Abstract
KCNJ11 null mutants, lacking Kir6.2 ATP-sensitive K+ (KATP) channels, exhibit a marked susceptibility towards hypertension (HTN)-induced heart failure. To gain insight into the molecular alterations induced by knockout of this metabolic sensor under hemodynamic stress, wild-type (WT) and Kir6.2 knockout (Kir6.2-KO) cardiac proteomes were profiled by comparative 2-DE and Orbitrap MS. Despite equivalent systemic HTN produced by chronic hyperaldosteronism, 114 unique proteins were altered in Kir6.2-KO compared to WT hearts. Bioinformatic analysis linked the primary biological function of the KATP channel-dependent protein cohort to energetic metabolism (64% of proteins), followed by signaling infrastructure (36%) including oxido-reductases, stress-related chaperones, processes supporting protein degradation, transcription and translation, and cytostructure. Mapped protein–protein relationships authenticated the primary impact on metabolic pathways, delineating the KATP channel-dependent subproteome within a nonstochastic network. Iterative systems interrogation of the proteomic web prioritized heart-specific adverse effects, i.e., “Cardiac Damage”, “Cardiac Enlargement”, and “Cardiac Fibrosis”, exposing a predisposition for the development of cardiomyopathic traits in the hypertensive Kir6.2-KO. Validating this maladaptive forecast, phenotyping documented an aggravated myocardial contractile performance, a massive interstitial fibrosis and an exaggerated left ventricular size, all prognostic indices of poor outcome. Thus, Kir6.2 ablation engenders unfavorable proteomic remodeling in hypertensive hearts, providing a composite molecular substrate for pathologic stress-associated cardiovascular disease.
Keywords: ATP-sensitive K+ channel, Bioinformatics, Heart failure, Kir6.2, Systems biology
1 Introduction
Systemic HTN is a leading risk factor in the development of cardiomyopathy and heart failure [1–3]. The adverse cardiac manifestations of HTN encompass ventricular enlargement, interstitial fibrosis, and ultimately organ dysfunction, significantly contributing to morbidity and mortality in the population at large [4–6]. Heart adaptation to hypertensive stress load is however highly variable, with only certain individuals progressing from a state of risk to one of overt disease [7]. Homeostatic processes that ensure stress tolerance are only partially understood [8, 9], warranting elucidation of individual cardioprotective components and their underlying systems organization within the hypertensive myocardium.
A prototypic stress-response element is the ATP-sensitive K+ (KATP) channel, formed through heteromeric assembly of the inwardly rectifying K+channel, Kir6.x, with the regulatory ATP-binding cassette sulfonylurea receptor protein [10–13]. The KCNJ11 gene encodes the pore-forming Kir6.2 isoform of cardiac KATP channels, particularly abundant in the sarcolemma of ventricular myocytes [14–16]. KATP channels act as high-fidelity molecular rheostats, adjusting membrane potential-dependent functions to match energetic demands of the working heart [17, 18]. Established in both experimental models and in humans, intact Kir6.2 is required for securing cardiac stress adaptation, with KATP channel malfunction implicated in the development of heart disease [19–21]. It has recently been recognized that a deficit in KATP channels impairs tolerance to systemic stressors, ranging from sympathetic surge [22–26] and endurance challenge [27] to hemodynamic load [28–30]. Moreover, genetic disruption of KATP channels compromises the protective benefits of preconditioning [31–33], while overexpression of channel subunits generates a resistant phenotype [34].
Cardioprotective properties of KATP channels have been linked to a tight integration of channel proteins with the cellular metabolic infrastructure, ensuring effective coupling of channel function with bioenergetic dynamics [35–41]. KATP channel subunits have been reported to physically associate and/or communicate with myocardial energy shuttles, i.e., creatine kinase, adenylate kinase, and lactate dehydrogenase, and energy production pathways catalyzed by glycolytic or fatty acid biosynthesis enzymes, including glyceraldehyde-3-phosphate dehydrogenase, triosephosphate isomerase, and long chain acyl-CoA dehydrogenase [42–46]. Privileged channel interactions with identified partners have been implicated in cellular protection, whereas dysfunction of – or uncoupling from – interacting partners predisposes to stress intolerance [47–49]. Although present knowledge indicates that the KATP channel is integrated with cell metabolism, contributing to stress tolerance, a broader understanding of the channel’s relationship with the cellular milieu and its implication on disease predisposition is currently lacking.
Indeed, biological functions can rarely be attributed to individual molecules, but rather arise through complex interactions between numerous cellular constituents [50–52]. Proteomic analysis provides a high-throughput, unbiased approach for large scale identification of proteins responsible for the execution of physiological processes [53]. As such, proteomic methodology is suited for dissecting the molecular fingerprint of KATP channel-dependent system associations. In the setting of HTN, the consequences of myocardial KATP channel deficiency were here examined by comparative 2-DE followed by nanoelectrospray linear IT tandem MS (nESI LC-MS/MS). Over one hundred identified proteins were differentially expressed within the cytosol of KATP channel knockouts compared to their WT counterparts. Bioinformatic examination of protein–protein relationships further mapped the primary impact on metabolic pathways, delineating that the KATP channel-dependent subproteome resides within a nonstochastic network. Stratification of the resolved interactome unmasked, in the absence of KATP channel function, a maladaptive predilection for cardiomyopathic traits under the stress of HTN. This proteomic approach thereby provides a systems-based diagnostic tool to chart the composite molecular substrate of KATP channel deficiency in the hypertensive heart.
2 Materials and methods
2.1 Volume overload hypertension model
Protocols were carried out in accordance with NIH guidelines, following approval of the Mayo Clinic Institutional Animal Care and Use Committee. KATP channel Kir6.2 knockout (Kir6.2-KO) mice, generated by disruption of the KCNJ11 gene encoding the Kir6.2 channel pore [54], were back-crossed for five generations to a C57BL/6 background. Targeted Kir6.2 disruption generates null mutants with ventricular myocytes lacking functional KATP channels [55]. Adult male C57BL/6 wild-type (WT) mice and age-matched male Kir6.2-KO counterparts underwent left nephrectomy through a retroperitoneal flank excision under isoflurane anesthesia to reduce urinary clearance. Mineralocorticoid-HTN was induced by subcutaneous implantation of a 50 mg 21 day release deoxycorticosterone-acetate tablet (Innovative Research of America) and supplementation with 1% NaCl plus 0.2% KCl in drinking water [28]. Mice were given standard rodent chow, and housed individually under a 12 h day/night cycle. Water and salt intake were measured weekly. Mice had similar nonfasting glycemic levels measured by tail sampling (OneTouch Ultra, Lifescan). Following one week of acclimatization training to restraint, blood pressure was measured by automated tail-cuff recording (Columbus Instruments) in awake restrained animals, two weeks post-nephrectomy. Blood pressure values were digitally derived from ten sequential recordings, documenting overt HTN in all WT and Kir6.2-KO by 14 days of mineralocorticoid/salt loading [28].
2.2 Protein extraction and quantitation
At 21 days, hypertensive WT and Kir6.2-KO animals were weighed, sacrificed under isoflurane anesthesia, and hearts excised and rinsed in PBS. Left ventricles including septum were removed, weighed ex vivo, snap-frozen in liquid N2, and stored at −80°C. Cytosolic tissue extracts were prepared by mechanical homogenization at 4°C in four volumes of extraction buffer, consisting of (in mM) HEPES 25 (pH 7.4), PMSF 0.25, and DTT 50, 1.25 μM pepstatin A, Mini-Complete™ protease inhibitor cocktail (Roche Applied Science), and 1% phosphatase inhibitor cocktails 1 and 2 (Sigma) [56]. Samples were centrifuged (16 000×g) at 4°C for 10 min, supernatants were transferred to fresh tubes, and protein was quantified in triplicate by BioRad protein assay using the microassay procedure with a bovine gamma globulin standard [57, 58].
2.3 2-DE and gel imaging
Protein extracts (100 μg) were resolved by IPG 2-DE following addition to IEF rehydration buffer (7 M urea, 2 M thiourea, 2% w/v CHAPS, 50 mM DTT, 1x BioRad pH 3–10 ampholytes). IPG Ready Strips™ (pH 3–10, 170 mm, BioRad) were actively rehydrated at 50 V for 10 h, followed by rapid voltage ramping with a series of 15 min steps at 100, 500, and 1000 V, and a final step at 10 000 V for 60 kVh at 20°C. Focused IPG strips were rinsed with distilled, deionized water, and incubated for 15 min in equilibration buffer (50 mM Tris-HCl, pH 8.8, 6 M urea, 30% v/v glycerol, 2% w/v SDS) containing 10 mg/mL DTT, followed by 15 min in equilibration buffer containing 25 mg/mL iodoacetamide. After horizontal positioning on 12.5% SDS–PAGE gels, strips were overlaid with SDS buffer (25 mM Tris, 192 mM glycine, 0.1% w/v SDS) containing 0.5% w/v agarose, and resolved orthogonally by SDS–PAGE in a Protean® II XL system (BioRad). Resolved 2-D gels were silver stained and digitized at 400 dpi for spot image analysis, including spot detection, matching, normalization, and quantification, conducted with BioRad PDQuest v.7.4.0 [58, 59]. Individual gel images were normalized by total intensity of valid spots. Fold change was calculated as the [mean Kir6.2-KO]:[mean WT] ratio for protein spots increasing, or the negative inverse for protein species decreasing, in KATP channel knockouts. For proteins identified in more than one spot, the sum of values for all spots was used to determine a weighted average treatment ratio, unless otherwise indicated.
2.4 Nanoelectrospray linear IT tandem MS
Significantly altered protein species were isolated, destained, and prepared for LC-MS/MS by reduction, alkylation, tryptic digestion, peptide extraction, and drying [59, 60]. Peptides were reconstituted in 0.15% formic acid, 0.05% TFA, and trap injected onto a 75 μm × 10 cm ProteoPep C18 PicoFrit™ nanoflow column (New Objective). Chromatography was performed using 0.2% formic acid in solvents A (99% water, 1% ACN) and B (80% ACN, 5% isopropanol, 15% water), with peptides eluted over 30 min with a 5–45% solvent B gradient using an Eksigent nanoHPLC system (MDS Sciex) coupled to an LTQ-Orbitrap mass spectrometer (Thermo Fisher Scientific). Continuous scanning of eluted peptide ions was carried out between 375–1600 m/z, automatically switching to MS/MS CID mode on ions exceeding an intensity of 8000. Raw MS/MS spectra were converted to.dta files using Bioworks 3.2 (Thermo Fisher Scientific), and merged files matching +1, 2, or 3 peptide charge states were correlated to theoretical tryptic fragments in Swiss-Prot (v.53.0) using Mascot™ v.2.2 [61]. Searches were conducted on mammalian sequences (53 539 entries), tolerating up to two missed cleavages, a mass tolerance of ±0.01 Da for precursor ions (including 13C peak detection) and ±0.6 Da for MS/MS product ions allowing for protein N-terminal acetylation, methionine oxidation, and cysteine carbamidomethylation. Protein identities were confirmed by matching multiple peptide spectra at p<0.05, with proteins accepted at p<0.01. Proteins identified by a single peptide were subjected to a higher stringency level (p<0.01), and were confirmed by manual spectrum inspection with detected fragment ions from the MS/MS spectrum required to be above baseline noise, have demonstrable continuity in b- or y-ion series, and proline residues yielding intense y-ions [62]. Protein assignments were further validated by congruence of observed versus predicted pI/Mr, using the ExPASy pI/Mr tool (http://us.expasy.org/tools/pi_tool.html), taking into consideration protein processing and PTMs [59].
2.5 Interactome network analysis
Differentially expressed proteins, with fold change ratios, were submitted as focus proteins for network analysis using Ingenuity Pathways Knowledge Base (Ingenuity® Systems, www.ingenuity.com) to identify associated functional networks. An overview of interactions was obtained by merging functional subnetworks into a composite interactome. The composite was depicted using the molecular interaction network visualization program Cytoscape 2.5.1, with topological properties characterized as an undirected network using Network Analyzer [63]. Computed properties included node degree (k), the number of links connected to the node, and node degree distribution (P[k]), the probability that a specified node has k links, defined as P[k] = X[k]/n, where X[k] is the number of nodes with degree k and n is the total number of network nodes [64, 65]. P[k] versus k discriminates between random and scale-free topographies, defined by normal and power law distributions, respectively [64]. The Anderson–Darling normality test [66] ruled out a normal distribution, so P[k] versus k was calculated as a power law relationship using a cumulative distribution function [67] to determine γ in the power law distribution (P[k] ~ k−γ) according to Eq. (1)
| (1) |
where γ is the power law exponent, n the number of networknodes, xi node degree, and xmin is the minimum node degree within the network, with statistical error σ [67] for Eq. (1) defined by Eq. (2)
| (2) |
To link expression data with predicted pathological outcomes, the resolved network was interrogated with Ingenuity Pathways Analysis by screening for associated toxicity functions.
2.6 Cardiac function and structure
Prior to and following the 21 day HTN protocol, trans-thoracic echocardiography (Model c256 with a 15L8 probe, Acuson) was performed in lightly anesthetized (1.25% isoflurane) WT and Kir6.2-KO mice. Images were digitally acquired and stored for offline blinded analysis. Stroke volume was determined by the product of aortic root cross-sectional area and the velocity time integral taken from peak trans-aortic Doppler tracings [29, 30]. The product of stroke volume and heart rate, expressed as mL/min, was used to calculate cardiac output [29]. Interstitial fibrosis was quantified by computer analysis (MetaMorph, Visitron Universal Imaging) of 0.5 μm thick, paraffin-embedded, Sirius red stained sections.
2.7 Statistical analysis
Comparison between groups was performed using a standard t-test of variables with 95% confidence intervals, with data expressed as mean±standard error.
3 Results
3.1 KATP channel knockout alters cardiac proteome in hypertension
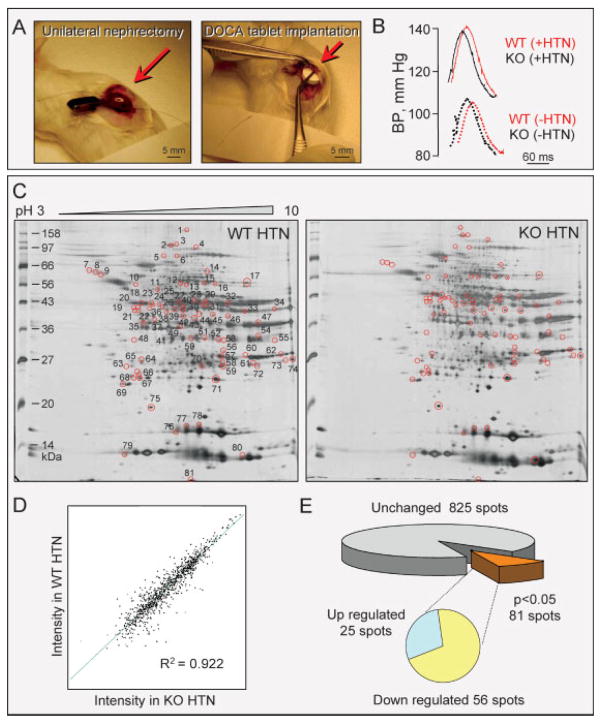
Systemic HTN is a common condition of stress faced by the heart [9] with chronic hyperaldosteronism a leading cause [68], replicated herein through unilateral nephrectomy and mineralocorticoid/salt challenge (Fig. 1A). Sustained hemodynamic stress produced comparable HTN in WT and KATP channel knockout (Kir6.2-KO) mice (Fig. 1B). At 14 days of follow-up, systolic blood pressure increased significantly from 112 ± 5 to 137 ± 5 mm Hg in WT (n = 14, p<0.05), and similarly from 110 ± 4 to 135 ± 5 mm Hg in age-matched mice lacking the KCNJ11-encoded Kir6.2 channel pore (n = 12, p<0.05 compared to baseline and p = 0.8 when compared to WT). To assess the molecular consequences of KATP channel disruption, the left ventricular cytosolic proteome was extracted from WT (n = 4) and Kir6.2-KO (n = 5) hearts and profiled by comparative 2-DE (Fig. 1C). In broad pH range (pH 3–10) silver-stained gels, more than 900 protein species were consistently resolved. Reproducibility was documented by a high degree of correlation (R2 = 0.922) between protein spot intensities from WT and Kir6.2-KO gels (Fig. 1D). Densitometric quantification demonstrated that a subset of 81 unique protein spots (9% of the total) was significantly altered in response to Kir6.2 deletion, 56 of which were downregulated and 25 upregulated (Fig. 1E). Thus, comparative expression profiling of WT and Kir6.2-KO myocardium revealed that, in response to equivalent hypertensive stress, the absence of functional KATP channels translates into a distinct, remodeled cardiac subproteome.
Figure 1.
Impact of Kir6.2 KO on the ventricular proteome in HTN. (A) Unilateral nephrectomy (left) and implantation of deoxy-corticosterone acetate (DOCA) releasing tablet (right) were implemented in WT and age/sex-matched Kir6.2-KO mice to induce HTN through volume overload. (B) Blood pressure measurements revealed significant yet comparable HTN at 14 days of volume overload in WTand Kir6.2-KO. (C) Representative 2-D gels of left ventricular cytoplasmic protein extracts (100 μg per gel) from hypertensive WT (WT HTN) and Kir6.2-KO (KO HTN) mice resolved by pH 3–10 IEF/12.5% SDS–PAGE. Differentially expressed spots are circled, and numbered on the WT HTN gel to cross-reference with the corresponding protein identities determined by LTQ-Orbitrap MS/MS analysis of excised tryptic digests (listed in Fig. 3 and Fig. 4 and Table S1 and S2 of Supporting Information). (D) Average normalized intensities of matching protein spots from WT HTN and Kir6.2-KO HTN gels indicated a high level of gel-to-gel reproducibility and correlation (R2 = 0.922) across cohorts. (E) Statistical analysis comparing WT to Kir6.2-KO gel densitometric spot quantitation revealed a subset of 81 spots that differed in Kir6.2-KO extracts (p<0.05), with 56 down-regulated and 25 up-regulated protein species.
3.2 Mapping the Kir6.2-dependent subproteome
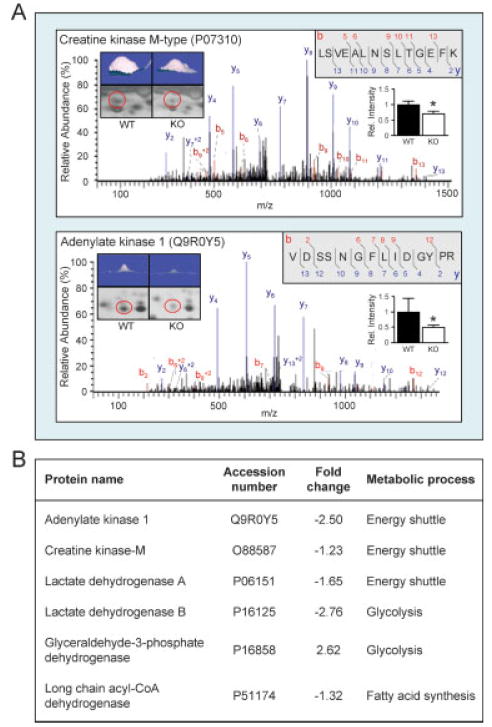
To characterize the KATP channel-dependent subproteome in the setting of HTN, a discriminatory analysis was carried out by LTQ-Orbitrap MS/MS contrasting WT versus Kir6.2-KO. As examples of this multistep identification approach, spots 40 and 67 (Fig. 1C and enlarged in Fig. 2A, left insets), both down-regulated in the Kir6.2-KO dependent subproteome (Fig. 2A, right insets), were identified by MS/MS spectral peptide assignments as creatine kinase M (Fig. 2A, upper spectrum and peptide sequence) and adenylate kinase 1 (Fig. 2A, lower spectrum and peptide sequence) isoforms. These prototypic phosphotransfer enzymes have been demonstrated to couple cellular bioenergetics with KATP channel activity [42, 43, 47]. Indeed, metabolic enzymes with previously recognized associations to KATP channel function were altered in response to Kir6.2 deletion (Fig. 2B), including long chain acyl-CoA dehydrogenase, lactate dehydrogenase A and B, glyceraldehyde-3-phosphate dehydrogenase, and triosephosphate isomerase [42–47], thereby validating the sensitivity and specificity of the employed proteomic approach.
Figure 2.
Metabolic enzymes associated with KATP channel function are among the differentially affected subproteome. (A) Representative LTQ-Orbitrap MS/MS product ion spectra obtained for spots #40 (upper) and #67 (lower), modified from BioWorks 3.2 to indicate detected b-ions in red and y-ions in blue, with corresponding peptide sequences of the identified proteins, creatine kinase M-type and adenylate kinase 1 (upper right insets). Spots are shown for comparison by 2-D gel region enlargement (circled in red) and 3-D rendering (upper left insets), with relative spot intensities indicated in the histogram (middle right insets). (B) A number of additional metabolic enzymes with known association to KATP channels were significantly altered in Kir6.2-KO hypertensive hearts.
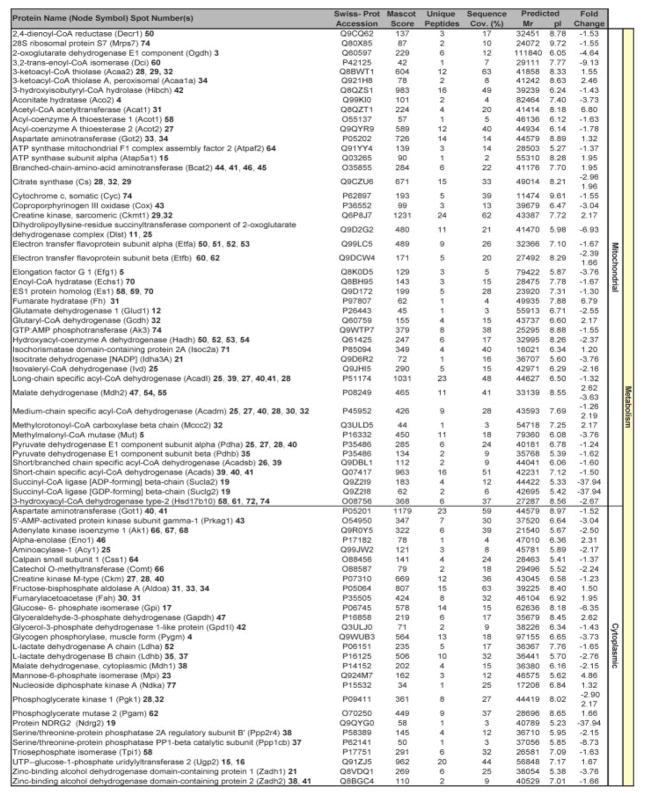
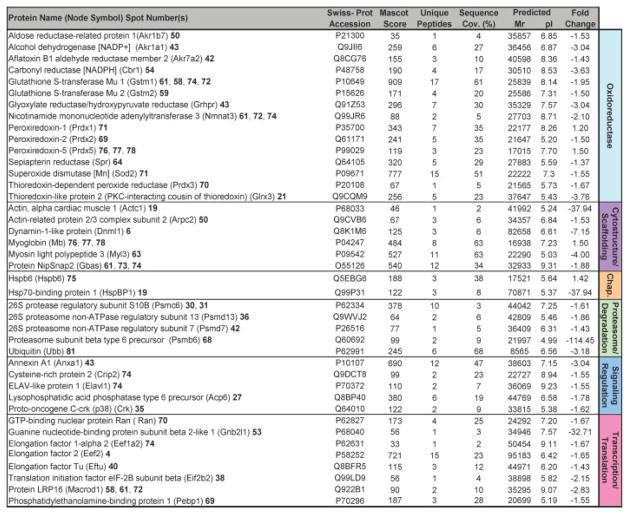
Beyond verifying these previously identified KATP channel interactions, the high-throughput approach utilized here expanded the metabolism-related KATP channel-dependent subproteome (Fig. 3). Specifically, a total of 114 unique proteins were found to be altered in Kir6.2-KO hearts, following assignments for 63 of the 81 differentially expressed protein species [see Table S1 and S2 of Supporting Information]. Cellular metabolism was the primary function associated with 64% of identified proteins, accounting for 45 mitochondrial and 28 cytoplasmic metabolic proteins within the resolved KATP channel-dependent subproteome (Fig. 3). Complete ontological annotation revealed that the remaining 41 identified proteins form a metabolism-related infrastructure encompassing oxidoreductases (15 proteins), stress related chaperones (2), components of the proteasome (6), signaling regulation (5), transcription and translation (8), as well as cellular structure and scaffolding (6) (Fig. 4). Thus, this comprehensive protein identification and ontological stratification approach mapped a definitive set of 114 proteins in the setting of HTN, expanding by over an order of magnitude the resolved elements of the Kir6.2 interactome.
Figure 3.
Metabolism is the predominant function of Kir6.2-KO proteins differentially altered in response to HTN. Significantly altered spots identified by LTQ-Orbitrap MS/MS analysis and functionally categorized by their respective Swiss-Prot ontological annotations were primarily associated with metabolism related processes, with 45 and 28 proteins involved in mitochondrial or cytoplasmic metabolism, respectively. Protein names are listed with corresponding spot numbers to locate their 2-D gel position(s) in Fig. 1, and with Swiss-Prot gene names for locating them in the protein interaction network in Fig. 5. Protein Mascot score, number of unique identified peptides, % sequence cov. (coverage), predicted Mr and pI for each protein (following expected post-translational processing, e.g., removal of a mitochondrial signal peptide), and fold change (KO vs. WT) are indicated. For proteins detected in more than one spot, the maximum score and corresponding number of unique peptides are reported. Fold change was calculated as described in experimental procedures, and for proteins detected in both increasing and decreasing spots, both values are indicated. Complete MS/MS data for all proteins is outlined in Table S1 and S2 of Supporting Information.
Figure 4.
Remaining Kir6.2-KO proteins differentially altered in response to HTN comprise a metabolism infrastructure related module. Significantly altered spots identified by LTQ-Orbitrap MS/MS analysis included 41 proteins, and functionally categorized by their respective Swiss-Prot ontological annotations as oxidoreductases, cytostructure, and scaffolding proteins, stress related chaperones, proteasome, and degradation components, and elements of signaling regulation, and transcription and translation. Protein names are listed with corresponding spot numbers to locate their 2-D gel position(s) in Fig. 1, and with Swiss-Prot gene names for locating them in the protein interaction network in Fig. 5. Protein Mascot score, number of unique identified peptides, % sequence cov. (coverage), predicted Mr and pI for each protein (following expected post-translational processing, e.g., removal of a mitochondrial signal peptide), and fold change (KO vs. WT) are indicated. Fold change was calculated as described in experimental procedures. Complete MS/MS data for all proteins is outlined in Table S1 and S2 of Supporting Information.
3.3 Kir6.2-dependent network assembly and topography
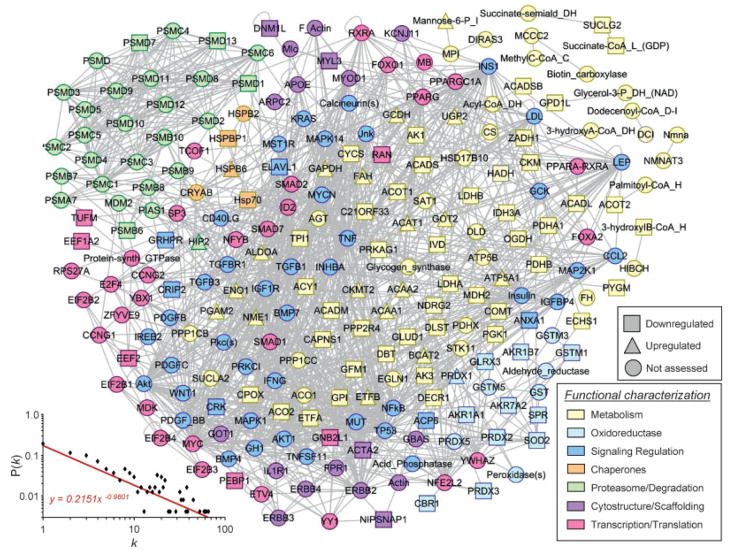
Network analysis was carried out to chart physiological and pathophysiological systems processes associated with the Kir6.2-dependent subproteome. The 114 identified proteins were integrated into a composite neighborhood comprised of 242 nodes linked by 1215 interactions or edges (Fig. 5). Network topology was not random, demonstrating a definitive nonstochastic property confirmed by examination of the interrelationship between node degree (k) and degree distribution (P[k]) (Fig. 5). The organized assemblage followed a power law distribution indicative of scale-free architecture, where P(k) ~ k−γ, with γ = 1.713 ± 0.046 (Fig. 5) falling within the predicted confidence range of biological networks [52, 53]. The resolved KATP channel dependent network unmasked ontological linkage to modules of bioenergetic and metabolism related processes, integrating multiple energy producing, consuming and distributing pathways (Fig. 5). By encompassing vital hallmarks of inter-linked cardiovascular energy metabolism [69], the composite non-stochastic, metabolism-centric neighborhood provides an unbiased systems framework for predictive interrogation of KATP channel criticality in the setting of HTN.
Figure 5.
Kir6.2-dependent protein–protein interaction network in HTN. The 114 differentially expressed proteins of the Kir6.2-dependent subproteome were submitted to Ingenuity Pathways Analysis as focus nodes, generating a 242 protein interaction network. Nodes are listed by Swiss-Prot gene names. Nodes are colored to correspond to functional categorization, while node shape indicates directionality of expression level change, unless the protein was not detected during proteomic analysis (legends, lower right). A plot of degree distribution (P[k]) versus degree (k) followed a power law distribution, indicating a scale free, nonstochastic network architecture (lower left).
3.4 Adverse outcomes predicted from the Kir6.2-dependent network
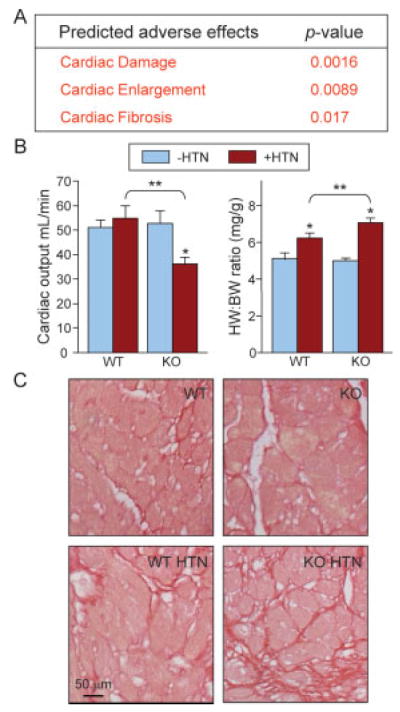
Criticality enables coordination of complex behaviors, balancing stability with adaptability [70]. Interrogation of the KATP channel-dependent network for links to a broad spectrum of 134 pathological conditions and toxicological pathways, curated in Ingenuity Pathways Knowledge Base, exclusively extracted three adverse effects: “Cardiac Damage”, “Cardiac Enlargement”, and “Cardiac Fibrosis” (Fig. 6A). This highly targeted cardiac-specific outcome with a likelihood of 1 in >2 000 000 of obtaining only these three outcomes at random (1/134×1/133×1/132 = 1/[2.35×106]), without other significantly over-represented pathological or toxicological link-ages, indicated a critical role for KATP channels in coordinating proper adaptive response to hypertensive stress load. The maladaptive response forecast was validated at both functional and structural levels. Specifically, in response to HTN, Kir6.2-KO (n = 8) compared to WT (n = 8) demonstrated poor cardiac muscle performance (Fig. 6B, left), an increase in heart to body weight ratios (Fig. 6B, right), and exaggerated collagen deposition (Fig. 6C), indicating damage, enlargement, and fibrosis, respectively. With HTN, heart contractile function was significantly impaired only in Kir6.2-KO, with a 32% decrease in cardiac output, in contrast to WT where no significant change was observed (Fig. 6B, left). Increased left ventricular mass in the setting of systemic HTN, a major predictor of poor outcome, was significantly greater in the Kir6.2-KO versus WT cohort (Fig. 6B, right). Moreover, Sirius red staining of left ventricular tissue revealed greater prevalence of noncontractile, collagen-rich fibrotic deposition in hypertensive Kir6.2-KO relative to WT in response to HTN (Fig. 6C). Thus, objective forecasting of severe adverse cardiac outcomes, predicted from the resolved Kir6.2-dependent sub-proteome network, establishes a role for KATP channels as molecular contributors to criticality, ensuring adaptability under pathophysiological hypertensive stress load.
Figure 6.
Functional validation of adverse outcomes predicted from the Kir6.2-dependent network. (A) Interrogation of the KATP channel-dependent proteome for existing toxicological pathways in Ingenuity Pathways Knowledge Base revealed three statistically significant adverse effects: “Cardiac Damage”, “Cardiac Enlargement”, and “Cardiac Fibrosis”. Each of these predicted adverse effects was confirmed in vivo by (B) marked reduction of cardiac function as measured by cardiac output and increased cardiac mass measured by normalized heart to body weight (HW:BW) and (C) cardiac fibrosis as illustrated by Sirius red staining of increased collagen deposition (* p<0.05, −HTN vs. +HTN; ** p<0.05, WT +HTN vs. KO +HTN).
4 Discussion
High blood pressure is a common cause of cardiovascular disease, yet there is marked variation in the susceptibility of hypertensive individuals to develop cardiomyopathy [1–9]. Why certain patients progress to overt heart failure while others do not, despite similar risk load, has been attributed to genomically predetermined homeostatic reserve [71]. In this regard, genetic divergence in stress-responsive KATP channels was recently implicated as a risk factor for transition from hypertensive stress to cardiac maladaptation [72], with knockout of the Kir6.2 channel pore precipitating severe heart failure when superimposed upon HTN [28]. However, the global molecular adaptation that occurs in response to myocardial KATP channel deficiency, and that underlies individual susceptibility for developing hypertensive cardiomyopathy, remains unknown.
The present study demonstrates that KATP channel deficit significantly remodels the cardiac proteome in HTN. Specifically, in a chronic hypertensive KATP channel knockout model, we identified a myocardial proteome of 114 altered proteins assembling into an interconnected, metabolism-centric KATP channel-dependent neighborhood. This resolved subproteome exposed a predilection for cardiomyopathy and onset of heart failure through unfavorable reorganization of the elucidated set of altered proteins and their expanded protein web. Dissecting the molecular fingerprint of KATP channel-dependent associations by comprehensive protein identification, ontological stratification, and predictive interrogation thus creates a systems framework tool to forecast outcome.
2-D gel separation coupled with nanoelectrospray linear IT tandem MS provided a high throughput, unbiased method to resolve and characterize the multiplicity of changes arising from the KATP channel-dependent subproteome in the hypertensive state [50–53, 59]. High specificity protein cartography of the hypertensive KATP channel knockout heart displayed pronounced re-arrangement in cellular energetics (73 of 114 proteins), consistent with the proposed role for KATP channels as metabolic checkpoints supporting the adaptive, cardioprotective response to sustained hemodynamic stress [21, 28]. In fact, over two-thirds of detected changes were down-regulated, in line with the notion that intact KATP channels contribute to functional metabolic integration within the heart [17, 49, 73]. Down-regulated proteins included creatine kinase, adenylate kinase, and lactate dehydrogenase isoforms, all previously recognized for their direct KATP channel linkages in support of cardiac homeostasis [35,39, 42, 43, 47, 48]. The remaining 41 altered members of the KATP channel-dependent subproteome encompassed oxido-reductases, stress-related chaperones, proteasome subunits, and proteins involved in signaling regulation, transcription and translation, as well as cellular structure and scaffolding components. Here, unbiased large scale proteomic mining establishes the comprehensive nature of the KATP channel related metabolic infrastructure of the hypertensive heart.
Mapping of protein–protein relationships authenticated the primary impact on metabolic pathways, delineating the KATP channel-dependent subproteome within a functional neighborhood comprised of 242 proteins. The resolved interactome fulfilled criteria of nonstochastic topography based on indices of connectedness, including node degree (k) and degree distribution (P[k]), with the log–log plot of P[k] versus k fitting a power law distribution indicative of scale-free network topography [52]. Most biological systems display a scale-free organization, with disease-perturbed regulatory networks differing from their normal counterparts in functional prioritization [74, 75]. Iterative systems interrogation of the KATP channel-dependent web prioritized only heart-specific adverse effects, i.e., “Cardiac Damage”, “Cardiac Enlargement”, and “Cardiac Fibrosis”, exposing a predisposition for development of cardiomyopathic traits specific to the hypertensive Kir6.2 KO proteome. Validating this maladaptive forecast, functional and structural phenotyping documented aggravated myocardial contractile performance associated with massive interstitial fibrosis and exaggerated left ventricular size, strong prognostic indices of poor outcome. Indeed, in experimental HTN, knockout of the KCNJ11 gene, encoding the Kir6.2 pore-forming subunit of KATP channels, causes defective decoding of HTN-induced metabolic distress signals setting in motion pathological calcium overload and aggravated structural remodeling, ultimately precipitating organ failure and survival disadvantage [28]. In this way, the networked integration of proteins provides a systems understanding of pathophysiological processes associated with development of hypertensive cardiomyopathy in the setting of KATP channel deficiency.
In summary, this study provides the first proteomic profiling of KATP channel-deficient hypertensive hearts, expanding the understanding of molecular alterations caused by lack of a critical metabolic sensor during hemodynamic stress challenge. In contrast to the WT that tolerates hypertensive load, genetically compromised hearts devoid of the KCNJ11-encoded Kir6.2 KATP channel function engender unfavorable proteomic remodeling, revealing the metabolism-centric molecular substrate for pathologic stress-associated cardiovascular disease. In conjunction with the use of bioinformatic interrogation to predict adverse effects [53, 76], the resolved subproteome unmasked risk for maladaptive cardiomyopathic outcome in the context of KATP channelopathy and environmental challenge. In view of KCNJ11 genetic variants conferring cardiac disease risk [72, 77], the present demonstration that KATP channels are critical for the proteomic response to hemodynamic load establishes a foundation to further investigate the role of these cardioprotective channels in predictive medicine.
Acknowledgments
The authors thank Dr. Satsuki Yamada for intra-surgical photography, Jonathan Nesbitt for technical expertise, and the Mayo Proteomics Research Center staff, particularly Benjamin Madden, Kenneth Johnson, Christopher Mason and Roman Zenka, for expert guidance in mass spectrometry and bioinformatic analysis. Support was provided by the National Institutes of Health, Marriott Heart Disease Research Program, Marriott Foundation, Ted Nash Long Life Foundation, and the Japanese Ministry of Education, Science, Sports, Culture and Technology. JZ holds a Mayo Graduate School fellowship. DKA is the recipient of a Marriott Individualized Medicine Career Development Award. AT holds the Marriott Family Professorship in Cardiovascular Research at Mayo Clinic.
Abbreviations
- HTN
hypertension
- KATP
ATP-sensitive K+
- Kir6.2-KO
Kir6.2 knockout
- WT
wild-type
Footnotes
The authors have declared no conflict of interest.
References
- 1.Kostis JB. From hypertension to heart failure: update on the management of systolic and diastolic dysfunction. Am J Hypertens. 2003;16:18S–22S. doi: 10.1016/s0895-7061(03)00966-x. [DOI] [PubMed] [Google Scholar]
- 2.Krum H, Gilbert RE. Demographics and concomitant disorders in heart failure. Lancet. 2003;362:147–158. doi: 10.1016/S0140-6736(03)13869-X. [DOI] [PubMed] [Google Scholar]
- 3.Tocci G, Sciarretta S, Volpe M. Development of heart failure in recent hypertension trials. J Hypertens. 2008;26:1477–1486. doi: 10.1097/HJH.0b013e3282fe1d3d. [DOI] [PubMed] [Google Scholar]
- 4.Rosamond W, Flegal K, Furie K, Go A, et al. Heart disease and stroke statistics -2008 update: a report from the American Heart Association statistics committee and stroke statistics subcommittee. Circulation. 2008;117:e25–e146. doi: 10.1161/CIRCULATIONAHA.107.187998. [DOI] [PubMed] [Google Scholar]
- 5.Towbin JA, Bowles NE. The failing heart. Nature. 2002;415:227–233. doi: 10.1038/415227a. [DOI] [PubMed] [Google Scholar]
- 6.Susic D, Frohlich ED. The aging hypertensive heart: a brief update. Nat Clin Pract Cardiovasc Med. 2008;5:104–110. doi: 10.1038/ncpcardio1091. [DOI] [PubMed] [Google Scholar]
- 7.Bell J. Predicting disease using genomics. Nature. 2004;429:453–456. doi: 10.1038/nature02624. [DOI] [PubMed] [Google Scholar]
- 8.Chien KR. Stress pathways and heart failure. Cell. 1999;98:555–558. doi: 10.1016/s0092-8674(00)80043-4. [DOI] [PubMed] [Google Scholar]
- 9.Lifton RP, Gharavi AG, Geller DS. Molecular mechanisms of human hypertension. Cell. 2001;104:545–556. doi: 10.1016/s0092-8674(01)00241-0. [DOI] [PubMed] [Google Scholar]
- 10.Miki T, Seino S. Roles of KATP channels as metabolic sensors in acute metabolic changes. J Mol Cell Cardiol. 2005;38:917–925. doi: 10.1016/j.yjmcc.2004.11.019. [DOI] [PubMed] [Google Scholar]
- 11.Nichols CG. KATP channels as molecular sensors of cellular metabolism. Nature. 2006;440:470–476. doi: 10.1038/nature04711. [DOI] [PubMed] [Google Scholar]
- 12.Ashcroft FM. ATP-sensitive K+ channels and disease: from molecule to malady. Am J Physiol Endocrinol Metab. 2007;293:E880–E889. doi: 10.1152/ajpendo.00348.2007. [DOI] [PubMed] [Google Scholar]
- 13.Bryan J, Muñoz A, Zhang X, Düfer M, et al. ABCC8 and ABCC9: ABC transporters that regulate K+ channels. Pflugers Arch. 2007;453:703–718. doi: 10.1007/s00424-006-0116-z. [DOI] [PubMed] [Google Scholar]
- 14.Inagaki N, Gonoi T, Clement JP, Namba N, et al. Reconstitution of IKATP: An inward rectifier subunit plus the sulfonylurea receptor. Science. 1995;270:1166–1170. doi: 10.1126/science.270.5239.1166. [DOI] [PubMed] [Google Scholar]
- 15.Inagaki N, Gonoi T, Clement JP, Wang CZ, et al. A family of sulfonylurea receptors determines the pharmacological properties of ATP-sensitive K+ channels. Neuron. 1996;16:1011–1017. doi: 10.1016/s0896-6273(00)80124-5. [DOI] [PubMed] [Google Scholar]
- 16.Lorenz E, Terzic A. Physical association between recombinant cardiac ATP-sensitive K+ channel subunits Kir6.2 and SUR2A. J Mol Cell Cardiol. 1999;31:425–434. doi: 10.1006/jmcc.1998.0876. [DOI] [PubMed] [Google Scholar]
- 17.Alekseev AE, Hodgson DM, Karger AB, Park S, et al. ATP-sensitive K+ channel channel/enzyme multimer: Metabolic gating in the heart. J Mol Cell Cardiol. 2005;38:895–905. doi: 10.1016/j.yjmcc.2005.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zingman LV, Alekseev AE, Hodgson-Zingman DM, Terzic A. ATP-sensitive potassium channels: metabolic sensing and cardioprotection. J Appl Physiol. 2007;103:1888–1893. doi: 10.1152/japplphysiol.00747.2007. [DOI] [PubMed] [Google Scholar]
- 19.Zingman LV, Hodgson DM, Alekseev AE, Terzic A. Stress without distress: homeostatic role for KATP channels. Mol Psychiatry. 2003;8:253–254. doi: 10.1038/sj.mp.4001323. [DOI] [PubMed] [Google Scholar]
- 20.Bienengraeber M, Olson TM, Selivanov VA, Kathmann EC, et al. ABCC9 mutations identified in human dilated cardiomyopathy disrupt catalytic KATP channel gating. Nat Genet. 2004;36:382–387. doi: 10.1038/ng1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kane GC, Liu XK, Yamada S, Olson TM, Terzic A. Cardiac KATP channels in health and disease. J Mol Cell Cardiol. 2005;38:937–943. doi: 10.1016/j.yjmcc.2005.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zingman LV, Hodgson DM, Bast PH, Kane GC, et al. Kir6.2 is required for adaptation to stress. Proc Natl Acad Sci USA. 2002;99:13278–13283. doi: 10.1073/pnas.212315199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu XK, Yamada S, Kane GC, Alekseev AE, et al. Genetic disruption of Kir6.2, the pore-forming subunit of ATP-sensitive K+ channel, predisposes to catecholamine-induced ventricular dysrhythmia. Diabetes. 2004;53:S165–S168. doi: 10.2337/diabetes.53.suppl_3.s165. [DOI] [PubMed] [Google Scholar]
- 24.Tong XY, Porter LM, Liu GX, Dhar-Chowdhury P, et al. Consequences of cardiac myocyte-specific ablation of KATP channels in transgenic mice expressing dominant negative Kir6 subunits. Am J Physiol Heart Circ Physiol. 2006;291:H543–H551. doi: 10.1152/ajpheart.00051.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Olson TM, Alekseev AE, Moreau C, Liu XK, et al. KATP channel mutation confers risk for vein of Marshall adrenergic atrial fibrillation. Nat Clin Pract Cardiovasc Med. 2007;4:110–116. doi: 10.1038/ncpcardio0792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reyes S, Kane GC, Miki T, Seino S, Terzic A. KATP channels confer survival advantage in cocaine overdose. Mol Psychiatry. 2007;12:1060–1061. doi: 10.1038/sj.mp.4002083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kane GC, Behfar A, Yamada S, Perez-Terzic C, et al. ATP-sensitive K+ channel knockout compromises the metabolic benefit of exercise training, resulting in cardiac deficits. Diabetes. 2004;53:S169–S175. doi: 10.2337/diabetes.53.suppl_3.s169. [DOI] [PubMed] [Google Scholar]
- 28.Kane GC, Behfar A, Dyer RB, O’Cochlain DF, et al. KCNJ11 gene knockout of the Kir6.2 KATP channel causes maladaptive remodeling and heart failure in hypertension. Hum Mol Genet. 2006;15:2285–2297. doi: 10.1093/hmg/ddl154. [DOI] [PubMed] [Google Scholar]
- 29.Kane GC, Lam CF, O’Cochlain F, Hodgson DM, et al. Gene knockout of the KCNJ8-encoded Kir6.1 KATP channel imparts fatal susceptibility to endotoxemia. FASEB J. 2006;20:2271–2280. doi: 10.1096/fj.06-6349com. [DOI] [PubMed] [Google Scholar]
- 30.Yamada S, Kane GC, Behfar A, Liu XK, et al. Protection conferred by myocardial ATP-sensitive K+ channels in pressure overload-induced congestive heart failure revealed in KCNJ11 Kir6.2-null mutant. J Physiol. 2006;577:1053–1065. doi: 10.1113/jphysiol.2006.119511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suzuki M, Sasaki N, Miki T, Sakamoto N, et al. Role of sarcolemmal KATP channels in cardioprotection against ischemia/reperfusion injury in mice. J Clin Invest. 2002;109:509–516. doi: 10.1172/JCI14270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gumina RJ, Pucar D, Bast P, Hodgson DM, et al. Knockout of Kir6.2 negates ischemic preconditioning-induced protection of myocardial energetics. Am J Physiol Heart Circ Physiol. 2003;284:H2106–H2113. doi: 10.1152/ajpheart.00057.2003. [DOI] [PubMed] [Google Scholar]
- 33.Gumina RJ, O’Cochlain DF, Kurtz CE, Bast P, et al. KATP channel knockout worsens myocardial calcium stress load in vivo and impairs recovery in stunned heart. Am J Physiol Heart Circ Physiol. 2007;292:H1706–H1713. doi: 10.1152/ajpheart.01305.2006. [DOI] [PubMed] [Google Scholar]
- 34.Du Q, Jovanovic S, Clelland A, Sukhodub A, et al. Overexpression of SUR2A generates a cardiac phenotype resistant to ischemia. FASEB J. 2004;20:1131–1141. doi: 10.1096/fj.05-5483com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Elvir-Mairena JR, Jovanovic A, Gomez LA, Alekseev AE, Terzic A. Reversal of the ATP-liganded state of ATP-sensitive K+ channels by adenylate kinase activity. J Biol Chem. 1996;271:31903–31908. doi: 10.1074/jbc.271.50.31903. [DOI] [PubMed] [Google Scholar]
- 36.Dzeja PP, Terzic A. Phosphotransfer reactions in the regulation of ATP-sensitive K+ channels. FASEB J. 1998;12:523–529. doi: 10.1096/fasebj.12.7.523. [DOI] [PubMed] [Google Scholar]
- 37.Bienengraeber M, Alekseev AE, Abraham MR, Carrasco AJ, et al. ATPase activity of the sulfonylurea receptor: A catalytic function for the KATP channel complex. FASEB J. 2000;14:1943–1952. doi: 10.1096/fj.00-0027com. [DOI] [PubMed] [Google Scholar]
- 38.Zingman LV, Alekseev AE, Bienengraeber M, Hodgson DM, et al. Signaling in channel/enzyme multimers: ATPase transitions in SUR module gate ATP-sensitive K+ conductance. Neuron. 2001;31:233–245. doi: 10.1016/s0896-6273(01)00356-7. [DOI] [PubMed] [Google Scholar]
- 39.Selivanov VA, Alekseev AE, Hodgson DM, Dzeja PP, Terzic A. Nucleotide-gated KATP channels integrated with creatine and adenylate kinases: amplification, tuning and sensing of energetic signals in the compartmentalized cellular environment. Mol Cell Biochem. 2004;256:243–256. doi: 10.1023/b:mcbi.0000009872.35940.7d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park S, Lim BB, Perez-Terzic C, Mer G, Terzic A. Interaction of asymmetric ABCC9-encoded nucleotide binding domains determines KATP channel SUR2A catalytic activity. J Proteome Res. 2008;7:1721–1728. doi: 10.1021/pr7007847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Burke MA, Mutharasan RK, Ardehali H. The sulfonyl-urea receptor, an atypical ATP-binding cassette protein, and its regulation of the KATP channel. Circ Res. 2008;102:164–176. doi: 10.1161/CIRCRESAHA.107.165324. [DOI] [PubMed] [Google Scholar]
- 42.Carrasco AJ, Dzeja PP, Alekseev AE, Pucar D, et al. Adenylate kinase phosphotransfer communicates cellular energetic signals to ATP-sensitive potassium channels. Proc Natl Acad Sci USA. 2001;98:7623–7628. doi: 10.1073/pnas.121038198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Crawford RM, Ranki HJ, Botting CH, Budas GR, Jovanovic A. Creatine kinase is physically associated with the cardiac ATP-sensitive K+ channel in vivo. FASEB J. 2002;16:102–104. doi: 10.1096/fj.01-0466fje. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dhar-Chowdhury P, Harrell MD, Han SY, Jankowska D, et al. The glycolytic enzymes, glyceraldehyde-3-phosphate dehydrogenase, triosephosphate isomerase, and pyruvate kinase are components of the KATP channel macromolecular complex and regulate its function. J Biol Chem. 2005;280:38464–38470. doi: 10.1074/jbc.M508744200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jovanovic S, Du Q, Crawford RM, Budas GR, et al. Glyceraldehyde 3-phosphate dehydrogenase serves as an accessory protein of the cardiac sarcolemmal KATP channel. EMBO Rep. 2005;6:848–852. doi: 10.1038/sj.embor.7400489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu GX, Hanley PJ, Ray J, Daut J. Long-chain acyl-coenzyme A esters and fatty acids directly link metabolism to KATP channels in the heart. Circ Res. 2001;88:918–924. doi: 10.1161/hh0901.089881. [DOI] [PubMed] [Google Scholar]
- 47.Abraham MR, Selivanov VA, Hodgson DM, Pucar D, et al. Coupling of cell energetics with membrane metabolic sensing: Integrative signaling through creatine kinase phosphotransfer disrupted by M-CK gene knock-out. J Biol Chem. 2002;277:24427–24434. doi: 10.1074/jbc.M201777200. [DOI] [PubMed] [Google Scholar]
- 48.Crawford RM, Budas GR, Jovanovic S, Ranki HJ, et al. M-LDH serves as a sarcolemmal KATP channel subunit essential for cell protection against ischemia. EMBO J. 2002;21:3936–3948. doi: 10.1093/emboj/cdf388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hodgson DM, Zingman LV, Kane GC, Perez-Terzic C, et al. Cellular remodeling in heart failure disrupts KATP channel-dependent stress tolerance. EMBO J. 2003;22:1732–1742. doi: 10.1093/emboj/cdg192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hartwell LH. From molecular to modular cell biology. Nature. 1999;402:C47–C52. doi: 10.1038/35011540. [DOI] [PubMed] [Google Scholar]
- 51.Kitano H. Computational systems biology. Nature. 2002;420:206–210. doi: 10.1038/nature01254. [DOI] [PubMed] [Google Scholar]
- 52.Barabási AL, Oltvai ZN. Network biology: understanding the cell’s functional organization. Nat Rev Genet. 2004;5:101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- 53.Weston AD, Hood L. Systems biology, proteomics, and the future of health care: toward predictive, preventative, and personalized medicine. J Proteome Res. 2004;3:179–196. doi: 10.1021/pr0499693. [DOI] [PubMed] [Google Scholar]
- 54.Miki T, Nagashima K, Tashiro F, Kotake K, et al. Defective insulin secretion and enhanced insulin action in KATP channel-deficient mice. Proc Natl Acad Sci USA. 1998;95:10402–10406. doi: 10.1073/pnas.95.18.10402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Suzuki M, Li RA, Miki T, Uemura H, et al. Functional roles of cardiac and vascular ATP-sensitive potassium channels clarified by Kir6.2-knockout mice. Circ Res. 2001;88:570–577. doi: 10.1161/01.res.88.6.570. [DOI] [PubMed] [Google Scholar]
- 56.Arrell DK, Neverova I, Fraser H, Marbán E, Van Eyk JE. Proteomic analysis of pharmacologically preconditioned cardiomyocytes reveals novel phosphorylation of myosin light chain 1. Circ Res. 2001;89:480–487. doi: 10.1161/hh1801.097240. [DOI] [PubMed] [Google Scholar]
- 57.Arrell DK, Elliott ST, Kane LA, Guo Y, et al. Proteomic analysis of pharmacological preconditioning: Novel protein targets converge to mitochondrial metabolism pathways. Circ Res. 2006;99:706–714. doi: 10.1161/01.RES.0000243995.74395.f8. [DOI] [PubMed] [Google Scholar]
- 58.Arrell DK, Niederländer NJ, Perez-Terzic C, Behfar A, Terzic A. Embryonic stem cell cardiac differentiation: A proteomic perspective. Adv Mol Med. 2006;2:149–156. [Google Scholar]
- 59.Arrell DK, Niederländer NJ, Faustino RS, Behfar A, Terzic A. Cardioinductive network guiding stem cell differentiation revealed by proteomic cartography of tumor necrosis factor alpha-primed endodermal secretome. Stem Cells. 2008;26:387–400. doi: 10.1634/stemcells.2007-0599. [DOI] [PubMed] [Google Scholar]
- 60.Behfar A, Perez-Terzic C, Faustino RS, Arrell DK, et al. Cardiopoietic programming of embryonic stem cells for tumor-free heart repair. J Exp Med. 2007;204:405–420. doi: 10.1084/jem.20061916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Perkins DN, Pappin DJC, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 62.Link AJ, Eng J, Schieltz DM, Carmack E, et al. Direct analysis of protein complexes using mass spectrometry. Nat Biotechnol. 1999;17:676–682. doi: 10.1038/10890. [DOI] [PubMed] [Google Scholar]
- 63.Assenov Y, Ramírez F, Schelhorn SE, Lengauer T, Albrecht M. Computing topological parameters of biological networks. Bioinformatics. 2008;24:282–284. doi: 10.1093/bioinformatics/btm554. [DOI] [PubMed] [Google Scholar]
- 64.Barabásí AL, Albert R. Emergence of scaling in random networks. Science. 1999;286:509–512. doi: 10.1126/science.286.5439.509. [DOI] [PubMed] [Google Scholar]
- 65.Jeong H, Tombor B, Albert R, Oltavi ZN, Barabásí AL. The large-scale organization of metabolic networks. Nature. 2000;407:651–654. doi: 10.1038/35036627. [DOI] [PubMed] [Google Scholar]
- 66.Anderson TW, Darling DA. Asymptotic theory of certain “goodness-of-fit” criteria based on stochastic processes. Ann Math Stat. 1952;23:193–212. [Google Scholar]
- 67.Newman MEJ. The structure and function of complex networks. SIAM Rev. 2003;45:167–256. [Google Scholar]
- 68.Weber KT. Aldosterone in congestive heart failure. N Engl J Med. 2001;345:1689–1697. doi: 10.1056/NEJMra000050. [DOI] [PubMed] [Google Scholar]
- 69.Weiss JN, Yang L, Qu Z. Network perspectives of cardiovascular metabolism. J Lipid Res. 2006;47:2355–2366. doi: 10.1194/jlr.R600023-JLR200. [DOI] [PubMed] [Google Scholar]
- 70.Nykter M, Price ND, Aldana M, Ramsey SA, et al. Gene expression dynamics in the macrophage exhibit criticality. Proc Natl Acad Sci USA. 2008;105:1897–1900. doi: 10.1073/pnas.0711525105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bleumink GS, Schut AF, Sturkenboom MC, Deckers JW, et al. Genetic polymorphisms and heart failure. Genet Med. 2004;6:465–474. doi: 10.1097/01.gim.0000144061.70494.95. [DOI] [PubMed] [Google Scholar]
- 72.Reyes S, Terzic A, Mahoney DW, Redfield MM, et al. KATP channel polymorphism is associated with left ventricular size in hypertensive individuals: a large-scale community-based study. Hum Genet. 2008;123:665–667. doi: 10.1007/s00439-008-0519-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dzeja PP, Terzic A. Phosphotransfer networks and cellular energetics. J Exp Biol. 2003;206:2039–2047. doi: 10.1242/jeb.00426. [DOI] [PubMed] [Google Scholar]
- 74.Hood L, Heath JR, Phelps ME, Lin B. Systems biology and new technologies enable predictive and preventative medicine. Science. 2004;306:640–643. doi: 10.1126/science.1104635. [DOI] [PubMed] [Google Scholar]
- 75.Loscalzo J, Kohane I, Barabási AL. Human disease classification in the postgenomic era: a complex systems approach to human pathobiology. Mol Syst Biol. 2007;3:124. doi: 10.1038/msb4100163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Arrell DK, Niederländer NJ, Perez-Terzic C, Chung S, et al. Pharmacoproteomics: advancing the efficacy and safety of regenerative therapeutics. Clin Pharmacol Ther. 2007;82:316–319. doi: 10.1038/sj.clpt.6100310. [DOI] [PubMed] [Google Scholar]
- 77.Sattiraju S, Reyes S, Kane GC, Terzic A. KATP channel pharmacogenomics: from bench to bedside. Clin Pharmacol Ther. 2008;83:354–357. doi: 10.1038/sj.clpt.6100378. [DOI] [PMC free article] [PubMed] [Google Scholar]