotide binding domains, NBD1 and NBD2, of the mammalian KATP channel SUR2A ABC protein were individually expressed in the E. coli C41(DE3) strain, using a solubility enhancing NusA tag (Figure 1A). NusA fusion protein (pI/MW = 4.6/66 kDa) increased protein solubility and stability, enabling reliable protein expression,50,51 while removal of the tag protein reinstated a high aggregation pattern (not illustrated). Binding to a Ni2+-NTA column was targeted by histidine tags, with imidazole-elution and additional size-exclusion purification yielding oligomeric NBD1 and NBD2 products at >94 ± 3% purity, detected at corresponding molecular weights on SDS-PAGE (n = 4; Figure 1A). Purified NBD1 and NBD2 displayed a monodisperse size distribution of dynamic light scattering intensity bars, excluding aggregation and indicating a homogeneous profile of purified proteins (Figure 1B). On average, dynamic light scattering revealed a diameter of 52.7 ± 1.0 nm (n = 16) and 32.8 ± 0.9 nm (n = 11) for NBD1 and NBD2, respectively, significantly different from 15.1 ± 3.5 nm (n = 10) recorded for NusA (P < 0.05; Figure 1C). Structural properties of NBD1 and NBD2 were validated by mapping the microarchitecture of purified proteins using atomic force microscopy which generates three-dimensional topographic maps of sample surface. At nanoscale resolution in solution, NBD1 and NBD2 demonstrated island-like, table-mountain topographies (Figure 1D,E). On average, NBD1 was 56.8 ± 3.7 nm long and 19.7 ± 2.2 nm wide (n = 43), dimensions distinct from the long and short axis of NBD2, 33.9 ± 2.3 nm and 13.4 ± 2.2 nm (n = 22), respectively (P < 0.05; Figure 1C–E). Thus, complementary analysis demonstrates the monodisperse and homogeneous properties of ABCC9-encoded SUR2A NBD1 and NBD2.
Figure 1.
Structure of SUR2A NBD1 and NBD2 proteins. Recombinant nucleotide binding domains, NBD1 and NBD2, were expressed using a solubility enhancing NusA tag, and purified from Ni-affinity and size exclusion chromatography. (A) SDS-PAGE of Coomassie blue-stained NBD1 (86 kDa) and NBD2 (89 kDa) fusion constructs or NusA (66 kDa) alone, loaded 5 μg/protein samples, migrating at corresponding molecular weights. (B) Homogeneous NBD1 and NBD2 display monodisperse size distribution profile on dynamic light scattering, distinct from the size of NusA. (C) NBD1 and NBD2 protein size independently determined by dynamic light scattering and atomic force microscopy. Structures of NBD1 (D) and NBD2 (E) mapped at nanoscale resolution by atomic force microscopy.
Assembly of NBD1/NBD2 Heterodomains
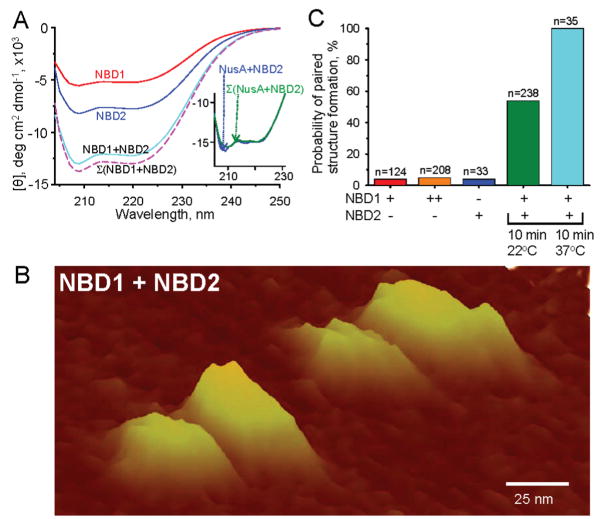
Recombinant NBD1 and NBD2 displayed contour depressions, or troughs, in far-UV circular dichroism spectroscopy spectra at 209 and 220 nm (Figure 2A), indicative of the distinctive α-helix structural profile.41,52 The conformational preference of NBD1 and NBD2 for the rigorous α-helix configuration, in the absence of a detectable random coil profile, demonstrated a well-folded structure for both recombinant proteins (Figure 2A). Coincubation of NBD1 with NBD2 produced a significant change in α-helicity (Figure 2A). Measured by circular dichroism, the experimentally determined conformational change was characteristic of an intimate protein–protein interaction,52,53 and the readout differed significantly from the theoretical sum of individual NBD (Σ(NBD1 + NBD2)) spectra (n = 4, P < 0.05), suggesting direct physical interaction between NBD1 and NBD2 (Figure 2A). In contrast to NBD1/NBD2 coincubation, NusA coincubation with NBD2 did not produce detectable change in α-helicity indicating lack of direct physical interaction with the solubility enhancing tag (Figure 2A, inset).
Figure 2.
Interaction of NBD1 and NBD2. (A) Circular dichroism spectra reveal the distinctive α-helix profile of NBD1, NBD2, and NBD1 + NBD2. Of note, the circular dichroism spectrum measured from coincubated NBD1 and NBD2 demonstrated a profile distinct from the theoretically predicted sum Σ(NBD1 + NBD2), indicating structural change upon protein–protein interaction. Inset: Coincubation of NusA with an NBD did not produce a significant change in α-helicity beyond the predicted sum of the two coincubated proteins. Units of the x and y axis are identical as in panel A. (B) Paired nanostructures of NBD1/NBD2 complexes, after incubation at 37 °C for 10 min, resolved by atomic force microscopy. (C) The probability for NBD1 and NBD2 to assemble exceeded the likelihood of NBD1 or NBD2, when incubated alone, to form nonspecific aggregates even with increase in protein concentration as determined by atomic force microscopy. Enhanced formation of NBD1/NBD2 complexes at physiological temperature. + and − indicate presence or absence of an NBD; ++ indicates presence of an NBD at a concentration >2-fold higher than in +.
Heterodomain interaction of NBDs was verified by topography mapping using atomic force microscopy (Figure 2B).54,55 Once NBD1 and NBD2 were free to interact, the ensuing microarchitecture revealed paired nanostructures, referred to as hetero-oligomers, with larger/smaller island sets corresponding to the larger NBD1 and smaller NBD2 doublet components (Figure 2B). Alone, neither NBD1 nor NBD2 displayed a significant probability for paired structures even following increase in the concentration of the added protein (Figure 2C). Indeed, the limited occurrence of NBD1 or NBD2 self-pairing (<10%) was in sharp contrast to massive pairing when NBD1 freely interacted with NBD2, in particular at physiological temperature (Figure 2C). In doublets, NBD1 was 53.8 ± 4.3 nm long and 20.1 ± 2.0 nm wide (n = 22), recognizable from the long and short axis of NBD2 at 37.9 ± 2.1 nm and 13.9 ± 1.8 nm (n = 22), respectively (P < 0.05; Figure 2B). These dimensions displayed significant, albeit small, changes compared to the topography of individual proteins (P < 0.05), establishing a change in secondary structure upon NBD1/NBD2 heterodomain formation.
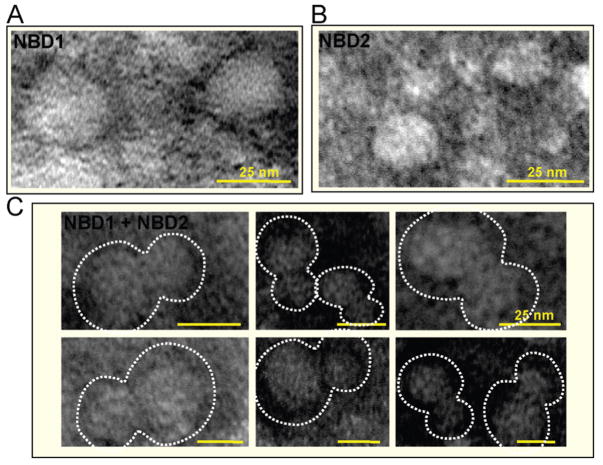
The change of molecular architecture upon assembly was independently validated by transmission electron microscopy (Figure 3). Electron micrographs of NBD1 or NBD2 revealed individual spherical particles (Figure 3A,B), whereas the NBD1/NBD2 mixture displayed paired particles with larger and smaller components (Figure 3C), corresponding to the larger NBD1 and smaller NBD2 doublets similarly observed by atomic force microscopy (Figure 2B). Because of a drying procedure, the observed dimensions of NBD1, NBD2, or paired NBDs in electron micrographs were slightly smaller than those obtained in atomic force microscopy operation in fluid, yet overall molecular architectures were comparable.
Figure 3.
Electron micrographs of negatively stained isolated NBD1 (A) or NBD2 (B) versus the NBD1 + NBD2 mixture (C). Note that pairing occurs only when NBD1 and NBD2 are free to interact forming heterodomain complexes as shown in (C). All bars = 25 nm.
NBD1/NBD2 Heterodomain Interaction Promotes ATPase Activity
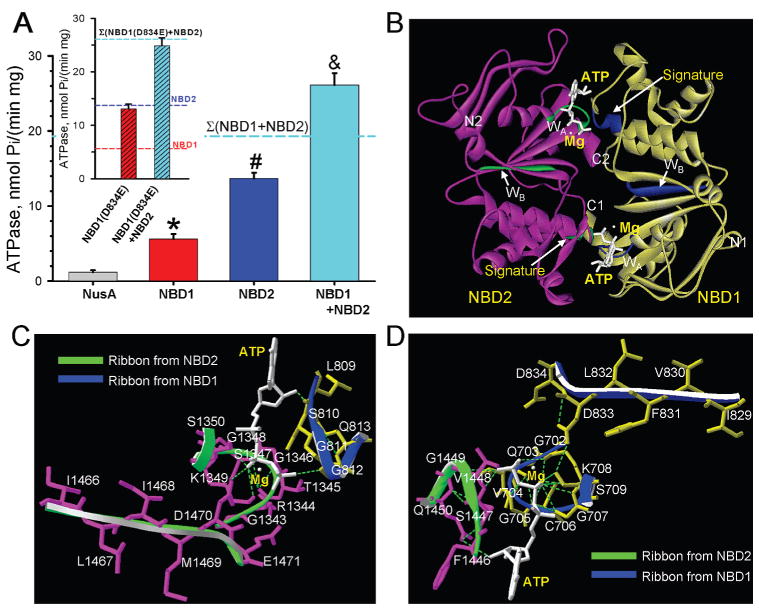
Purified recombinant NBD1 and NBD2 proteins generated a specific ATPase activity of 5.6 ± 0.7 nmol Pi/(min mg) (n = 7) and 13.7 ± 0.8 nmol Pi/(min mg) (n = 7), respectively, values significantly higher than the background Vmax activity of 1.2 ± 0.3 nmol Pi/(min mg) (n = 5) measured in samples containing the NusA tag protein alone (P < 0.05; Figure 4A). The catalytic function of NBD1 and NBD2 was sensitive to treatment with the γ-phosphate analogue, beryllium fluoride, which reduced ATPase activity to 48 ± 3% (n = 4) and 93 ± 2% (n = 4) of control, respectively (not illustrated). Site-directed mutagenesis of the NBD2 Walker A motif (K1349A) reduced ATPase activity by 32 ± 12% (n = 4), which was further diminished by 60 ± 8% (n = 4) in the double NBD2 mutant where the Walker A lysine residue was mutated along the aspartate residue (D1470A) in the Walker B motif, which by itself has only a marginal effect, suggesting the importance of tandem residue-based coordination on regulation of ATP catalysis. Notably, the ATPase activity was markedly enhanced when NBD1 was allowed to interact with NBD2 (Figure 4A). The Vmax of 26.1 ± 1.6 nmol Pi/(min mg) (n = 7) measured in the NBD1/NBD2 complex was significantly higher than the individual catalytic activity of either NBD1 or NBD2, or their arithmetic sum (Figure 4A). The functional consequence of NBD1/NBD2 interaction was further verified by site-directed mutagenesis at the distal residue of the Walker B motifs (D/E), which serves as a catalytic base critical in defining ATPase properties in prokaryotic ABC proteins.56 Here, the corresponding SUR2A NBD1 D834 was mutated into a glutamate residue normally present at the equivalent Walker B position (E1471) in NBD2 (Figure 4B). The engineered NBD1(D834E) mutant exhibited a specific activity of 13.1 ± 0.7 nmol Pi/(min mg) (n = 4), significantly higher than native NBD1 but similar to wildtype NBD2 (Figure 4A, inset). Mutant NBD1 (D834E) in the presence of wildtype NBD2 generated an ATPase activity of 24.9 ± 0.8 nmol Pi/(min mg) (n = 3). This value was not significantly different from the arithmetic sum of their respective catalytic activities (Figure 4A, inset), suggesting that although the individual ATPase activity can be altered within an NBD it is the complementarity of native NBD1 and NBD2 that secure enhanced intrinsic ATPase activity. Taken together, these data indicate that the interaction of asymmetric NBD1 and NBD2 heterodomains has a direct functional implication on the catalytic behavior of the SUR2A protein.
Figure 4.
NBD1/NBD2 assembly promotes intrinsic ATPase activity. (A) The ATPase activity of NBD1 or NBD2 was significantly higher than background ascribed to the NusA tag. Upon NBD1/NBD2 interaction, ATPase activity significantly increased, exceeding the sum of individual NBD1 + NBD2 catalytic activities. *, #, and & indicate significantly greater (P < 0.05) than NusA, than NusA or NBD1, and NusA, NBD1, or NBD2, respectively. Inset: The D834E mutation increased NBD1 ATPase activity approximating wildtype NBD2 catalysis. NBD1 (D834E) and NBD2, free to interact, produced a total ATPase that did not exceed the arithmetic sum of individual catalytic activities. (B) Homology structural model of the NBD1/NBD2 heterodimer, developed in the “head to tail” orientation, indicated that Walker A (WA) and Walker B (WB) motifs along with a signature motif delineate nucleotide docking sites. NBD1 in yellow, NBD2 in purple, and the ATPase cofactor Mg2+ in white. Hydrogen bonds shown as dotted lines. N- and C-termini of individual NBDs are labeled. Close-up view of ATP-binding sites at the “head” (C) and “tail” (D) of the heterodimer.
Favorable Structural Environment for Catalysis at the Interface of NBD1 and NBD2
Supported by present nanoscale protein mapping and enzymatic analysis, atomic structural modeling predicted the residues at the NBD1/NBD2 interface, in the presence of the molecular substrate ATP, using a “head to tail” orientation with respective amino termini diagonally positioned reflecting crystallographic findings in ABC proteins.48,57–61 The generated heterodimer SUR2A NBD1/NBD2 model revealed that discrete domains of the NBDs sandwiched ATP molecules, facilitating protein–protein interaction in the nucleotide-bound state (Figure 4B). The ATP-binding pocket in the “head” location was composed of the NBD2 Walker A (GRTGSGKS) and Walker B (ILIMDE) motifs in conjunction with the signature LSGGQ motif provided by NBD1 (Figure 4B,C). Conversely, the ATP-binding site at the “tail” was delineated by NBD1 Walker A (GQVGCGKS) and B (IVFLDD) and the NBD2 signature FSVGQ motif (Figure 4B,D). The asymmetric ATPase activity of ABCC9 NBDs experimentally measured in recombinant proteins (Figure 4A) would suggest that a limited ATPase activity is generated at the “tail” site (Figure 4C), indicating that a primary site for ATP hydrolysis is associated with the active catalytic pocket at the “head” delineated by NBD2 Walker motifs and the accompanying NBD1 LSGGQ amino acid residues (Figure 4C), in line with the role of the LSGGQ motif in accelerating ATP hydrolysis.62 Thus, physical assembly of ABCC9-encoded NBD1 with NBD2 proteins creates a favorable structural environment for nucleotide docking, and aligns nucleotide binding domains into a conformational state that secures optimal ATP hydrolysis within the heterodimer.
Discussion
Cardiac KATP channels harness energetic decoding capabilities, harbored in the ABCC9-encoded SUR2A regulatory protein, and provide a high-fidelity feedback mechanism that adjusts membrane excitability to match fluctuating demands of cellular homeostasis.24,26 The operation of KATP channels requires functional SUR2A nucleotide binding domains, NBD1 and NBD2.8,34,63,64 Applying a panel of proteomic methods, we here provide direct evidence for physical interaction between asymmetric NBD1 and NBD2 SUR2A domains, establishing a conformational platform for tandem function in this unconventional ABC protein of the KATP channel.
Interdomain interaction of NBD1 with NBD2 produced a conformational change in the secondary protein structure, validated by nanoscale resolution imaging of the paired structure. The cooperative interaction, further delineated by in silico modeling, translated into enhanced maximal velocity of catalysis, implying favorable structural rearrangements at the interface of coupled NBD1 and NBD2 proteins. The identified molecular substrate for optimal catalysis in the SUR2A NBD1/NBD2 complex sets a basis for a structure–function relationship within the ABCC9-encoded protein. Although the mechanistic foundation coupling structural adaptation with catalytic activity has not been previously reported for a non-transporter ABC protein, the recently resolved crystal structure of the ABC transporter HlyB NBD implicates enhanced structural asymmetry within the dimer interface,65 underscoring molecular NBD complementarity during the ATP hydrolysis cycle.
Expression of mammalian ABCC proteins is challenging as these membrane-associated proteins display significant predisposition for generation of inclusion bodies and aggregation, limiting solubility and purification.66,67 Application of the NusA solubility enhancing tag50 reduced the propensity for aggregation in recombinant ABCC9 NBD1 and NBD2. On purification, fusion proteins demonstrated distinct α-helices, indicating a well-folded structure for both NBD1 and NBD2.41,52 The size distribution of purified NBD1 and NBD2 was typical of a monodisperse distribution profile consistent with the mapped monolithic microarchitecture of the proteins, and clearly distinct from NusA. Although purification of monomeric SUR NBDs has not been attained,68,69 purified oligomeric NBD1 and NBD2 retained catalytic activity, while homogeneity was demonstrated by dynamic light scattering and atomic force microscopy. Moreover, point mutations introduced in the conserved Walker motifs of SUR2A NBD affected ATPase activity ruling out possible contamination. In this way, the uniform nature of the soluble, nonaggregated, and stable ABCC9 NBD1 and NBD2 proteins secured molecules amenable for structural probing.
Hydrodynamic dimensions determined by dynamic light scattering, a method for sizing proteins in solution based on extracting diffusion coefficients from the autocorrelation function,40 were validated by the topographic nanoscale approach of atomic force microscopy.42,44,45 Protein–protein interaction predicted by measuring the secondary structural change in circular dichroism52,53 was further captured on atomic force microscopy scans and electron micrographs.42,44,45,70 Assembly of SUR2A NBD1 and NBD2 was accelerated at physiological temperature, a condition reported to favor protein–protein interaction.71 Combined biophysical and spectroscopic methods, along with nanoimaging, thereby provided the first opportunity to resolve the physical properties of ABC protein domains.
Interdomain interactions of isolated recombinant ABCC9-encoded SUR2A NBD1/NBD2 protein translated into enhanced catalytic activity, exceeding the sum of individual NBDs ATPase activities. Enhanced ATPase activity is in line with reports from related ABC proteins, such as the cystic fibrosis transmembrane conductance regulator (CFTR),72–74 although the outcome of NBD interaction on intrinsic catalytic activity may differ between members of the ABC protein family.67,69,75 Site-directed mutagenesis in the predicted catalytic base here altered ATPase activity, and disrupted the potentiation in catalytic behavior. This is of significance as the overall state of the catalytic function within the SUR2A regulatory module critically determines the behavior of the KATP channel complex with ATPase-driven conformations linked to channel pore gating.7,8,34 Indeed, the cooperative interaction of SUR2A NBD1 and NBD2 implies a favorable structural rearrangement at the NBD1/NBD2 interface conferring optimal operation of the regulatory KATP channel subunit. This dynamic structural paradigm of catalytic regulation establishes a molecular substrate with functional consequences for the ABCC proteome.
Acknowledgments
Supported by National Institutes of Health, American Heart Association, Marriott Heart Disease Research Program, and Mayo Clinic FUTR Career Development Award.
Abreviations
- ABC
ATP-binding cassette
- KATP channel
ATP-sensitive potassium channel
- NBD(s)
nucleotide binding domain(s)
- SUR
sulfonylurea receptor
- TMD
transmembrane domain
References
- 1.Dean M. The genetics of ATP-binding cassette transporters. Methods Enzymol. 2005;400:409–429. doi: 10.1016/S0076-6879(05)00024-8. [DOI] [PubMed] [Google Scholar]
- 2.Linton KJ. Structure and function of ABC transporters. Physiology (Bethesda) 2007;22:122–130. doi: 10.1152/physiol.00046.2006. [DOI] [PubMed] [Google Scholar]
- 3.Higgins CF, Linton KJ. The ATP switch model for ABC transporters. Nat Struct Mol Biol. 2004;11:918–926. doi: 10.1038/nsmb836. [DOI] [PubMed] [Google Scholar]
- 4.Biemans-Oldehinkel E, Doeven MK, Poolman B. ABC transporter architecture and regulatory roles of accessory domains. FEBS Lett. 2006;580:1023–1035. doi: 10.1016/j.febslet.2005.11.079. [DOI] [PubMed] [Google Scholar]
- 5.Oram JF, Vaughan AM. ATP-Binding cassette cholesterol transporters and cardiovascular disease. Circ Res. 2006;99:1031–1043. doi: 10.1161/01.RES.0000250171.54048.5c. [DOI] [PubMed] [Google Scholar]
- 6.Solbach TF, Konig J, Fromm MF, Zolk O. ATP-binding cassette transporters in the heart. Trends Cardiovasc Med. 2006;16:7–15. doi: 10.1016/j.tcm.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 7.Zingman LV, Alekseev AE, Bienengraeber M, Hodgson D, Karger AB, Dzeja PP, Terzic A. Signaling in channel/enzyme multimers: ATPase transitions in SUR module gate ATP-sensitive K+ conductance. Neuron. 2001;31:233–245. doi: 10.1016/s0896-6273(01)00356-7. [DOI] [PubMed] [Google Scholar]
- 8.Alekseev AE, Hodgson DM, Karger AB, Park S, Zingman LV, Terzic A. ATP-sensitive K+ channel channel/enzyme multimer: metabolic gating in the heart. J Mol Cell Cardiol. 2005;38:895–905. doi: 10.1016/j.yjmcc.2005.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walker JE, Saraste M, Runswick MJ, Gay NJ. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Higgins CF. ABC transporters: physiology, structure and mechanism–an overview. Res Microbiol. 2001;152:205–210. doi: 10.1016/s0923-2508(01)01193-7. [DOI] [PubMed] [Google Scholar]
- 11.Ashcroft FM. From molecule to malady. Nature. 2006;440:440–447. doi: 10.1038/nature04707. [DOI] [PubMed] [Google Scholar]
- 12.Mikhailov MV, Campbell JD, de Wet H, Shimomura K, Zadek B, Collins RF, Sansom MS, Ford RC, Ashcroft FM. 3-D structural and functional characterization of the purified KATP channel complex Kir6.2-SUR1. EMBO J. 2005;24:4166–4175. doi: 10.1038/sj.emboj.7600877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Inagaki N, Gonoi T, Clement JP, Wang CZ, Aguilar-Bryan L, Bryan J, Seino S. A family of sulfonylurea receptors determines the pharmacological properties of ATP-sensitive K+ channels. Neuron. 1996;16:1011–1017. doi: 10.1016/s0896-6273(00)80124-5. [DOI] [PubMed] [Google Scholar]
- 14.Bienengraeber M, Alekseev AE, Abraham MR, Carrasco AJ, Moreau C, Vivaudou M, Dzeja PP, Terzic A. ATPase activity of the sulfonylurea receptor: a catalytic function for the KATP channel complex. FASEB J. 2000;14:1943–1952. doi: 10.1096/fj.00-0027com. [DOI] [PubMed] [Google Scholar]
- 15.Masia R, Enkvetchakul D, Nichols CG. Differential nucleotide regulation of KATP channels by SUR1 and SUR2A. J Mol Cell Cardiol. 2005;39:491–501. doi: 10.1016/j.yjmcc.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 16.Zingman LV, Hodgson DM, Bienengraeber M, Karger AB, Kathmann EC, Alekseev AE, Terzic A. Tandem function of nucleotide binding domains confers competence to sulfonylurea receptor in gating ATP-sensitive K+ channels. J Biol Chem. 2002;277:14206–14210. doi: 10.1074/jbc.M109452200. [DOI] [PubMed] [Google Scholar]
- 17.Inagaki N, Gonoi T, Clement JP, Namba N, Inazawa J, Gonzalez G, Aguilar-Bryan L, Seino S, Bryan J. Reconstitution of IKATP: an inward rectifier subunit plus the sulfonylurea receptor. Science. 1995;270:1166–1170. doi: 10.1126/science.270.5239.1166. [DOI] [PubMed] [Google Scholar]
- 18.Hambrock A, Loffler-Walz C, Kloor D, Delabar U, Horio Y, Kurachi Y, Quast U. ATP-Sensitive K+ channel modulator binding to sulfonylurea receptors SUR2A and SUR2B: opposite effects of MgADP. Mol Pharmacol. 1999;55:832–840. [PubMed] [Google Scholar]
- 19.Lorenz E, Terzic A. Physical association between recombinant cardiac ATP-sensitive K+ channel subunits Kir6.2 and SUR2A. J Mol Cell Cardiol. 1999;31:425–434. doi: 10.1006/jmcc.1998.0876. [DOI] [PubMed] [Google Scholar]
- 20.Matsushita K, Kinoshita K, Matsuoka T, Fujita A, Fujikado T, Tano Y, Nakamura H, Kurachi Y. Intramolecular interaction of SUR2 subtypes for intracellular ADP-Induced differential control of KATP channels. Circ Res. 2002;90:554–561. doi: 10.1161/01.res.0000012666.42782.30. [DOI] [PubMed] [Google Scholar]
- 21.Bryan J, Munoz A, Zhang X, Dufer M, Drews G, Krippeit-Drews P, Aguilar-Bryan L. ABCC8 and ABCC9: ABC transporters that regulate K+ channels. Pflugers Arch. 2006;453:703–718. doi: 10.1007/s00424-006-0116-z. [DOI] [PubMed] [Google Scholar]
- 22.Zingman LV, Hodgson DM, Bast PH, Kane GC, Perez-Terzic C, Gumina RJ, Pucar D, Bienengraeber M, Dzeja PP, Miki T, Seino S, Alekseev AE, Terzic A. Kir6.2 is required for adaptation to stress. Proc Natl Acad Sci US A. 2002;99:13278–13283. doi: 10.1073/pnas.212315199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ashcroft FM. ATP-sensitive potassium channelopathies: focus on insulin secretion. J Clin Invest. 2005;115:2047–2058. doi: 10.1172/JCI25495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kane GC, Liu XK, Yamada S, Olson TM, Terzic A. Cardiac KATP channels in health and disease. J Mol Cell Cardiol. 2005;38:937–943. doi: 10.1016/j.yjmcc.2005.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moreau C, Prost AL, Derand R, Vivaudou M. SUR, ABC proteins targeted by KATP channel openers. J Mol Cell Cardiol. 2005;38:951–963. doi: 10.1016/j.yjmcc.2004.11.030. [DOI] [PubMed] [Google Scholar]
- 26.Nichols CG. KATP channels as molecular sensors of cellular metabolism. Nature. 2006;440:470–476. doi: 10.1038/nature04711. [DOI] [PubMed] [Google Scholar]
- 27.Hambrock A, de Oliveira Franz CB, Hiller S, Grenz A, Ackermann S, Schulze DU, Drews G, Osswald H. Resveratrol binds to the sulfonylurea receptor (SUR) and induces apoptosis in a SUR subtype-specific manner. J Biol Chem. 2007;282:3347–3356. doi: 10.1074/jbc.M608216200. [DOI] [PubMed] [Google Scholar]
- 28.Kane GC, Behfar A, Dyer RB, O’Cochlain DF, Liu XK, Hodgson DM, Reyes S, Miki T, Seino S, Terzic A. KCNJ11 gene knockout of the Kir6.2 KATP channel causes maladaptive remodeling and heart failure in hypertension. Hum Mol Genet. 2006;15:2285–2297. doi: 10.1093/hmg/ddl154. [DOI] [PubMed] [Google Scholar]
- 29.Yamada S, Kane GC, Behfar A, Liu XK, Dyer RB, Faustino RS, Miki T, Seino S, Terzic A. Protection conferred by myocardial ATP-sensitive K+ channels in pressure overload-induced congestive heart failure revealed in KCNJ11 Kir6.2-null mutant. J Physiol. 2006;577:1053–1065. doi: 10.1113/jphysiol.2006.119511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Du Q, Jovanovic S, Clelland A, Sukhodub A, Budas G, Phelan K, Murray-Tait V, Malone L, Jovanovic A. Overexpression of SUR2A generates a cardiac phenotype resistant to ischemia. FASEB J. 2006;20:1131–1141. doi: 10.1096/fj.05-5483com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carrasco AJ, Dzeja PP, Alekseev AE, Pucar D, Zingman LV, Abraham MR, Hodgson D, Bienengraeber M, Puceat M, Janssen E, Wieringa B, Terzic A. Adenylate kinase phosphotransfer communicates cellular energetic signals to ATP-sensitive potassium channels. Proc Natl Acad Sci US A. 2001;98:7623–7628. doi: 10.1073/pnas.121038198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abraham MR, Selivanov VA, Hodgson DM, Pucar D, Zingman LV, Wieringa B, Dzeja PP, Alekseev AE, Terzic A. Coupling of cell energetics with membrane metabolic sensing. Integrative signaling through creatine kinase phosphotransfer disrupted by M-CK gene knock-out. J Biol Chem. 2002;277:24427–24434. doi: 10.1074/jbc.M201777200. [DOI] [PubMed] [Google Scholar]
- 33.Selivanov VA, Alekseev AE, Hodgson DM, Dzeja PP, Terzic A. Nucleotide-gated KATP channels integrated with creatine and adenylate kinases: amplification, tuning and sensing of energetic signals in the compartmentalized cellular environment. Mol Cell Biochem. 2004;256–257:243–256. doi: 10.1023/b:mcbi.0000009872.35940.7d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bienengraeber M, Olson TM, Selivanov VA, Kathmann EC, O’Cochlain F, Gao F, Karger AB, Ballew JD, Hodgson DM, Zingman LV, Pang YP, Alekseev AE, Terzic A. ABCC9 mutations identified in human dilated cardiomyopathy disrupt catalytic KATP channel gating. Nat Genet. 2004;36:382–387. doi: 10.1038/ng1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ueda K, Komine J, Matsuo M, Seino S, Amachi T. Cooperative binding of ATP and MgADP in the sulfonylurea receptor is modulated by glibenclamide. Proc Natl Acad Sci US A. 1999;96:1268–1272. doi: 10.1073/pnas.96.4.1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matsuo M, Kimura Y, Ueda K. KATP channel interaction with adenine nucleotides. J Mol Cell Cardiol. 2005;38:907–916. doi: 10.1016/j.yjmcc.2004.11.021. [DOI] [PubMed] [Google Scholar]
- 37.Miroux B, Walker JE. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J Mol Biol. 1996;260:289–298. doi: 10.1006/jmbi.1996.0399. [DOI] [PubMed] [Google Scholar]
- 38.Chifflet S, Torriglia A, Chiesa R, Tolosa S. A method for the determination of inorganic phosphate in the presence of labile organic phosphate and high concentrations of protein: application to lens ATPases. Anal Biochem. 1988;168:1–4. doi: 10.1016/0003-2697(88)90002-4. [DOI] [PubMed] [Google Scholar]
- 39.Park S, Ajtai K, Burghardt TP. Inhibition of myosin ATPase by metal fluoride complexes. Biochim Biophys Acta. 1999;1430:127–140. doi: 10.1016/s0167-4838(98)00262-3. [DOI] [PubMed] [Google Scholar]
- 40.Gakh O, Adamec J, Gacy AM, Twesten RD, Owen WG, Isaya G. Physical evidence that yeast frataxin is an iron storage protein. Biochemistry. 2002;41:6798–6804. doi: 10.1021/bi025566+. [DOI] [PubMed] [Google Scholar]
- 41.Greenfield NJ. Using circular dichroism spectra to estimate protein secondary structure. Nat Protocols. 2006;1:2876–2890. doi: 10.1038/nprot.2006.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Perez-Terzic C, Gacy AM, Bortolon R, Dzeja PP, Puceat M, Jaconi M, Prendergast FG, Terzic A. Structural plasticity of the cardiac nuclear pore complex in response to regulators of nuclear import. Circ Res. 1999;84:1292–1301. doi: 10.1161/01.res.84.11.1292. [DOI] [PubMed] [Google Scholar]
- 43.Kuznetsov VY, Ivanov YD, Bykov VA, Saunin SA, Fedorov IA, Lemeshko SV, Hoa HB, Archakov AI. Atomic force microscopy detection of molecular complexes in multiprotein P450cam containing monooxygenase system. Proteomics. 2002;2:1699–1705. doi: 10.1002/1615-9861(200212)2:12<1699::AID-PROT1699>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 44.Frederix PL, Akiyama T, Staufer U, Gerber C, Fotiadis D, Muller DJ, Engel A. Atomic force bio-analytics. Curr Opin Chem Biol. 2003;7:641–647. doi: 10.1016/j.cbpa.2003.08.010. [DOI] [PubMed] [Google Scholar]
- 45.Nakazawa K, Yamakoshi Y, Tsuchiya T, Ohno Y. Purification and aqueous phase atomic force microscopic observation of recombinant P2X2 receptor. Eur J Pharmacol. 2005;518:107–110. doi: 10.1016/j.ejphar.2005.06.018. [DOI] [PubMed] [Google Scholar]
- 46.Perez-Terzic C, Faustino RS, Boorsma BJ, Arrell DK, Niederlander NJ, Behfar A, Terzic A. Stem cells transform into a cardiac phenotype with remodeling of the nuclear transport machinery. Nat Clin Pract Cardiovasc Med. 2007;4:S68–76. doi: 10.1038/ncpcardio0763. [DOI] [PubMed] [Google Scholar]
- 47.Perez-Terzic C, Behfar A, Mery A, van Deursen JM, Terzic A, Puceat M. Structural adaptation of the nuclear pore complex in stem cell-derived cardiomyocytes. Circ Res. 2003;92:444–452. doi: 10.1161/01.RES.0000059415.25070.54. [DOI] [PubMed] [Google Scholar]
- 48.Zaitseva J, Jenewein S, Jumpertz T, Holland IB, Schmitt L. H662 is the linchpin of ATP hydrolysis in the nucleotide-binding domain of the ABC transporter HlyB. EMBO J. 2005;24:1901–1910. doi: 10.1038/sj.emboj.7600657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 50.Davis GD, Elisee C, Newham DM, Harrison RG. New fusion protein systems designed to give soluble expression in Escherichia coli. Biotechnol Bioeng. 1999;65:382–388. [PubMed] [Google Scholar]
- 51.De Marco V, Stier G, Blandin S, de Marco A. The solubility and stability of recombinant proteins are increased by their fusion to NusA. Biochem Biophys Res Commun. 2004;322:766–771. doi: 10.1016/j.bbrc.2004.07.189. [DOI] [PubMed] [Google Scholar]
- 52.Sreerama N, Woody RW. Computation and analysis of protein circular dichroism spectra. Methods Enzymol. 2004;383:318–351. doi: 10.1016/S0076-6879(04)83013-1. [DOI] [PubMed] [Google Scholar]
- 53.Greenfield NJ. Circular dichroism analysis for protein-protein interactions. Methods Mol Biol. 2004;261:55–78. doi: 10.1385/1-59259-762-9:055. [DOI] [PubMed] [Google Scholar]
- 54.Wang H, Yang Y, Schofield MJ, Du C, Fridman Y, Lee SD, Larson ED, Drummond JT, Alani E, Hsieh P, Erie DA. DNA bending and unbending by MutS govern mismatch recognition and specificity. Proc Natl Acad Sci US A. 2003;100:14822–14827. doi: 10.1073/pnas.2433654100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fotiadis D, Jastrzebska B, Philippsen A, Muller DJ, Palczewski K, Engel A. Structure of the rhodopsin dimer: a working model for G-protein-coupled receptors. Curr Opin Struct Biol. 2006;16:252–259. doi: 10.1016/j.sbi.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 56.Moody JE, Millen L, Binns D, Hunt JF, Thomas PJ. Cooperative, ATP-dependent association of the nucleotide binding cassettes during the catalytic cycle of ATP-binding cassette transporters. J Biol Chem. 2002;277:21111–21114. doi: 10.1074/jbc.C200228200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hopfner KP, Karcher A, Shin DS, Craig L, Arthur LM, Carney JP, Tainer JA. Structural biology of Rad50 ATPase: ATP-driven conformational control in DNA double-strand break repair and the ABC-ATPase superfamily. Cell. 2000;101:789–800. doi: 10.1016/s0092-8674(00)80890-9. [DOI] [PubMed] [Google Scholar]
- 58.Smith PC, Karpowich N, Millen L, Moody JE, Rosen J, Thomas PJ, Hunt JF. ATP binding to the motor domain from an ABC transporter drives formation of a nucleotide sandwich dimer. Mol Cell. 2002;10:139–149. doi: 10.1016/s1097-2765(02)00576-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Campbell JD, Sansom MS, Ashcroft FM. Potassium channel regulation. EMBO Rep. 2003;4:1038–1042. doi: 10.1038/sj.embor.7400003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen J, Lu G, Lin J, Davidson AL, Quiocho FA. A tweezers-like motion of the ATP-binding cassette dimer in an ABC transport cycle. Mol Cell. 2003;12:651–661. doi: 10.1016/j.molcel.2003.08.004. [DOI] [PubMed] [Google Scholar]
- 61.Procko E, Ferrin-O’Connell I, Ng SL, Gaudet R. Distinct structural and functional properties of the ATPase sites in an asymmetric ABC transporter. Mol Cell. 2006;24:51–62. doi: 10.1016/j.molcel.2006.07.034. [DOI] [PubMed] [Google Scholar]
- 62.Tombline G, Bartholomew L, Gimi K, Tyndall GA, Senior AE. Synergy between conserved ABC signature Ser residues in P-glycoprotein catalysis. J Biol Chem. 2004;279:5363–5373. doi: 10.1074/jbc.M311964200. [DOI] [PubMed] [Google Scholar]
- 63.Zingman LV, Hodgson DM, Alekseev AE, Terzic A. Stress without distress: homeostatic role for KATP channels. Mol Psychiatry. 2003;8:253–254. doi: 10.1038/sj.mp.4001323. [DOI] [PubMed] [Google Scholar]
- 64.Olson TM, Alekseev AE, Moreau C, Liu XK, Zingman LV, Miki T, Seino S, Asirvatham SJ, Jahangir A, Terzic A. KATP channel mutation confers risk for vein of Marshall adrenergic atrial fibrillation. Nat Clin Pract Cardiovasc Med. 2007;4:110–116. doi: 10.1038/ncpcardio0792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zaitseva J, Oswald C, Jumpertz T, Jenewein S, Wiedenmann A, Holland IB, Schmitt L. A structural analysis of asymmetry required for catalytic activity of an ABC-ATPase domain dimer. EMBO J. 2006;25:3432–3443. doi: 10.1038/sj.emboj.7601208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Roosbeek S, Caster H, Liu QZ, Berne PF, Duverger N, Christiaens B, Vandekerckhove J, Peelman F, Labeur C, Rosseneu M. Expression and activity of the nucleotide-binding domains of the human ABCA1 transporter. Protein Expression Purif. 2004;35:102–110. doi: 10.1016/j.pep.2003.12.015. [DOI] [PubMed] [Google Scholar]
- 67.Ramaen O, Sizun C, Pamlard O, Jacquet E, Lallemand JY. Attempts to characterize the NBD heterodimer of MRP1: transient complex formation involves Gly771 of the ABC signature sequence but does not enhance the intrinsic ATPase activity. Biochem J. 2005;391:481–490. doi: 10.1042/BJ20050897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mikhailov MV, Ashcroft SJ. Interactions of the sulfonylurea receptor 1 subunit in the molecular assembly of beta-cell KATP channels. J Biol Chem. 2000;275:3360–3364. doi: 10.1074/jbc.275.5.3360. [DOI] [PubMed] [Google Scholar]
- 69.de Wet H, Mikhailov MV, Fotinou C, Dreger M, Craig TJ, Venien-Bryan C, Ashcroft FM. Studies of the ATPase activity of the ABC protein SUR1. FEBS J. 2007;274:3532–3544. doi: 10.1111/j.1742-4658.2007.05879.x. [DOI] [PubMed] [Google Scholar]
- 70.Sokolova AV, Kreplak L, Wedig T, Mucke N, Svergun DI, Herrmann H, Aebi U, Strelkov SV. Monitoring intermediate filament assembly by small-angle x-ray scattering reveals the molecular architecture of assembly intermediates. Proc Natl Acad Sci US A. 2006;103:16206–16211. doi: 10.1073/pnas.0603629103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Palleros DR, Welch WJ, Fink AL. Interaction of hsp70 with unfolded proteins: effects of temperature and nucleotides on the kinetics of binding. Proc Natl Acad Sci US A. 1991;88:5719–5723. doi: 10.1073/pnas.88.13.5719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sharma S, Davis JA, Ayvaz T, Traxler B, Davidson AL. Functional reassembly of the Escherichia coli maltose transporter following purification of a MalF-MalG subassembly. J Bacteriol. 2005;187:2908–2911. doi: 10.1128/JB.187.8.2908-2911.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang Z, Stalcup LD, Harvey BJ, Weber J, Chloupkova M, Dumont ME, Dean M, Urbatsch IL. Purification and ATP hydrolysis of the putative cholesterol transporters ABCG5 and ABCG8. Biochemistry. 2006;45:9929–9939. doi: 10.1021/bi0608055. [DOI] [PubMed] [Google Scholar]
- 74.Kidd JF, Ramjeesingh M, Stratford F, Huan LJ, Bear CE. A heteromeric complex of the two nucleotide binding domains of cystic fibrosis transmembrane conductance regulator (CFTR) mediates ATPase activity. J Biol Chem. 2004;279:41664–41669. doi: 10.1074/jbc.M407666200. [DOI] [PubMed] [Google Scholar]
- 75.Biswas-Fiss EE. Interaction of the nucleotide binding domains and regulation of the ATPase activity of the human retina specific ABC transporter, ABCR. Biochemistry. 2006;45:3813–3823. doi: 10.1021/bi052059u. [DOI] [PubMed] [Google Scholar]