Abstract
We have developed a version of our surface generalized Born (SGB) model that employs a Gaussian surface, as opposed to the van der Waals surface used previously. The Gaussian surface is smooth and its properties are analytically differentiable with respect to the positions of atoms. A significant advantage of a solvent model based on this analytically differentiable surface is the availability of analytical gradients of the surface and solvation forces. An efficient and robust algorithm is designed to construct and triangulate the Gaussian surface for large biomolecules with arbitrary shapes, and to compute the various terms required for energy gradients. The Gaussian surface is shown to better mimic the boundary between the solute and solvent by properly addressing solvent accessibility, as is demonstrated by comparisons with standard Poisson–Boltzmann calculations for proteins of different sizes. These results also demonstrate that surface definition is a dominant contribution to differences between GB and PB calculations, especially if the system is large. Application of the new surface to prediction of long loop regions is presented, and significant improvement in the energetics is seen compared with results obtained using the van der Waals surface, even in the absence of optimized empirical correction terms that were used in the latter calculations.
Keywords: Born model, novel Gaussian surface, biomolecules, implicit solvent model, solvation energy, protein structure prediction, loop structure prediction, energy minimization, molecular surface
Introduction
Solvation is a fundamental phenomenon in biomolecular systems. It plays a significant role in energetics, dynamics, and structures of biomolecules, such as proteins, in solution. The development of accurate and efficient solvation models is thus crucial to understanding the chemical and physical properties that underlie many biomolecular processes. Explicit solvent models treat solvent molecules at an atomic level of detail. The high computational cost associated with explicit solvent simulations has been a bottleneck in their application to complex computational modeling tasks such as protein–ligand docking and protein folding. The inclusion of a large number of explicit solvent molecules significantly increases the computational expense, especially due to the extensive equilibration and averaging required to converge the solvation free energy.
Driven by these computational exigencies, a great deal of effort has been put into the development of implicit solvation models over the past 2 decades. Poisson–Boltzmann (PB) methods, which solve the PB equation to calculate solvation free energies, have been established as a standard approach. The PB equation can be derived using well-defined approximations, is tractable for large systems, and is amenable to parameterization against experimental data. However, the computational expense involved in solving the PB equation, whether by means of finite-difference,1–4 finite-element,5,6 or boundary element methods,7–10 is significantly larger than that of calculating the energy of the solute with a molecular mechanics force field, and it is also nontrivial to compute gradients of the solvation free energy for large systems.
Simpler implicit solvation models such as effective-dielectric models or solvent-exposed surface area models do not provide a robust description of electrostatics, and hence, have great difficulty in achieving the same accuracy as the PB methods.11 Among a variety of semianalytical approximations to the PB methods, the generalized Born (GB) model proposed by Still and coworkers12 appears quite promising because of its simplicity and ability to at least semiquantitatively reproduce PB results. In the GB model, the pair-interaction energy is described by an analytical function of atomic positions, which makes it easier to compute atom-based effective solvation forces. The major computational expense arises from calculating the self-interaction energy, or the effective Born radius, for each atom. A recent study shows that with “perfect” effective Born radii, defined as those obtained by solving the PB equation with only one atom charged at a time, the GB model gives a very good approximation to the underlying Poisson Equation (PE) theory for a variety of biomacromolecular structures and conformations.13 A variety of approaches to calculate the self-interaction energies have been developed by various laboratories such as the overlapping spheres approach by the groups of Cramer, Truhlar, Case, and Brooks,14–19 the asymptotic approach by the Still group,20 the analytical continuum electrostatics (ACE) model of the Karplus group,21 and the surface GB (SGB) model developed in the Friesner group.22
There are two main approximations associated with the calculation of the effective Born radii. The first approximation is the Coulomb field approximation (CFA). In the CFA, the contribution to the electric field from the induced polarization charge distribution on the dielectric boundary is neglected and the electric field is approximated as only the Coulombic field generated by internal atomic charges. The resultant electrostatic solvation energy has an inverse fourth-power distance dependence coming from the classical monopole-induced dipole interactions. This treatment is rigorously correct for a source charge lying at the center of a sphere (i.e., a single atom). However, the deviation can be very large when the shape of the macromolecule is far from spherical and the source charge approaches the surface. Parameter optimization can reduce the errors in the CFA, depending on the size of the system. Our group has introduced several empirical corrections terms to this, which have considerably improved the agreement between the GB model and the PB model.22 Recently, the Brooks group has developed an empirical correction term that involves the square root of a fifth-order distance dependence.18
The other approximation lies in the description of the boundary that delimits solute and solvent. One of the simplest approaches, which has dominated in work on GB models, is to represent each atom by a hard sphere with chosen radius. The whole biomolecule is hence a superposition of all atomic spheres. This surface is termed the van der Waals (VDW) surface. This simple surface is easy to implement and display graphically, but it overestimates the volume of space near molecules that water can access. As the size of the solute increases, many narrow invaginations or even cavities form, which are treated as having a high dielectric constant. However, in many cases such regions are physically inaccessible to solvent molecules because they are too narrow or enclosed.
To address this problem, the solvent-accessible surface was introduced by Lee and Richards.23 It is defined as the surface traced by the locus of a probe sphere rolling over the VDW surface. The radius of the probe sphere is chosen to approximate that of a water molecule. The obtained surface is displaced outward from the VDW surface by a distance equal to the radius of the probe sphere. Later, the definition of the molecular surface was presented by Richards.24 This surface is defined to be the smooth surface traced by the inward facing part of the probe sphere rolling over the VDW surface, and consists of two parts: the contact surface, the part of the VDW surface that is accessible to the probe sphere, and the reentrant surface, the part of the inward-facing surface of the probe sphere when it is simultaneously in contact with more than one atom. A variety of implementations of the molecular surface have been reported.25–28 Probably the most important of these algorithms is the efficient analytical implementation given by Connolly.28–30
Each of these definitions represents component atoms as hard spheres, which leads to either the unavailability of analytical electrostatic forces or discontinuities of analytical electrostatic solvation forces at the intersections of the atomic spheres.
An alternative description of the shape of molecules is the isocontour of electron density of molecules, which can be approximated using Gaussian functions. The Gaussian technology has been adopted as a simple solution to calculate the electron density in quantum chemistry.31 In this technology, the total electron density at a point in space is calculated as the sum of each atomic contribution, which is a three-dimensional Gaussian function.
Both the Connolly surface and a Gaussian surface of the type discussed above can be designed to exclude water molecules from regions where they obviously are unable to penetrate. The two surfaces will differ in their details, particularly in complex regions involving multiple points of contact for a probe water molecule. In such regions, it is far from clear which surface provides a more accurate description of the physical system, and in any case, both methods require extensive parameterization to properly describe first shell interactions of water molecules with polar or charged groups. However, the Gaussian surface does possess a significant practical advantage; it is smooth, that is, continuous and analytically differentiable. This property enables us to calculate the electrostatic solvation force analytically. Furthermore, a Gaussian function’s derivative is itself a Gaussian, which facilitates efficient computation of the electrostatic solvation force.
There have been some attempts to adopt the idea of the Gaussian surface, that is, isocontour of overlapping Gaussian functions, in molecular modeling applications. Walker et al.32 employed Gaussian-type functions to calculate electron density and describe molecular shape. Gabdoulline and Wade33 used Gaussian functions to calculate isocontour points to analyze molecular interaction properties by visually inspecting shape complementarities of the isocontours. Both of these studies generated only a discretized set of surface points, which is insufficient for carrying out volume or surface integration for calculating GB self-interaction energies. Friedrichs et al.34 constructed a molecular surface, but then used a Gaussian function to assign dielectric constants to grid points in their PB method. Duncan and Olson35 used Connolly’s triangulated molecular surface as a starting point and gradually moved these surface points towards the contour value along the direction of the gradient of the density until all surface points were within a predefined error tolerance of the chosen contour value. In principle, it would be possible to apply their approach to our goal of calculating GB self-energies, but we describe here a more efficient and robust surface generation method.
Another gaussian non-isocontour description of molecular shape has been used to calculate efficiently the volume or surface area of molecules, using analytical expressions.36 Recently, this Gaussian volume exclusion approach has been applied to generating a smooth dielectric function for PB methods37 and the development of a shape-based docking scoring function.38 It has also been used by Schaefer and Karplus21 to carry out the volume integral in the calculation of self-interaction energies in their GB method. However, these approaches cannot be used to carry out surface integrals because of the lack of local surface properties. Furthermore, the results were compared with the results from the VDW surface and excellent agreements were achieved. However, the VDW surface is not a good benchmark to choose because it is well known for its weakness in addressing solvent-accessibility, as was discussed above.
In this article, we explore how the surface definition impacts the calculated electrostatic solvation energies. In particular, we present an efficient and robust method to directly construct the Gaussian surface and develop a new-generation SGB model based on this surface. The electrostatic solvation force is subsequently derived and successfully applied to protein structure minimization. We choose the SGB approach to calculate the self-interaction energy of each atom in the GB model. A noteworthy point is that we do not include the correction terms from our previous SGB model22 in the new SGB model. The correction terms are specific to the underlying model, and will need to be reparameterized; we will pursue this direction in future publications. However, in the absence of any empirical terms of this type, the Gaussian SGB model appears to perform well in the initial tests we report here.
The article is organized as follows. In the next section we summarize the theory involved in the PB method and the SGB model. Then we describe the algorithm to construct the Gaussian surface and deduce the formulae to calculate the gradient of electrostatic solvation free energy with respect to atomic coordinates, that is, the electrostatic solvation force (details are presented in the Appendix). Then we present the new-generation Gaussian surface-based SGB model, comparing with the PB and prior SGB model results, for a number of proteins. After validating the new method by applying it to loop prediction cases where the prior SGB model has been successful, we then apply it to a number of long loop prediction problems for which the prior SGB model exhibited clear energy errors, using the gradient optimization capabilities to generate optimized structures. We then discuss the implications of the results, and conclude by summarizing our findings and presenting future directions.
Theory
Finite-Difference PB Method
The PB equation treats the aqueous solvent as a dielectric continuum. One solves eq. (1) below to find the resulting electrostatic potential, φ(r⃑), at point r⃑ from the charge density, ρ(r⃑), and the dielectric constant, ε(r⃑), of the system:
| (1) |
A PB model is specified by the charge distribution (arising from the molecular mechanics potential function of the solute) and the functional form of the dielectric of the system. Typically, this is defined as a dielectric surface; all points in the system falling inside the surface (i.e., in the interior of the protein) are defined to have dielectric of unity (or whatever the internal dielectric is specified as—this itself is a subject of considerable controversy), whereas points falling outside of the surface are defined to have dielectric 80 (given that we are modeling aqueous solution).
To solve the PB equation, we use the DelPhi program of Honig and coworkers.2,39 – 41 DelPhi uses a finite difference approach to solving the PB equation, with a number of specialized numerical techniques that ensure both accuracy and high efficiency. We have tested the grid resolution in Delphi for the present problem and obtained converged results using 0.5 Å/grid (that is, finer grids minimally change the results presented below). For maximum consistency, we have input the OPLS-AA charges and van der Waals radii into DelPhi.
Surface-Generalized Born Model
The SGB model is an approximation to the boundary element formulation of the PB equation, in which the polarization effects throughout the entire volume of the system can be exactly reproduced by an appropriate distribution of induced polarization charge at the surface of solute molecules. A derivation of this approximation has been presented previously,22 and was shown to be equivalent to the volume formulation of the GB method by Still and coworkers,12 assuming equivalent dielectric surfaces.
The total electrostatic solvation free energy can be divided into two terms—a self-term and a pair term:
| (2) |
The formulas to calculate the self-term and the pair term are given as
| (3) |
| (4) |
where qk is the charge on atom k, r⃑k is its coordinate, ε is the dielectric constant, R⃑ is the coordinate of a point on the dielectric boundary, n⃑(R⃑) is the normal at the point, αk is the Born alpha radius for atom k, rkl is the distance between a pair of atom k and l,
Computational Algorithm and Methods
Construction and Triangulation of the Gaussian Surface
Calculation of the Gaussian Density Function for the System on a Discrete Grid
We define the dielectric boundary of the molecule using a Gaussian density function, constructed as a superposition of atomic functions. First, the biomolecular system is mapped onto a cubic grid. Although the grid resolution is specified as a parameter in the new SGB model, we have chosen for the calculations presented in this article the resolution of 0.5 Å/grid, which is more than enough to obtain converged results (that is, finer grids minimally change the results presented below). The Gaussian density D at each grid point is defined by summing the atomic Gaussian contributions:
| (5) |
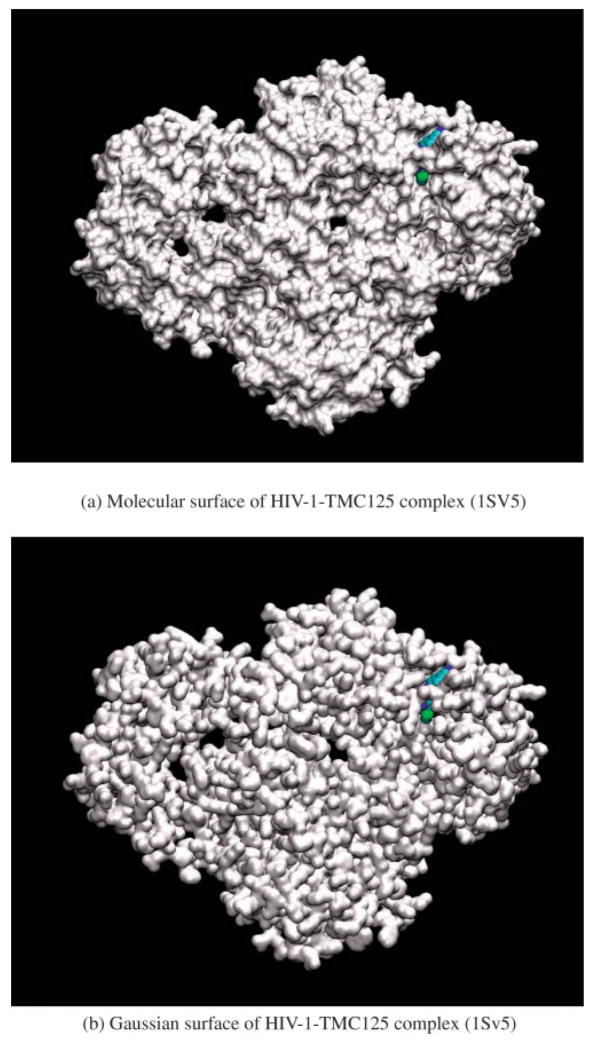
where the sum is over all atoms i of the solute, r⃑i is the coordinate of atom i, r⃑ is the coordinate of the grid point, Ri is the radius of atom i, and a is a parameter defining the smoothness of the surface. The radii we use are taken from the Lennard–Jones parameters of the OPLS-AA force field.42 We have experimented with the optimal value of a, which controls the “smoothness” of the surface, by comparing the shape and curvature of the Gaussian surface of a three-atom system (Fig. 3) and another 56-atom system (results not shown) with the molecular surface and chosen the value of 2.5. This represents a compromise between a surface that is too smooth to accurately define the shape (value of a too small), and a surface that is too close to a VDW surface, with the associated problem of inappropriately high solvent accessibility, for example, in the macromolecular interior (value of a too large). The choice of the water probe radius used for the molecular surface will affect our choice of a as well. The larger the water probe radius, the smoother the molecular surface, an effect that can be mimicked by decreasing the value of a. Here we have used the standard water probe radius of 1.4 Å. More quantitative tests can be imagined to determine the value of a, such as comparing the surface areas of the Gaussian surface and molecular surface on a representative set of structures. However, it is clear in our much simpler tests that the value of 2.5 for a is nearly optimal, and further refinement, if any is required, will take place in the context of parameter optimization with respect to experimental solvation free energies. As shown in Figure 4, the Gaussian surface of the large HIV-1-TMC125 complex (two chains, 979 residues in total) created using a = 2.5 mimics the molecular surface. In addition, our application of the new solvation model based on this Gaussian surface to protein loop prediction further validates that this choice of a is a reasonable one.
Figure 3.
A comparison of the VDW surface (a), the molecular surface (b), and the Gaussian surface (c) and (d) for a simple system of three atoms, each pair of which are tangent to each other; (c) represents the Gaussian surface in a triangulated surface mesh representation and (d) is solidified Gaussian surface rendered by VMD.54,55
Figure 4.
The Gaussian surface and molecular surface for 1Sv5, which has 979 residues. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com]
With a = 2.5, the contribution to D from atoms that are located more than twice their radius from the grid point in question is negligible, and neglected in our algorithm. The gradients of D with respect to the x component xi, y component yi, and z component zi of the r⃑i are calculated for the later computation of the effective electrostatic solvating force:
| (6) |
where Di, the electron density contribution from atom i, can be obtained from eq. (5) and need not to be computed repeatedly, which makes the computation of eq. (6) efficient.
Interpolation of Gaussian Surface Points
Having calculated the density D at the grid points, we can judge whether a grid point is located inside or outside of the surface by comparing its electron density with the chosen contour value for the Gaussian surface. If it is larger than (or equal to) the preset value, the grid point is inside (or on) the surface; if it is smaller than the preset value, the grid point is outside the surface. When one of two adjacent grid points has density larger than the preset value and the other has density smaller than the preset value, the surface must intersect the cube edge connecting these two points since the density is continuous.
The contour cutoff value Dcutoff for the Gaussian surface cannot be chosen arbitrarily once the value of a is chosen. Their relationship can be deduced from the case of a single atom, for which the Gaussian surface should be identical to the hard-sphere surface. Applying eq. (5) to the surface of this single atom, we have
| (7) |
Because a = 2.5, the value of Dcutoff is exp(−2.5), approximately 0.082085.
The position of the intersecting surface point can be approximated by linear interpolation:
| (8) |
where r⃑in and Din are the coordinate and the density of the grid point inside the surface, respectively, and r⃑out and Dout are the coordinate and the density of the grid point outside the surface. Actually, two coordinate components of r⃑in − r⃑out are 0 and the third component is l or −l, where l is the size of the grid, which reduces the computational cost by two-thirds. The partial derivative of r⃑ with respect to xi is
| (9) |
Similarly, we have
| (10) |
| (11) |
Triangulation of the Gaussian Surface
There are eight grid points in each cube, and there are two states for each grid point, either outside or inside the surface. Thus, there are in all 28 = 256 ways in which a surface can intersect the cube. It is possible to triangulate the entire Gaussian surface by independently triangulating the intersecting surface component in each cube. However, enumerating a triangulation pattern for each of the 256 cases is tedious. Two different symmetries of the cube reduce the problem from 256 cases to 15 cases, as proposed in the “marching cube” algorithm by Lorensen et al.43 First, the topology of the triangulated surface is unchanged if the states (being inside or outside the Gaussian surface as mentioned above) of all eight vertices in a cube are inverted together. The only thing that changes is the surface normal. This inversion symmetry reduces the number of the cases from 256 to 128. The second symmetry, rotational symmetry, further reduces the problem to 15 basic cases, as shown in Figure 1a. However, it is found by Dürst that these 15 triangulation patterns are not self-consistent, that is, the obtained surface is not always enclosed.44 Figure 2 illustrates an example where an artificial gap can be observed. Heiden et al.45 proposed an additional triangulation pattern for the inverted case of basic case 3, 6, 7, and 4, respectively, to solve this problem. Actually, the additional triangulation pattern for the inverted case of basic case 4 is unnecessary from the point of view of self-consistency. With a slight modification of Heiden and coworkers’ patterns by eliminating the unnecessary inverted case of basic case 4, we have our improved marching cube triangulation patterns, 18 in all, as presented in Figure 1a and b. This algorithm always converges and is very accurate and efficient.
Figure 1.
Patterns for three-dimensional grid triangulation: 15 basic patterns from the original marching cube algorithm in (a), and three additional patterns in (b) to achieve the closing of the surface. The prime to the right of the pattern numbers in (b) denotes inversion of the corresponding basic pattern in (a).
Figure 2.

Examples of producing artificial gaps. With the original marching cube algorithm, the cube in (b), adjacent to the cube in (a), is triangulated in such a way that the surface points on the common face of the cube in (a) and (b) are not connected by the same two edges. The surface is thus not enclosed, and an artificial gap is produced. With the improved marching cube algorithm, this problem is solved. The surface points are connected by the same two edges.
Characterization of Gaussian Surface Triangles
The center R⃑j,, area Aj, and unit normal n⃑j (pointing outward) of each surface triangle j, which are needed in the later computation of the effective Born radius in the SGB model, are subsequently calculated by
| (12) |
| (13) |
and
| (14) |
where N⃑j is the surface normal before normalization, r⃑I, r⃑II, and r⃑III are the positions of the three vertices of the surface triangle, numbered so that the direction of the unit normal n⃑j complies with the right-hand rule. With the knowledge of vector differentiation ∇r = r⃑/r,, where r⃑ = i⃑ x + j⃑ y + k⃑z and r = |r⃑|, the gradients for the center, normal, and surface area of a surface triangle can be easily derived:
| (15) |
| (16) |
| (17) |
| (18) |
| (19) |
where
| (20) |
| (21) |
| (22) |
Derivation of the Effective Electrostatic Solvation Force
Because of its length, this derivation has been placed in an Appendix.
Results
We first investigate the question of whether the Gaussian surface as defined above properly addresses issues of solvent accessibility, from a qualitative point of view. To answer this question, first we study a simple system of three identical atoms, where each pair of atomic spheres is tangent to each other. The atomic radius is set to 2.0 Å, close to that of a heavy atom (C, N, O) in a biomolecular system. The VDW and Gaussian surfaces for this system are presented in Figure 3. In the VDW surface, as shown in Figure 3a, an enclosing cavity forms in the middle of the three atomic spheres. This cavity is assigned the high dielectric constant of the solvent in implicit solvent models. However, it is in practice solvent inaccessible and should be assigned a low dielectric constant. In the molecular surface, as shown in Figure 3b, this cavity no longer exists. The region occupied by the cavity is now treated as part of the solute. Furthermore, the solvent-inaccessible invagination area between each pair of atoms is corrected in the molecular surface by a reentrant surface. Similarly, in the Gaussian surface, the cavity in the middle of the system is eliminated, as shown in Figures 3c and 3d, and the narrow invagination areas between each pair of atoms are also corrected. Actually Figures 3b and 3d show that the Gaussian surface behaves very similarly to the molecular surface. The physical fact that the density at a point is a sum of the contributions from adjacent atoms ensures the correct treatment of solvent accessibility in the Gaussian surface, leading to the formation of this kind of reentrant surface. On the other hand, the Gaussian surface is smooth, as expected, which is vital for its application to the development of implicit solvent models with analytical electrostatic solvation forces.
To examine the same question for a large system, in Figure 4 we compare the molecular surface and Gaussian surface for the complex of HIV-1 and nonnucleoside reverse transcriptase inhibitor (NNRTI) TMC125-R165335 (etravirine), 1SV5 in the protein data bank (PDB),46 which has 979 residues. Clearly, both surfaces show similar encapsulation of the ligand and reveal three key cavities in HIV-1. In addition, while the shape and curvature of the Gaussian surface are similar to the molecular surface, the Gaussian surface appears to be smoother, and tends to remove the artificial sharp boundaries that occur at the intersections of atomic spheres in the molecular surface.
Quantitative comparisons are more difficult to evaluate in a rigorous fashion. Many previous workers have set as an objective for approximate continuum methods such as generalized Born approaches the reproduction of the solvation free energy obtained from accurate numerical solution of the PB equation. However, agreement with PB results is not by any means equivalent to agreement with experimental data. The PB results themselves are strongly dependent upon the definition of the dielectric surface (with regard to both functional form and the specific atomic parameters used to build the surface). It is far from clear, for example, whether the Connolly surface is a better or worse representation of physical reality than the Gaussian surface defined above, yet the numerical values of solvation free energy obtained from each can be quite different. Furthermore, we have recently shown that both PB and GB models deviate substantially from explicit solvent potentials of mean force when a single bridging water separates two charged groups.47 Hence, one cannot simply use detailed comparisons of a GB model with any particular PB calculation as a measure of quantitative accuracy of the predicted solvation free energy of the GB model in question. Direct comparison with experimental data (e.g., predicting the conformations of loops and side chains in protein structures) is ultimately the only important measure of the accuracy and relevance of the model.
However, we can extend the analysis initiated above with regard to eliminating qualitatively incorrect features of the dielectric surface (which are clearly wrong based on straightforward physical arguments), and correlate elimination of these features with closer agreement of PB and GB results. That is, we expect that when we use the Gaussian surface rather than the VDW surface in an SGB calculation, solvation free energy differences between different conformations become closer on average to the PB values as a result.
We denote our new-generation of SGB model based on the Gaussian surface the Gaussian surface-based SGB model (GSGB). Because the error in solvent-accessibility in the VDW surface becomes more serious as the size of the biomolecular system becomes large, we apply the new GSGB model to a series of conformations, generated by a minimization on a native structure, of three systems ranging from a small peptide to a large protein, and compare the GSGB results obtained with the results from molecular surface-based DelPhi and the original VDW surface-based SGB (VSGB) model. The absolute value of the electrostatic solvation energies for the same conformation differs across these three methods, and we do not expect to see the absolute results of VSGB and GSGB match well with DelPhi because we do not incorporate any correction terms to improve the errors introduced by the Coulomb field approximation used to calculate the effective Born radii. The differences are highly dependent upon parameterization, and, as argued above, it is not necessarily clear that the DelPhi values are correct compared with experimental results. Moreover, in most applications, only the relative energies are important. Therefore, we also set to zero in all three approaches the electrostatic solvation energy for the conformation with the lowest solvation energy in DelPhi and present the relative energies.
The absolute and relative results for the small peptide, PDB code 1ct6, which has 124 atoms, or nine residues, are shown in Figures 5 and 6. Both VSGB and GSGB agree reasonably well with DelPhi (noting again that below some threshold, quantitative differences are difficult to interpret). This system is so small that most of its atoms are substantially exposed to water. Few cavities form, and few atoms are buried in the interior of the system. In this case, the VDW surface does not overestimate the solvent-accessible surface, and thus the VSGB solvent model does rather well. Apparently the small deviation of VSGB and GSGB from DelPhi comes much more from the Coulomb field approximation in the GB formulation to PB rather than from the surface definition. As a matter of fact, VSGB is somewhat closer to the PB results than GSGB, which is quantified by the smaller RMSD of the VSGB points from DelPhi. However, the difference between the two is quite small.
Figure 5.

The absolute VSGB and GSGB solvation free energies for a series of conformations, generated by a minimization of the peptide 1ct6, as compared with DelPhi results. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com]
Figure 6.

The VSGB and GSGB results for a series of conformations, generated by a minimization on the peptide 1ct6, compared with DelPhi results. For comparison, the electrostatic solvation free energy of the conformation with the lowest solvation energy in DelPhi is set to zero in all three models, as is the same in Figure 8 and Figure 10. The DelPhi trend line is y = x, for comparison. The VSGB trend line is y = 0.81x + 0.76 with the correlation coefficient R = 0.993 and the GSGB trend line is y = 0.75x − 1.23 with R = 0.994. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com]
As we increase the size of the system to that of 1cbh, 499 atoms or 36 residues, the VSGB results begin to deviate erratically from the DelPhi results, while the GSGB results stay reasonably close to the DelPhi results, as shown by the VSGB’s poorer correlation with DelPhi in Figure 7 and its larger RMSD in Figure 8. In this case, there are some cavities formed inside the protein and some atoms are buried in the interior, and the error in the definition of surface begins to surpass the error in the Coulomb field approximation in the GB formulation.
Figure 7.

The absolute VSGB and GSGB results for a series of conformations, generated by a minimization on the PDB structure 1cbh, compared with DelPhi results. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com]
Figure 8.

The VSGB and GSGB results for a series of conformations of a small protein 1cbh, compared with DelPhi results. The VSGB trend line is y = 0.81x − 14.23 with R = 0.971 and the GSGB trend line is y = 0.70x − 1.14 with R = 0.995. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com]
Last, we study the protein 1mo9. This is a large system, which consists of two protein chains with a total of 16,012 atoms or 1046 residues. The results are shown in Figures 9 and 10. It is clear that the VSGB results now deviate erratically from the DelPhi results while the GSGB results still agree well. When the system gets this large, the error in the definition of surface becomes dominant while that in the Coulomb field approximation becomes less important. In such a large system, many cavities exist inside the protein. Furthermore, many atoms, including charged atoms, are entirely buried in the interior of the protein. The self-interaction energies for these buried atoms are small, and their effective Born radius can be as large as 10 Å or more. However, in the VDW surface, all these cavities are treated as high dielectric, and many of the buried atoms are still treated as exposed to water to different extents. Hence, in the VSGB model the calculated self-interaction energies of buried atoms tend to be larger than their corresponding PB values and their effective Born radii tend to be systematically smaller than the effective PB values. In fact, the effective Born radii rarely exceed 10 Å in the VSGB model, as shown in Figure 11. On the other hand, the cavities and buried atoms are realistically treated in the Gaussian surface, which leads to good qualitative agreement of the relative energies in the GSGB model with DelPhi. In addition, the plot in Figure 11 indicates that the relationship between the effective Born radii of GSGB and DelPhi is systematically linear (on average; the deviation of individual atoms’ effective Born radii mostly comes from the Coulomb field approximation, which will cancel to a great extent in the calculation of relative energies, especially for large systems) while the relationship between VSGB and DelPhi is roughly linear for small Born radii but then deviates sharply, with a bias towards smaller radii for atoms in the interior of the system (Born radii >7 Å), as we expected. The success of the GSGB model on such a large system also highlights the robustness of our Gaussian surface construction algorithm.
Figure 9.

The absolute VSGB and GSGB results for a series of conformations, generated by a minimization on the PDB structure 1mo9, compared with DelPhi results. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com]
Figure 10.

The VSGB and GSGB results for a series of conformations of a large protein 1mo9, compared with DelPhi results. The VSGB trend line is y = 0.69x − 34.43 with R = 0.717 and the GSGB trend line is y = 0.85x − 6.69 with R = 0.971. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com]
Figure 11.

Comparison of effective Born radii of the VSGB and GSGB models with DelPhi. The VSGB trend line is y = 0.45x + 2.53 with R = 0.776 and the GSGB trend line is y = 0.78x + 2.45 with R = 0.811. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com]
To test the analytical electrostatic solvation forces, and obtain a preliminary assessment of the energetics in the GSGB model, we have integrated the model with the protein local optimization program (PLOP),48–53 a protein structure prediction package developed by the Jacobson and Friesner groups. The sampling and minimization algorithms in PLOP have been described previously, and this description will not be repeated here.50–52 Replacement of forces and energies from the previously used VSGB model with their GSGB equivalents was straightforward, as no changes were made in the underlying algorithms.
We have previously reported the use of PLOP for predicting the conformations of loops with lengths ranging from 4 residues to 12 residues, starting from the native protein structure.50 In general, the performance of PLOP and the VSGB model in these tests has been excellent, representing a clear advance in the state of the art compared to alternative results in the published literature. However, there are on occasion loops that exhibit substantial deviations from the native structure, and some of these outliers can be identified as failures of the energy model (as opposed to errors caused by incomplete sampling, which represent an entirely different class of problems). Such failures become more frequent as the length of the loop increases; if long loop prediction is to be made robust, the sources of these errors must be identified and eliminated.
We have hypothesized that problems associated with the VSGB dielectric surface are responsible in part for the long loop energy errors that we have observed. One can imagine that in long loop prediction, incorrect structures with small pockets or invaginations are created, and that these are incorrectly assigned more solvation free energy than they physically deserve by the VSGB surface. If this is indeed the case, we would expect that when such false positives are presented to the GSGB surface, their energy relative to the native structure should become less favorable. Note that we do not necessarily expect that the current version of the GSGB model will unerringly select the native conformation as best; previous experience indicates that some degree of empirical parameterization to experimental data is needed to obtain highly accurate and robust results, and we have not yet carried out such parameterization for the GSGB model (which has to be started from scratch). However, reduction of the large energy gaps seen in the VSGB false positives is a reasonable expectation.
As a first test suite, we selected two set of “loop decoys”: series of conformations for protein loops of various lengths ranging from native-like to highly nonnative structures. These decoy sets consist of all loop conformations generated during the testing of a previously published loop prediction algorithm that utilized the VSGB solvent model.50 The first decoy set contains 25 four-residue loops, each with around 300 decoys as well as relaxed native structures by minimization and by side-chain optimization followed by minimization. The second decoy set contains 12 12-residue loops, each with around 1100 decoys as well as relaxed native structures. We then compute the VSGB and GSGB energy of each conformation of each loop. In these calculations (and the following loop calculations as well), crystal packing effects are considered, that is, the simulated systems include multiple copies of the conformation described by the corresponding PDB file with the exact number of copies depending on the space group of protein unit cell. As we have reported previously, VSGB on average does an excellent job: the average RMSD of lowest-energy conformations for the four-residue decoy set is 0.27 Å with a standard deviation of 0.33 Å, and the average RMSD 0.60 Å for the 12-residue decoy set with a standard deviation of 0.45 Å. The GSGB does even slightly better. The GSGB average RMSD for the four-residue decoy set is 0.20 Å with standard deviation 0.13 Å, and 0.58 ± 0.39 Å for the 12-residue decoy set. The detailed results are shown in Tables 1 and 2. These results are encouraging because the VSGB model required extensive parameterization (adjustable atomic radii and several types of correction terms) to obtain these excellent results, while the GSGB model has not yet been subjected to parameter optimization.
Table 1.
Details of the Results of PLOP/VSGB and PLOP/GSGB Models for the Four-Residue Decoy Set.
| PDB ID | Beginning residue of loop | Ending residue of loop | Number of decoys | RMSDVSGB (Å) | RMSDGSGB (Å) |
|---|---|---|---|---|---|
| 1ads | 99 | 102 | 308 | 0.15 | 0.15 |
| 1bam | 92 | 95 | 260 | 0 | 0 |
| 1bgc | 40 | 43 | 340 | 0.22 | 0.17 |
| 1cbs | 21 | 24 | 318 | 0.22 | 0.19 |
| 1fkf | 42 | 45 | 308 | 0.29 | 0.52 |
| 1frd | 59 | 62 | 274 | 0.17 | 0.17 |
| 1gpr | 123 | 126 | 286 | 0.23 | 0.23 |
| 1iab | 100 | 103 | 281 | 0.17 | 0.17 |
| 1mba | 97 | 100 | 308 | 0.42 | 0.42 |
| 1nfp | 37 | 40 | 280 | 0.16 | 0.16 |
| 1pbe | 117 | 120 | 306 | 0.13 | 0.13 |
| 1pda | 139 | 142 | 316 | 0.2 | 0.2 |
| 1pgs | 226 | 229 | 330 | 0.14 | 0.14 |
| 1plc | 74 | 77 | 330 | 0.17 | 0.17 |
| 1ppn | 42 | 45 | 320 | 0.42 | 0.42 |
| 1prn | 66 | 69 | 324 | 0.11 | 0.11 |
| 1rcf | 111 | 114 | 306 | 0.14 | 0.22 |
| 1tca | 287 | 290 | 290 | 0.1 | 0.1 |
| 1thw | 194 | 197 | 332 | 0.26 | 0.26 |
| 1tib | 46 | 49 | 310 | 0.2 | 0.16 |
| 1tml | 42 | 45 | 316 | 0.48 | 0.47 |
| 1tys | 131 | 134 | 306 | 0.21 | 0.09 |
| 1xif | 82 | 85 | 326 | 0.25 | 0.11 |
| 1xnb | 30 | 33 | 300 | 0.09 | 0.1 |
| 2cmd | 163 | 166 | 276 | 1.78 | 0.11 |
| Average | 0.27 | 0.20 | |||
| STDEV | 0.33 | 0.13 |
The crystal packing environment is considered in calculations.
Table 2.
Details of the Results of PLOP/VSGB and PLOP/GSGB Models for the 12-Residue Decoy Set.
| PDB ID | Beginning residue of loop | Ending residue of loop | Number of decoys | RMSDVSGB (Å) | RMSDGSGB (Å) |
|---|---|---|---|---|---|
| 1ixh_1 | 160 | 171 | 1152 | 0.49 | 0.49 |
| 1cex_1 | 40 | 51 | 1043 | 0.27 | 0.27 |
| 1arb_1 | 74 | 85 | 1148 | 0.22 | 0.22 |
| 7rsa_1 | 13 | 24 | 1152 | 0.44 | 0.43 |
| 1akz_1 | 181 | 192 | 552 | 0.42 | 0.41 |
| 153l_1 | 98 | 109 | 1152 | 0.28 | 0.28 |
| 1bkf_1 | 9 | 20 | 1152 | 1.16 | 1.16 |
| 1dad_1 | 204 | 215 | 1147 | 1.7 | 1.38 |
| 1dim_1 | 213 | 224 | 1152 | 0.24 | 0.24 |
| 1kuh_1 | 90 | 101 | 1102 | 0.81 | 0.81 |
| 2ayh_1 | 21 | 32 | 1151 | 0.38 | 0.38 |
| 351c_1 | 15 | 26 | 1146 | 0.74 | 0.9 |
| Average | 0.60 | 0.58 | |||
| STDEV | 0.45 | 0.39 |
The crystal packing environment is considered in the calculations.
Then we selected seven 13-residue loops (chosen from our test suite of 40 such loops) for which the VSGB solvent model scored nonnative conformations as having significantly lower energy than the native. These failures, which may reflect deficiencies of the energy function, are much more common in longer, more flexible loops. Details of each test case are listed in Table 3. We compute the GSGB energy of the 52 lowest energy conformations generated by VSGB. We also compute these energies for the relaxed native structures. We then minimize each conformation using the GSGB analytical gradient methods described above, which results in a substantial reduction in the GSGB gradient and energy.
Table 3.
Details of Each 13-Residue Loop, and Their Energy Errors with PLOP/VSGB and New PLOP/GSGB Models.
| PDB ID | No. of residues | Beginning residue of loop | Ending residue of loop | PLOP/VSGB energy error (kcal/mol) | PLOP/GSGB energy “error” (kcal/mol) |
|---|---|---|---|---|---|
| 1cnv | 283 | _:110 | _:122 | −1.35 | 24.04 |
| 1dpg | 970 | A:352 | A:364 | −18.84 | 14.45 |
| 1mo9 | 1044 | A:107 | A:119 | −17.18 | −1.91 |
| 1ock | 824 | A:43 | A:55 | −30.95 | −21.20 |
| 1ojq | 212 | A:167 | A:179 | −9.91 | 4.56 |
| 1yge | 839 | _:452 | _:464 | −41.24 | −34.00 |
| 2hlc | 460 | A:91 | A:103 | −13.96 | −10.86 |
The crystal packing environment is considered in the calculations.
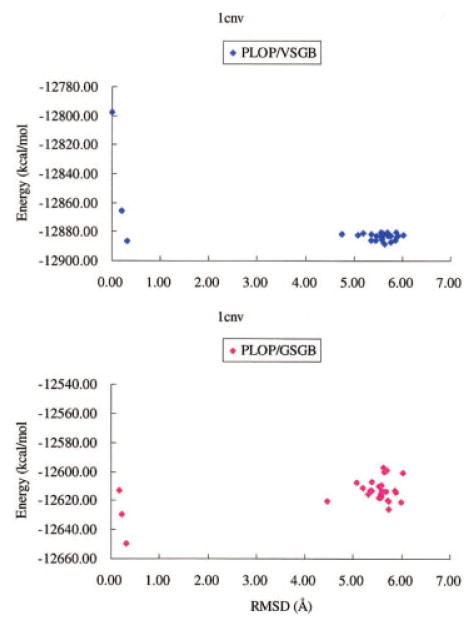
The results for the various test cases are summarized in Table 3. The “energy error” in Table 3 is defined by taking the lowest energy native-like structure (choosing whichever of the minimized native structure, or side-chain optimized native structure, is lowest in energy, typically the latter) and comparing its energy with the lowest energy loop generated by PLOP. For the VSGB model, the energy errors are all negative; these relatively large negative values are, in fact, why the specific test cases were selected. We also plot the GSGB and VSGB energy vs. RMSD for the 1cnv test case in Figure 12 for all of the native-like and false positive conformations, to provide some illustrative detailed results.
Figure 12.
Comparison between PLOP/VSGB energy and PLOP/GSGB energy vs. RMSD for native-like and nonnative conformations of a 13-residue loop (residues 110–122) in 1cnv. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com]
It is clear that, as was hypothesized above, the GSGB model has substantially reduced the energy gaps between the false positive VSGB conformations and the native or native-like structures. In the cases of 1cnv, 1dpg, 1ojq, a low RMSD loop conformation is actually lowest in energy. In the case of 1mo9, a low RMSD structure is almost lowest in energy, with the energy error being reduced to only −1.91 kcal/mol. In the remaining cases of 1yge and 1ock, the energy gap is quantitatively reduced, opening the possibility that further parameter optimization will, in fact, enable the native structure to be selected as lowest in energy (although it should be noted that for 1yge and 1ock, the gap is still quite large, potentially indicating other significant problems with the model). Note that the present test cases are selected to have an extremely high degree of difficulty with regard to recognition of the native structure (compared to the most loop prediction test cases we have investigated, where VSGB works very well, or randomly generated structural perturbations, which do not seek out and maximize flaws in the energy model, and hence, are often trivial to rank as higher in energy than the native structure). Finally, it is possible that one or more of these cases is failing for reasons unrelated to the solvent model, or even the force field, for example, incorrect assignment of the protonation states of titrable residues on the loop or on neighboring side chains. Further investigation will be required to more rigorously assess the quality of the current model, and to discriminate among the various possible reasons for the remaining energy errors.
All of the aforementioned calculations were carried out on Intel PIII 1.40-GHz processors, and a summary of the computational performance of the new GSGB model compared to the VSGB model is presented in Table 4. In general, a single-point total energy calculation with the GSGB model takes about 1.4 times as long as with VSGB, and a single-point gradient calculation with GSGB takes about 2.1 times as long as the VSGB, which does not compute derivatives of the Born alphas. As the primary purpose of this article is to introduce and validate the new GSGB model, we used a resolution of 0.5 Å/grid, which may be finer than is required for many applications. In practice, a coarser resolution or multilevel resolution can be used to speed up the new model significantly. It turns out that when we used a resolution of 1.0 Å/grid, the GSGB model became as fast as or even faster than the VSGB model in single-point energy calculation and the results are within +2% of the results of 0.5 Å/grid resolution, as shown in Table 5.
Table 4.
Computational Performance of the PLOP/GSGB Model, Compared to That of PLOP/VSGB.
| PDB ID | No. of residues | CPUPLOP/VSGB for a single-point energy calc. (s) | CPUPLOP/GSGB for a single-point energy calc. (s) | CPUPLOP/VSGB for a single-point energy and gradient calc. (s) | CPUPLOP/GSGB for a single-point energy and gradient calc. (s) |
|---|---|---|---|---|---|
| 1cnv | 283 | 110 | 158 | 193 | 405 |
| 1dpg | 970 | 455 | 580 | 827 | 1786 |
| 1mo9 | 1044 | 534 | 687 | 987 | 2232 |
| 1ock | 824 | 400 | 528 | 738 | 1575 |
| 1ojq | 212 | 96 | 149 | 165 | 369 |
| 1yge | 839 | 423 | 555 | 782 | 1639 |
| 2hlc | 460 | 182 | 256 | 327 | 661 |
The resolution of the GSGB model is 0.5 Å/grid. The crystal packing environment is considered in the calculations.
Table 5.
Computational Performance and Accuracy of the PLOP/GSGB Model with Resolutions of 1.0 Å/Grid and 0.5 Å/Grid.
| PDB ID | No. of residues | CPUPLOP/GSGB, 1.0 Å/grid for a single-point energy calc. (s) | EPLOP/GSGB, 0.5 Å/grid (kcal/mol) | EPLOP/GSGB, 1.0 Å/grid (kcal/mol) | ΔE% |
|---|---|---|---|---|---|
| 1cnv | 283 | 113 | −2066.69 | −2111.48 | 2.17% |
| 1dpg | 970 | 447 | −9863.37 | −9948.29 | 0.86% |
| 1mo9 | 1044 | 529 | −5751.50 | −5829.03 | 1.35% |
| 1ock | 824 | 399 | −4029.33 | −4069.05 | 0.99% |
| 1ojq | 212 | 100 | −2470.08 | −2474.59 | 0.18% |
| 1yge | 839 | 421 | −4779.18 | −4886.95 | 2.26% |
| 2hlc | 460 | 186 | −6526.00 | −6613.41 | 1.34% |
The crystal packing environment is considered in the calculations.
Discussion and Conclusion
It is clear from the results presented above that the definition of the boundary between solvent and solute in GB models has a large impact on their abilities to estimate the electrostatic solvation energy accurately. The Gaussian surface has been shown to qualitatively reduce the problem of spurious cavities and invaginations relative to a VDW surface. A new-generation SGB model based on the Gaussian surface, without any correction terms to the first Coulombic approximation, nor with any force field parameter optimizations, is demonstrated to qualitatively match PB results, even for a system with more than 1000 residues. Our results demonstrate that the surface definition dominates the qualitative differences between GB and PB results when the VDW surface is used for the former when the system gets large. The significance of the remaining differences in relative energies of different conformations requires further investigation, as does the question of how the details of surface definition (e.g., Connolly surface vs. Gaussian surface) affects accuracy compared to physical reality.
The present model does not incorporate any correction terms or parameter optimization designed to improve agreement with experimental data. Once a surface has been defined that qualitatively exhibits the correct behavior with regard to excluding water from unphysical regions, it is our view that other aspects of the model should be calibrated by direct fitting to experiment and/or explicit solvent simulations, as opposed to the use of a particular approximation (e.g., PB solutions using the Connolly surface) as a benchmark. We have recently shown that both GB and PB continuum solvation methods display serious errors in modeling ion pairs separated by a single bridging water; there was no evidence from these calculations that PB results are at all superior to GB results in comparison with potentials of mean force obtained using explicit solvent models.47 One productive direction to follow is optimization of the model to maximize the accuracy of protein side-chain prediction. We previously optimized the VSGB model using this approach, dramatically reducing overprediction of salt bridges via the use of empirical correction terms. However, such correction terms are specific to the underlying continuum model, so a new round of optimization will have to be carried out for the present GSGB model. The combination of correction terms and the use of the Gaussian surface should result in a model that is an improvement over previous efforts.
Significant progress can also be made with regard to computational performance. The efficiency of the GSGB model can be further improved by utilizing variable grid spacings, where a finer grid is used in the neighborhood of charged or polar atoms and a coarser grid is used in the neighborhood of uncharged or nonpolar atoms. Such an approach would speed up both the surface construction and subsequent surface integration to compute effective Born radii. Also, there is no need to compute the GSGB solvation force for every minimization step because the force does not change significantly within such a short period. With careful bookkeeping, we only need to update it once every several minimization steps. This way, the computational cost of minimization can be cut to a fraction of what it is now.
Acknowledgments
Contract/grant sponsor: the NSF; contract/grant number: MCB-0346399, to M.P.J.
Contract/grant sponsor: NIH; contract/grant numbers: GM071790 and GM56531
Contract/grant sponsor: HHMI Biomedical Research Support Program; contract/grant number: 5300246 to the UCSF School of Medicine
Contract/grant sponsor: NIH; contract/grant number: GM-52018, to R.A.F.
Z.Y. thanks Dr. David Pincus for providing the seven test cases of long loop conformations, Dr. David Rinaldo for helping with figure preparation, and Dr. Yixiang Cao at Schrodinger for helpful calculations.
Appendix Derivation of the Effective Electrostatic Solvation Force
The electrostatic solvation energy consists of two components: self-interaction energy and pair-interaction energy. Correspondingly, the electrostatic solvation force has two components: one from the gradient of the self-interaction energy, and the other from the gradient of the pair-interaction energy:
| (A.1) |
The gradients of the total self-interaction energy are the sum of the gradients of the individual self-interaction energy for each atom:
| (A.2) |
From the formula for the self-interaction energy of each atom, eq. (3), we have
| (A.3) |
where
| (A.4) |
In eq. (A.4), δ is the Delta function. Similarly, we have the formulae for the partial derivatives with regard to the y, z components
| (A.5) |
and
| (A.6) |
where
| (A.7) |
| (A.8) |
Then the gradient of the effective Born radius αk can be derived based on the gradient of the self-interaction energy:
| (A.9) |
| (A.10) |
| (A.11) |
Last, the gradient of the pair-interaction energy is derived based on the gradient of the effective Born radius:
| (A.12) |
| (A.13) |
Similarly, we have the gradients with regard to the other two coordinate components:
| (A.14) |
| (A.15) |
where
| (A.16) |
| (A.17) |
References
- 1.Warwicker J, Watson HC. J Mol Biol. 1982;157:671. doi: 10.1016/0022-2836(82)90505-8. [DOI] [PubMed] [Google Scholar]
- 2.Nicholls A, Honig B. J Comp Chem. 1991;12:435. [Google Scholar]
- 3.Davis ME, McCammon JA. J Comp Chem. 1989;10:387. [Google Scholar]
- 4.Lapidus L, Pinder GF. Numerical Solution of Partial Differential Equations in Science and Engineering. Wiley; New York: 1982. [Google Scholar]
- 5.Cortis CM, Friesner RA. J Comput Chem. 1997;18:1570. [Google Scholar]
- 6.Cortis CM, Friesner RA. J Comput Chem. 1997;18:1591. [Google Scholar]
- 7.Zauhar RJ, Morgan RS. J Mol Biol. 1985;186:815. doi: 10.1016/0022-2836(85)90399-7. [DOI] [PubMed] [Google Scholar]
- 8.Zauhar RJ, Morgan RS. J Comp Chem. 1988;9:171. [Google Scholar]
- 9.Pascual-Ahuir JL, Silla E, Tomasi J. J Comp Chem. 1987;8:778. [Google Scholar]
- 10.Aguilar MA, Olivares FJ, Tomasi J. J Comp Chem. 1993;98:7375. [Google Scholar]
- 11.Edinger SR, Cortis CM, Shenkin PS, Friesner RA. J Phys Chem B. 1997;1997:1190. [Google Scholar]
- 12.Still WC, Tempczyk A, Hawley RC, Hendrickson T. J Am Chem Soc. 1990;112:6127. [Google Scholar]
- 13.Onufriev A, Case DA, Bashford D. J Comp Chem. 2002;23:1297. doi: 10.1002/jcc.10126. [DOI] [PubMed] [Google Scholar]
- 14.Hawkins GD, Cramer CJ, Truhlar DG. Chem Phys Lett. 1995;246:122. [Google Scholar]
- 15.Hawkins GD, Cramer CJ, Truhlar DG. J Phys Chem. 1996;100:19824. [Google Scholar]
- 16.Srinivasan J, Trevathan MW, Beroza P, Case DA. Theor Chem Acc. 1999;101:426. [Google Scholar]
- 17.Tsui V, Case DA. J Am Chem Soc. 2000;122:2489. [Google Scholar]
- 18.Lee MS, Salsbury FRJ, Brooks CL., III J Chem Phys. 2002;116:10606. [Google Scholar]
- 19.Im W, Lee MS, Brooks CLI. J Comp Chem. 2003;24:1691. doi: 10.1002/jcc.10321. [DOI] [PubMed] [Google Scholar]
- 20.Qiu D, Shenkin PS, Hollinger FP, Still WC. J Phys Chem A. 1997;101:3005. [Google Scholar]
- 21.Schaefer M, Karplus M. J Phys Chem. 1996;100:1578. [Google Scholar]
- 22.Ghosh A, Rapp CS, Friesner RA. J Phys Chem B. 1998;102:10983. [Google Scholar]
- 23.Lee B, Richards FM. J Mol Biol. 1971;55:379. doi: 10.1016/0022-2836(71)90324-x. [DOI] [PubMed] [Google Scholar]
- 24.Richards FM. Annu Rev Biophys Bioeng. 1977;6:151. doi: 10.1146/annurev.bb.06.060177.001055. [DOI] [PubMed] [Google Scholar]
- 25.Greer J, Bush BL. Proc Natl Acad Sci. 1978;75:303. doi: 10.1073/pnas.75.1.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pearl LH, Honegger A. J Mol Graphics. 1983;1:9. [Google Scholar]
- 27.Pavlov MY, Fedorov BA. Biopolymer. 1983;22:1507. [Google Scholar]
- 28.Connolly ML. J Appl Crystallogr. 1983;16:548. [Google Scholar]
- 29.Connolly ML. J Appl Crystallogr. 1985;18:499. [Google Scholar]
- 30.Connolly ML. J Mol Graphics. 1993;11:139. doi: 10.1016/0263-7855(93)87010-3. [DOI] [PubMed] [Google Scholar]
- 31.Boys SF. Proc R Soc Lond. 1950;A200:542. [Google Scholar]
- 32.Walker DP, Arteca GA, Mezey PG. J Comput Chem. 1991;12:220. [Google Scholar]
- 33.Gabdoulline RR, Wade RC. J Mol Graphics. 1996;14:341. doi: 10.1016/s0263-7855(97)00008-8. [DOI] [PubMed] [Google Scholar]
- 34.Friedrichs M, Zhou R, Edinger SR, Friesner RA. J Phys Chem B. 1999;103:3057. [Google Scholar]
- 35.Duncan BS, Olson AJ. Biopolymer. 1993;33:231. doi: 10.1002/bip.360330205. [DOI] [PubMed] [Google Scholar]
- 36.Grant JA, Pickup BT. J Phys Chem. 1995;99:3503. [Google Scholar]
- 37.Grant JA, Pickup BT, Nicholls A. J Comp Chem. 2001;22:608. [Google Scholar]
- 38.McGann MR, Almond HR, Nicholls A, Grant JA, Brown FK. Biopolymer. 2003;68:76. doi: 10.1002/bip.10207. [DOI] [PubMed] [Google Scholar]
- 39.Gilson MK, Sharp K, Honig B. J Comp Chem. 1987;9:327. [Google Scholar]
- 40.Honig B, Nicholls A. Science. 1995;268:1144. doi: 10.1126/science.7761829. [DOI] [PubMed] [Google Scholar]
- 41.Rocchia W, Sridharan S, Nicholls A, Alexov E, Chiabrera A, Honig B. J Comp Chem. 2002;23:128. doi: 10.1002/jcc.1161. [DOI] [PubMed] [Google Scholar]
- 42.Jorgensen WL, Maxwell DS, Tirado-Rives J. J Am Chem Soc. 1996;118:11225. [Google Scholar]
- 43.Lorensen WE, Cline HE. Comput Graphics. 1987;21:163. [Google Scholar]
- 44.Dürst M. Comput Graphics. 1988;22:72. [Google Scholar]
- 45.Heiden W, Goetze T, Brickman J. J Comp Chem. 1992;14:246. [Google Scholar]
- 46.Das K, Clark AD, Jr, Lewi PJ, Heeres J, de Jonge MR, Koymans LMH, Vinkers HM, Daeyaert F, Ludovici DW, Kukla MJ, De Corte B, Kavash RW, Ho CY, Ye H, Lichtenstein MA, Andries K, Pauwels R, de Bethune M-P, Boyer PL, Clark P, Hughes SH, Janssen PAJ, Arnold E. J Med Chem. 2004;47:2550. doi: 10.1021/jm030558s. [DOI] [PubMed] [Google Scholar]
- 47.Yu Z, Jacobson MP, Josovitz J, Rapp CS, Friesner RA. J Phys Chem B. 2004;108:6643. [Google Scholar]
- 48.Jacobson MP, Friesner RA, Xiang Z, Honig B. J Mol Biol. 2002;320:597. doi: 10.1016/s0022-2836(02)00470-9. [DOI] [PubMed] [Google Scholar]
- 49.Jacobson MP, Kaminski GA, Friesner RA, Rapp CS. J Phys Chem B. 2002;106:11673. [Google Scholar]
- 50.Jacobson MP, Pincus DL, Rapp CS, Day TJF, Honig B, Shaw DE, Friesner RA. Proteins. 2004;55:351. doi: 10.1002/prot.10613. [DOI] [PubMed] [Google Scholar]
- 51.Li X, Jacobson MP, Friesner RA. Proteins. 2004;55:368. doi: 10.1002/prot.20014. [DOI] [PubMed] [Google Scholar]
- 52.Andrec M, Harano Y, Jacobson MP, Friesner RA, Levy RM. J Struct Funct Genom. 2002;2:103. doi: 10.1023/a:1020435630054. [DOI] [PubMed] [Google Scholar]
- 53.Guallar V, Jacobson MP, McDermott A, Friesner RA. J Mol Biol. 2004;337:227. doi: 10.1016/j.jmb.2003.11.016. [DOI] [PubMed] [Google Scholar]
- 54.Sanner M, Olsen A, Spehner J. In Proceedings of the 11th ACM Symposium on Computational Geometry; New York: ACM; 1995. p. C6. [Google Scholar]
- 55.Humphrey W, Dalke A, Schulten K. J Mol Graphics. 1996;14:33. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]