Abstract
We have generated clones (L2.3 and RG3.6) of neural progenitors with radial glial properties from rat E14.5 cortex that differentiate into astrocytes, neurons and oligodendrocytes. Here we describe a different clone (L2.2) that gives rise exclusively to neurons, but not to glia. Neuronal differentiation of L2.2 cells was inhibited by BMP2 and enhanced by SHH similar to cortical interneuron progenitors. Compared to L2.3, differentiating L2.2 cells expressed significantly higher levels of mRNAs for glutamate decarboxylases (GAD), DLX transcription factors, calretinin, calbindin, neuropeptide Y (NPY) and somatostatin. Increased levels of DLX-2, GAD and calretinin proteins were confirmed upon differentiation. L2.2 cells differentiated into neurons that fired action potentials in vitro, and their electrophysiological differentiation was accelerated and more complete when co-cultured with developing astroglial cells but not with conditioned medium from these cells. The combined results suggest that clone L2.2 resembles GABAergic interneuron progenitors in the developing forebrain.
Keywords: neuronal progenitor, v-myc, L2.2, RG3.6, GABA
INTRODUCTION
Multipotent neural stem/progenitor cells (NSPC) undergo lineage restrictions before they become mature cell types in the central nervous system (CNS) (Lu et al., 2002; Rowitch et al., 2002; Noble et al., 2004). CNS radial glial cells (RG) are NSPC (Hartfuss et al., 2001; Gotz et al., 2002; Noctor et al., 2002) that acquire glial restricted precursor (GRP) marker A2B5/4D4 or neuronal restricted precursor (NRP) marker 5A5/PSA-NCAM both in vivo and in vitro as they transition to different lineage-restricted precursors (RP) (Li et al., 2004). These RP markers have been used to isolate antigenically-defined populations of cells from developing CNS that have different capacities for differentiation (Mayer-Proschel et al., 1997; Rao and Mayer-Proschel, 1997; Rao et al., 1998). RP may be particularly advantageous for transplantation since they can migrate widely into tissues before they differentiate into mature cells that become relatively immobile. However, cells that behave as NSPC in culture, giving rise to neurons, astrocytes and oligodendrocytes, exhibit more limited differentiation in certain tissues such as the adult spinal cord where they give rise mainly to glia but few neurons (Cao et al., 2001; Han et al., 2004; Hasegawa et al., 2005). Nevertheless, NRP cells maintain their lineage restriction after transplantation into adult spinal cord in that only neuronal phenotypes were observed after 4 weeks in vivo (Han et al., 2002).
Because NSPC and RP exhibit heterogeneities and have been difficult to stabilize in culture, retroviral vectors containing oncogenes have been used to immortalize immature CNS cells (Cepko, 1988; Cepko, 1989). Non-transforming oncogenes have been used to immortalize primary CNS NSPC cells from mouse (Bernard et al., 1989; Ryder et al., 1990; Frisa et al., 1994), rat (Frederiksen et al., 1988) and human (Villa et al., 2000; De Filippis et al., 2007). Most of these cell lines can differentiate into neurons and glia but one clone differentiates predominantly into glia and may represent a GRP, however, it can give rise to some neurons (Wu et al., 2002). Although NRP cells have been described (Noble et al., 2003), to our knowledge, only one cloned NRP that differentiates exclusively into CNS neurons has been reported (Bosch et al., 2004), but it has not been well characterized molecularly.
We immortalized NSPC from rat embryonic cortices with v-myc and obtained clones that proliferate in serum-free medium containing FGF2. Clones L2.3 (Li et al., 2004) and RG3.6 (Hasegawa et al., 2005) isolated from rat E14.5 cortex display properties of radial glia and NSPC that differentiate into astrocytes, neurons and oligodendrocytes. Here we describe in detail a different clone, L2.2, which proliferates in defined medium with FGF2, and upon withdrawal of FGF2 differentiates exclusively into neurons but not into astrocytes (GFAP+) or oligodendrocytes (GalC+). L2.2 cells and their differentiated progenies express cortical interneuron markers including DLX transcription factors and GADs. In co-culture with GFP-labeled radial glial RG3.6 cells, L2.2 cells developed mature action potentials much faster than L2.2 cells alone. Therefore, L2.2 represents an immortalized NRP clone derived from rat developing forebrain that gives rise to GABAergic neurons in a process that can be modulated by astroglia.
METHODS
Cell culture, differentiation and factor treatments
Generation of precursor clones (L2.2 and L2.3) from embryonic rat cortical cultures and their culturing conditions were described previously (Li et al., 2004). Briefly, E14.5 cortices were dissected without meningeal membranes. The mechanically dissociated cells were cultured as neurospheres for 3 to 4 days in DMEM/F12 (Invitrogen) supplemented with 25 mM glucose (Sigma), 2 mM glutamine (Invitrogen), penicillin/streptomycin (Invitrogen), 10 ng/ml FGF2 (BD Biosciences), 2 µg/ml heparin (Sigma) and 1× B27 (Invitrogen). FGF2 was refreshed daily in these cultures to promote proliferation. The neurospheres were then passaged by mild trypsinization (0.025% for 5 min) and cultured as adherent cells on laminin-coated dishes in the presence of FGF2 (10 ng/ml) and leukemia inhibitory factor (LIF, 10 ng/ml, Chemicon) for 2 days before infected with PK-VM-2 retrovirus contain v-myc (Villa et al., 2000). For immortalization, the cells were then incubated with 5 ml of serum-free infection medium including 50% viral supernatant, 25% fresh DMEM/F12 medium and 25% neurosphere-conditioned medium with 8 µg/ml polybrene (Sigma) for 8 h. This infection procedure was repeated twice during a 24-h period. Infected cells were then selected by their resistance to 200 µg/ml G418 (Invitrogen). After 4–5 days in selection, individual colonies were expanded and immunostained. Colonies contained BLBP+ cells with a polarized morphology and epithelioid-shaped cells that were BLBP−. Several colonies including L2 were recloned by limiting dilution, yielding clones that were BLBP+ (e.g. L2.3) and others that were BLBP− (e.g. L2.2). Two cell differentiation protocols were followed in this study. The first protocol has minor modifications from that described previously (Li et al., 2004). Briefly, immortalized clones (e.g. L2.2, L2.3) were cultured overnight on laminin-coated substrates in FGF2 containing DMEM/F12 culture medium (described above), the medium was then removed and replaced with DMEM/F12 culture medium lacking FGF2 (DMEM/F12). This is a default differentiation protocol for most experiments performed in this study unless stated otherwise. In some experiments, 0.5% fetal bovine serum (FBS) was added to promote cell survival as indicated. The second protocol utilized media containing N2 supplement (N2B27) (Ying et al., 2003), which promotes neuronal cell survival and differentiation. We used N2B27 differentiation medium for measuring neuronal action potentials. For factor treatment experiments, we added BMP2 (25 ng/ml, R&D, human recombinant), LIF (10 ng/ml, Chemicon, ESGRO), or SHH agonist (100 nM, Curis, Cur-0199567) to DMEM/F12 differentiation medium. After maintenance for the number of days indicated, cultures were then fixed and stained with cell type specific markers.
Gene expression analysis
L2.2 and L2.3 cells were cultured on laminin-coated 35 mm dishes in DMEM/F12 serum free medium containing FGF2 (10 ng/ml) at 3×105 cells per dish. The next day, differentiation was initiated by changing to medium lacking FGF2 and including 0.5% FBS. Triplicate cultures were harvested at day 0 (prior to FGF2 withdrawal), and 1 or 3 after differentiation. RNA was prepared from L2.2 and L2.3 cultures using the mirVana miRNA Isolation kit (Ambion/Applied Biosystems), which isolates and separates low molecular weight (LMW) from high molecular weight (HMW) RNA. 0.5 µg of HMW RNA was labeled using the NanoAmp™ RT-IVT Labeling Kit (Applied Biosystems) and hybridized to AB 1700 Rat Genome Survey Microarrays following the manufacturer’s protocols.
Array data were quality-assessed, aggregated, quantile-normalized, and analyzed using the ABarray Package for R (http://www.r-project.org/) and Bioconductor (http://www.bioconductor.org). Probes exhibiting a signal to noise ratio (S/N) < 3 were excluded from further analysis. A two-way ANOVA was performed on remaining probes using cell clone and time as factors. Significant probes were determined to have an acceptable FDR of 5% using the Benjamini-Hochberg method. Significant probes were k-means clustered (k=6) to identify similar expression patterns, as we described previously (Pan et al., 2004). Cluster centers, along with the hierarchically clustered heatmap, were plotted using R. Gene-level interpretation of probe data was determined using annotation previously described (Goff et al., 2007).
Magnetic Bead Cell Separation
Ventral forebrain tissue was obtained from day 14 rat embryos isolated from timed-pregnant Sprague Dawley rats (Hilltop Lab Animals, Inc). Each tissue sample (n=3; each from a separate litter) was pooled from nine embryos. After dissociation, cells were labeled with A2B5 antibody and separated from other cells following the MACS protocol (Miltenyi Biotecs) using anti-immunoglobulin (IgM) microbeads. The negative fraction (A2B5−) was then labeled with monoclonal 5A5 antibody and separated from other cells following the MACS protocol. The cells retained in the column were eluted as the A2B5−/5A5+ fraction. To verify that both the A2B5+ and A2B5−/5A5+ fractions were positive for these cell surface markers, we counterstained a portion of each fraction with goat anti-mouse FITC (Molecular Probes, Invitrogen). Immunocytochemistry of both negative and positive fractions for A2B5 and 5A5 displayed separation of the A2B5+ and 5A5+ fractions from other cell types. The remaining portion of each fraction was saved for further analysis. HMW RNA was isolated from A2B5+ cells and A2B5−/5A5+ cells using Qiagen’s miRNeasy Mini Kit.
Quantitative RT-PCR (qPCR)
1 µg of HMW RNA was reverse-transcribed into cDNA using oligo-dT primer and SuperScript II reverse transcriptase (Invitrogen). The qPCR reactions were carried out on an ABI PRISM 7900HT Sequence Detection System (Applied Biosystems) as described previously (Li et al., 2003). GAPDH was used to normalize the expression levels of each sample. Primers for detecting genes are listed in Table I.
Table 1.
Primers for qPCR analysis
| Gene names | Forward | Reverse |
|---|---|---|
| BLBP/Fabp7 | GGAAGCTGACAGACAGCCAGA | CGCCCAGAGCCTTCATGTAC |
| nestin | CAAGCAGCAGGGTCACTTCC | AGGTTTGTGGCTAAGGAGGTCA |
| GAD-1 | GGGTGTGCTGCTCCAGTGTT | GCTTGTCTGGCTGGAAGAGG |
| GAD-2 | CTCCAACATGTACGCCATGC | CTGACGTGAATGCGATGAGC |
| DLX-1 | CCTACGTCCCCAGCTACACG | GAAGCGGGTGAGTGCGAA |
| DLX-2 | CATGGGCTCCTACCAGTACCA | CGTAGGAAGTGTACGCGGC |
| DLX-5 | GGAGAACTCGGCTTCCTGGT | TGGGAATTGATTGAGCTGGC |
| DLX-6 | GGGAAATCAGGTTCAACGGA | AGTCTGCTGAAAGCGGTGGT |
| calbindin | GCGCTCTCTCAAACTAGCCG | TGACTGCAGGTGGGATTCTG |
| calretinin | TGACTGCATCCCAGTTCCTG | TTCCGTCAGCATCAAAGTGC |
| parvalbumin | TCTGGTGGCCGAAAGCTAAG | GAGAGGTGGGAGACCCAAGC |
| NPY | CCCGCCATGATGCTAGGTAA | GAGGGTCAGTCCACACAGCC |
| neurotensin | TGTGCTTTCTTGGATGGGATT | ATTGCTTCCAGCTTGCATGA |
| somatostatin | GAGCAGGACGAGATGAGGCT | TGGGTTCGAGTTGGCAGAC |
| TH | GTACCCATGTTGGCTGACCG | TCCAATGTCCTGGGAGAACTG |
| DARPP-32 | ACAGCACAAAAGCCTGCAGA | ACCACGCTGCTCCTGAGTCT |
| Olg-1 | GGGCTTCGTTGTACGAGCTG | ATGACGAGATGGGTGGCTG |
| Olig-2 | GAAGCAGATGACTGAGCCCG | CTGTTGATCTTCAGGCGCAG |
| Pax6 | TCTAACCGAAGGGCCAAGTG | GAGGAGACAGGTGTGGTGGG |
| Nkx2.1 (Titf1) | CGGCCCTGAACTCTGAAGC | CTGGCAGAGTGCATCCACAG |
| Nkx6.2 | GGCTTGCCTACTCTCTGGGC | GGAACCACACCTTCACCTGG |
| Lhx6 | GTCAGGAAAGGCAAATTCCG | CCACAGGTGAAGGAGGGACA |
| Emx1 | GCATCGGGACCCTCTTCAC | AAGAAGCGATTCCGAAGCAC |
Immunocytochemistry
The methods for immunocytochemistry were described previously (Li et al., 2004) except that 2% glutaradelhyde was included in fixative solution where cellular glutamate was to be detected. Antibodies used in this study were mouse IgMs: A2B5 (1:200, Chemicon), 5A5 (anti-PSA-NCAM, 1:200, DSHB), O4 (1:50, McKinnon lab); mouse IgGs: anti-vimentin (1:10, DSHB), anti-nestin (1:20, DSHB), anti-β-III tubulin (1:500, TuJ1, Covance), anti-GalC (1:50, McKinnon lab), anti-parvalbumin (1:200, Chemicon), and anti-calbindin (1:200, Sigma); rabbit IgGs: anti- BLBP (1:1000, Chemicon), anti-NCAM (1:50) (Friedlander et al., 1994), anti-GFAP (1:200, Dako), anti-NG2 (1:500, Levine lab), anti-DLX-2 (1:200, Chemicon), anti-glutamate (1:500, Chemicon), anti-GAD65/67 (1:200, Chemicon), anti-calretinin (1:1000, Chemicon), antineuropeptide Y (1:500, Chemicon), and anti-somatostatin (1:200). Secondary antibodies included Oregon-Green- or Rhodamine-Red-conjugated against appropriate species (1:200, Molecular Probes). DAPI (10 µg/ml, Sigma) was included in the secondary antibody incubations to label nuclei.
Western blot analysis
Cultured cells were harvested in SDS lysis buffer (62.5 mM Tris-HCl, pH 6.8, 2% SDS, 10% glycerol, 5% β-mercaptoethanol, 12.5 mM EDTA, 0.05 % bromophenol blue), and heat-denatured at 95°C for 5 min. Proteins were separated in 10% SDS-PAGE and transferred onto PVDF membranes. The membranes were then blotted with primary antibodies indicated, followed by horseradish peroxidase (HRP)-conjugated secondary antibodies (1:5000, Jackson lab). The blots were developed using ECL plus detection system (GE Healthcare Amersham). Anti-GAPDH (mouse IgG, 1:1000, Chemicon) was used to normalize the sample loading.
Electrophysiological techniques
Whole-cell patch-clamp recordings were performed on L2.2 cells after incubation in N2B27 differentiation culture. The external neuronal recording solution (NRS) contained 1.67 mM CaCl2, 1 mM MgCl2, 5.36 mM KCl, 137 mM NaCl, 17 mM glucose, 10 mM HEPES and 13.15 mM sucrose. The pipette solution contained 105 mM K-methanesulfonate, 17.5 mM KCl, 10 mM HEPES, 0.2 mM EGTA, 8 mM NaCl, 2 mM Mg-ATP, 2 mM Na-ATP, 0.3 mM Na-GTP, and 20 mM phosphocreatinine. The pH of the pipette solution was set to 7.3 with KOH. The typical range of pipette resistance and access resistance was 3–5 MΩ and 7–20 MΩ, respectively. Currents were recorded under voltage clamp mode with an Axoclamp 200 amplifier, digitized at 2.9 kHz and filtered at 5 kHz. Potentials were recorded under current clamp mode, digitized at 2.9 kHz and filtered at 1 kHz. Signals were digitized with a CED Power1401 interface. Series resistance was monitored throughout the recording and an experiment was terminated if it increased by more than 10%. Data acquisition parameters and voltage pulse generation were under the control of custom software written in the Plummer lab.
RESULTS
L2.2 is a neuronal restricted progenitor (NRP) clone
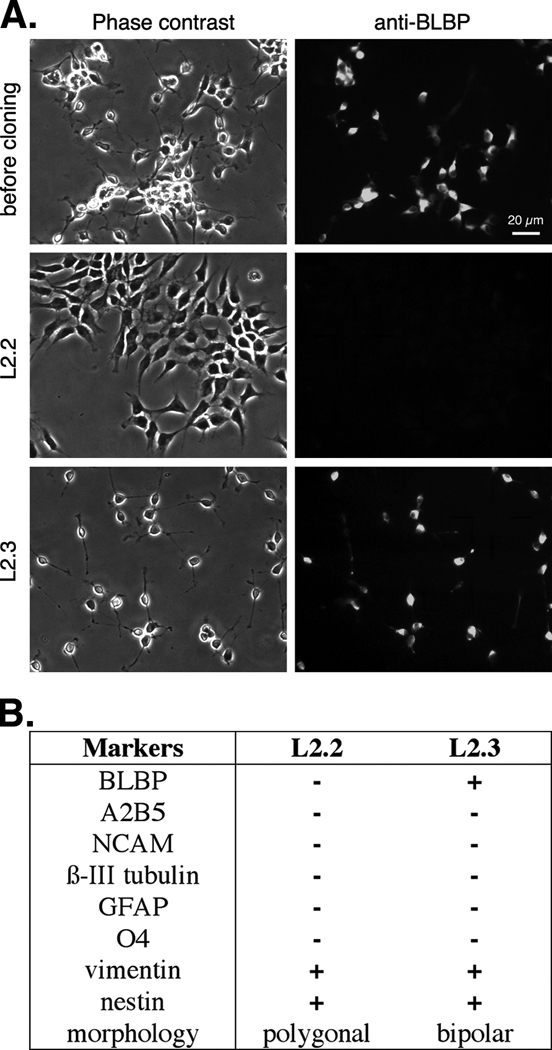
To isolate progenitor clones, we immortalized neurosphere cultures that were derived from rat E14.5 cortex (Li et al., 2004). The clones derived were initially screened for expression of the NSPC markers nestin and vimentin (Li et al., 2004). After they were recloned, about half of these also expressed the RG marker BLBP and a representative clone L2.3 that was analyzed in detail was found to support neuronal migration, a key property of RG (Li et al., 2004). Among those that did not express BLBP, a representative clone L2.2 expressed progenitor markers including nestin and vimentin (Fig. 1A, 1B). In contrast to the bipolar radial morphology of L2.3 cells, L2.2 cells had polygonal morphology when cultured on laminin-coated substrates (Fig. 1A). Upon differentiation, L2.2 cells exhibited morphological changes sending out neuronal processes (Fig. 2A). Long neuronal-like fibers and fasciculated bundles were seen in 4- and 6- day differentiated L2.2 cultures. L2.3 cells gave rise to neurons and glia, while L2.2 cells differentiated exclusively into TuJ1+ neurons within 6 days (Fig. 2B). No glial cell types were detected by immunostaining for GFAP (Fig. 2B) and GalC (data not shown) in the L2.2 differentiation cultures even in the presence of FBS (Li et al., 2004). Western blot analysis showed that after 6 days of differentiation, L2.2 cells expressed neuron-specific β-III tubulin, but not the glial marker GFAP, whereas L2.3 differentiated cells expressed both proteins (Fig. 2C). The multipotential differentiation of L2.3 cells indicates their neural stem cell nature. Thus, the L2.2 and L2.3 clones that we isolated in parallel from E14.5 rat cortical cultures both proliferate in the presence of FGF2 but they show contrasting differentiation upon FGF2 withdrawal. In addition, the L2.3 cells can be induced to differentiate primarily into GFAP+ astrocytes in the presence of LIF or 10% FBS (Li and Grumet, 2007) but these conditions did not promote differentiation of L2.2 cells (data not shown). Whereas L2.2 cells are neurogenic in various differentiation conditions, L2.3 cells display multipotential properties of NSPC but they are almost exclusively gliogenic under certain differentiation conditions (Li and Grumet, 2007).
Fig. 1. Generation and characterization of the cortical neuronal progenitor clone L2.2.
A. L2.2 and L2.3 exhibited distinct morphology on laminin substrates. L2.3 cells expressed the radial glial marker, BLBP, while L2.2 cells did not. B. Summary showing expression of cell type specific markers detected by immunofluorescence in the two clones without differentiation.
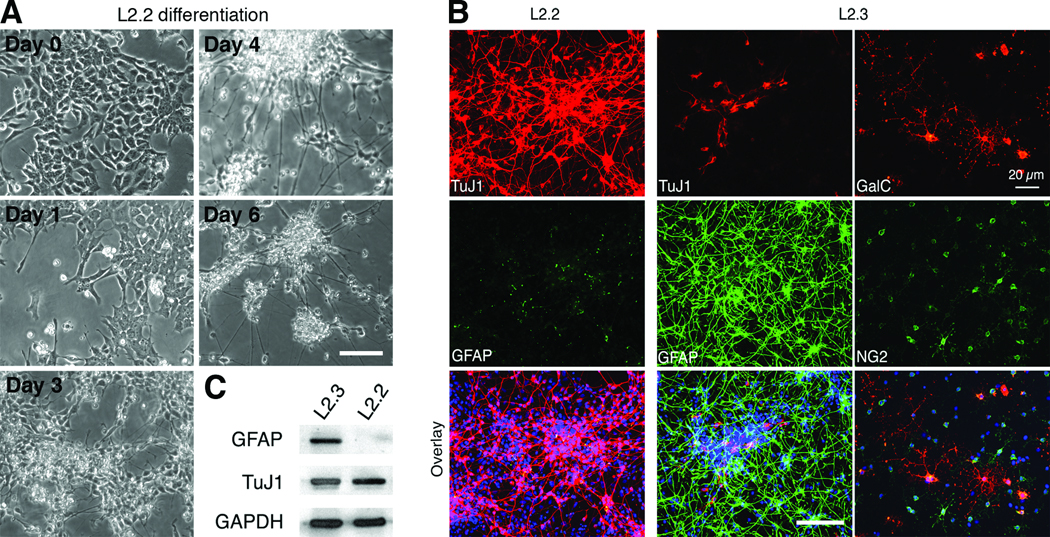
Fig. 2. Restricted neuronal differentiation of L2.2 clone.
L2.2 and L2.3 progenitor cells were maintained in FGF2 containing serum free medium. To initiate differentiation, FGF2 was withdrawn from the culture medium, and 0.5% of FBS was added to promote cell survival. A. Phase-contrast images showing morphological changes of L2.2 cells during differentiation after FGF2 withdrawal. B. L2.2 cells differentiated exclusively into TuJ1+ neurons, while L2.3 cells differentiate into both GFAP+ astrocytes and TuJ1+ neurons within 6 days. C. Western blot analysis confirmed the restricted neuronal differentiation of L2.2 cells with only β-III tubulin (TuJ1) expression and no detectable GFAP expression. Differentiated L2.3 cells expressed both markers. GAPDH was used to normalize the sample loading. Scale bars, 50 µm.
The expression of several key transcription factors (e.g. Pax6 for neurogenic and Olig-1, -2 for gliogenic) were found to differ in these two clones. To verify and expand the comparison, we did a comprehensive microarray analysis on L2.2 and L2.3 in a time course differentiation study (Supplemental Table 1). Proliferating cells in the presence of FGF2 were harvested as the first time point (0 day). In sister cultures the medium was changed to remove FGF2 and add 0.5% FBS. One or three days after changing the medium, additional cells were harvested for 1 and 3 day points. On the AB1700 Rat Genome Survey arrays, 3,181 probes (11.8%) were significantly regulated between cell clones and/or during the course of differentiation, as measured by two-way ANOVA and corrected for multiple testing (Supplemental Table 1). These significant probes were K-means clustered (k=6; selected to maximize explained variability) to identify grouped, interpretable expression patterns (Fig. 3A). Although a large group of genes only showed moderate changes in both cell types (clusters 6 and 4, where the group means appear unchanged but individual genes, shown in grey, are regulated), several other clusters appear to represent biologically interpretable groups of mRNAs. Genes in cluster 4 were expressed at relatively higher levels in undifferentiated L2.3 than in L2.2 cells and included the neural stem cell markers nestin and prominin (Supplemental Table 1), supporting the idea that L2.3 are NSPC. Clusters 2 and 5 contain genes expressed at relatively higher levels in L2.2 compared to L2.3 cells including Tubb3, Pax6, DLX-5, NeuroD-1 and -3, supporting the notion that clone L2.2 is neurogenic (Anderson et al., 1997; Lee, 1997; Heins et al., 2002; Katsetos et al., 2003). In contrast, genes in cluster 1 showed relatively higher levels in undifferentiated L2.3 vs. L2.2 cells including Olig-1 and –2, and BLBP/Fabp7 (Supplemental Table 1), which are associated with glial differentiation (Lu et al., 2001; Lu et al., 2002; Anthony et al., 2005), and these genes were down-regulated in L2.3 cells during differentiation (Figure 3A). Genes in cluster 3 including GFAP, S100β, and tenascin-R showed much higher up-regulation in L2.3 than in L2.2 cells and represent markers of astroglial differentiation (Supplemental Table 1). These results provide additional support for the idea that clone L2.2 is neurogenic whereas clone L2.3 is both neurogenic and gliogenic. The relatively high levels of DLX-5 gene expression found in undifferentiated L2.2 cells (cluster 5) suggested that it may be derived from an interneuron progenitor since the DLX family of transcription factors are expressed in these ventrally derived cortex-invading interneurons (Anderson et al., 1997).
Fig. 3. Differences in gene expression patterns in L2.2 and L2.3 cells and during differentiation.
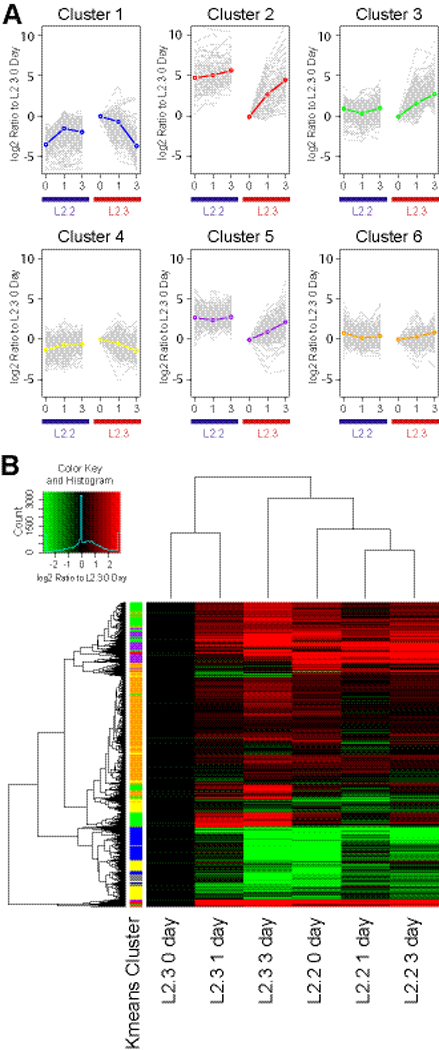
Upon FGF2 withdrawal, total RNA samples of L2.2 and L2.3 cultures were harvested after 0, 1 and 3 days and analyzed on rat genome survey chip (AB 1700). Significant genes were k-means clustered (k=6) to identify similar expression patterns (A), and were also shown as the hierarchically clustered heatmap plotted using R (B). For each k-means cluster, the calculated mean of all cluster members is plotted and individual genes are plotted in grey to show variability within the cluster. Assignment of a gene to a specific cluster is defined by an 80% membership in that cluster upon repeated fittings. Detailed gene expression profiles and cluster memberships are in Supplemental Table 1.
A heatmap was constructed and juxtaposed next to the k-means clusters to visualize the contributions of probe groups to the cluster centers (Figure 3B). Clustering confirmed close relationships between the individual clones during differentiation and some relationship between L2.3 at 3-days and L2.2, consistent with their capacities for neuronal differentiation.
BMP2 suppresses neuronal differentiation of L2.2 cells
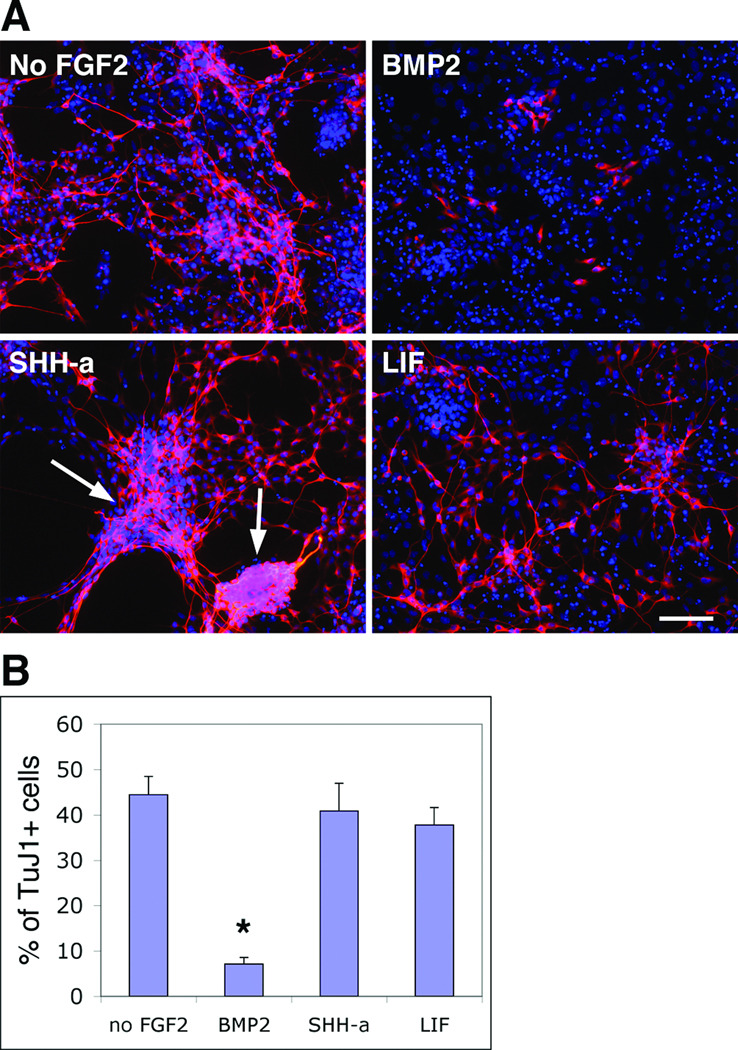
The neuronal subtype differentiation of cortical progenitors can be influenced by dorsoventral morphogens such as BMPs and SHH (Gulacsi and Lillien, 2003). We therefore tested whether L2.2 cells respond to these factors similarly to their counterparts in vivo. Proliferating L2.2 cells were treated for 3 days with BMP2, LIF and SHH-agonist in the absence of FGF2 on laminin-coated substrates. In contrast to withdrawal of FGF2, treatment with BMP2 (25 ng/ml) yielded a lower percentage of TuJ1+ cells (Fig. 4), suggesting an inhibition of differentiation. Furthermore, the few TuJ1+ cells in BMP2 treated cultures showed simpler cell morphologies by comparison to control cells, which were more branched and process bearing (Fig. 4A). FBS (10%), which can mimic BMP differentiation (Kondo and Raff, 2000), also inhibited L2.2 differentiation (data not shown). SHH-agonist did not significantly increase the number of TuJ1+ cells with a 3-day treatment, but larger bundles of neuronal processes and aggregates of cell bodies were observed (Fig. 4A, arrows), which indicates more extensive neuronal maturation. LIF treatment also did not inhibit L2.2 differentiation (Fig. 4). Thus clone L2.2 exhibited responsiveness to dorsoventral morphogens in culture that influence neuronal development in the brain. In contrast, the RG clone L2.3 responded to these factors quite differently than the L2.2 clone; BMP2 increased the TuJ1+ neuronal population while LIF caused most L2.3 cells to become GFAP+ astrocytes upon differentiation (Li and Grumet, 2007).
Fig. 4. BMP2 suppressed L2.2 neuronal differentiation.
A. L2.2 cells were allowed to differentiate after FGF2 withdrawal and in the presence of the indicated factors for 3 days. Immunostaining with neuron specific antibody, TuJ1 (red), showed that BMP2 (25 ng/ml) in comparison with control (No FGF2) suppressed neuronal differentiation, while SHH-agonist (100 nM) and LIF (10 ng/ml) did not have significant effects. Enhanced fasciculation and aggregation of the neuronal processes were seen in SHH-a treated cultures (arrows). DAPI (blue) was used to label nuclei. B. Percentage of TuJ1 positive cells among total cells (DAPI+) were calculated for different treatments, and the values were shown as mean with standard errors from triplicate experiments (n = 604 for No FGF2; 465 for BMP2; 605 for SHH-a and 477 for LIF). The large neuronal aggregates or bundles where individual cells could not be distinguished were excluded from counting. *, p < 0.05 (Student t-test). Scale bar, 50 µm.
Co-culture of L2.2 cells with radial glia promotes development of action potentials
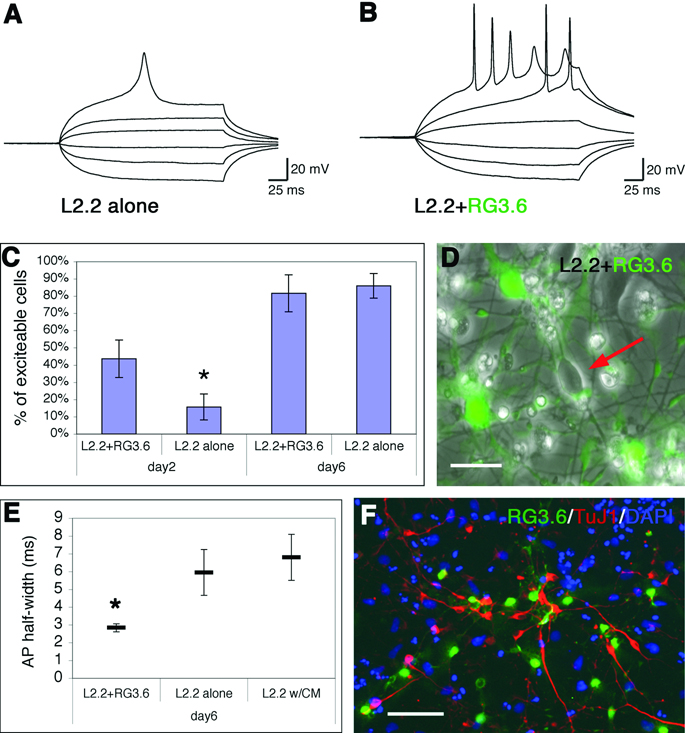
To determine whether L2.2 cells can differentiate into functional neurons in culture, we made whole-cell current clamp recordings. Cells that were selected for recording had oval cell bodies and at least two processes resembling mature neurons. Roughly 10–50% of cells showed this morphology depending on culture conditions and the duration of differentiation. At day 2 after FGF2 withdrawal, 15.7±7.5% of the recorded L2.2 cells exhibited action potential (AP) firing and the percentage of excitable cells increased to 86±7.1% at day 6 (Fig. 5A, C). At both times in vitro, the APs were eliminated by the Na channel blocker tetrodotoxin.
Fig. 5. Co-culturing L2.2 cells with RG3.6 cells promoted development of AP.
A. Example of current-clamp recording from an L2.2 cell 6 days after FGF2 withdrawal. Responses to six depolarizing current injections of increasing magnitude are shown, with the largest one evoking action potential firing. B. Example of current-clamp recording from a L2.2 cell co-cultured with RG3.6 cells 6 days after FGF2 withdrawal. Responses to six depolarizing current injections are shown, with the largest two evoking action potential firing. C. Whole cell current-clamp recordings made from L2.2 derived cells under four different conditions, L2.2 alone and L2.2+RG3.6 co-cultures at day 2 and day 6 after FGF2 withdrawal. The percentage of L2.2 cells that fired action potentials was significantly higher on day 2 for cells in co-cultures (n = 25) compared to being cultured alone (n = 23, p < 0.05); data show standard errors from 6 independent experiments. There was no difference in the percentage of excitable cells on day 6 (p > 0.95, n = 21 for both), however, nearly all recorded cells were excitable. D. An example of GFP and phase-contrast overlaid images showing L2.2 (GFP-negative, arrow) differentiation in co-culture with RG3.6 cells (GFP-positive). Only GFP-negative L2.2 cells were recorded. Scale bar, 10 µm. E. The half-width (in milliseconds) of AP in day 6 cultures of L2.2+RG3.6 (n=17), L2.2 alone (n=18) and L2.2 with RG3.6-conditioned medium (n=12). The average half-width was significantly lower in L2.2+RG3.6 co-cultures than in either L2.2 alone or L2.2 with RG3.6 condition medium, p <0.05. F. Six days after FGF2 withdrawal, immunostaining showed that TuJ1 positive (red) L2.2 cells were often seen in close proximity with GFP positive RG3.6 cells. DAPI (blue) was used to label the nuclei. Scale bar, 50 µm.
Although L2.2 cultures had cells that were electrically excitable, many of the cells died during the differentiation induced by FGF2 withdrawal. Given that neurons normally differentiate in the presence of developing glia, we tested effects of co-culturing with other cells. When L2.2 cells were co-cultured with GFP-astrocytes, many survived and expressed TuJ1 within 2 days, and the astrocytes changed morphologically from flat to assume more spindly shapes (Supplemental Fig. 1A–C). The morphological response of the astrocytes was observed in cocultures with L2.2 cells but not with the L2.3 or non-neuronal Hela cells (Supplemental Fig. 1B), suggesting a specific interaction between the neurogenic L2.2 and astroglia. Moreover, only the L2.2 cells induced up-regulation of BLBP in the astroglia (Supplemental Fig. 1C), a response that has been observed for radial glia when they interact with neurons (Feng and Heintz, 1995). However, since the astroglia are grown in medium containing FBS, which inhibits differentiation of the L2.2 cells, we tested whether the GFP-expressing RG3.6, a radial glial-like clone that differentiates primarily into astroglia, also could support L2.2 differentiation in the absence of serum. Indeed, such co-cultures resulted in good survival and extensive process extension of L2.2 cells as well as induction of GFAP in the RG3.6 cells (Supplemental Fig. 1D)
To assess effects of glia on L2.2 functional differentiation, we therefore co-cultured L2.2 cells with the GFP-labeled RG3.6 in defined medium. The L2.2 cells were identified in these cultures by the absence of GFP (Fig, 5D, arrow). Interestingly, under these conditions (L2.2+RG3.6), L2.2 cells developed AP firing more rapidly than when cultured alone (Fig. 5A vs B). The percentage of excitable cells was 43.7±10.9% at day 2, which was significantly higher than with L2.2 alone (Fig. 5C). Six days after FGF withdrawal, the percentage of excitable cells was not significantly different between L2.2+RG3.6 (81.7±10.6%) and L2.2 alone. Importantly, however, the APs fired by L2.2 cells cultured alone for 6-days were less mature than those fired by L2.2 cells in the RG3.6 co-cultures. In the former condition, APs were relatively broad and were often fired singly (Fig, 5A), whereas, in the latter condition, APs were sharp and frequently fired in bursts (Fig. 5B). Quantification of AP half-width showed that this difference was statistically significant (Fig. 5E). Delayed firing was often seen in both conditions (Fig. 5A, 5B). In co-cultures, about 50% of L2.2 cells (non-GFP) expressed TuJ1+ after 6 days of differentiation (Fig. 5F), while RG3.6 cells (GFP+) differentiated mainly into GFAP+ astrocytes (Hasegawa et al., 2005).
To explore the mechanism of the effect of RG3.6 on L2.2 functional differentiation, we also made current clamp recordings from L2.2 cells cultured in medium supplemented with RG3.6 conditioned medium (CM). The CM did not mimic the promoting effect of RG3.6 co-culture on the percentage of excitable cells, or AP half-width (Fig. 5E). In fact, all the measurements showed that L2.2 cells supplemented with RG3.6 CM were very similar to L2.2 cells cultured alone, indicating that the effects of RG3.6 on L2.2 differentiation are not mediated through secreted factors, but through cell-cell contact. Indeed, neuronal marker TuJ1 positive L2.2 cells were often seen in close contact with GFP-positive RG3.6 cells in L2.2+RG3.6 co-cultures (Fig. 5F).
L2.2 cells express cortical interneuron markers
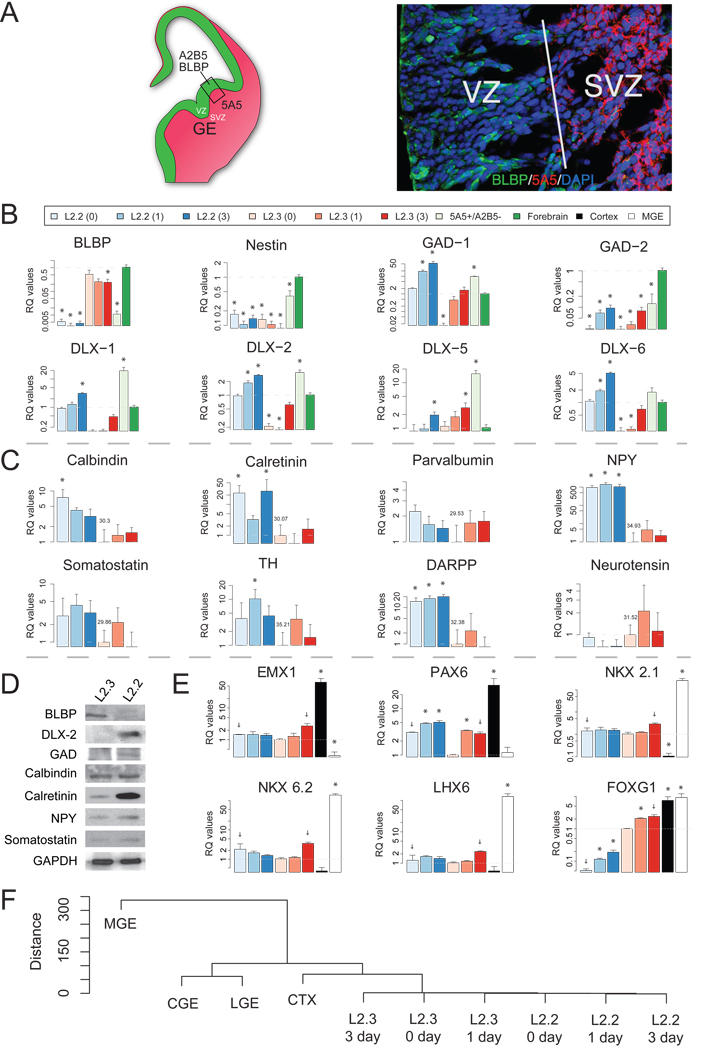
Microarray analysis of clone L2.2 provided strong evidence that it is neurogenic, and selective expression of DLX-5 and somatostatin (Supplemental Table 1) provided hints that it may represent a ventrally derived interneuron precursor. Considering that these cells responded to dorsal-ventral morphogens, we compared expression of interneuron markers between the two clones and with primary cortical interneurons and their precursors. Cortical interneurons are derived mostly from the ganglionic eminence (GE) in the developing ventral forebrain in rodents (Anderson et al., 1997) (Fig. 6A). We showed previously that the neuronal surface marker PSA-NCAM (recognized by 5A5 antibody) strongly labels cells in the SVZ, whereas the VZ contains BLBP+ radial glia (Fig. 6A), which are A2B5 positive in the lateral region of the cortex (Li et al., 2004; Li and Grumet, 2007). To separate these different types of cells, we micro-dissected rat E14.5 ventral forebrains and then removed most of the radial glia by magnetic bead sorting for 5A5+/A2B5− cells. Q-PCR analysis confirmed the success of the sorting in that BLBP expression was >50 fold lower in 5A5+/A2B5− cells than the unsorted forebrain (Fig. 6B). Thus, the 5A5+/A2B5− cells are likely to be enriched in GE NSPC. Similarly, we found drastic differences in BLBP expression between L2.2 and L2.3 (Fig. 6B, Fig. 1B, 1C). As expected, both L2.2 and L2.3 cells expressed the NSPC marker nestin, which decreased with differentiation in both clones (Fig. 6B). In addition, nestin was expressed robustly in 5A5+/A2B5− cells as well as in the unsorted forebrains (Fig. 6B), indicating the abundance of neural precursors.
Fig. 6. Clone L2.2-derived cultures express cortical interneuron markers similarly to 5A5- enriched primary interneurons.
(A, left panel) Schematic drawing showing expression patterns of RP markers (A2B5 for GRPs; 5A5 for NRPs) and RG marker (BLBP) on coronal sections of E14.5 forebrain. Strong 5A5 staining in the GE labels SVZ progenitors and newly formed neurons (red), whereas the VZ is not labeled but is positive for A2B5 and BLBP (green) (Li et al., 2004). (A, right panel) Confocol image of the boxed region in A showing immunostaining of the VZ/SVZ interface marked by BLBP (green) and 5A5 (red). qPCR analysis of undifferentiated (0 day) and differentiated (1 or 3 days) L2.2 and L2.3 cells was compared to 5A5+/A2B5− acutely isolated, MACS sorted primary cells from E14.5 ventral forebrains in their expression of RG markers and interneuron markers (B), and for interneuron subtype markers (C). The gene expression levels in whole forebrain were set as 1 for reference in (B), and those in L2.3 0 day were set as 1 in (C) where the cycle threshold (Ct) values are also indicated. (D) Western blot analysis showed differential expression of proteins between L2.2 and L2.3 clones after 6-day differentiation in culture. GAPDH was used to normalize the sample loading. (E) qPCR analysis on L2.2 and L2.3 samples and micro-dissected E14.5 cortex and GEs. The gene expression levels in L2.3 0 day were set as 1. “+”s indicate calculations were based on only duplicated samples. (F) Dendrogram showing correlation of different samples was drawn based on the expression of genes in panel E. * indicates expression levels were significantly different from those of references (p<0.05, Students T-test).
We then compared other markers in all these samples that may distinguish different types of precursors. The 5A5+/A2B5− cells expressed higher levels of GAD-1, DLX-1, -2, -5 and –6 than unsorted forebrain indicating that 5A5+/A2B5− sorting enriched for genes associated with cortical interneuron precursors. Importantly, L2.2 cells showed higher expression of these genes than L2.3 cells, and the levels increased further with differentiation (Fig. 6B). L2.3 cells expressed relatively low levels of these precursor markers that increased with differentiation, suggesting that at least some L2.3 cells can become ventral cell types after differentiation (Fig. 6B). Compared to L2.3 cells, L2.2 also expressed higher levels of calretinin, calbindin, NPY and somatostatin, which are associated with interneurons, and other ventral neuronal markers including DARPP32 and tyrosine hydroxylase (TH) (Fig. 6C). Moreover, increased expression of DLX-2, GAD and calretinin proteins was confirmed in L2.2 cells by western blot analysis (Fig. 6D). For NPY and somatostatin, clear differences between L2.2 and L2.3 were not observed upon differentiation perhaps because they also are expressed by neurons in the L2.3 cultures.
The expression of ventral cell type markers in L2.2 culture suggests it may be of ventral origin. However, while these studies were in progress, it was reported that dorsal progenitors expressing EMX1 and DLX5/6 generate a subpopulation of olfactory bulb GABAergic interneurons (Kohwi et al., 2007). To explore further the possibility that L2.2 clone may be related to these dorsal precursors, we performed additional qPCR analyses using primers for additional transcription factors that exhibit spatio-temporal restricted expression. Cortex-specific genes EMX1 and PAX6 showed higher expression levels in cortex samples, whereas NKX2.1, NKX6.2 and LHX6 showed higher expression levels in MGE (Fig. 6E) confirming the success of our tissue dissection. Although we were able to detect some levels of both sets of genes in L2.2 and L2.3, correlations among the samples based on the expression of these genes, suggested that L2.2 and L2.3 are more closely related to cortex than to GE (Fig. 6F). This suggests that clones L2.2 and L2.3 are more likely to be derived from dorsal cortex than GE. FOXG1, as a pan-telencephalic marker, showed similar expression between cortex and MGE (Fig. 6E). L2.2 showed lower expression levels than the tissue samples in the proliferating stage (L2.2, 0 day) that increased upon differentiation (L2.2 1 and 3 days, Fig. 6E).
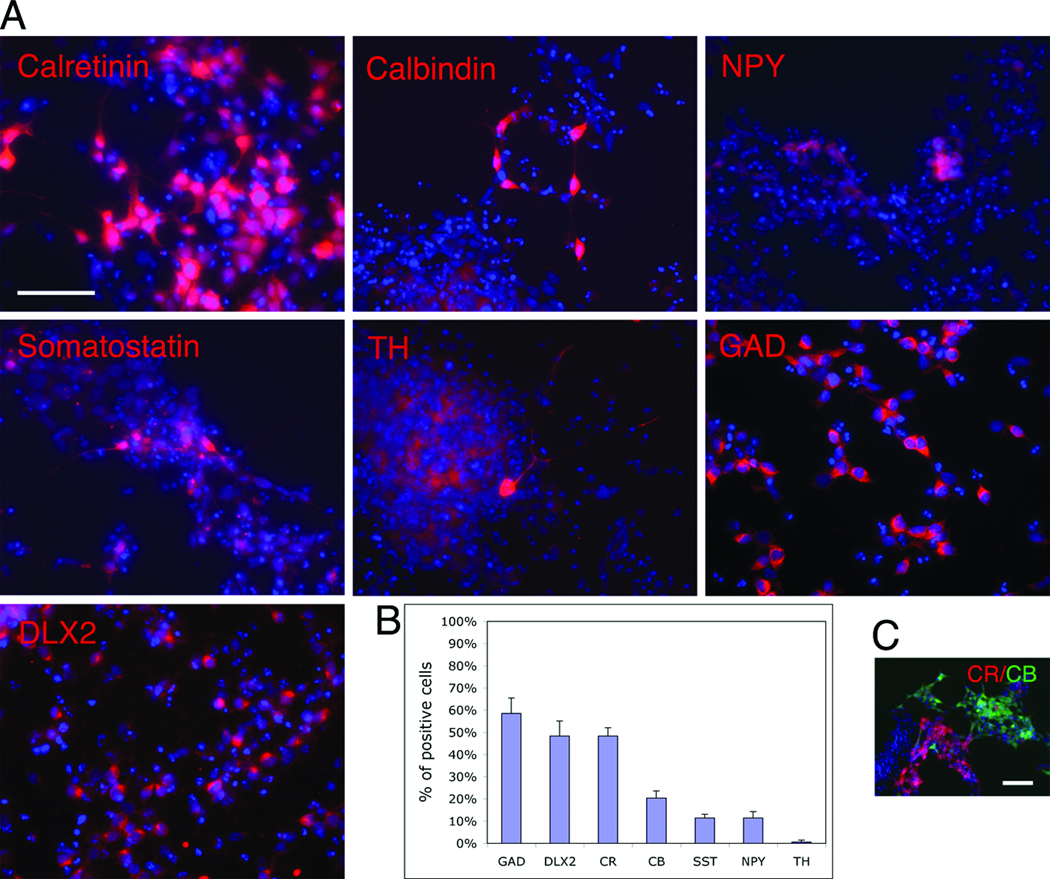
To confirm expression of interneuron subtype markers on cells, we immunostained 6-day differentiated L2.2 cultures. The results showed that nearly half (48 ± 6.8%) of these cells expressed calretinin and smaller percentages of cells expressed other interneuronal markers including calbindin (20.4 ± 3.2 %), NPY (11.5 ± 2.8 %) and somatostatin (11.5 ± 1.6 %) (Fig. 7A, 7B). Calretinin staining did not overlap on calbindin+ cells (Fig. 7C), suggesting that the majority of cells that differentiated from clone L2.2 are different types of neurons. The combined results suggest that L2.2 is a precursor that differentiates into several types of cells including those resembling DARPP32+ striatal projection neurons, TH+ olfactory bulb interneurons, and interneurons of the striatum and cortex expressing calretinin, calbindin, NPY and somatostatin.
Fig. 7. L2.2 differentiated neurons expressed cortical interneuron markers by immunostaining.
(A) After 6-day differentiation in N2B27 medium, L2.2 cells showed immunoreactivity for cortical interneuron markers including GAD, DLX-2, calretinin, calbindin, NPY, somatostatin and TH. DAPI (blue) was used to label nuclei. The percentages of marker positive cells among total cells (DAPI+) in L2.2 culture are shown as mean with standard errors from triplicate experiments in (B). Double immunostaining showed non-overlapping staining pattern between calretinin and calbindin (C). Scale bars, 50 µm.
DISCUSSION
We describe the generation and characterization of an immortalized NRP clone L2.2 that was derived from E14.5 rat cortical culture. L2.2 cells express markers typically found on NSPC, proliferate and can be expanded in the presence of FGF2, and differentiate upon FGF2 withdrawal into TuJ1+ neurons, which exhibit GABAergic interneuron phenotypes, but not into glia. Differentiated L2.2 cells express markers including DLX transcription factors and the calcium-binding protein calretinin. Importantly, differentiated L2.2 cells exhibited properties of functional neurons (i.e. firing action potentials), and acquisition of this property was accelerated when they were differentiated in co-cultures with glial cells. Thus L2.2 is the first clonal cell reported to differentiate exclusively into neurons that exhibit GABAergic properties.
Radial glia play a major role in contributing to neurogenesis during mammalian forebrain development (Malatesta et al., 2000; Miyata et al., 2001; Noctor et al., 2002; Anthony et al., 2004). Besides RG, the VZ of developing embryonic forebrain give rise to neuronal restricted precursors, also called intermediate progenitors or short neural progenitors, in both rodents (Haubensak et al., 2004; Gal et al., 2006) and humans (Piper et al., 2001; Howard et al., 2006; Mo et al., 2007). These two cell types have different gene expression profiles, and can be distinguished by the status of Notch signaling (Mizutani et al., 2007). Using v-myc immortalization, we have generated clones that may represent these two cell types, i.e. L2.2 behaving as intermediate progenitors, and L2.3 and RG3.6 behaving as RG (Li et al., 2004; Hasegawa et al., 2005). Thus, these clones may be useful models for characterizing two major progenitor cell types.
GABAergic inhibitory interneurons form synapses with glutamatergic projection neurons and modulate their functions in the forebrain. During development, projection neurons are generated mostly from radial glial cells in the VZ of the forebrain, and migrate radially out towards the pial surface (Englund et al., 2005; Hevner, 2006). Although progenitors in the dorsal cortex have the potential to give rise to interneurons (Gulacsi and Lillien, 2003), they are believed to originate mainly from the subpallium of the forebrain, and migrate tangentially into the developing cortex at least in rodents (Anderson et al., 1997; Lavdas et al., 1999; Pleasure et al., 2000; Anderson et al., 2001; Wichterle et al., 2001; Xu et al., 2004). Ventrally derived interneurons and their precursors express DLX transcription factors, which are essential for their tangential migration into the dorsal cortex (Anderson et al., 1997). The expression of DLX mRNAs in the L2.2 clone is consistent with the ventral origin in the developing forebrain of cortex-invading interneuron precursors. Although our E14.5 forebrain dissection protocol eliminated the ventral GE where most interneurons originate, we could not exclude the possibility of some contaminating ventral tissues in the culture. However, the isolation of many nestin+/BLBL− clones morphologically resembling L2.2 cells makes it unlikely that this highly represented phenotype was due to ventral contamination during dissection. Alternatively, L2.2 and related clones may be derived dorsally, given that Emx1-expressing cortical progenitors can give rise to calretinin-positive olfactory bulb GABAergic interneurons, which generate calretinin-positive interneurons (Pappas and Parnavelas, 1998). Our qPCR support the latter possibility (Fig. 6E and 6F).
Mature interneurons exhibit variable features and can be subclassified by their morphologies (Woo and Lu, 2006) and their expression of calcium-binding proteins (calretinin, calbindin and parvalbumin) and neuropeptides (neuropeptide Y, somatostatin, neurotensin and vasoactive intestinal peptide) (Marin et al., 2000; Xu et al., 2004). Upon differentiation, clone L2.2 gives rise primarily to calretinin+ neurons and to small percentages of calbindin-, NPY- and somatostatin-positive cells providing an indication of its plasticity in vitro. Parvalbumin is another calcium-binding protein but it was never detected by immunostaining or western blot in differentiated L2.2 cultures. Parvalbumin+ interneurons largely originate from medial GE (Wonders and Anderson, 2006), which was excluded in our dissection. In addition, parvalbumin+ cortical interneurons exhibit a fast-spiking firing pattern with short latency of AP in cultured slices (Butt et al., 2005). We rarely observed a fast-spiking firing pattern in L2.2 derived neurons; rather, most of our recordings showed delayed firing pattern with latencies of more than 100 ms (Fig 5D).
The GABAergic differentiation of the L2.2 clone provides a model to study differentiation and maturation of interneurons in vitro. Neural progenitor cell lines that are capable of generating GABAergic neurons have been described (Lundberg et al., 1996; Yamada et al., 1999; Tominaga et al., 2005). Among them, ST14A is most similar to clone L2.2. Despite the use of different immortalizing reagents (temperature-sensitive large T-antigen for ST14A vs. v-myc for L2.2) and the possible difference in their origins (E14.5 striatum for ST14A vs. E14.5 dorsal cortex for L2.2), L2.2 and ST14A are remarkably similar in that they both give rise to GABAergic neurons but not to glial cells. The success of immortalizing neuronal restricted interneuron precursors from both embryonic cortex (L2.2) and striatum (ST14A) suggests the existence of multiple sources for such cells in vivo.
Oncogene immortalized NSPC do not necessarily form tumors and many did not when transplanted into adult tissues (C17.2, RG3.6, ST14A) (Bosch et al., 2004; Hasegawa et al., 2005; Macias et al., 2006). Such clones may be particularly useful for testing hypotheses about cell transplantation in animal models of disease and traumatic injury. Furthermore, these studies may provide key information to develop protocols for generating useful cells from embryonic stem cells to facilitate translational research for human therapeutics. Moreover, potential concerns using v-myc expressing cells may be mitigated by the observation that v-myc expression dramatically decreased after growth factor withdrawal in immortalized human NSC (IhNSC) in culture (De Filippis et al., 2007) and was undetectable 24–48 hours after orthotopic engraftment into rodent brains (Flax et al., 1998).
In this study, we showed that L2.2 cells acquired excitability more rapidly in the RG3.6 coculture than when grown alone. In addition, the L2.2 cells in the co-cultures fired multiple APs that were sharper and had shorter half-widths than APs fired by L2.2 cells grown alone. This effect of RG3.6 cells on L2.2 functional differentiation is apparently mediated by cell-cell contact rather than secreted factors. This is in contrast to other neuronal responses induced by glial cells (i.e. astrocytes and Schwann cells) including synapse formation (Pfrieger and Barres, 1997; Ullian et al., 2001; Peng et al., 2003; Ullian et al., 2004; Ullian et al., 2004; Christopherson et al., 2005; Cao and Ko, 2007) that are mediated by secreted factors including BDNF (Levine et al., 1995; Levine et al., 1998). RT-PCR measurements indicate that RG3.6 cells express BDNF and other factors including NT3 and GDNF (Y.W. Chang and M. Grumet, unpublished data), but they do not appear to be sufficient to promote AP development in our cultures as CM failed to induce L2.2 differentiation. We tested for effects of BDNF (20 ng/ml) on L2.2 differentiation, but we did not observe accelerated AP development (Y. Han, H. Li and M. Plummer, unpublished data). Thus, development of action potentials and synapse formation may be separable events in neuronal maturation (Johnson et al., 2007), and glial cells may promote both of these steps but via different mechanisms.
Cell-cell contact plays important roles during neural development. For example, cerebellar Bergmann glial cells promote survival of granule neurons in culture (Hatten, 1985; Hatten, 1987; Edmondson et al., 1988), and reciprocally, granule neurons inhibit proliferation of Bergmann glial cells through Notch-mediated cell contact mechanisms (Hatten et al., 1997; Goldowitz and Hamre, 1998). Microarray analysis showed that L2.2 cells express higher levels of Notch ligands such as delta and Jagged, while RG3.6 and L2.3 cells express higher levels of the Notch receptors (Supplemental Table 1 and data not shown). Ventrally derived cortical interneurons migrate tangentially into dorsal cortex, but they also can switch to migrate radially along radial glial (Nadarajah and Parnavelas, 2002; Poluch and Juliano, 2007; Yokota et al., 2007). Our results that co-culture of RG3.6 with L2.2 promotes their differentiation, suggest that cell-cell interactions between interneuron progenitors and radial glia as they migrate tangentially across the cortex may promote their functional maturation to develop AP. Although we did not detect obvious synaptic activity in L2.2 cultures even two weeks after differentiation (data not shown), this could result from the lack of specific targets in culture, e.g. glutamatergic projection neurons. Co-cultures of L2.2 cells with primary neurons such as embryonic hippocampal neurons may allow them to form functional synapses, and this would be an important property facilitate functional integration after transplantation into the injured CNS in future studies (Marsala et al., 2004; Hasegawa et al., 2005).
In summary, we have described molecular and electrophysiological features of a novel clone L2.2 that proliferates in FGF2-containing defined medium, behaves as a NRP, and exhibits differentiation into GABAergic interneurons. Although additional studies may be needed to define better the most likely origin of clone L2.2, it is one of the best characterized GABAergic precursors that can now be used for additional in vitro studies including drug discovery.
Supplementary Material
http://cord.rutgers.edu/appendix/Li/Supplemental_Table_1.html
ACKNOWLEDGEMENTS
We thank Dr. Lee Rubin for SHH agonist. This work was supported by grants from NIH, the New Jersey Commission on Spinal Cord Research, the New Jersey Commission on Science & Technology, and Invitrogen, Inc. JLD is a graduate fellow and CC is a postdoctoral fellow of the New Jersey Commission on Spinal Cord Research. CLR is an NSF-IGERT fellow.
REFERENCES
- Anderson SA, Eisenstat DD, Shi L, Rubenstein JL. Interneuron migration from basal forebrain to neocortex: dependence on Dlx genes. Science. 1997;278:474–476. doi: 10.1126/science.278.5337.474. [DOI] [PubMed] [Google Scholar]
- Anderson SA, Marin O, Horn C, Jennings K, Rubenstein JL. Distinct cortical migrations from the medial and lateral ganglionic eminences. Development. 2001;128:353–363. doi: 10.1242/dev.128.3.353. [DOI] [PubMed] [Google Scholar]
- Anthony TE, Klein C, Fishell G, Heintz N. Radial glia serve as neuronal progenitors in all regions of the central nervous system. Neuron. 2004;41:881–890. doi: 10.1016/s0896-6273(04)00140-0. [DOI] [PubMed] [Google Scholar]
- Anthony TE, Mason HA, Gridley T, Fishell G, Heintz N. Brain lipid-binding protein is a direct target of Notch signaling in radial glial cells. Genes Dev. 2005;19:1028–1033. doi: 10.1101/gad.1302105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard O, Reid HH, Bartlett PF. Role of the c-myc and the N-myc proto-oncogenes in the immortalization of neural precursors. J Neurosci Res. 1989;24:9–20. doi: 10.1002/jnr.490240104. [DOI] [PubMed] [Google Scholar]
- Bosch M, Pineda JR, Sunol C, Petriz J, Cattaneo E, Alberch J, Canals JM. Induction of GABAergic phenotype in a neural stem cell line for transplantation in an excitotoxic model of Huntington's disease. Exp Neurol. 2004;190:42–58. doi: 10.1016/j.expneurol.2004.06.027. [DOI] [PubMed] [Google Scholar]
- Butt SJ, Fuccillo M, Nery S, Noctor S, Kriegstein A, Corbin JG, Fishell G. The temporal and spatial origins of cortical interneurons predict their physiological subtype. Neuron. 2005;48:591–604. doi: 10.1016/j.neuron.2005.09.034. [DOI] [PubMed] [Google Scholar]
- Cao G, Ko CP. Schwann cell-derived factors modulate synaptic activities at developing neuromuscular synapses. J Neurosci. 2007;27:6712–6722. doi: 10.1523/JNEUROSCI.1329-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao QL, Zhang YP, Howard RM, Walters WM, Tsoulfas P, Whittemore SR. Pluripotent stem cells engrafted into the normal or lesioned adult rat spinal cord are restricted to a glial lineage. Exp Neurol. 2001;167:48–58. doi: 10.1006/exnr.2000.7536. [DOI] [PubMed] [Google Scholar]
- Cepko C. Immortalization of neural cells via oncogene transduction. Trends Neurosci. 1988;11:6–8. doi: 10.1016/0166-2236(88)90039-2. [DOI] [PubMed] [Google Scholar]
- Cepko CL. Immortalization of neural cells via retrovirus-mediated oncogene transduction. Annu Rev Neurosci. 1989;12:47–65. doi: 10.1146/annurev.ne.12.030189.000403. [DOI] [PubMed] [Google Scholar]
- Christopherson KS, Ullian EM, Stokes CC, Mullowney CE, Hell JW, Agah A, Lawler J, Mosher DF, Bornstein P, Barres BA. Thrombospondins are astrocyte-secreted proteins that promote CNS synaptogenesis. Cell. 2005;120:421–433. doi: 10.1016/j.cell.2004.12.020. [DOI] [PubMed] [Google Scholar]
- De Filippis L, Lamorte G, Snyder EY, Malgaroli A, Vescovi AL. A novel, immortal, and multipotent human neural stem cell line generating functional neurons and oligodendrocytes. Stem Cells. 2007;25:2312–2321. doi: 10.1634/stemcells.2007-0040. [DOI] [PubMed] [Google Scholar]
- Edmondson JC, Liem RK, Kuster JE, Hatten ME. Astrotactin: a novel neuronal cell surface antigen that mediates neuron-astroglial interactions in cerebellar microcultures. J Cell Biol. 1988;106:505–517. doi: 10.1083/jcb.106.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englund C, Fink A, Lau C, Pham D, Daza RA, Bulfone A, Kowalczyk T, Hevner RF. Pax6, Tbr2, and Tbr1 are expressed sequentially by radial glia, intermediate progenitor cells, and postmitotic neurons in developing neocortex. J Neurosci. 2005;25:247–251. doi: 10.1523/JNEUROSCI.2899-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng L, Heintz N. Differentiating neurons activate transcription of the brain lipid-binding protein gene in radial glia through a novel regulatory element. Development. 1995;121:1719–1730. doi: 10.1242/dev.121.6.1719. [DOI] [PubMed] [Google Scholar]
- Flax JD, Aurora S, Yang C, Simonin C, Wills AM, Billinghurst LL, Jendoubi M, Sidman RL, Wolfe JH, Kim SU, Snyder EY. Engraftable human neural stem cells respond to developmental cues, replace neurons, and express foreign genes. Nat Biotechnol. 1998;16:1033–1039. doi: 10.1038/3473. [DOI] [PubMed] [Google Scholar]
- Frederiksen K, Jat PS, Valtz N, Levy D, McKay R. Immortalization of precursor cells from the mammalian CNS. Neuron. 1988;1:439–448. doi: 10.1016/0896-6273(88)90175-4. [DOI] [PubMed] [Google Scholar]
- Friedlander DR, Milev P, Karthikeyan L, Margolis RK, Margolis RU, Grumet M. The neuronal chondroitin sulfate proteoglycan neurocan binds to neural adhesion molecules Ng-CAM/L1/NILE and N-CAM, and inhibits neuronal adhesion and neurite outgrowth. Journal of Cell Biology. 1994;125:669–680. doi: 10.1083/jcb.125.3.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisa PS, Goodman MN, Smith GM, Silver J, Jacobberger JW. Immortalization of immature and mature mouse astrocytes with SV40 T antigen. J Neurosci Res. 1994;39:47–56. doi: 10.1002/jnr.490390107. [DOI] [PubMed] [Google Scholar]
- Gal JS, Morozov YM, Ayoub AE, Chatterjee M, Rakic P, Haydar TF. Molecular and morphological heterogeneity of neural precursors in the mouse neocortical proliferative zones. J Neurosci. 2006;26:1045–1056. doi: 10.1523/JNEUROSCI.4499-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff L, Davila J, Jörnsten R, Keles S, Hart R. Bioinformatic Analysis of Neural Stem Cell. Journal of Biomolecular Techniques. 2007 In press. [PMC free article] [PubMed] [Google Scholar]
- Goldowitz D, Hamre K. The cells and molecules that make a cerebellum. Trends Neurosci. 1998;21:375–382. doi: 10.1016/s0166-2236(98)01313-7. [DOI] [PubMed] [Google Scholar]
- Gotz M, Hartfuss E, Malatesta P. Radial glial cells as neuronal precursors: a new perspective on the correlation of morphology and lineage restriction in the developing cerebral cortex of mice. Brain Res Bull. 2002;57:777–788. doi: 10.1016/s0361-9230(01)00777-8. [DOI] [PubMed] [Google Scholar]
- Gulacsi A, Lillien L. Sonic hedgehog and bone morphogenetic protein regulate interneuron development from dorsal telencephalic progenitors in vitro. J Neurosci. 2003;23:9862–9872. doi: 10.1523/JNEUROSCI.23-30-09862.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han SS, Kang DY, Mujtaba T, Rao MS, Fischer I. Grafted lineage-restricted precursors differentiate exclusively into neurons in the adult spinal cord. Exp Neurol. 2002;177:360–375. doi: 10.1006/exnr.2002.7995. [DOI] [PubMed] [Google Scholar]
- Han SS, Liu Y, Tyler-Polsz C, Rao MS, Fischer I. Transplantation of glial-restricted precursor cells into the adult spinal cord: survival, glial-specific differentiation, and preferential migration in white matter. Glia. 2004;45:1–16. doi: 10.1002/glia.10282. [DOI] [PubMed] [Google Scholar]
- Hartfuss E, Galli R, Heins N, Gotz M. Characterization of CNS precursor subtypes and radial glia. Dev Biol. 2001;229:15–30. doi: 10.1006/dbio.2000.9962. [DOI] [PubMed] [Google Scholar]
- Hasegawa K, Chang Y-W, H L, Berlin Y, Ikeda O, Kane-Goldsmith, Grumet M. Embryonic radial glia bridge spinal cord lesions and promote functional recovery following spinal cord injury. Exp Neurol. 2005;193:394–410. doi: 10.1016/j.expneurol.2004.12.024. [DOI] [PubMed] [Google Scholar]
- Hasegawa K, Chang YW, Li H, Berlin Y, Ikeda O, Kane-Goldsmith N, Grumet M. Embryonic radial glia bridge spinal cord lesions and promote functional recovery following spinal cord injury. Exp Neurol. 2005;193:394–410. doi: 10.1016/j.expneurol.2004.12.024. [DOI] [PubMed] [Google Scholar]
- Hatten ME. Neuronal regulation of astroglial morphology and proliferation in vitro. J Cell Biol. 1985;100:384–396. doi: 10.1083/jcb.100.2.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatten ME. Neuronal inhibition of astroglial cell proliferation is membrane mediated. J Cell Biol. 1987;104:1353–1360. doi: 10.1083/jcb.104.5.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatten ME, Alder J, Zimmerman K, Heintz N. Genes involved in cerebellar cell specification and differentiation. Curr Opin Neurobiol. 1997;7:40–47. doi: 10.1016/s0959-4388(97)80118-3. [DOI] [PubMed] [Google Scholar]
- Haubensak W, Attardo A, Denk W, Huttner WB. Neurons arise in the basal neuroepithelium of the early mammalian telencephalon: a major site of neurogenesis. Proc Natl Acad Sci U S A. 2004;101:3196–3201. doi: 10.1073/pnas.0308600100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heins N, Malatesta P, Cecconi F, Nakafuku M, Tucker KL, Hack MA, Chapouton P, Barde YA, Gotz M. Glial cells generate neurons: the role of the transcription factor Pax6. Nat Neurosci. 2002;5:308–315. doi: 10.1038/nn828. [DOI] [PubMed] [Google Scholar]
- Hevner RF. From radial glia to pyramidal-projection neuron: transcription factor cascades in cerebral cortex development. Mol Neurobiol. 2006;33:33–50. doi: 10.1385/MN:33:1:033. [DOI] [PubMed] [Google Scholar]
- Howard B, Chen Y, Zecevic N. Cortical progenitor cells in the developing human telencephalon. Glia. 2006;53:57–66. doi: 10.1002/glia.20259. [DOI] [PubMed] [Google Scholar]
- Johnson MA, Weick JP, Pearce RA, Zhang SC. Functional neural development from human embryonic stem cells: accelerated synaptic activity via astrocyte coculture. J Neurosci. 2007;27:3069–3077. doi: 10.1523/JNEUROSCI.4562-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsetos CD, Legido A, Perentes E, Mork SJ. Class III beta-tubulin isotype: a key cytoskeletal protein at the crossroads of developmental neurobiology and tumor neuropathology. J Child Neurol. 2003;18:815–866. doi: 10.1177/088307380301801205. discussion 867. [DOI] [PubMed] [Google Scholar]
- Kohwi M, Petryniak MA, Long JE, Ekker M, Obata K, Yanagawa Y, Rubenstein JL, Alvarez-Buylla A. A subpopulation of olfactory bulb GABAergic interneurons is derived from Emx1- and Dlx5/6-expressing progenitors. J Neurosci. 2007;27:6878–6891. doi: 10.1523/JNEUROSCI.0254-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T, Raff M. Oligodendrocyte precursor cells reprogrammed to become multipotential CNS stem cells. Science. 2000;289:1754–1757. doi: 10.1126/science.289.5485.1754. [DOI] [PubMed] [Google Scholar]
- Lavdas AA, Grigoriou M, Pachnis V, Parnavelas JG. The medial ganglionic eminence gives rise to a population of early neurons in the developing cerebral cortex. J Neurosci. 1999;19:7881–7888. doi: 10.1523/JNEUROSCI.19-18-07881.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JE. NeuroD and neurogenesis. Dev Neurosci. 1997;19:27–32. doi: 10.1159/000111182. [DOI] [PubMed] [Google Scholar]
- Levine ES, Crozier RA, Black IB, Plummer MR. Brain-derived neurotrophic factor modulates hippocampal synaptic transmission by increasing N-methyl-D-aspartic acid receptor activity. Proc Natl Acad Sci U S A. 1998;95:10235–10239. doi: 10.1073/pnas.95.17.10235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine ES, Dreyfus CF, Black IB, Plummer MR. Brain-derived neurotrophic factor rapidly enhances synaptic transmission in hippocampal neurons via postsynaptic tyrosine kinase receptors. Proc Natl Acad Sci U S A. 1995;92:8074–8077. doi: 10.1073/pnas.92.17.8074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Babiarz J, Woodbury J, Kane-Goldsmith N, Grumet M. Spatiotemporal heterogeneity of CNS radial glial cells and their transition to restricted precursors. Dev Biol. 2004;271:225–238. doi: 10.1016/j.ydbio.2004.02.028. [DOI] [PubMed] [Google Scholar]
- Li H, Berlin Y, Hart RP, Grumet M. Microtubules are critical for radial glial morphology: possible regulation by MAPs and MARKs. Glia. 2003;44:37–46. doi: 10.1002/glia.10267. [DOI] [PubMed] [Google Scholar]
- Li H, Grumet M. BMP and LIF signaling coordinately regulate lineage restriction of radial glia in the developing forebrain. Glia. 2007;55:24–35. doi: 10.1002/glia.20434. [DOI] [PubMed] [Google Scholar]
- Lu QR, Cai L, Rowitch D, Cepko CL, Stiles CD. Ectopic expression of Olig1 promotes oligodendrocyte formation and reduces neuronal survival in developing mouse cortex. Nat Neurosci. 2001;4:973–974. doi: 10.1038/nn718. [DOI] [PubMed] [Google Scholar]
- Lu QR, Sun T, Zhu Z, Ma N, Garcia M, Stiles CD, Rowitch DH. Common developmental requirement for Olig function indicates a motor neuron/oligodendrocyte connection. Cell. 2002;109:75–86. doi: 10.1016/s0092-8674(02)00678-5. [DOI] [PubMed] [Google Scholar]
- Lundberg C, Field PM, Ajayi YO, Raisman G, Bjorklund A. Conditionally immortalized neural progenitor cell lines integrate and differentiate after grafting to the adult rat striatum. A combined autoradiographic and electron microscopic study. Brain Res. 1996;737:295–300. doi: 10.1016/0006-8993(96)00923-7. [DOI] [PubMed] [Google Scholar]
- Macias MY, Syring MB, Pizzi MA, Crowe MJ, Alexanian AR, Kurpad SN. Pain with no gain: allodynia following neural stem cell transplantation in spinal cord injury. Exp Neurol. 2006;201:335–348. doi: 10.1016/j.expneurol.2006.04.035. [DOI] [PubMed] [Google Scholar]
- Malatesta P, Hartfuss E, Gotz M. Isolation of radial glial cells by fluorescent-activated cell sorting reveals a neuronal lineage. Development. 2000;127:5253–5263. doi: 10.1242/dev.127.24.5253. [DOI] [PubMed] [Google Scholar]
- Marin O, Anderson SA, Rubenstein JL. Origin and molecular specification of striatal interneurons. J Neurosci. 2000;20:6063–6076. doi: 10.1523/JNEUROSCI.20-16-06063.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsala M, Kakinohana O, Yaksh TL, Tomori Z, Marsala S, Cizkova D. Spinal implantation of hNT neurons and neuronal precursors: graft survival and functional effects in rats with ischemic spastic paraplegia. Eur J Neurosci. 2004;20:2401–2414. doi: 10.1111/j.1460-9568.2004.03702.x. [DOI] [PubMed] [Google Scholar]
- Mayer-Proschel M, Kalyani AJ, Mujtaba T, Rao MS. Isolation of lineage-restricted neuronal precursors from multipotent neuroepithelial stem cells. Neuron. 1997;19:773–785. doi: 10.1016/s0896-6273(00)80960-5. [DOI] [PubMed] [Google Scholar]
- Miyata T, Kawaguchi A, Okano H, Ogawa M. Asymmetric inheritance of radial glial fibers by cortical neurons. Neuron. 2001;31:727–741. doi: 10.1016/s0896-6273(01)00420-2. [DOI] [PubMed] [Google Scholar]
- Mizutani K, Yoon K, Dang L, Tokunaga A, Gaiano N. Differential Notch signaling distinguishes neural stem cells from intermediate progenitors. Nature. 2007;449:351–355. doi: 10.1038/nature06090. [DOI] [PubMed] [Google Scholar]
- Mo Z, Moore AR, Filipovic R, Ogawa Y, Kazuhiro I, Antic SD, Zecevic N. Human cortical neurons originate from radial glia and neuron-restricted progenitors. J Neurosci. 2007;27:4132–4145. doi: 10.1523/JNEUROSCI.0111-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadarajah B, Parnavelas JG. Modes of neuronal migration in the developing cerebral cortex. Nat Rev Neurosci. 2002;3:423–432. doi: 10.1038/nrn845. [DOI] [PubMed] [Google Scholar]
- Noble M, Arhin A, Gass D, Mayer-Proschel M. The cortical ancestry of oligodendrocytes: common principles and novel features. Dev Neurosci. 2003;25:217–233. doi: 10.1159/000072270. [DOI] [PubMed] [Google Scholar]
- Noble M, Proschel C, Mayer-Proschel M. Getting a GR(i)P on oligodendrocyte development. Dev Biol. 2004;265:33–52. doi: 10.1016/j.ydbio.2003.06.002. [DOI] [PubMed] [Google Scholar]
- Noctor SC, Flint AC, Weissman TA, Wong WS, Clinton BK, Kriegstein AR. Dividing precursor cells of the embryonic cortical ventricular zone have morphological and molecular characteristics of radial glia. J Neurosci. 2002;22:3161–3173. doi: 10.1523/JNEUROSCI.22-08-03161.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan JZ, Jornsten R, Hart RP. Screening anti-inflammatory compounds in injured spinal cord with microarrays: a comparison of bioinformatics analysis approaches. Physiol Genomics. 2004;17:201–214. doi: 10.1152/physiolgenomics.00177.2003. [DOI] [PubMed] [Google Scholar]
- Papas IS, Parnavelas JG. Basic fibroblast growth factor promotes the generation and differentiation of calretinin neurons in the rat cerebral cortex in vitro. Eur J Neurosci. 1998;10:1436–1445. doi: 10.1046/j.1460-9568.1998.00147.x. [DOI] [PubMed] [Google Scholar]
- Peng HB, Yang JF, Dai Z, Lee CW, Hung HW, Feng ZH, Ko CP. Differential effects of neurotrophins and schwann cell-derived signals on neuronal survival/growth and synaptogenesis. J Neurosci. 2003;23:5050–5060. doi: 10.1523/JNEUROSCI.23-12-05050.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfrieger FW, Barres BA. Synaptic efficacy enhanced by glial cells in vitro. Science. 1997;277:1684–1687. doi: 10.1126/science.277.5332.1684. [DOI] [PubMed] [Google Scholar]
- Piper DR, Mujtaba T, Keyoung H, Roy NS, Goldman SA, Rao MS, Lucero MT. Identification and characterization of neuronal precursors and their progeny from human fetal tissue. J Neurosci Res. 2001;66:356–368. doi: 10.1002/jnr.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleasure SJ, Anderson S, Hevner R, Bagri A, Marin O, Lowenstein DH, Rubenstein JL. Cell migration from the ganglionic eminences is required for the development of hippocampal GABAergic interneurons. Neuron. 2000;28:727–740. doi: 10.1016/s0896-6273(00)00149-5. [DOI] [PubMed] [Google Scholar]
- Poluch S, Juliano SL. A normal radial glial scaffold is necessary for migration of interneurons during neocortical development. Glia. 2007;55:822–830. doi: 10.1002/glia.20488. [DOI] [PubMed] [Google Scholar]
- Rao MS, Mayer-Proschel M. Glial-restricted precursors are derived from multipotent neuroepithelial stem cells. Dev Biol. 1997;188:48–63. doi: 10.1006/dbio.1997.8597. [DOI] [PubMed] [Google Scholar]
- Rao MS, Noble M, Mayer-Proschel M. A tripotential glial precursor cell is present in the developing spinal cord. Proc Natl Acad Sci U S A. 1998;95:3996–4001. doi: 10.1073/pnas.95.7.3996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowitch DH, Lu QR, Kessaris N, Richardson WD. An 'oligarchy' rules neural development. Trends Neurosci. 2002;25:417–422. doi: 10.1016/s0166-2236(02)02201-4. [DOI] [PubMed] [Google Scholar]
- Ryder EF, Snyder EY, Cepko CL. Establishment and characterization of multipotent neural cell lines using retrovirus vector-mediated oncogene transfer. J Neurobiol. 1990;21:356–375. doi: 10.1002/neu.480210209. [DOI] [PubMed] [Google Scholar]
- Tominaga M, Honda S, Okada A, Ikeda A, Kinoshita S, Tomooka Y. A bipotent neural progenitor cell line cloned from a cerebellum of an adult p53-deficient mouse generates both neurons and oligodendrocytes. Eur J Neurosci. 2005;21:2903–2911. doi: 10.1111/j.1460-9568.2005.04119.x. [DOI] [PubMed] [Google Scholar]
- Ullian EM, Christopherson KS, Barres BA. Role for glia in synaptogenesis. Glia. 2004;47:209–216. doi: 10.1002/glia.20082. [DOI] [PubMed] [Google Scholar]
- Ullian EM, Harris BT, Wu A, Chan JR, Barres BA. Schwann cells and astrocytes induce synapse formation by spinal motor neurons in culture. Mol Cell Neurosci. 2004;25:241–251. doi: 10.1016/j.mcn.2003.10.011. [DOI] [PubMed] [Google Scholar]
- Ullian EM, Sapperstein SK, Christopherson KS, Barres BA. Control of synapse number by glia. Science. 2001;291:657–661. doi: 10.1126/science.291.5504.657. [DOI] [PubMed] [Google Scholar]
- Villa A, Snyder EY, Vescovi A, Martinez-Serrano A. Establishment and properties of a growth factor-dependent, perpetual neural stem cell line from the human CNS. Exp Neurol. 2000;161:67–84. doi: 10.1006/exnr.1999.7237. [DOI] [PubMed] [Google Scholar]
- Wichterle H, Turnbull DH, Nery S, Fishell G, Alvarez-Buylla A. In utero fate mapping reveals distinct migratory pathways and fates of neurons born in the mammalian basal forebrain. Development. 2001;128:3759–3771. doi: 10.1242/dev.128.19.3759. [DOI] [PubMed] [Google Scholar]
- Wonders CP, Anderson SA. The origin and specification of cortical interneurons. Nat Rev Neurosci. 2006;7:687–696. doi: 10.1038/nrn1954. [DOI] [PubMed] [Google Scholar]
- Woo NH, Lu B. Regulation of cortical interneurons by neurotrophins: from development to cognitive disorders. Neuroscientist. 2006;12:43–56. doi: 10.1177/1073858405284360. [DOI] [PubMed] [Google Scholar]
- Wu YY, Mujtaba T, Han SS, Fischer I, Rao MS. Isolation of a glial-restricted tripotential cell line from embryonic spinal cord cultures. Glia. 2002;38:65–79. doi: 10.1002/glia.10049. [DOI] [PubMed] [Google Scholar]
- Xu Q, Cobos I, De La Cruz E, Rubenstein JL, Anderson SA. Origins of cortical interneuron subtypes. J Neurosci. 2004;24:2612–2622. doi: 10.1523/JNEUROSCI.5667-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K, Hisatsune T, Uchino S, Nakamura T, Kudo Y, Kaminogawa S. NMDA receptor mediated Ca2+ responses in neurons differentiated from p53−/− immortalized Murine neural stem cells. PLoS ONE. 1999;264:165–167. doi: 10.1016/s0304-3940(99)00134-2. [DOI] [PubMed] [Google Scholar]
- Ying QL, Stavridis M, Griffiths D, Li M, Smith A. Conversion of embryonic stem cells into neuroectodermal precursors in adherent monoculture. Nat Biotechnol. 2003;21:183–186. doi: 10.1038/nbt780. [DOI] [PubMed] [Google Scholar]
- Yokota Y, Gashghaei T, Han C, Watson H, Campbell KJ, Anton ES. Radial glial dependent and independent dynamics of interneuronal migration in the developing cerebral cortex. PLoS ONE. 2007;2:e794. doi: 10.1371/journal.pone.0000794. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
http://cord.rutgers.edu/appendix/Li/Supplemental_Table_1.html