Summary
Since the discovery of the 26S proteasome, much progress has been made in determining the structure of this large dynamic protein complex. Until now, a vast amount of structural information of the proteasome has been obtained from all kinds of structure determination techniques, and the function of the protease core is well understood at atomic detail. Yet our understanding of the entire 26S proteasome structure, particularly its 19S regulatory complex, is still limited at a low-resolution blob-ology level. In this review, we highlight the recent progress made in understanding the mechanism of 20S gate opening by the proteasomal activators. We also emphasized the recent methodological advances, particularly in achieving the near atomic resolution by single particle electron cryomicroscopy, and the possible approaches that will enable more detailed structural analysis of the entire 26S proteasome.
Introduction
In all eukaryotic cells, the 26S proteasome catalyzes most of the ATP-dependent intracellular protein degradation [1]. The protease core of this large protein machine is a barrel-shaped 20S core particle (CP) that is capped at each end by a 19S regulatory particle (RP). Ubiquitinated proteins targeted for degradation must first attach to the 19S RP where they are unfolded, then translocated into the 20S CP for degradation. This ubiquitin-proteasome pathway plays a critical role in governing cellular processes that range from normal protein turnover, to detoxification by degrading misfolded and damaged proteins, and to cell cycle control [2].
Ever since the first picture of a 26S proteasome was captured by using an electron microscope in 1991 [3], structural biologists have been fascinated with the goal of determining the structure of this large protein complex, or its components, by all possible structural tools, from x-ray crystallography [4–6], electron cryomicroscopy (cryoEM) [7,8*,9], NMR spectroscopy [10] to atomic force microscopy [11]. Over the years, much progress has been made in revealing the structures and understanding the functions of this dynamic protein machinery in a divide-and-conquer process. The 20S CP, which is the most stable part of 26S proteasome, was the first and still is the biggest proteasomal component to be crystallized and to have its structure determined by the x-ray crystallography [4–6]. This achievement was followed by more structural studies focused on the functions of the 20S CP, such as the structure and function of its gate [12*], its assembly [13,14], and its complexes with various proteasomal activators that are simpler than the 19S RP [15–17**,18,19]. As for the 19S RP, only a few components of this dynamic multi-subunit complex have their structures determined, such as the ubiquitin receptor Rpn13 [20,21]. The “holy grail” of proteasome structural biology is, of course, to obtain an atomic structure of the entire 26S proteasome. This feat has not yet been achieved despite remarkable progress made in the methodologies of all structure determination techniques. It is understandable that crystallizing the 26S proteasome is very difficult, due to its size, number of components and dynamic nature. As an alternative approach, the 26S proteasome has also been a target of single particle EM studies since its discovery. Although still at a level of blob-ology, the single particle cryoEM has produced a number of three-dimensional (3D) reconstructions of this large protein complex to the modest resolution of near 20 Å and resolved individual domains [7,8*,9]. In light of the recent progresses in single particle cryoEM methodology, and achievements such as subnanometer resolution structures of large assemblies like the ribosome [22] and the clathrin coats [23] and even near atomic resolution of icosahedral viruses [24–26], we could expect that a 3D reconstruction of the 26S proteasome beyond subnanometer resolution and a pseudo atomic model of this large protein complex will soon be produced.
The 20S core particle
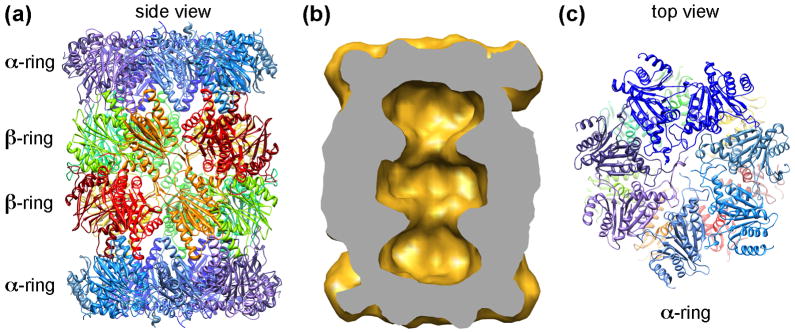
The 20S CP carries out the protease activity of the 26S proteasome. Until now, the structures of the 20S CP from archaea, yeast and mammals were all determined by the x-ray crystallography [4–6]. Their structures are highly conserved, consisting of four axially-stacked heptameric rings, two inner β-rings and two outer α-rings (Fig. 1a), forming three continuous chambers inside the 20S CP (Fig 1b). Such a “self-compartmentalized” structure keeps the proteolytic active sites, located at the N-termini of three of the seven β-subunits, sequestered in the central chamber and prevents unregulated substrate degradation [27]. Access to the central degradation chamber is through a channel of only 13 Å in diameter in the middle of the outer α-rings, known as the α-annulus. Only unfolded substrates can pass through this channel to enter the central chamber for degradation. Furthermore, the N-termini of α-subunits in eukaryotic 20S CP extend across the central channel and function as a gate that blocks substrates from entering the inner degradation chamber (Fig. 1c) [12*]. Archaea has a simpler 20S CP, its α- and β-rings are formed by seven identical α- and β-subunits [5]. The N-termini of archaeal 20S α-subunits are less ordered but still function as a gate [28]. Without an activator, the 20S gate remains closed to block the free access of even unfolded substrates from entering the 20S CP.
Figure 1.
Architecture of the 20S CP. (a): A ribbon diagram of the yeast 20S proteasome (pdb accession code: 1RYP) [4–6]. Two inner β-rings and two outer α-rings are marked. (b) Volume of the yeast 20S CP, calculated from atomic coordinates and filtered to 20 Å. The volume is cut in half, showing three chambers within the 20S CP. (c) The top surface of the α-ring. The N-termini of the α-subunits form a closed gate block the entrance to the inner chamber of the 20S CP.
In addition to the functions of the 20S CP, the x-ray crystallographic studies have also revealed its assembly process, including the crystal structure of 20S α-ring alone [13], and the more recent studies of proteasome assembly chaperones (PACs) [2,14,29].
The 20S CP – proteasomal activator complexes
The 20S CP alone has a closed gate and requires an activator to regulate its protease activity. Three different types of proteasomal activators have been identified so far. They are the 19S RP, 11S activator (PA28/PA26/REG) [30], and PA200 (or Blm10 in yeast) [31]. All three types of proteasome activators can associate with the 20S CP and stimulate its peptidase activity by inducing the gate opening by similar yet distinct mechanisms. Among them, only the 19S RP contains the ATPase subunits that can also unfold globular proteins. The base subunit of the 19S RP contains six ATPases, Rpt1–6. These ATPases interact directly with the α-ring in the 20S CP and controls its gate opening.
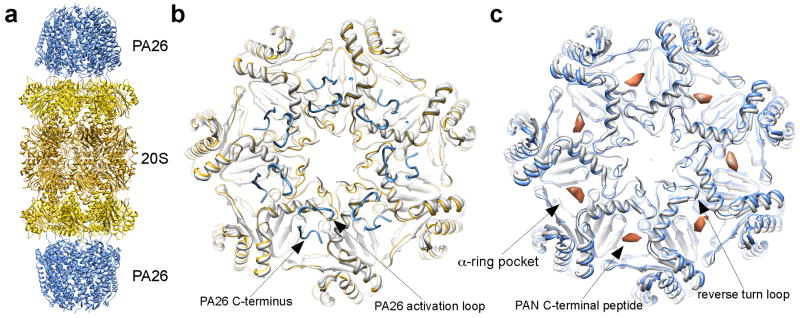
The first clue of proteasomal activator induced gate opening in the 20S CP came from the atomic structures of 20S CP in complex with the PA26 from Trypanosoma brucei (Fig. 2a) [15–17]. PA26 is a heptameric 11S activator (or PA28 in most organisms) [32]. It is not an ATPase and does not unfold globular proteins, but rather stimulates the degradation of unfolded peptides by the 20S by inducing its gate opening [15]. The exact function of PA26 or PA28 in the protein degradation pathway is not quite clear. Nevertheless, the atomic structure of its complex with the 20S CP provided the atomic details of how a proteasomal activator interacts with the 20S CP and induces the gate opening. Different from the hexameric proteasomal ATPases, which waggle in the heptameric 20S CP [28], the heptameric PA26 forms a stable complex with the 20S CP facilitating the crystallization of the complex for structure determination. The crystal structure showed that the PA26 binds to the 20S CP by inserting its C-termini into the 20S α-ring pockets located between adjacent α-subunits. But such interaction does not induce significant conformational changes in the body of the α-ring, except a reverse turn loop in the α-subunit very subtly moves away from the central pore. Apparently it is moved by a loop in the bottom of the PA26 (named the activation loop) (Fig. 2b). Surprisingly, this less than 2Å movement of the reverse turn loop, which connects the N-terminus that forms the 20S gate to the helix H0, is sufficient to disrupt the closed gate formed by the N-termini of α-subunits and stabilize the open-gate conformation.
Figure 2.
Gate opening of the 20S CP by proteasomal activators. (a) Ribbon diagram of 20S-PA26 complex (PDB accession code: 1YA7) [17]. (b) Top view of the 20S CP’s α-ring, showing both a closed gate (light gray, PDB accession code: 3C92) and an open gate (yellow) in the presence of the PA26 (blue). The locations of the PA26 C-terminus and activation loop are marked. Notice that the 20S reverse turn loop (pointed in (c)) is moved by the PA26 activation loop. (c) CryoEM structure of 20S-PAN peptides, with an open-gate conformation (light blue, PDB accession code: 3C91). The structure of a closed gate is shown in light gray (PDB access code: 3C92) [33]. The densities correspond to the PAN C-terminal peptide, indicates the binding site of PAN’s C-termini in the 20S CP.
The relocation of the reverse turn loop in the α-subunits is apparently a mechanism that is shared by the other proteasomal activators, such as proteasomal ATPases, to open the 20S gate [33**]. The difference between these activators is how each activator moves this reverse turn loop. The PA26 moves it by two interactions with the 20S CP. One interaction is the binding of its C-termini to the 20S α-ring pocket, and the other is its activation loop directly interacts with the reverse turn loop of the α-subunit [15–17**]. However, the proteasomal ATPases uses a distinct mechanism to move this reverse turn loop [33]. The C-termini of proteasomal ATPases contain a conserved hydrophobic-tyrosine-X motif named HbYX motif [34**]. This HbYX motif binds to the same pocket in the 20S α-ring as the C-termini of PA26 do, but such binding is sufficient to open the 20S gate without an activation loop interacting with the reverse turn loop. This result was shown by experiments in which the synthetic peptides of only 7 or 8 residues corresponding to the C-terminus of proteasomal ATPases stimulate the 20S gate opening [34]. This mechanism has also been observed by the single particle cryoEM, where these peptides bound to the same α-ring pockets as the C-termini of PA26. Binding of these peptides induces a rotation of the individual α-subunits by ~ 4°. The reverse turn loop of the 20Sα-subunit moves along to the position that the closed gate is disrupted and open gate is stabilized. Thus, without the activation loop, the C-termini of the proteasomal ATPases with the HbYX motif is sufficient to induce the gate opening in the 20S CP [33] (Fig. 2c).
The third class of proteasomal activator, the PA200 (or Blm10 in yeast), also forms complex with the 20S CP and opens the 20S gate [18,19,35], but the exact mechanism is not yet clear. One can certainly hypothesize that it may use a similar mechanism as either the proteasomal ATPases or the PA26. There must be some differences because the PA200 functions as a monomer, unlike the hexameric proteasomal ATPases or the heptameric PA26. With only one C-terminus, the PA200 must have multiple interactions with the 20S CP. Structural studies at higher resolution are definitely needed to answer this question.
The eukaryotic 26S proteasome
Although all three types of proteasomal activators regulate the gate opening in the 20S CP, only the ATPases unfold substrates and translocate unfolded substrates into the 20S CP for degradation. No atomic structure of any proteasomal ATPases has been determined yet, thus we know very little about how they unfold and translocate the substrates. We know even less about how structurally the entire 19S RP function as a whole machinery in the process of ATP dependent degradation of ubiquitinated substrates, except a few subunits such as the ubiquitin receptors in the 19S RP [20,21].
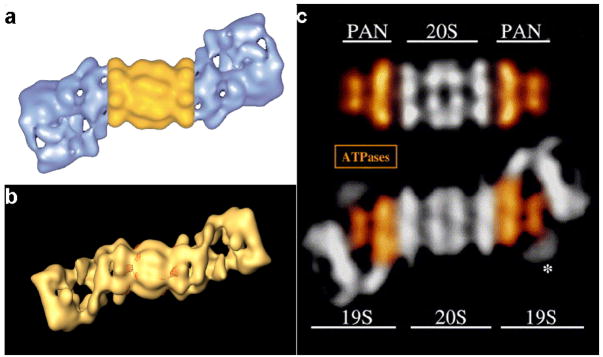
Ever since the 26S proteasome was identified as a protein degradation machine, single particle cryoEM has been the method of choice to determine the structure of this large and dynamic complex [36]. The first 3D reconstruction of the 26S proteasome was reported ten years ago in 1998, at a resolution of about 28 Å (Fig. 3a) [7]. It was then discovered that the conformation of the19S RP is rather dynamic. This dynamic nature of the 19S RP seemed to be the major limiting factor preventing the 3D reconstruction of this large complex to reach higher resolution. Over the last ten years, the resolution improvement of the 26S proteasome 3D reconstruction did not match the pace of the methodological development of the single particle cryoEM. The most recent 3D reconstructions of the human [9] as well as the Drosophila 26S proteasome (Fig. 3b) [8] are still at a resolution above 20 Å. Such an unsatisfying resolution only resolves large domains such as the 20S CP and the base and the lid of the 19S RP but not any individual subunits. Even for a simpler model system in archaea, where the PAN ATPase is the closest homolog of the eukaryotic proteasomal ATPases in the 19S RP and structurally similar with part of the 19S base subunit (Fig. 3a) [28], the resolution is unsatisfying (unpublished result). This situation is quite different from that of the protein synthesis machinery, the ribosome, whose 3D reconstruction by single particle cryoEM has been improved to subnanometer resolution as the methodologies of this technique have developed.
Figure 3.
Structure of the 26S proteasome. (a) and (b) 3D reconstructions of the 26S proteasome, by single particle EM (reprinted with permission from [7] and [9]). (c) Comparison of the 20S-PAN complex with the 26S proteasome shows the similarity between the PAN ATPase complex (colored in gold) and the 19S base subunit (colored in gold) (reprinted with permission from [28]). The 20S-PAN complex may be used as a simpler model system to dissect the mechanism of substrates unfolding and translocation by the eukaryotic proteasomal ATPases.
In 2008, the single particle cryoEM has experienced an unusual successful year, in which a number of laboratories have cracked the 4 Å resolution barrier [24,25]. Although all of these near-atomic resolution structures are obtained from protein assemblies with high symmetry, such as virus particles with icosahedral symmetry, the methodological development itself brought new hope into the battle of determining the near-atomic structure of the 26S proteasome. Achieving a high-resolution 3D reconstruction by single particle cryoEM requires averaging a large number of images of structurally/conformationally homogeneous (identical) particles. Thus, the challenge of determining a high-resolution structure of the 26S proteasome lies in the conformational and/or compositional heterogeneities presented in the particles of 26S proteasome, particularly within the 19S RP. The methods used to generate the near-atomic resolution structures of the icosahedral virus may not be directly applicable to the 26S proteasome. Therefore, different approaches will need to be developed to achieve a high-resolution structure of the 26S proteasome by single particle cryoEM.
Strategies towards an atomic structure of the 26S proteasome
X-ray crystallography is still the most successful method of determining atomic structures of large complexes. During the past ten years, a number of large macromolecular assemblies have been crystallized, such as ribosome [37] and fatty acid synthase [38]. Therefore, we still hope that at one day the 26S proteasome will be crystallized. Until then, an alternative approach is to continue the effort to improve the resolution of 3D reconstructions of the 26S proteasome by single particle cryoEM. If one can achieve the subnanometer resolution 3D reconstruction of the 26S proteasome and identify every subunit in the 19S RP, then, it is possible to generate a complete atomic model by docking the atomic structures of individual components into the 3D density map. In the past, such an approach has generated a complete atomic model of clathrin coat [23].
The major obstacle in achieving the subnanometer single particle reconstruction of 26S proteasome seems to be the structural/conformational heterogeneities in the particles used for 3D reconstruction. Recently, progresses in dealing with such heterogeneity were made by using a number of methods, such as maximum likelihood classification [39,40], bootstrap resampling [41] and statistical analysis of 2D images [42] to computationally separate the conformationally or compositional heterogeneous protein complexes into different classes. These methods were demonstrated by a number of examples where particles were separated to calculated 3D reconstructions of different conformations or compositions [43–46]. We can expect that similar approaches will be applied to the 26S proteasome for the purpose of not only to separate its different conformations, but also to improve the resolution of its 3D reconstructions. To make such approach work, it is necessary to have a very large dataset so that the number of particles in each conformational class is sufficiently large to generate 3D reconstructions toward high resolution. Generating such a large dataset could be tedious, if not impossible, with the traditional manual data acquisition methods. But this task becomes more feasible with the automated data acquisition procedures [8,47]. By combining these two recent developments, automated data acquisition and sorting heterogeneous particles, we can anticipate the effort of determining the structure of eukaryotic 26S proteasome by single particle cryoEM will soon break the limit of 20 Å resolution.
It is worthwhile to note the progress made with another approach used to determine the 3D architecture of nuclear pore complex [48]. Different from the traditional approaches of structure determination, a computational method was developed to model the 3D architecture of this large macromolecular assembly by combining all available proteomic data with the relatively low-resolution 3D reconstruction from EM [49]. This same approach could be applied to the 26S proteasome to generate a more detailed 3D arrangement of each subunit within the 26S proteasome, particularly the 19S RP, than our current understanding of this large complex [50].
Conclusions
Since the first image of the 26S proteasome, tremendous structural information has already been obtained about this large protein machine, including the atomic structure of 20S CP and its interaction with the non-ATPase activators. However, we are still far from understanding at atomic detail how the 26S proteasome functions as an integrated machine. Achieving a complete atomic structure of the entire 26S proteasome is still a major goal in structural studies of this large complex. It seems that neither the x-ray crystallography nor the single particle cryoEM will be sufficient to achieve this goal. Instead, the combination of these two methods will provide insight into the structure of the protein degradation machine. With the recent developments in structural biology methodologies, particularly in single particle cryoEM and computational modeling, achieving this goal becomes very feasible.
Acknowledgments
This work was supported by an NIH grant R01 GM082893 and a Sandler Family Supporting Foundation grant, the Sandler Opportunity Award in Basic Science. The TF20 electron microscope system at UCSF was established in part with grants from the Sandler New Technology Award and a UCSF Academic Senate Shared Equipment Grant.
Footnotes
Dedication
This work is dedicated to Professor Wang Renhui, who taught me electron microscopy.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
Papers of particular interest, published within the period of review, have been highlighted as:
* of special interest
** of outstanding interest
- 1.Goldberg AL. Nobel committee tags ubiquitin for distinction. Neuron. 2005;45:339–344. doi: 10.1016/j.neuron.2005.01.019. [DOI] [PubMed] [Google Scholar]
- 2.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 3.Ikai A, Nishigai M, Tanaka K, Ichihara A. Electron microscopy of 26 S complex containing 20 S proteasome. FEBS Lett. 1991;292:21–24. doi: 10.1016/0014-5793(91)80824-m. [DOI] [PubMed] [Google Scholar]
- 4.Groll M, Ditzel L, Lowe J, Stock D, Bochtler M, Bartunik HD, Huber R. Structure of 20S proteasome from yeast at 2.4 A resolution. Nature. 1997;386:463–471. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- 5.Lowe J, Stock D, Jap B, Zwickl P, Baumeister W, Huber R. Crystal structure of the 20S proteasome from the archaeon T. acidophilum at 3.4 A resolution. Science. 1995;268:533–539. doi: 10.1126/science.7725097. [DOI] [PubMed] [Google Scholar]
- 6.Unno M, Mizushima T, Morimoto Y, Tomisugi Y, Tanaka K, Yasuoka N, Tsukihara T. The structure of the mammalian 20S proteasome at 2.75 A resolution. Structure. 2002;10:609–618. doi: 10.1016/s0969-2126(02)00748-7. [DOI] [PubMed] [Google Scholar]
- 7.Walz J, Erdmann A, Kania M, Typke D, Koster AJ, Baumeister W. 26S proteasome structure revealed by three-dimensional electron microscopy. J Struct Biol. 1998;121:19–29. doi: 10.1006/jsbi.1998.3958. [DOI] [PubMed] [Google Scholar]
- 8*.Nickell S, Beck F, Korinek A, Mihalache O, Baumeister W, Plitzko JM. Automated cryoelectron microscopy of “single particles” applied to the 26S proteasome. FEBS Lett. 2007;581:2751–2756. doi: 10.1016/j.febslet.2007.05.028. An automated data acquisition procedure was described here and used to collect a dataset of frozen hydrated 26S proteasome. Although the resolution of the 26S proteasome 3D reconstruction is still at over 20 Å, it represents a promising approach to reach higher resoltuion 3D reconstructions of the 26S proteasome. [DOI] [PubMed] [Google Scholar]
- 9.da Fonseca PC, Morris EP. Structure of the human 26S proteasome: subunit radial displacements open the gate into the proteolytic core. J Biol Chem. 2008;283:23305–23314. doi: 10.1074/jbc.M802716200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sprangers R, Kay LE. Quantitative dynamics and binding studies of the 20S proteasome by NMR. Nature. 2007;445:618–622. doi: 10.1038/nature05512. [DOI] [PubMed] [Google Scholar]
- 11.Rosenzweig R, Osmulski PA, Gaczynska M, Glickman MH. The central unit within the 19S regulatory particle of the proteasome. Nat Struct Mol Biol. 2008;15:573–580. doi: 10.1038/nsmb.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12*.Groll M, Bajorek M, Kohler A, Moroder L, Rubin DM, Huber R, Glickman MH, Finley D. A gated channel into the proteasome core particle. Nat Struct Biol. 2000;7:1062–1067. doi: 10.1038/80992. This work described the formation and the function of 20S gate. This was followed by many other studies characterizing the activator induced gate opening. [DOI] [PubMed] [Google Scholar]
- 13.Groll M, Brandstetter H, Bartunik H, Bourenkow G, Huber R. Investigations on the maturation and regulation of archaebacterial proteasomes. J Mol Biol. 2003;327:75–83. doi: 10.1016/s0022-2836(03)00080-9. [DOI] [PubMed] [Google Scholar]
- 14.Yashiroda H, Mizushima T, Okamoto K, Kameyama T, Hayashi H, Kishimoto T, Niwa S, Kasahara M, Kurimoto E, Sakata E, et al. Crystal structure of a chaperone complex that contributes to the assembly of yeast 20S proteasomes. Nat Struct Mol Biol. 2008;15:228–236. doi: 10.1038/nsmb.1386. [DOI] [PubMed] [Google Scholar]
- 15.Whitby FG, Masters EI, Kramer L, Knowlton JR, Yao Y, Wang CC, Hill CP. Structural basis for the activation of 20S proteasomes by 11S regulators. Nature. 2000;408:115–120. doi: 10.1038/35040607. [DOI] [PubMed] [Google Scholar]
- 16.Forster A, Whitby FG, Hill CP. The pore of activated 20S proteasomes has an ordered 7-fold symmetric conformation. Embo J. 2003;22:4356–4364. doi: 10.1093/emboj/cdg436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17**.Forster A, Masters EI, Whitby FG, Robinson H, Hill CP. The 1.9 A structure of a proteasome-11S activator complex and implications for proteasome-PAN/PA700 interactions. Mol Cell. 2005;18:589–599. doi: 10.1016/j.molcel.2005.04.016. The high-resolution structure of 20S-PA26 compelx presented in this paper described how a proteasomal activator interacts with the 20S CP and how to induce the gate opening. The mechanism described here is partly shared by the other proteasomal activators. [DOI] [PubMed] [Google Scholar]
- 18.Ortega J, Heymann JB, Kajava AV, Ustrell V, Rechsteiner M, Steven AC. The axial channel of the 20S proteasome opens upon binding of the PA200 activator. J Mol Biol. 2005;346:1221–1227. doi: 10.1016/j.jmb.2004.12.049. [DOI] [PubMed] [Google Scholar]
- 19.Iwanczyk J, Sadre-Bazzaz K, Ferrell K, Kondrashkina E, Formosa T, Hill CP, Ortega J. Structure of the Blm10–20 S Proteasome Complex by Cryo-electron Microscopy. Insights into the Mechanism of Activation of Mature Yeast Proteasomes. J Mol Biol. 2006;363:648–659. doi: 10.1016/j.jmb.2006.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Husnjak K, Elsasser S, Zhang N, Chen X, Randles L, Shi Y, Hofmann K, Walters KJ, Finley D, Dikic I. Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature. 2008;453:481–488. doi: 10.1038/nature06926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schreiner P, Chen X, Husnjak K, Randles L, Zhang N, Elsasser S, Finley D, Dikic I, Walters KJ, Groll M. Ubiquitin docking at the proteasome through a novel pleckstrin-homology domain interaction. Nature. 2008;453:548–552. doi: 10.1038/nature06924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chandramouli P, Topf M, Menetret JF, Eswar N, Cannone JJ, Gutell RR, Sali A, Akey CW. Structure of the mammalian 80S ribosome at 8.7 A resolution. Structure. 2008;16:535–548. doi: 10.1016/j.str.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fotin A, Cheng Y, Sliz P, Grigorieff N, Harrison SC, Kirchhausen T, Walz T. Molecular model for a complete clathrin lattice from electron cryomicroscopy. Nature. 2004;432:573–579. doi: 10.1038/nature03079. [DOI] [PubMed] [Google Scholar]
- 24.Jiang W, Baker ML, Jakana J, Weigele PR, King J, Chiu W. Backbone structure of the infectious epsilon15 virus capsid revealed by electron cryomicroscopy. Nature. 2008;451:1130–1134. doi: 10.1038/nature06665. [DOI] [PubMed] [Google Scholar]
- 25.Yu X, Jin L, Zhou ZH. 3.88 Å structure of cytoplasmic polyhedrosis virus by cryo-electron microscopy. Nature. 2008;453:415–419. doi: 10.1038/nature06893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang X, Settembre E, Xu C, Dormitzer PR, Bellamy R, Harrison SC, Grigorieff N. Near-atomic resolution using electron cryomicroscopy and single-particle reconstruction. Proc Natl Acad Sci U SA. 2008;105:1867–1872. doi: 10.1073/pnas.0711623105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baumeister W, Walz J, Zuhl F, Seemuller E. The proteasome: paradigm of a self-compartmentalizing protease. Cell. 1998;92:367–380. doi: 10.1016/s0092-8674(00)80929-0. [DOI] [PubMed] [Google Scholar]
- 28.Smith DM, Kafri G, Cheng Y, Ng D, Walz T, Goldberg AL. ATP binding to PAN or the 26S ATPases causes association with the 20S proteasome, gate opening, and translocation of unfolded proteins. Mol Cell. 2005;20:687–698. doi: 10.1016/j.molcel.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 29.Hirano Y, Hayashi H, Iemura S, Hendil KB, Niwa S, Kishimoto T, Kasahara M, Natsume T, Tanaka K, Murata S. Cooperation of multiple chaperones required for the assembly of mammalian 20S proteasomes. Mol Cell. 2006;24:977–984. doi: 10.1016/j.molcel.2006.11.015. [DOI] [PubMed] [Google Scholar]
- 30.Knowlton JR, Johnston SC, Whitby FG, Realini C, Zhang Z, Rechsteiner M, Hill CP. Structure of the proteasome activator REGalpha (PA28alpha) Nature. 1997;390:639–643. doi: 10.1038/37670. [DOI] [PubMed] [Google Scholar]
- 31.Ustrell V, Hoffman L, Pratt G, Rechsteiner M. PA200, a nuclear proteasome activator involved in DNA repair. EMBO J. 2002;21:3516–3525. doi: 10.1093/emboj/cdf333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dubiel W, Pratt G, Ferrell K, Rechsteiner M. Purification of an 11 S regulator of the multicatalytic protease. J Biol Chem. 1992;267:22369–22377. [PubMed] [Google Scholar]
- 33**.Rabl J, Smith DM, Yu Y, Chang SC, Goldberg AL, Cheng Y. Mechanism of gate opening in the 20S proteasome by the proteasomal ATPases. Mol Cell. 2008;30:360–368. doi: 10.1016/j.molcel.2008.03.004. In this paper, the single particle cryoEM was used to study how the C-termini of proteasomal ATPases induce the 20S gate opening. It described the similarity as well as the difference of the gate-opening mechanism between the an ATPase and a non-ATPase proteasomal activator. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34**.Smith DM, Chang SC, Park S, Finley D, Cheng Y, Goldberg AL. Docking of the proteasomal ATPases’ carboxyl termini in the 20S proteasome’s alpha ring opens the gate for substrate entry. Mol Cell. 2007;27:731–744. doi: 10.1016/j.molcel.2007.06.033. This paper identified a conserved the C-terminal motif in the proteasomal ATPases that is required for induce the 20S gate opening. It showed that the short synthetic peptides with the gate-opening motif is sufficient to induce the 20S gate opening. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmidt M, Haas W, Crosas B, Santamaria PG, Gygi SP, Walz T, Finley D. The HEAT repeat protein Blm10 regulates the yeast proteasome by capping the core particle. Nat Struct Mol Biol. 2005;12:294–303. doi: 10.1038/nsmb914. [DOI] [PubMed] [Google Scholar]
- 36.Peters JM, Cejka Z, Harris JR, Kleinschmidt JA, Baumeister W. Structural features of the 26 S proteasome complex. J Mol Biol. 1993;234:932–937. doi: 10.1006/jmbi.1993.1646. [DOI] [PubMed] [Google Scholar]
- 37.Bingel-Erlenmeyer R, Kohler R, Kramer G, Sandikci A, Antolic S, Maier T, Schaffitzel C, Wiedmann B, Bukau B, Ban N. A peptide deformylase-ribosome complex reveals mechanism of nascent chain processing. Nature. 2008;452:108–111. doi: 10.1038/nature06683. [DOI] [PubMed] [Google Scholar]
- 38.Maier T, Leibundgut M, Ban N. The crystal structure of a mammalian fatty acid synthase. Science. 2008;321:1315–1322. doi: 10.1126/science.1161269. [DOI] [PubMed] [Google Scholar]
- 39.Scheres SH, Nunez-Ramirez R, Gomez-Llorente Y, San Martin C, Eggermont PP, Carazo JM. Modeling experimental image formation for likelihood-based classification of electron microscopy data. Structure. 2007;15:1167–1177. doi: 10.1016/j.str.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scheres SH, Valle M, Carazo JM. Fast maximum-likelihood refinement of electron microscopy images. Bioinformatics. 2005;21(Suppl 2):ii243–244. doi: 10.1093/bioinformatics/bti1140. [DOI] [PubMed] [Google Scholar]
- 41.Penczek PA, Yang C, Frank J, Spahn CM. Estimation of variance in single-particle reconstruction using the bootstrap technique. J Struct Biol. 2006;154:168–183. doi: 10.1016/j.jsb.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 42.Elad N, Clare DK, Saibil HR, Orlova EV. Detection and separation of heterogeneity in molecular complexes by statistical analysis of their two-dimensional projections. J Struct Biol. 2008;162:108–120. doi: 10.1016/j.jsb.2007.11.007. [DOI] [PubMed] [Google Scholar]
- 43.Clare DK, Bakkes PJ, van Heerikhuizen H, van der Vies SM, Saibil HR. Chaperonin complex with a newly folded protein encapsulated in the folding chamber. Nature. 2009;457:107–110. doi: 10.1038/nature07479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Elad N, Farr GW, Clare DK, Orlova EV, Horwich AL, Saibil HR. Topologies of a substrate protein bound to the chaperonin GroEL. Mol Cell. 2007;26:415–426. doi: 10.1016/j.molcel.2007.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scheres SH, Gao H, Valle MTHG, Eggermont PP, Frank J, Carazo JM. Disentangling conformational states of macromolecules in 3D-EM through likelihood optimization. Nat Methods. 2007;4:27–29. doi: 10.1038/nmeth992. [DOI] [PubMed] [Google Scholar]
- 46.Zhang W, Kimmel M, Spahn CM, Penczek PA. Heterogeneity of large macromolecular complexes revealed by 3D cryo-EM variance analysis. Structure. 2008;16:1770–1776. doi: 10.1016/j.str.2008.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stagg SM, Lander GC, Pulokas J, Fellmann D, Cheng A, Quispe JD, Mallick SP, Avila RM, Carragher B, Potter CS. Automated cryoEM data acquisition and analysis of 284742 particles of GroEL. J Struct Biol. 2006;155:470–481. doi: 10.1016/j.jsb.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 48.Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, et al. The molecular architecture of the nuclear pore complex. Nature. 2007;450:695–701. doi: 10.1038/nature06405. [DOI] [PubMed] [Google Scholar]
- 49.Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, et al. Determining the architectures of macromolecular assemblies. Nature. 2007;450:683–694. doi: 10.1038/nature06404. [DOI] [PubMed] [Google Scholar]
- 50.Robinson CV, Sali A, Baumeister W. The molecular sociology of the cell. Nature. 2007;450:973–982. doi: 10.1038/nature06523. [DOI] [PubMed] [Google Scholar]