Abstract
Elevated BAFF (TNFSF13B) levels have been found in patients with B-cell malignancies and autoimmune diseases suggesting that it may play a pathogenic role. We previously found that a SNP in the TNFSF13B promoter resulted in increased transcription suggesting that genetic variation in TNFSF13B may influence its expression. We therefore wanted to determine if genetic variation in TNFSF13B is associated with high BAFF levels and non-Hogkin lymphoma (NHL) risk. We genotyped 9 tagSNPs within TNFSF13B in a clinic-based study of 441 NHL cases and 475 matched controls and evaluated the association of individual SNPs with risk of NHL, 3 tagSNPs were significant (p<0.05). When categorized into low, moderate, and high risk groups based on risk alleles, we found the permutation-corrected odds ratio (OR) for the trend to be 1.43 (p=0.0019) for risk of B-cell NHL, 1.69 (p=0.0093) for diffuse large B cell lymphoma (DLBCL), 1.43 (p=0.029) for follicular lymphoma, and 1.06 (p=0.21) for CLL/SLL. The mean serum BAFF level in those who carried the low risk alleles was 2 ng/ml compared to 4.3 ng/ml in those with the high risk alleles (p=0.02). Taken together, our data suggest that genetic variation in the TNFSF13B gene is significantly associated with NHL risk and elevated serum BAFF levels.
Introduction
B cell activating factor, BAFF,(1) also called BLyS,(2) is a TNF family member shown to be critical for B cell development and homeostasis (3). Overexpression of BAFF results in elevated numbers of mature B cells in the spleen and periphery and development of autoimmune-like manifestations reminiscent of systemic lupus erythematosus (4) As BAFF transgenic mice age, they develop a secondary pathology that resembles Sjögren's syndrome (5), which is associated with intense B cell activity and a predisposition to development of B cell non-Hodgkin lymphoma (NHL) (6). Together, these results implicate BAFF as a critical component of both normal and pathologic B lymphocyte biology
The similarities between the phenotype of BAFF transgenic mice and that of patients with B-cell malignancies, e.g., elevated Bcl-2 levels, apoptosis resistance, and prevalence of autoimmune disorders, raises the question of whether or not BAFF is involved in the pathogenesis of malignancy. The biologic impact of BAFF on NHL B-cells has been found to be quite similar to that seen in normal human B cells in that it co-stimulates IgM mediated proliferation and promotes survival (7-10). Both autocrine and paracrine expression of BAFF in NHL results in degradation and phosphorylation of Ik-Bα and activation of NF-κb, p50-p65 and p50-c-rel (9). Incubation of NHL B cells with BAFF also results in up-regulation of anti-apoptotic proteins, Bcl-2 and Bcl-xl, and downregulation of the anti-apoptotic protein Bax (9). Additional studies have also shown that BAFF mediated induction of NF-κb and NFAT upregulates BAFF expression resulting in a positive feedback loop that contributes NHL B cell survival (8). BAFF also enhances the survival of other malignant B cells including those from patients with chronic lymphocytic leukemia (CLL) (11, 12), multiple myeloma (MM) (13, 14), and Hodgkin lymphoma (HL) (15). Similar to what has been found in autoimmune disease, serum BAFF levels are elevated in patients with NHL and high BAFF levels correlate with patient outcome and response to therapy (10).
The finding that BAFF levels are elevated in NHL patients may lend insight as to how this disease arises and how malignant B cells maintain their growth potential. However, little is known about the mechanistic details underlying control of BAFF expression, in either the normal or malignant scenario. In previous studies, we found that BAFF levels were elevated in CLL patients with a family history of lymphoproliferative disorders and that single nucleotide polymorphisms (SNP) in the promoter of the TNFSF13B gene (−871C→T, rs9514828) resulted in increased BAFF transcription (16). Taken together these data suggest that dysregulation of BAFF may occur at the genetic level and that genetic variation in TNFSF13B may influence its expression. We therefore set out to determine if additional SNPs in TNFSF13B are associated with high serum BAFF levels and risk of developing NHL.
Methods
Study population and data collection
The Institutional Review Board at the Mayo Clinic has reviewed and approved this study. Details of this on-going clinic-based case-control study at the Mayo Clinic are available elsewhere (17). Briefly, we offered enrollment to patients with newly diagnosed (within nine months), histologically-confirmed Hodgkin and Non-Hodgkin lymphoma who were aged 20 years or older, HIV negative, and a resident of Minnesota, Iowa or Wisconsin at the time of diagnosis. A Mayo hematopathologist reviewed materials for each case and assigned a diagnosis according to the WHO classification criteria. Clinic-based controls were selected from patients visiting the Mayo Clinic for a pre-scheduled general medical exam. Eligibility requirements included no prior diagnoses of lymphoma or leukemia, HIV negative, aged 20 or older and a resident of Minnesota, Iowa or Wisconsin. Controls were randomly selected and frequency matched to our cases by 5-year age group, gender, and county of residence. All cases and controls were Caucasian. Serum samples were available from all cases and controls used in the study. Additional frozen serum specimens from untreated follicular lymphoma and DLBCL patients from Mayo Clinic who were not part of the clinic-based case-control study were also used for determination of serum BAFF levels.
Genotyping
Nine SNPs from TNFSF13B were genotyped as part of a larger genotyping project that merged data from the ParAllele (now Affymetrix) Immune-Inflammation panel (9,412 SNPs from 1,253 genes) and a custom Illumina genotyping panel (384 SNPs from 100 genes), as previously described in detail (18). Briefly, both platforms used tagging SNPs from the HapMap, covered 5 kb up and downstream of each gene, had a minor allele frequency (MAF) > 0.05, and had pairwise r2 values of 0.8 or greater. Tagged SNPs were selected using LDSelect (19). Quality controls were included in the design of each panel, including duplicate study samples and CEPH trios (18). For the Immune and Inflammation panel, the sample success rate was 98.75%, the assay call rate was 99.13%, and the concordance rate was 98.95% (based on 48 duplicates), while on the Illumina panel the sample success rate was 99.93%, the assay call rate was 93.49%, and the concordance rate was 99.99% (based on 48 CEPH duplicates and 79 study sample duplicates). A total of 916 people (475 controls and 441 cases) were genotyped in both assays and passed all quality control measures.
BAFF ELISA
ELISA plates were coated with anti-BAFF (ZymoGenetics Inc, Seattle, WA) and BAFF was detected with biotinylated anti-BAFF mAb (ZymoGenetics) followed by strepavidin-HRP and TMB substrate (10). Patient serum samples from untreated cases and controls that had been genotyped were diluted 1:5 in triplicate and BAFF levels were calculated from a standard curve generated using recombinant human BAFF (ZymoGenetics) in 20% human sera. The detection limit of purified BAFF was 300 pg/ml. Samples that fell below the detection limit were assigned a value of 0.001 ng/ml. To address inter and intra assay variability a standard curve and an internal control with a known BAFF concentration (5 ng/ml) was included on every ELISA assay plate. The mean BAFF value was 5.9 ng/ml (+/− 1.2 ng/ml, range=4.69−8.50 ng/ml) with a coefficient variance of 20% for the internal control for all assays used in this study.
Detection of BAFF splice variants
RNA was isolated from patient peripheral blood samples using TRIzol (Invitrogen). RNA from available untreated controls and cases that had been genotyped and analyzed for serum BAFF levels were used for the analysis. cDNA was generated with SuperScript® III First-Strand Synthesis SuperMix (Invitrogen). PCR amplification of BAFF was done using HotStarTaq Master Mix (Qiagen) and the following primers; 5'- TCC AGG AGA AGG CAA CTC-3' (forward) and 5'GCA GGA ATT ATT GGG TAG TGT-3' (reverse). PCR products were separated on a 3% agarose gel and visualized with ethidium bromide staining. The ratio of ΔBAFF to BAFF was calculated using densitometry with AlphaImager software (Alpha Innotech).
Statistical analysis
Allele frequencies from cases and controls were estimated using observed genotype frequencies. The frequencies in the controls were compared to allele frequencies expected under Hardy-Weinberg Equilibrium (HWE) using a Pearson goodness-of-fit test or Fisher's exact test (MAF<0.05). All SNPs were in HWE. The independent effects of the matching variables age, gender, and geographic region were examined in unconditional logistic regression models; geographic region was not significant and therefore was dropped from further consideration. We used unconditional logistic regression analysis to examine associations between each SNP and the risk of NHL, adjusting for the effects of age and gender. The most prevalent homozygous genotype was used as the reference group. Each polymorphism was modeled individually as having a log-additive effect, and odds ratios (ORs) and corresponding 95% confidence intervals (CIs) were estimated. For gene-level haplotype analyses, we used the global score test of Schaid et al (20) as implemented in the S-plus program Haplo.stats
For SNPs that had p<0.05, a composite risk variable was created that combined high-risk alleles across the three SNPs. This variable was created as follows: (1) subjects homozygous for the high-risk allele for all three SNPs were classified as highest risk; (2) those not homozygous for any of the three SNPs were classified as lowest risk; and (3) subjects with the remaining combinations were classified as intermediate risk (i.e., they are homozygous high risk allele for at least one of the SNPs, but not all). This grouping allows the highest- and lowest-risk groups to be as homogenous as possible. An OR and 95% CI were then estimated for this composite risk variable, which was modeled in a log-additive manner. Because our approach in forming this composite risk variable is data driven,(21) we obtained ORs, 95%CI, and empirical p-values through permutations. We simulated the case-control status 10000 times. Within each permutation, each SNP within the gene was analyzed and its significance was determined. Then, a high-risk variable was created in the same manner as described above using the SNPs with a p<0.05. This high-risk variable was examined in a logistic regression model and the parameter estimate and Wald p-value was recorded. Our empirical p-value is the number of permuted p-values that are more extreme than the one in the observed data. Our empirical parameter estimate is the observed log-odds ratio minus the mean of the permuted log-odds ratios, and our upper (lower) 95% CI is the observed OR divided by the 2.5th (97.5th) percentile of the permuted ORs.
Statistical comparisons of BAFF levels between patient groups were compared using Mann-Whitney tests. All analyses were implemented using SAS (SAS Institute, Cary, NC, Version 8, 1999) and S-Plus (Insightful Corp, Seattle, WA, Version 7.05, 2005) software systems. Two-sided P-values <0.05 were considered statistically significant.
Results
Genetic variation in the BAFF gene is associated with NHL Risk
Descriptive characteristics of the 441 cases and 475 matched controls from the clinic-based case-control study are shown in Table 1. We genotyped 9 tagSNPs within TNFSF13B and the tagSNPs covered 70% of the gene based on the most recent version of HapMap (Build 36, version 126) based on an r2 of 0.8 and a MAF ≥0.05. In the single SNP analysis (Table 2), 3 of the 9 tagSNPs were significant at p<0.05; rs1224141 (p=0.03), rs12583006 (p=0.01), and rs12428930 (p=0.01) when comparing all NHL cases to controls. ORs for the minor allele in the ordinal model for these SNPs ranged from 0.75 to 0.77. The global p-value for haplotype analysis of all 9 SNPs was not significant (p=0.12). As shown in Table 2, none of the SNPs were individually associated with risk of the three common histolgic subtypes, follicular lymphoma (FL), diffuse large B cell lymphoma (DLBCL), or small/chronic lymphocytic leukemia (SLL/CLL). Similarly, no association was found with the haplotype results.
Table 1.
Patient Characteristics
| Variable | Level | Controls | Cases | p-Value |
|---|---|---|---|---|
| Age category | <40 | 28 (5.9%) | 30 (6.8%) | 0.05 |
| 40−49 | 53 (11.2%) | 73 (16.6%) | ||
| 50−59 | 100 (21.1%) | 82 (18.6%) | ||
| 60−69 | 136 (28.6%) | 138 (31.3%) | ||
| 70+ | 158 (33.3%) | 118 (26.8%) | ||
| Mean age | Mean (N, SD) | 61.7 (475,12.84) | 60.1 (441,13.45) | 0.07 |
| Gender | Male | 260 (54.7%) | 255 (57.8%) | 0.35 |
| Female | 215 (45.3%) | 186 (42.2%) | ||
| Race | Caucasian | 475 (100%) | 441 (100%) | |
| Disease type | ||||
| DLBCL | 0 | 86 (19.5%) | ||
| FL | 0 | 98 (22.2%) | ||
| CLL | 0 | 123 (27.9%) | ||
| Other B-NHL | 0 | 91 (20.6%) | ||
| Other T-NHL | 0 | 23 (5.2%) | ||
| Unknown | 0 | 20 (4.5%) | ||
| Family history | 66 | 102 | 0.06 | |
| None | 136 (33.3%) | 106 (31.3%) | ||
| NHL: ICD9=202 | 12 (2.9%) | 15 (4.4%) | ||
| Leukemia: ICD9=208 | 9 (2.2%) | 19 (5.6%) | ||
| Other | 252 (61.6%) | 199 (58.7%) | ||
*Numbers presented are N (%), unless otherwise noted and p-values were generated with Chi-square tests for categorical variables or Student's t-tests for continuous variables
Table 2.
Single SNP analysis of TNFSF13B SNPs and risk of NHL and NHL subtypes
| ALL NHL (n=441) |
CLL/SLL (n=123) |
Follicular NHL (n= 113) |
DLBCL (n=69) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Position (bp) |
Gene region |
Major/Minor Alleles |
MAF control |
OR (95% CI) |
P-value | OR (95% CI) |
P-value | OR (95% CI) |
P-value | OR (95% CI) |
P-value |
| rs9514828 | 107719374 | promoter | C/T | 0.49 | 1.01 (0.84, 1.21) | 0.93 | 1.08 (0.82, 1.43) | 0.57 | 1.00 (0.75, 1.34) | 0.99 | 0.99 (0.70, 1.40) | 0.95 |
| rs8181791 | 107732046 | intron | A/G | 0.37 | 0.91 (0.75, 1.10) | 0.34 | 1.06 (0.78, 1.43) | 0.71 | 1.01 (0.74, 1.37) | 0.96 | 0.79 (0.54, 1.17) | 0.24 |
| rs1224141 | 107733724 | intron | T/G | 0.234 | 0.77 (0.61, 0.97) | 0.03 | 0.82 (0.57, 1.18) | 0.28 | 0.81 (0.56, 1.16) | 0.25 | 0.78 (0.49, 1.24) | 0.29 |
| rs12583006 | 107735453 | intron | T/A | 0.25 | 0.75 (0.60, 0.94) | 0.01 | 0.86 (0.61, 1.21) | 0.39 | 0.80 (0.55, 1.14) | 0.22 | 0.65 (0.41, 1.04) | 0.07 |
| rs12428930 | 107737706 | intron | A/C | 0.25 | 0.75 (0.60, 0.94) | 0.01 | 0.88 (0.63, 1.24) | 0.48 | 0.79 (0.55, 1.14) | 0.20 | 0.65 (0.40, 1.04) | 0.07 |
| rs10508198 | 107740789 | intron | G/C | 0.30 | 1.21 (0.99, 1.47) | 0.06 | 0.97 (0.71, 1.33) | 0.85 | 1.19 (0.88, 1.62) | 0.25 | 1.39 (0.96, 2.03) | 0.08 |
| rs1224147 | 107754759 | intron | T/C | 0.234 | 1.18 (0.96, 1.46) | 0.12 | 0.88 (0.62, 1.25) | 0.47 | 1.20 (0.87, 1.67) | 0.27 | 1.13 (0.74, 1.71) | 0.58 |
| rs9520836 | 107755064 | intron | A/G | 0.26 | 1.01 (0.82, 1.25) | 0.91 | 1.03 (0.75, 1.42) | 0.86 | 1.01 (0.72, 1.42) | 0.94 | 1.02 (0.67, 1.53) | 0.94 |
| rs7982100 | 107756366 | intron | C/T | 0.007 | 1.94 (0.77, 4.93) | 0.16 | 2.12 (0.60, 7.41) | 0.24 | 1.76 (0.45, 6.94) | 0.42 | 1.02 (0.12, 8.47) | 0.99 |
*Odds ratio and 95% confidence interval assuming log-additive genetic model adjusted for gender, and age.
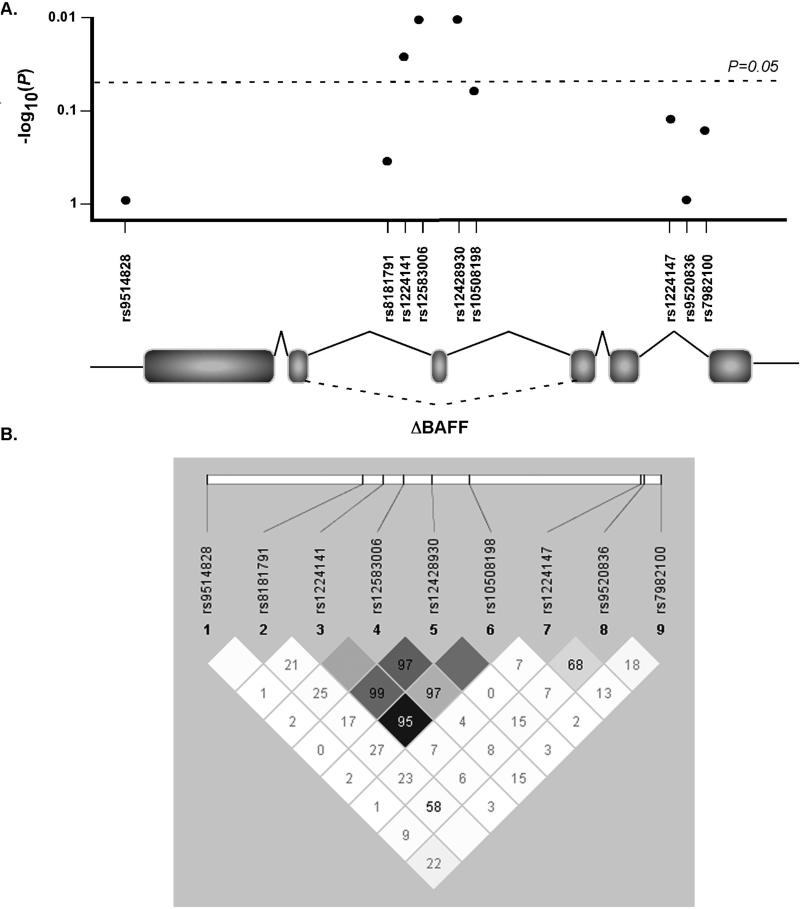
The human TNFSF13B gene encodes for 282 amino acid type II transmembrane protein, BAFF, and consists of 6 exons (Fig. 1A and B). In addition to full length BAFF, an alternate splice isoform called ΔBAFF has also been described. ΔBAFF has a 57 basepair deletion and therefore lacks exon 3. The 3 SNPs that were found to significantly correlate with lymphoma risk are in a non-coding region of the TNFSF13B gene, flank exon 3 (Fig. 1A), and are in strong linkage disequilibrium with each other (Fig 1B).
Figure 1. TNFSF13B gene structure and tagSNP mapping.
A. Three SNPs, rs1224141, rs12583006, and rs12428930 in TNFSF13B are significantly associated with NHL risk and are located in the non-coding region of TNFSF13B flanking exon 3. B. Linkage disequilibrium (LD) chart showing strong LD among rs1224141, rs12583006, and rs12428930. Numbers represent |D’| values and darker shading represents higher r^2 values of correlation between SNPs.
Combined Association of BAFF SNPs with Follicular Lymphoma and DLBCL
As a secondary analysis (Table 3), for those SNPs showing significant results, we evaluated a composite risk variable with risk of disease by classifying participants into low, moderate, and high risk groups based on the risk alleles at the significant SNPs (high risk alleles: rs1224141T, rs12583006 T , and rs12428930 A). We found a permutation-adjusted OR of 1.43 (p=0.0019) for risk of B-cell NHL. When the analysis was restricted by histologic subtype, we found that diffuse large B cell lymphoma (DLBCL) had an OR of 1.69 (p=0.0093), follicular lymphoma (FL) had an OR= 1.43 (p=0.029), and chronic/small lymphocytic leukemia (CLL/SLL) had an OR=1.06 (p=0.21). Because there was not a significant correlation of the high risk alleles with CLL/SLL, we performed an additional analysis in which we included all B-cell NHL cases excluding CLL/SLL, and the OR was 1.61 (p=0.0001). These results suggest that combined genetic variation in TNFSF13B strongly predisposes to development of both FL and DLBCL, but has little association with risk of CLL/SLL.
Table 3.
Association of high/moderate/low risk variable in BLyS SNPs rs1224141, rs12583006, and rsl 2428930 with NH
| Samples | Number of subjects | OR (95% C.I.)* | P-value* | Permutation-adjusted OR (95% CI)+ | ||
|---|---|---|---|---|---|---|
| Low Risk | Moderate Risk | High Risk | ||||
| Controls | 52 | 315 | 108 | REF | NA | NA |
| All NHL cases | 35 | 244 | 155 | 1.57 (1.25, 1.97) | 0.0001 | 1.43 (0.95, 1.54) |
| DLBCL cases | 4 | 38 | 27 | 1.90 (1.21, 3.00) | 0.006 | 1.69 (1.10, 2.07) |
| Follicular cases | 7 | 68 | 38 | 1.59 (1.10, 2.29) | 0.014 | 1.43 (0.90, 1.60) |
| CLL/SLL cases | 14 | 66 | 36 | 1.24 (0.87, 1.77) | 0.23 | 1.06 (0.68, 1.18) |
| B-cell NHL excluding CLL/SLL | 11 | 106 | 65 | 1.74 (1.33, 2.27) | 0.0001 | 1.61 (1.06, 1.74) |
Odds ratios, 95% confidence intervals, and p-values generated assuming a log-additive effect between low-moderate-high risk coding for each case type independently versus the control subjects. All analyses adjusted for region, gender, and age.
Permutation results based upon 10,000 random permutations. Adjusted OR and p-values calculated using permutation methods.
Elevated BAFF levels are associated with high risk TNFSF13B gene variants and NHL
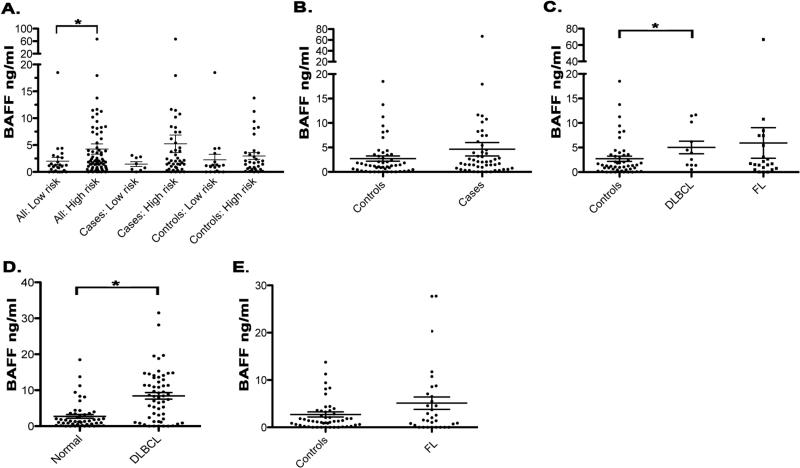
We next wanted to determine if the TNFSF13B high or low risk alleles were associated with elevated BAFF levels. Serum from cases (excluding SLL/CLL) and controls who had been genotyped and carried either the high risk or low risk alleles were tested by ELISA to determine BAFF levels (Fig. 2A). For this analysis, we used only those cases that were previously untreated at the time of serum collection, as we have shown that B-cell depleting therapies effect serum BAFF levels in NHL patients (22). The mean serum BAFF level in those individuals who carried the low risk alleles at all three SNPs was significantly lower (p=0.02) at 2 ng/ml (n=26, range: undetectable-18.5 ng/ml) compared to 4.3 ng/ml in those with the high risk alleles (n=74, range: undetectable- 66.8 ng/ml). When separated into cases and controls, the mean serum BAFF level in NHL cases who carry the low risk alleles was again lower (p=0.16) at 1.45 ng/ml (n=8, range: undetectable-3.1 ng/ml) compared to 5.2 ng/ml (n=42, range: undetectable-66.8 ng/ml) in cases with high risk alleles. In the controls, the mean serum BAFF level in those individuals who carry the low risk alleles was 2.3 ng/ml (n=18, range: undetectable-18.5 ng/ml) compared to 3.0 ng/ml (n=32, range: undetectable-13.8 ng/ml) in controls with high risk alleles (p=0.19).
Figure 2. Association of elevated serum BAFF levels with the high risk TNFSF13B gene alleles and NHL.
Serum BAFF levels were tested by ELISA as described in Materials and Methods. A. Serum from untreated cases who had been genotyped and carried either the TNFSF13B low risk alleles (n=8) or high risk alleles (n=42) and untreated controls that carried either the TNFSF13B low risk alleles (n=18) or high risk alleles (n=32) B. Serum BAFF levels in controls (n=50) versus cases (n=50). C. Serum BAFF levels by histologic subtype (DLBCL n=11, FL n=21). D Serum BAFF levels in DLBCL patients (n=57). E. Serum BAFF levels in FL patients (n=32). *p<0.05
To determine if BAFF levels were elevated in our NHL cases, serum BAFF levels (analyzed in Figure 2A) were compared to the controls (Fig. 2B). The mean seum BAFF level was lower (p=0.13) in the controls at 2.7 ng/ml (n=50, range: undetectable-18.5 ng/ml) compared to 4.63 ng/ml (n=50, range: undetectable-66.8 ng/ml) in cases. When broken down by histologic subtype we found that the mean serum BAFF level was higher, compared to controls, in patients with DLBCL (5.0 ng/ml, n=11, range: 0.5−11.7 ng/ml, p=0.02) and FL (5.9 ng/ml, n=21, range: undetectable-66.8 ng/ml, p=0.37), although not significantly (Fig. 2C). Serum BAFF levels in patients with marginal zone B-cell lymphoma (4.3 ng/ml, n=9, range: undetectable-18.0 ng/ml, p=0.26), mantle cell lymphoma (1.6 ng/ml, n=6, range: undetectable-4.0 ng/ml, p=0.6), lymphoplasmacytic lymphoma (0.3 ng/ml, n=1), or other B-cell NHLs (1.8 ng/ml, n=2, range: 0.8−2.8 ng/ml) were similar to or less than that seen in the controls, although the numbers of specimens were small.
To confirm our findings in FL and DLBCL we analyzed BAFF levels in an independent set of serum samples collected from previously untreated DLBCL (Fig. 2D) and FL (Fig. 2E) patients and matched controls. Compared to controls, BAFF levels were significantly elevated in patients with DLBCL (8.43 ng/ml, n=57, range: undetectable-31.5 ng/ml, p<0.0001). In the FL patients, we also saw higher BAFF levels, although as seen in Fig. 2C, this difference was not statistically significant (5.1 ng/ml, n=32, range: undetectable-27.73 ng/ml, p=0.4). Taken together, these results suggest that serum BAFF levels are elevated in NHL patients, particularly DLBCL, and that indviduals who carry the TNFSF13B high risk alleles have higher BAFF levels compared to those with low risk alleles.
High risk TNFSF13B gene alleles do not influence BAFF alternate splicing
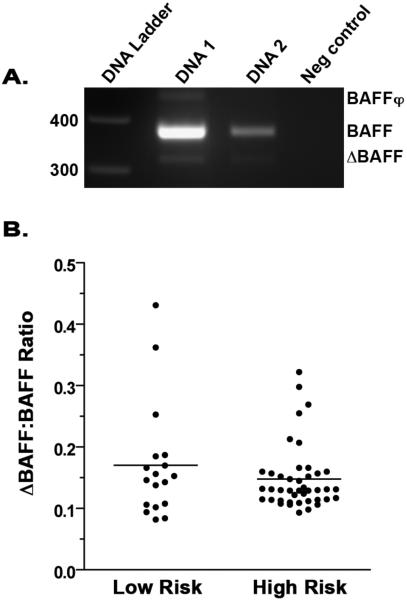
The high risk SNPs that are associated with NHL risk are not located in a consensus regulatory region of the BAFF gene, but they do flank exon 3, which can be alternately spliced to encode for ΔBAFF. Functionally this may be significant, as ΔBAFF has been reported to homerdimerize with BAFF and oppose its activity (23, 24). To determine if there were differences in ΔBAFF mRNA levels in individuals who carry the high risk versus low risk alleles, we employed a PCR approach. Using available matched specimens from the genotyping and serum BAFF ELISA studies, we isolated mRNA from PBMCs for amplification. Primers were designed to amplfy both full-length BAFF as well as ΔBAFF (Fig. 3) ΔBAFF levels were quantified and normalized to full-length BAFF. Our results suggest that ΔBAFF levels were not found to be significantly different (p=0.5) between individuals who carried the high risk (n=42, mean=0.148) versus the low risk alleles (n=18, mean =0.173). We also measured the level of BAFFφ mRNA expression, a BAFF transcript identified in human cell lines that is nonfunctional due to incomplete splicing of intronic sequences leading to premature stop codons (23), and saw no significant differences in expression. Taken together, these data suggest that the high risk alleles associated with NHL risk do not influence BAFF alternate splicing and ΔBAFF mRNA levels.
Figure 3. Expression of ΔBAFF is not associated with the high risk TNFSF13B gene alleles.
A. Expression of BAFF, ΔBAFF, and BAFFφ mRNA was determined by RT-PCR. Two DNA specimens and a no DNA template control are shown. B. ΔBAFF mRNA levels in individuals (untreated cases and controls) who carry the high risk alleles (n=42) versus low risk alleles (n=18) were determined by PCR. Primers were designed to amplify full-length BAFF as well as ΔBAFF and data were quantified and normalized to full-length BAFF as described in Materials and Methods. Values represent the ratio of the densitometric value of ΔBAFF/BAFF.
Discussion
Early results from association studies (25-33) show a promising role for common genetic alleles in immune and inflammatory response genes in NHL etiology. Our studies further contribute to these findings and suggest that genetic variation in the TNFSF13B gene is associated with NHL overall, and FL and DLBCL in particular. To date, the strongest etiology finding is from the InterLymph consortium (33), a pooled analysis of 3586 cases of NHL and 4018 controls (representing 8 studies), which reported that the tumor necrosis factor (TNF) −308G→A polymorphism was associated with increased NHL risk, particularly for DLBCL. Additionally, the interleukin 10 (IL-10) 3575T→A and the lymphotoxin-α (LTA) 252A→G polymorphisms were also found to be associated with increased risk, particularly for DLBCL. A role for immune SNPs has been further supported by the finding from Cerhan et al (17) showing that genetic variation in genes associated with the immune response, mitogen-activated protein kinase signaling, lymphocyte trafficking and migration, and coagulation pathways are important in the etiology of NHL. Previous to our current study, we have shown that the presence of a SNP in the promoter of the TNFSF13B gene (−871C→T, rs9514827) resulted in increased BAFF transcription and that the −871T allele was more prevalent in familial CLL patients (16). Analysis of the −871C→T SNP in SLE (34), Sjogren's Syndrome (35), and idiopathic thrombocytopenic purpura (36) also found that this SNP correlated with elevated BAFF levels, however, it did not correlate with disease development. In our current study this same SNP was not found to significantly associate with NHL risk (Table 2) nor with CLL subtype. This lack of finding may be due to the small number of our subjects reporting a family history of leukemia (n=19) or a family history of NHL (n=15), and it remains to be explored whether or not this will hold true in patients with familial NHL. However, our current studies clearly suggest that additional TNFSF13B SNPs contribute to the development of FL and DLBCL and further support the idea that genetic variation in genes involved in inflammation is associated with NHL risk.
In addition to correlating with NHL etiology, the high risk TNFSF13B gene alleles are also associated with elevated serum BAFF levels (Fig. 2). This finding suggests that SNPs in TNFSF13B may contribute to dysregulated BAFF expression. Confirming previous studies in DLBCL (10) we also show that serum BAFF levels are elevated in a clinic-based study of NHL patients and in an independent set of DLBLC patients. Previous reports have shown that BAFF levels are elevated in FL (7), and while we did not see a statistically significant increase in serum BAFF levels in the latter group of FL patients, there was a subset of FL patients that did have elevated BAFF. In support of our findings, numerous groups have measured serum BAFF levels and it has been found to be elevated in patients with NHL (10, 37, 38), CLL (11, 16), and MM (14). The mechanisms that control the homeostatic level of BAFF are not defined, but they may directly affect the number and composition of the of B cell pool. In support of this, Lesley et al showed in a murine system that elevated BAFF levels resulted in rescue of auto-antigen engaged B cells from elimination, predisposing to autoimmune disease (39). This data suggests that alterations in BAFF levels can have significant effects on B cells, and in addition to autoimmune disorders, may contribute to the development of malignant disease. In mouse models, transgenic overexpression of BAFF results in elevated numbers of mature B cells and development of autoimmune disorders, but these mice do not develop B cell tumors (4). However, when BAFF transgenic mice are crossed with TNF−/− mice, 35% of the mice develop B cell lymphoma (40). This suggests that BAFF alone may not be oncogenic, rather, it may work in combination with other inflammatory cytokines to promote development of B cell tumors. It will be of interest to further explore whether genetic variation in the TNFSF13B gene in combination with variation in TNF, IL-10, or LTA will have compounding effects on NHL risk.
Characterization of the functional significance of non-coding SNPs has proven to be technically challenging (41). Because the high risk allele SNPs we identified as being risk factors for NHL were located in a non-coding region of the BAFF gene it was unlikely that they would directly influence BAFF mRNA sequence or function. However, their location within the BAFF gene, flanking exon 3, was intriguing in light of recent reports suggesting a role for the alternate BAFF splice product ΔBAFF. Transgenic overexpression of ΔBAFF results in reduced B cell numbers and T-dependent antibody responses, opposite to what has been observed in BAFF transgenic mice. Initially we hypothesized that the high risk BAFF gene alleles were associated with alternate splicing of BAFF resulting in reduced expression of ΔBAFF. However, our data suggest that there was little variation in ΔBAFF levels between individuals who carried the low or high risk alleles (Fig. 3). Then again, because our PCR experiments amplified BAFF and ΔBAFF from whole blood, our results do not fully exclude the possibility that there is variability in ΔBAFF expression in lymphocyte subsets or in tumor cells. Ideally, a ΔBAFF specific ELISA could be used to determine if this alternate form of BAFF is expressed in the serum of patients and if its levels are varied in individuals who carry the TNFSF13B gene risk alleles, however this is currently not feasible. Another potential hypothesis is that the SNPs of interest are located in an miRNA coding sequence and could therefore influence RNA or protein expression. However, based on data from the Sanger Institute miRBase (http://microrna.sanger.ac.uk/) (42), no known miRNAs are found in this region if the TNFSF13B gene.
Our approach to identifying associations between the TNFSF13B gene and NHL risk was an indirect approach using tagSNPs. While this method is extremely useful for initial identification of genes associated with disease risk, tagSNPs may not alter protein expression or function. As our results have shown, it is likely that the SNPs identified in this study either indirectly effect expression of TNFSF13B or they are linked to additional SNPs in the region that may have a regulatory function. In ongoing studies, we are resequencing the human TNFSF13B gene in hopes of identifying and characterizing novel SNPs that are involved in BAFF regulation and NHL risk.
Although these studies highlight the significance of BAFF in NHL, it will also be of interest to determine if genetic variation or altered expression of BAFF receptors is associated with NHL risk. Three receptors, B cell maturation antigen (BCMA, TNFRSF17) (43), transmembrane activator and CAML interactor (TACI, TNFRSF13B) (44), and BAFF-R (TNFRSF13C) (45) have been identified as receptors for BAFF and are predominantly expressed on B cells. The expression profile of BAFF receptors on NHL B cells typically mirrors that of their normal B cell counter-part, however a large-scale analysis of their expression and SNP content in NHL is yet to be performed. Initially we observed that NHL B cells express BAFF-R, and have variable expression of TACI and BCMA (10). Deregulated expression of BAFF receptors combined with high expression of BAFF may have compounding effects and clinical significance. Moreaux et al has recently shown, using a hierarchical clustering of gene expression profile from 65 multiple myeloma patients, that the level of TACI gene expression is associated with a specific gene expression profile; TACIhi myeloma cells displayed a mature plasma cell gene signature while the TACIlo group had a gene signature of plasmablasts (46). Additionally two groups have recently identified mutations in TACI as an important factor in common variable immunodeficiency (CVID) and IgA deficiency (IgAD), both of which predispose to development of lymphoma (47, 48). Continued genetic analysis of BAFF and its receptors will hopefully lend further insight into their role in development of NHL.
There are several strengths to our study, including the careful characterization of cases and controls, central pathology review, and extensive genotyping quality controls. We have previously evaluated the potential for population stratification in our study population, and found this is not likely (49). Limitations of our study include the relatively small sample size for NHL subtypes, and an all white population, limiting generalizablity. While our study was not population-based, by restricting to regional cases and controls (i.e., Minnesota, Iowa and Wisconsin), we decrease referral bias and ensure that our cases and controls are derived from the same underlying population. Finally, we cannot rule out that our findings are false positives. Although only nine SNPs were evaluated, we did not correct for multiple testing. As such further replication will be needed to validate our findings.
In summary, we have found that genetic variation in the TNFSF13B gene is significantly associated with an increased risk of developing B-cell NHL, particularly FL and DLBCL. Additionally, the increased lymphoma risk is associated with an increase in serum BAFF levels. Taken together, our data further highlight the role of the TNFSF13B gene in B cell malignancies. Further characterization of the precise genetic and environmental mechanisms that control BAFF expression will hopefully provide insight into how this gene is regulated or dysregulated in both the normal and malignant scenario.
Funding
This work was supported in part by; The National Institutes of Health [CA097274 to J.C., CA92153 to J.C., and CA121569 to S.A.], The Leukemia and Lymphoma Society [S.A] and ZymoGenetics Inc and Merck Serono International S.A., an affiliate of Merck KGaA, Darmstadt, Germany. Lymphoma Research Foundation Follicular Lymphoma Research Grantee [A.N.]
References
- 1.Schneider P, MacKay F, Steiner V, et al. BAFF, a novel ligand of the tumor necrosis factor family, stimulates B cell growth. J Exp Med. 1999;189:1747–1756. doi: 10.1084/jem.189.11.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moore PA, Belvedere O, Orr A, et al. BLyS: member of the tumor necrosis factor family and B lymphocyte stimulator. Science. 1999;285:260–263. doi: 10.1126/science.285.5425.260. [DOI] [PubMed] [Google Scholar]
- 3.Thompson JS, Schneider P, Kalled SL, et al. BAFF binds to the tumor necrosis factor receptor-like molecule B cell maturation antigen and is important for maintaining the peripheral B cell population. J Exp Med. 2000;192:129–135. doi: 10.1084/jem.192.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mackay F, Woodcock SA, Lawton P, et al. Mice transgenic for BAFF develop lymphocytic disorders along with autoimmune manifestations. J Exp Med. 1999;190:1697–1710. doi: 10.1084/jem.190.11.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Groom J, Kalled SL, Cutler AH, et al. Association of BAFF/BLyS overexpression and altered B cell differentiation with Sjogren's syndrome. J Clin Invest. 2002;109:59–68. doi: 10.1172/JCI14121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zulman J, Jaffe R, Talal N. Evidence that the malignant lymphoma of Sjogren's syndrome is a monoclonal B-cell neoplasm. N Engl J Med. 1978;299:1215–1220. doi: 10.1056/NEJM197811302992204. [DOI] [PubMed] [Google Scholar]
- 7.Briones J, Timmerman JM, Hilbert DM, Levy R. BLyS and BLyS receptor expression in non-Hodgkin's lymphoma. Exp Hematol. 2002;30:135–141. doi: 10.1016/s0301-472x(01)00774-3. [DOI] [PubMed] [Google Scholar]
- 8.Fu L, Lin-Lee YC, Pham LV, et al. Constitutive NF-{kappa}B and NFAT Activation Leads to Stimulation of The BLyS Survival Pathway in Aggressive B Cell Lymphomas. Blood. 2006;107:4540–4548. doi: 10.1182/blood-2005-10-4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.He B, Chadburn A, Jou E, et al. Lymphoma B cells evade apoptosis through the TNF family members BAFF/BLyS and APRIL. J Immunol. 2004;172:3268–3279. doi: 10.4049/jimmunol.172.5.3268. [DOI] [PubMed] [Google Scholar]
- 10.Novak AJ, Grote DM, Stenson M, et al. Expression of BLyS and its receptors in B-cell non-Hodgkin lymphoma: correlation with disease activity and patient outcome. Blood. 2004;104:2247–2253. doi: 10.1182/blood-2004-02-0762. [DOI] [PubMed] [Google Scholar]
- 11.Kern C, Cornuel JF, Billard C, et al. Involvement of BAFF and APRIL in the resistance to apoptosis of B-CLL through an autocrine pathway. Blood. 2004;103:679–688. doi: 10.1182/blood-2003-02-0540. [DOI] [PubMed] [Google Scholar]
- 12.Novak AJ, Bram RJ, Kay NE, Jelinek DF. Aberrant expression of B-lymphocyte stimulator by B chronic lymphocytic leukemia cells: a mechanism for survival. Blood. 2002;100:2973–2979. doi: 10.1182/blood-2002-02-0558. [DOI] [PubMed] [Google Scholar]
- 13.Novak AJ, Darce JR, Arendt BK, et al. Expression of BCMA, TACI, and BAFF-R in multiple myeloma: a mechanism for growth and survival. Blood. 2004;103:689–694. doi: 10.1182/blood-2003-06-2043. [DOI] [PubMed] [Google Scholar]
- 14.Moreaux J, Legouffe E, Jourdan E, et al. BAFF and APRIL protect myeloma cells from apoptosis induced by interleukin 6 deprivation and dexamethasone. Blood. 2004;103:3148–3157. doi: 10.1182/blood-2003-06-1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chiu A, Xu W, He B, et al. Hodgkin lymphoma cells express TACI and BCMA receptors and generate survival and proliferation signals in response to BAFF and APRIL. Blood. 2007;109:729–739. doi: 10.1182/blood-2006-04-015958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Novak AJ, Grote DM, Ziesmer SC, et al. Elevated Serum B-Lymphocyte Stimulator Levels in Patients With Familial Lymphoproliferative Disorders. J Clin Oncol. 2006;24:983–987. doi: 10.1200/JCO.2005.02.7938. [DOI] [PubMed] [Google Scholar]
- 17.Cerhan JR, Ansell SM, Fredericksen ZS, et al. Genetic variation in 1253 immune and inflammation genes and risk of non-Hodgkin lymphoma. Blood. 2007;110:4455–4463. doi: 10.1182/blood-2007-05-088682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cerhan JR, Liu-Mares W, Fredericksen ZS, et al. Genetic variation in tumor necrosis factor and the nuclear factor-kappaB canonical pathway and risk of non-Hodgkin's lymphoma. Cancer Epidemiol Biomarkers Prev. 2008;17:3161–3169. doi: 10.1158/1055-9965.EPI-08-0536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carlson CS, Eberle MA, Rieder MJ, et al. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am J Hum Genet. 2004;74:106–120. doi: 10.1086/381000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schaid DJ, Rowland CM, Tines DE, Jacobson RM, Poland GA. Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am J Hum Genet. 2002;70:425–434. doi: 10.1086/338688. Epub 2001 Dec 2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee WC, Huang HY. Data-dredging gene-dose analyses in association studies: biases and their corrections. Cancer Epidemiol Biomarkers Prev. 2005;14:3004–3006. doi: 10.1158/1055-9965.EPI-05-0605. [DOI] [PubMed] [Google Scholar]
- 22.Ansell SM, Novak AJ, Ziesmer S, et al. Serum Cytokine Levels Predict Time to Progression after Rituximab as Initial Therapy in Patients with Follicular Grade 1 Non-Hodgkin Lymphoma. ASH Annual Meeting Abstracts. 2007;118:999A. [Google Scholar]
- 23.Gavin AL, Ait-Azzouzene D, Ware CF, Nemazee D. DeltaBAFF, an alternate splice isoform that regulates receptor binding and biopresentation of the B cell survival cytokine, BAFF. J Biol Chem. 2003;278:38220–38228. doi: 10.1074/jbc.M306852200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gavin AL, Duong B, Skog P, et al. deltaBAFF, a splice isoform of BAFF, opposes full-length BAFF activity in vivo in transgenic mouse models. J Immunol. 2005;175:319–328. doi: 10.4049/jimmunol.175.1.319. [DOI] [PubMed] [Google Scholar]
- 25.Lan Q, Zheng T, Rothman N, et al. Cytokine polymorphisms in the Th1/Th2 pathway and susceptibility to non-Hodgkin lymphoma. Blood. 2006;107:4101–4108. doi: 10.1182/blood-2005-10-4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Monne M, Piras G, Palmas A, et al. Cytotoxic T-lymphocyte antigen-4 (CTLA-4) gene polymorphism and susceptibility to non-Hodgkin's lymphoma. Am J Hematol. 2004;76:14–18. doi: 10.1002/ajh.20045. [DOI] [PubMed] [Google Scholar]
- 27.Lech-Maranda E, Baseggio L, Bienvenu J, et al. Interleukin-10 gene promoter polymorphisms influence the clinical outcome of diffuse large B-cell lymphoma. Blood. 2004;103:3529–3534. doi: 10.1182/blood-2003-06-1850. [DOI] [PubMed] [Google Scholar]
- 28.Demeter J, Porzsolt F, Ramisch S, et al. Polymorphism of the tumour necrosis factor-alpha and lymphotoxin-alpha genes in chronic lymphocytic leukaemia. Br J Haematol. 1997;97:107–112. doi: 10.1046/j.1365-2141.1997.9912636.x. [DOI] [PubMed] [Google Scholar]
- 29.Chouchane L, Ahmed SB, Baccouche S, Remadi S. Polymorphism in the tumor necrosis factor-alpha promotor region and in the heat shock protein 70 genes associated with malignant tumors. Cancer. 1997;80:1489–1496. doi: 10.1002/(sici)1097-0142(19971015)80:8<1489::aid-cncr17>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 30.Warzocha K, Ribeiro P, Bienvenu J, et al. Genetic polymorphisms in the tumor necrosis factor locus influence non-Hodgkin's lymphoma outcome. Blood. 1998;91:3574–3581. [PubMed] [Google Scholar]
- 31.Juszczynski P, Kalinka E, Bienvenu J, et al. Human leukocyte antigens class II and tumor necrosis factor genetic polymorphisms are independent predictors of non-Hodgkin lymphoma outcome. Blood. 2002;100:3037–3040. doi: 10.1182/blood-2002-02-0654. [DOI] [PubMed] [Google Scholar]
- 32.Mainou-Fowler T, Dickinson AM, Taylor PR, et al. Tumour necrosis factor gene polymorphisms in lymphoproliferative disease. Leuk Lymphoma. 2000;38:547–552. doi: 10.3109/10428190009059274. [DOI] [PubMed] [Google Scholar]
- 33.Rothman N, Skibola CF, Wang SS, et al. Genetic variation in TNF and IL10 and risk of non-Hodgkin lymphoma: a report from the InterLymph Consortium. Lancet Oncol. 2006;7:27–38. doi: 10.1016/S1470-2045(05)70434-4. [DOI] [PubMed] [Google Scholar]
- 34.Kawasaki A, Tsuchiya N, Fukazawa T, Hashimoto H, Tokunaga K. Analysis on the association of human BLYS (BAFF, TNFSF13B) polymorphisms with systemic lupus erythematosus and rheumatoid arthritis. Genes Immun. 2002;3:424–429. doi: 10.1038/sj.gene.6363923. [DOI] [PubMed] [Google Scholar]
- 35.Gottenberg JE, Sellam J, Ittah M, et al. No evidence for an association between the −871 T/C promoter polymorphism in the B-cell-activating factor gene and primary Sjogren's syndrome. Arthritis Res Ther. 2006;8:R30. doi: 10.1186/ar1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Emmerich F, Bal G, Barakat A, et al. High-level serum B-cell activating factor and promoter polymorphisms in patients with idiopathic thrombocytopenic purpura. Br J Haematol. 2007;136:309–314. doi: 10.1111/j.1365-2141.2006.06431.x. [DOI] [PubMed] [Google Scholar]
- 37.Briones J, Timmerman JM, Panicalli DL, Levy R. Antitumor immunity after vaccination with B lymphoma cells overexpressing a triad of costimulatory molecules. J Natl Cancer Inst. 2003;95:548–555. doi: 10.1093/jnci/95.7.548. [DOI] [PubMed] [Google Scholar]
- 38.Oki Y, Georgakis GV, Migone TS, Kwak LW, Younes A. Elevated serum BLyS levels in patients with non-Hodgkin lymphoma. Leuk Lymphoma. 2007;48:1869–1871. doi: 10.1080/10428190701549562. [DOI] [PubMed] [Google Scholar]
- 39.Lesley R, Xu Y, Kalled SL, et al. Reduced competitiveness of autoantigen-engaged B cells due to increased dependence on BAFF. Immunity. 2004;20:441–453. doi: 10.1016/s1074-7613(04)00079-2. [DOI] [PubMed] [Google Scholar]
- 40.Batten M, Fletcher C, Ng LG, et al. TNF deficiency fails to protect BAFF transgenic mice against autoimmunity and reveals a predisposition to B cell lymphoma. J Immunol. 2004;172:812–822. doi: 10.4049/jimmunol.172.2.812. [DOI] [PubMed] [Google Scholar]
- 41.Hudson TJ. Wanted: regulatory SNPs. Nat Genet. 2003;33:439–440. doi: 10.1038/ng0403-439. [DOI] [PubMed] [Google Scholar]
- 42.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Madry C, Laabi Y, Callebaut I, et al. The characterization of murine BCMA gene defines it as a new member of the tumor necrosis factor receptor superfamily. Int Immunol. 1998;10:1693–1702. doi: 10.1093/intimm/10.11.1693. [DOI] [PubMed] [Google Scholar]
- 44.von Bulow GU, Russell H, Copeland NG, et al. Molecular cloning and functional characterization of murine transmembrane activator and CAML interactor (TACI) with chromosomal localization in human and mouse. Mamm Genome. 2000;11:628–632. doi: 10.1007/s003350010125. [DOI] [PubMed] [Google Scholar]
- 45.Thompson JS, Bixler SA, Qian F, et al. BAFF-R, a newly identified TNF receptor that specifically interacts with BAFF. Science. 2001;293:2108–2111. doi: 10.1126/science.1061965. [DOI] [PubMed] [Google Scholar]
- 46.Moreaux J, Cremer FW, Reme T, et al. The level of TACI gene expression in myeloma cells is associated with a signature of microenvironment dependence versus a plasmablastic signature. Blood. 2005;106:1021–1030. doi: 10.1182/blood-2004-11-4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Castigli E, Wilson SA, Garibyan L, et al. TACI is mutant in common variable immunodeficiency and IgA deficiency. Nat Genet. 2005;37:829–834. doi: 10.1038/ng1601. [DOI] [PubMed] [Google Scholar]
- 48.Salzer U, Chapel HM, Webster AD, et al. Mutations in TNFRSF13B encoding TACI are associated with common variable immunodeficiency in humans. Nat Genet. 2005;37:820–828. doi: 10.1038/ng1600. [DOI] [PubMed] [Google Scholar]
- 49.Cerhan JR, Wang S, Maurer MJ, et al. Prognostic significance of host immune gene polymorphisms in follicular lymphoma survival. Blood. 2007;109:5439–5446. doi: 10.1182/blood-2006-11-058040. [DOI] [PMC free article] [PubMed] [Google Scholar]