Abstract
Following an initial report, there have been multiple replications of an association of alcohol dependence (AD) to markers within a haplotype block that includes the 3′-half of the gene encoding the GABAA α-2 subunit (GABRA2), on chromosome 4p. We examined the intergenic extent of this haplotype block and the association to AD of markers in the adjacent 5′ haplotype block in GABRG1, which encodes the GABAA receptor γ-1 subunit. We genotyped 15 SNPs in the GABRG1-GABRA2 interval as well as at 34 ancestry informative markers in three samples: 435 AD and 635 screened control subjects from Connecticut and 812 participants from a multi-center AD treatment trial. We observed two large haplotype blocks in the GABRG1-GABRA2 intergenic interval with a region of increased recombination midway between the two genes. Markers in the two haplotype blocks were in moderate linkage disequilibrium. Compared with markers in the GABRA2 haplotype block, markers in the 5′ GABRG1 haplotype showed greater allelic, genotypic, and haplotypic association with AD in European Americans from both AD samples. Logistic regression analysis indicated that genetic elements in the GABRG1 haplotype block likely contribute to AD risk in an additive fashion while those in the GABRA2 haplotype block may act in a dominant fashion in relation to risk of AD.
Keywords: psychiatric genetics, GABA(A) receptor, GABRG1, GABRA2, alcohol dependence, polymorphism
INTRODUCTION
Alcohol dependence [AD (MIM 103780)] is a common psychiatric disorder, recently estimated to affect 3.8% of the adult US population during a one-year period (Grant et al. 2004). A variety of adverse consequences are associated with AD, including medical, social, and legal problems (Caetano&Cunradi 2002). Based on heritability estimates ranging from 0.52 to 0.64 (Kendler 2001), considerable efforts have been made to identify genes that increase risk for the disorder.
Genomewide linkage scans implicated a region on chromosome 4p12 that harbors a clusterof four genes encoding GABAA receptor subunits (γ-1, α-2, α-4, and β-1) (Long et al. 1998; Reich et al. 1998). Because GABAA receptors have been implicated in biological processes related to the acute and chronic effects of alcohol (Koob 2004; Krystal et al. 2006), the GABAA receptor genes at 4p12 are both positional and functional candidates for AD risk. Fine mapping of markers in the chromosome 4p GABAA receptor gene cluster in relation to AD involved a study of 69 single nucleotide polymorphisms (SNPs) in multiplex families from the Collaborative Study on the Genetics of Alcoholism (COGA) (Edenberg et al. 2004). These investigators found significant associations for multiple markers in the gene encoding the GABAA α-2 subunit [GABRA2 (MIM 137140)] and for a single marker in the adjacent GABRG1 (MIM 137166) gene, which encodes the GABAA γ-1 subunit. There was no evidence of association with other members of the gene cluster. This association to GABRA2 was subsequently evaluated in three independent samples of subjects of European ancestry (Covault et al. 2004; Lappalainen et al. 2005; Fehr et al. 2006), and in each sample an association of AD with a haplotype block spanning the central and 3′-regions of GABRA2 was observed.
Two other clusters of genes encoding GABAA subunits, located on chromosomes 5 and 15, have been examined for association to AD in other studies. Results for markers in the GABAA gene cluster containing genes for β2, α6, α1 and γ2 subunits on chromosome 5q have been mixed, with association reported in some samples (Loh et al. 2000; Radel et al. 2005), but not others (Sander et al. 1999; Dick et al. 2005). Fine mapping of a GABAA gene cluster containing genes for the α5, β3 and γ3 subunits on chromosome 15q showed modest evidence of haplotypic association to AD for SNPs in GABRG3, encoding the γ3 subunit (Dick et al. 2004).
The present study extends the findings of association of AD to the chromosome 4 GABAA gene cluster by examining the intergenic extent of the GABRA2 3′ region haplotype block associated with AD and by examining markers in the adjacent haplotype block in the 5′-region of GABRG1. We observed a stronger association of GABRG1 5′-upstream markers with AD in both study samples compared with markers in the GABRA2 haplotype block.
MATERIALS and METHODS
Subjects
Connecticut AD subjects [372 non-Hispanic Caucasians of European decent (EA) and 63 African-Americans (AA)] were recruited as part of ongoing studies of the genetics of AD or from clinical trials for the treatment of AD at the University of Connecticut Health Center (UCHC), Farmington, CT and the VA Connecticut Healthcare Center (VA-CT), West Haven, CT. Controls from CT (535 EA and 100 AA) were recruited by advertisement in the greater Hartford, CT area. Psychiatric diagnoses were made using the Structured Clinical Interview for DSM-III-R or DSM-IV (SCID) (First et al. 1997), or the Semi-Structured Assessment for Drug Dependence and Alcoholism (SSADDA) (Pierucci-Lagha et al. 2005). All controls were screened using the SCID or the SSADDA to exclude individuals with an alcohol or drug use disorder, or other major Axis I psychiatric disorder. Subjects were paid for their participation and all provided written, informed consent to participate in study protocols that were approved by the institutional review boards at UCHC, Yale University School of Medicine, and/or VA-CT. The diagnosis of AD for Project MATCH subjects (727 EA and 85 AA) was made using the Computerized Diagnostic Interview for DSM-IV (Blouin et al. 1988; American Psychiatric Association 1994). For both the CT and Project MATCH samples, analysis was limited to self-identified AA and EA subjects. For analysis of AA subjects, we pooled AD subjects from the CT (n=63) and Project MATCH (n=85) samples.
Demographic and clinical characteristics of the participant sample are listed in Table 1. For both EA and AA samples, the control groups were significantly younger than the AD groups and included more females. Similar to other samples, AD subjects had a moderate prevalence of affective/anxiety disorders, lifetime diagnosis of cocaine or opioid dependence (lifetime drug dependence diagnoses were not available for the Project MATCH sample) or antisocial personality disorder. Among the CT EA subjects, 294 controls (55%) and 264 alcoholics (71%) were examined in our prior association study of GABRA2 SNPs A-H (Covault et al. 2004).
Table 1.
Demographic and Clinical Characteristics of Sample.
| EA - Connecticut sample | EA - Project Match | AA Subjects | |||
|---|---|---|---|---|---|
| Controls (n=535) | AD (n=372) | AD (n=727) | Controls (n=100) | AD (n=148) | |
| Age - yrs (SD) | 30.0 (10.6) | 43.8 (9.1)*** | 40.6 (11.1)*** | 31.7 (10.4) | 41.6 (8.5)*** |
| % male | 42.8 | 75.3*** | 73.2*** | 34.0 | 66.9*** |
| % co-morbid diagnosis (lifetime): | |||||
| affective/anxiety disorder | 0 | 32 | 25 | 0 | 22 |
| cocaine or opioid dependence | 0 | 14.8 | n.d. | 0 | n.d. |
| antisocial personality disorder | 0 | 6.5 | 9.5 | 0 | 7.0 |
p< 0.001 vs. controls (by race);
n.d. – incomplete or no data. For the AA AD subjects in the CT sample (n=63), 52% reported lifetime cocaine or opioid dependence.
Genotyping
GABRG1 and GABRA2 SNPs were genotyped using a closed-tube fluorescent TaqMan 5′-nuclease allelic discrimination assay using MGB-probes and primers designed using Primer Express v2.0 software [Applied Biosystems Inc. (ABI) Foster City, CA]. Fluorescence plate reads and genotype calls were made using ABI 7700 and 7500 Sequence Detection Systems. Tenng of genomic DNA was PCR amplified in 96-well plates using a 10 μl reaction volume for 40 cycles at 94°C for 15s followed by 60°C for 60s. Repeat genotyping was carried out for 16% of samples with an observed error rate of 0.5%. PCR amplifications failed or provided ambiguous genotype results from 1.5% of reactions (1.7% controls, 3.4% CT cases and 0.4% Project MATCH cases). To estimate genetic ancestry proportions for each subject, DNA samples were also genotyped using a panel of 34 short tandem repeat ancestry informative markers: CSF1PO, D2S1338, D3S1358, D5S818, D7S820, D8S1179, D13S317, D16S539, D18S51, D19S433, D21S11, FGA, TH01, TPO-9, vWA, D17S799, D1S196, D7S640, D8S1827, D7S657, D22S274, D5S407, D2S162, D10S197, D11S935, D9S175, D5S410, D7S2469, D16S3017, D10S1786, D15S1002, D6S1610, D1S2628, D12S352 as described previously (Stein et al. 2004; Luo et al. 2005; Yang et al. 2005).
Genotype distributions were in Hardy-Weinberg equilibrium (HWE) for all 15 SNPs for each of the three EA groups (controls: mean p=0.55, range 0.22–1.0; CT AD subjects: mean p=0.48, range 0.06–1.0; Project MATCH AD subjects: mean p=0.48, range 0.15–1.0). For the AA groups, genotype distributions were consistent with HWE expectations for all but two markers in the control group [mean p=0.41, range 0.04–1.0; rs1440133 (SNP7), p=0.04 and rs529826 (SNP C), p=0.04] and two markers in the AD group [mean p=0.63, range 0.04–1.0; rs534459 (SNP B), p=0.04 and rs529826 (SNP C), p=0.04].
Data analysis
Diagnostic groups were compared on age using ANOVA and on sex and allele frequencies using 2X2 contingency tables and the χ2 test. Linkage disequilibrium (LD) plots, haplotype blocks, allele frequencies, tests of HWE, and haplotype frequencies for each population and diagnostic group were generated using the software program Haploview v3.32 (Barrett et al. 2005). Best-estimate haplotype pairs for each subject were generated using PHASE v2.1.1 software, which incorporates a Bayesian statistical method for reconstructing estimated haplotypes from population data (Stephens et al. 2001; Stephens&Donnelly 2003). Haplotype pairs were generated separately for AA and EA populations. The software program STRUCTURE v2.1 (Pritchard et al. 2000; Falush et al. 2003) was used to generate estimates of the proportion of African vs. European genetic ancestry for each subject based on genotype results from the 34 ancestry informative markers. Simulations used 100,000 burn-ins followed by 500,000 runs and a population parameter K=2. Estimated haplotype frequencies were compared for alcoholics vs. controls and AA vs. EA controls using a series of 2X2 contingency tables for each haplotype compared to the sum of all other haplotypes. The odds ratio (OR) is reported as a measure of effect size. Binary logistic regression analysis was used to control for age, sex, and the proportion of EA genetic ancestry in determining a corrected OR for AD as a function risk haplotype. Statistical analysis was carried out using SPSS software v14.0.
RESULTS
We genotyped 15 SNPs (average inter-marker interval of 20,530 bp, range 6,598–39,849 bp) extending 287,400 bp in the GABRG1-GABRA2 region of chromosome 4 in a sample of 1,634 self-identified EA and 248 AA subjects, Table 1, comprising AD and control subjects from Connecticut (CT) and a sample of AD subjects from Project MATCH, an NIAAA-sponsored, multi-center clinical trial conducted at 10 sites throughout the US. That study sought to identify predictors of response to three psychotherapeutic treatments for alcoholism (Project MATCH Research Group. 1998). Table 2 lists the primers, MGB-probes and annealing-extension temperatures used to examine the 15 GABRG1-GABRA2 region SNPs. The trivial names for the SNPs examined in GABRA2 (SNPs A-H) are the same as were used previously (Covault et al. 2004); the additional 7 SNPs in the intergenic and 5′-region of GABRG1 are numbered 1–7.
Table 2.
GABRA2 SNPs examined in samples of alcohol-dependent and control subjects
| SNP trivial name (this paper) |
NCBI SNP (dbSNP build 126) |
Genome assembly position1 |
Location relative to GABRA2 |
Location relative to GABRG1 |
PCR amplicon primers | Taqman MGB probes |
|---|---|---|---|---|---|---|
| 1 | rs2350439 | 45779174 | intron 3 | TTCTCTACTCCCATACTCTTATCTCATCTTC GGGAGTGAGTGGGAATGTAGATTTAA |
Vic-CCATGTTTCTCAAAATAGTAGTT 6FAM-CATGTTTCTCAAAATGGTAGTT |
|
| 2 | rs1497577 | 45788470 | intron 2 | TCCACTTGCGGGTATGTTTGA AATAAAGAGCAGATTGAAACACAAATAGG |
Vic-TGCTATGTCTTTACATCTC2 6FAM- TGCTATGTCATTACATCTC2 |
|
| 3 | rs1391166 | 45800624 | intron 1 | GTTTTGCTATCCCTATTAGTGCTCATTTA CTTGAATTTGGCTCCTGATTGC |
Vic- TGGTTAGAATGAAACAAAAA 6FAM- TGGTTAGAATGATACAAAAA |
|
| 4 | rs7654165 | 45821824 | 125 kb 3′ | 1 kb 5′ | GCTTTCTCTGCCTAGCCATCTTC GGTGGATTAACAGCTAAAGACTCCAA |
Vic-TCTTAAAACGCCCTCCC 6FAM- TCTTAAAACGCCTTCCC |
| 5 | rs10033451 | 45847089 | 99 kb 3′ | 26 kb 5′ ′ | CACTGACACATTACCCCGAAATT AAGAAAAGGGAAATCAGCGTACAAT |
Vic- CTTCTTAAACTTGTAATCAATT 6FAM- CTTAAACTTGCAATCAAT |
| 6 | rs4280776 | 45883288 | 63 kb 3′ | 62 kb 5′ | CTTTAAATTTTCATCTCTCTTACCATTCAG GAAAAATCACCTATAATCCCTCCTCTCTA |
Vic-AGTTTTGTTTCCCAATCCA 6FAM- AGTTTTGTTTCCTAATCCA |
| 7 | rs1440133 | 45896677 | 50 kb 3′ | 76 kb 5′ | CACAATGTGCTTTCTCATGAATTCA TCCTAATAGTCCAGGGAACTGTAATGTT |
Vic- CTACACAACCTCTTTTTC2 6FAM- CTACACAACCTCCTTTTC2 |
| A | rs567926 | 45936526 | 10 kb 3′ | 116 kb 5′ | TTTTGTCCATGCTCCTGTCATTT TTGAACTGTGTTGCCTGTTCTATAGAA |
Vic- TCTGTCTGACGTTACATTT 6FAM- ATTTCTGTCTGACATTACATTT |
| B | rs534459 | 45951562 | intron 9 | CCGGAATTCTTAACCACTAAATTTCA GCACTTCTGGACATTTTTTTGCTT |
Vic- TCTACCTACACATCCC 6FAM- TCTACCTACGCATCCC |
|
| C | rs529826 | 45966409 | intron 8 | ACACCCAGCTGGATTTATATATCAATAT ACCACATTCTTCCCCAAAATGA |
Vic- CAGAAAAGAAACACCTACAGTTA 6FAM- CAGAAAAGAAACATCTACAGTTA |
|
| D | rs279869 | 46002752 | intron 6 | TGAAAGGTGAAGAGCAAATTTCTTG TGGCCTAAAGGAGATGTTATAATCATCT |
Vic- AGAGTCTGTTAGAATCACT 6FAM- AGAGTCTGTTATAATCACTG |
|
| E | rs279858 | 46009350 | exon 5 (K132K) | GAAGCAACTTATTTGGCATTGTCA TCTGGACTCCAGATACCTTTTTTCA |
Vic-TGAGCTACTGATTTCTTCCCAT 6FAM-TGAGCTACTGATTTTTTCCCAT |
|
| F | rs279844 | 46024412 | intron 4 | GAAGCTACTGGGATATTAATTAGTTCAGTAGTTA CAAATGGCAACTTCTGATAAAGGA |
Vic- AGTTGTGAGTTTTAATATCT2 6FAM-: AGTTGTGAGATTTAATATCT2 |
|
| G | rs279837 | 46034080 | intron 3 | CAATGGTCCTGAGCATCCTCTAAT GTGGGACTTGACTTTTGCACAA |
Vic- CAAAGAGCAAAGTAAATAT 6FAM- CAAAGAGCAGAGTAAATAT |
|
| H | rs9291283 | 46066590 | intron 3 | ABI SNP assay 8262290 | TTAGAGTTATATTAAATATAGTAAC[A/G] GGAGATTTGTTCCAAAGGAAATGTG |
Nucleotide position relative to GABRA2 ATG using the March 2006 version of the human genome chromosome 4 annotation (www.genome.ucsc.edu).
Probe sequence from chromosome 4 negative strand.
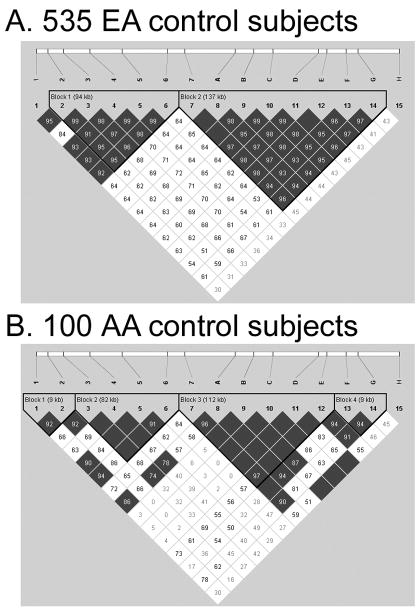
Two main haplotype blocks were observed in EA controls (Figure 1A). One was a 94 kb region that included SNP markers 2–6 and extended from GABRG1 intron 2 to 62,000 bp 5′ of the GABRG1 transcript start site. The second haplotype block extended 137 kb and included SNP marker 7 (50,000 bp 3′ from the 3′UTR of the GABRA2 transcript) and the 7 adjacent SNP markers A-G (ending in GABRA2 intron 3), which we had previously identified as being in high LD with one another (Covault et al. 2004). The LD plot and haplotype block structure for AA subjects was similar to EA subjects regarding the presence of a region of increased recombination in the 14 kb interval between SNPs 6 and 7 (Figure 1B). Among AAs, SNPs 3–6 were in high LD with one another in the GABRG1 block and in the GABRA2 haplotype block region SNPs 7 and A-E showed high LD. The LD plot and haplotype block structure for AD subjects paralleled those for the controls of their respective racial group (data not shown).
Figure 1.
LD plot from Haploview 3.32 for EA control subjects (Panel A) compared with that for AA control subjects (Panel B). Pair-wise SNP |D′| values (x100) of linkage are shown together with haplotype blocks identified using the four-gamete rule (Wang et al. 2002). Marker pairs in complete LD are indicated by an empty box. Darkened blocks indicate SNP pairs without evidence of extensive recombination (i.e. 4-gamete rule for haplotype block characterization with one (or two) 2-SNP haplotypes having a frequency < 0.02).
GABRG1 allele and genotype frequencies for the three EA groups and the two AA groups are shown in Table 3. Among EAs, all SNPs in the GABRG1 haplotype block (SNPs 2–6) showed significant association (nominal p<0.05) with AD for both allele and genotype frequencies in both the CT and Project MATCH samples. The strongest association (p=0.001) was observed for SNPs 4 and 5 in the 5′-upstream region of the gene. GABRA2 allele and genotype frequencies (SNP7 and SNPs A-H) are shown in Table 4. For the GABRA2 haplotype block region, the 3 SNPs closest to GABRG1 (i.e., SNPs 7, A, and B) showed modest allelic and genotypic association with AD in the CT EA sample (p=0.009–0.016 for allelic association), while SNPs D-G showed less evidence of association (p=0.021–0.032). In parallel with the CT sample, MATCH EA alcoholics showed a non-significantly higher prevalence than controls of the minor allele for each SNP in the 3′ GABRA2 haplotype block (p=0.083 – 0.167). None of the 15 markers showed allelic association with AD in the AA sample. Allele frequencies in the controls differed significantly by racial population for 13 of the 15 SNPs (Tables 3 and 4, right-hand column).
Table 3.
GABRG1 SNP allele and genotype frequencies for self-identified EA and AA subjects.
| EA - Connecticut sample | EA - Project Match | AA Subjects | EA c/w AA controls |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| SNP rs# location |
† Allele/ genotype |
Controls (n=535) |
AD (n=372) |
aχ2;p- value |
AD (n=727) |
bχ2; p-value |
Controls (n=100) |
AD (n=148) |
cχ2; p-value |
dχ2; p-value |
| rs2350439-1 | G | .508 | .583 | 9.72; | .555 | 3.37; | .535 | .555 | 0.19 | 0.50 |
| GABRG1-intron 3 | A | .492 | .417 | .002 | .455 | .066 | .465 | .445 | .665 | .479 |
| G/G | .261 | .373 | .297 | .290 | .308 | |||||
| G/A | .494 | .421 | 12.54; | .496 | 3.40; | .490 | .493 | 0.20 | ||
| A/A | .245 | .206 | .002 | .207 | .183 | .220 | .200 | .906 | ||
| rs1497577-2 | T | .544 | .473 | 8.46; | .493 | 6.28; | .636 | .583 | 1.41 | 5.78 |
| GABRG1-intron 2 | A | .456 | .527 | .004 | .507 | .012 | .364 | .417 | .234 | .016 |
| T/T | .305 | .258 | .243 | .424 | .345 | |||||
| T/A | .477 | .430 | 10.03; | .499 | 6.62; | .424 | .476 | 1.56 | ||
| A/A | .218 | .312 | .007 | .257 | .037 | .152 | .179 | .45 | ||
| rs1391166-3 | A | .505 | .563 | 5.40; | .564 | 8.41; | .696 | .728 | 0.52 | 21.3 |
| GABRG1-intron 1 | T | .495 | .437 | .020 | .436 | .004 | .304 | .272 | .470 | <.001 |
| A/A | .254 | .351 | .312 | .512 | .537 | |||||
| A/T | .502 | .425 | 9.24; | .505 | 8.71; | .369 | .381 | .87 | ||
| T/T | .244 | .225 | .010 | .184 | .013 | .119 | .119 | .650 | ||
| rs7654165-4 | C | .534 | .456 | 10.56; | .467 | 11.09; | .321 | .270 | 1.50 | 29.9 |
| GABRG1-1 kb 5′ | T | .466 | .544 | .001 | .533 | .001 | .679 | .730 | .221 | <.001 |
| C/C | .289 | .233 | .209 | .112 | .095 | |||||
| C/T | .490 | .446 | 11.76; | .515 | 11.97; | .418 | .351 | 1.69 | ||
| T/T | .221 | .321 | .003 | .276 | .003 | .469 | .554 | 0.43 | ||
| rs10033451-5 | T | .559 | .479 | 10.89; | .496 | 9.69; | .703 | .639 | 2.17 | 13.9 |
| GABRG1-26 kb 5′ | C | .441 | .521 | .001 | .504 | .002 | .297 | .361 | .140 | <.001 |
| T/T | .323 | .251 | .244 | .521 | .405 | |||||
| T/C | .472 | .457 | 10.68; | .503 | 10.28; | .365 | .466 | 3.22 | ||
| C/C | .205 | .292 | .005 | .253 | .006 | .115 | .128 | .200 | ||
| rs4280776-6 | T | .554 | .490 | 6.86; | .495 | 8.39; | .717 | .652 | 2.22 | 17.0 |
| GABRG1-62 kb 5′ | C | .446 | .510 | .009 | .505 | .004 | .283 | .348 | .136 | <.001 |
| T/T | .317 | .257 | .252 | .522 | .414 | |||||
| GABRA2-63 kb 3′ | T/C | .475 | .464 | 6.71; | .506 | 9.08; | .391 | .476 | 2.65 | |
| C/C | .208 | .278 | .035 | .242 | .011 | .087 | .110 | .266 | ||
Allele nucleotide designation refers to the chromosome plus strand sequence.
EA Controls compared with EA Connecticut Alcoholics
EA Controls compared with EA MATCH Alcoholics
AA Controls compared with AA Alcoholics (Connecticut and MATCH samples combined)
EA Controls compared with AA Controls
Table 4.
GABRA2 SNPs allele and genotype frequencies for self-identified EA and AA subjects.
| EA - Connecticut sample | EA - Project Match | AA Subjects | EA vs. AA controls |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| SNP rs# location |
† Allele/ genotype |
Controls (n=535) |
AD (n=372) |
aχ2; p-value |
AD (n=727) |
bχ2; p-value |
Controls (n=100) |
AD (n=148) |
cχ2; p-value |
dχ2; p-value |
| rs1440133-7 | G | .607 | .549 | 5.81; | .580 | 1.91 | .773 | .755 | 0.21 | 19.4 |
| GABRA2-50 kb 3′ | A | .393 | .451 | .016 | .420 | .167 | .227 | .245 | .648 | <.001 |
| G/G | .380 | .291 | .324 | .639 | .559 | |||||
| G/A | .453 | .515 | 7.30; | .512 | 4.97 | .268 | .393 | 5.03 | ||
| A/A | .166 | .194 | .026 | .165 | .083 | .093 | .048 | .08 | ||
| rs567926-A | A | .609 | .546 | 6.56; | .575 | 2.94; | .755 | .741 | 0.12 | 14.4 |
| GABRA2-9 kb 3′ | G | .391 | .454 | .010 | .425 | .087 | .245 | .259 | .727 | <.001 |
| A/A | .384 | .302 | .320 | .587 | .532 | |||||
| A/G | .451 | .488 | 6.55; | .510 | 5.88 | .337 | .417 | 1.83 | ||
| G/G | .165 | .210 | .038 | .170 | .053 | .075 | .050 | .40 | ||
| rs534459-B | G | .597 | .534 | 6.81; | .570 | 1.93; | .281 | .303 | 0.27 | 66.9 |
| GABRA2-intron 9 | A | .403 | .466 | .009 | .430 | .165 | .719 | .697 | .601 | <.001 |
| G/G | .365 | .283 | .311 | .112 | .051 | |||||
| G/A | .466 | .503 | 7.03; | .517 | 4.18; | .337 | .504 | 7.76 | ||
| A/A | .170 | .214 | .030 | .172 | .123 | .551 | .445 | .02 | ||
| rs529826-C | T | .600 | .557 | 3.02; | .576 | 1.39; | .279 | .298 | 0.18 | 61.4 |
| GABRA2-intron 8 | C | .400 | .443 | 0.82 | .424 | .238 | .721 | .702 | 0.669 | <.001 |
| T/T | .365 | .306 | .319 | .128 | .050 | |||||
| T/C | .496 | .502 | 3.24; | .514 | 3.09; | .302 | .496 | 10.33 | ||
| C/C | .166 | .192 | .197 | .166 | .213 | .570 | .454 | .006 | ||
| rs279869-D | G | .590 | .536 | 4.91; | .568 | 1.21; | .281 | .292 | 0.08 | 63.8 |
| GABRA2-intron 6 | T | .410 | .464 | .027 | .432 | .272 | .719 | .708 | .782 | <.001 |
| G/G | .357 | .285 | .310 | .112 | .049 | |||||
| G/T | .467 | .503 | 5.23; | .517 | 3.57; | .337 | .486 | 6.96 | ||
| T/T | .176 | .212 | .073 | .174 | .168 | .557 | .465 | .031 | ||
| rs279858-E | T | .598 | .542 | 5.29; | .570 | 2.00; | .760 | .762 | 0.00 | 18.6 |
| GABRA2-exon 5 | C | .402 | .458 | .021 | .430 | .157 | .240 | .236 | .956 | <.001 |
| T/T | .365 | .297 | .312 | .612 | .574 | |||||
| T/C | .465 | .490 | 5.21; | .515 | 4.16; | .296 | .376 | 2.76 | ||
| C/C | .169 | .213 | .074 | .173 | .125 | .092 | .050 | .252 | ||
| rs279844-F | A | .579 | .525 | 4.85; | .546 | 2.67 | .478 | .525 | 0.97 | 6.50 |
| GABRA2-intron 4 | T | .421 | .475 | .028 | .454 | .103 | .522 | .475 | .324 | .011 |
| A/A | .341 | .288 | .284 | .280 | .281 | |||||
| A/T | .476 | .472 | 4.87; | .524 | 4.71; | .398 | .489 | 2.81 | ||
| T/T | .183 | .239 | .088 | .192 | .095 | .323 | .230 | .246 | ||
| rs279837-G | A | .600 | .548 | 4.60; | .568 | 2.67; | .732 | .736 | 0.01 | 12.4 |
| GABRA2-intron 3 | G | .400 | .452 | .032 | .432 | .102 | .268 | .264 | .930 | <.001 |
| A/A | .367 | .301 | .312 | .566 | .535 | |||||
| A/G | .467 | .494 | 4.62; | .512 | 4.19; | .333 | .401 | 1.87 | ||
| G/G | .166 | .205 | .099 | .177 | .123 | .101 | .063 | .393 | ||
| rs9291283-H | G | .744 | .718 | 1.36; | .732 | 0.45 | .693 | .691 | 0.00 | 2.21 |
| GABRA2-intron 3 | A | .256 | .282 | .244 | .268 | .501 | .307 | .309 | .977 | .138 |
| G/G | .558 | .518 | .533 | .479 | .482 | |||||
| A/G | .372 | .399 | 1.35; | .398 | 0.91; | .427 | .418 | .03 | ||
| A/A | .070 | .083 | .510 | .069 | .636 | .094 | .099 | .985 | ||
Allele nucleotide designation refers to the chromosome plus strand sequence.
EA Controls compared with EA Connecticut Alcoholics
EA Controls compared with EA MATCH Alcoholics
AA Controls compared with AA Alcoholics (Connecticut and MATCH combined)
EA Controls compared with AA Control
Haplotype frequencies for the two major haplotype blocks in the GABRG1-GABRA2 interval were estimated using markers defining the slightly shorter core of each block based on the haplotype structure for AAs (Figure 1). Four markers (SNPs 3–6) were used to define the GABRG1 haplotype block and six markers (SNPs 7 and A-E) for the GABRA2 haplotype block. Estimated haplotype frequencies were compared to the sum of all other haplotypes for alcoholics vs. controls and for AA vs. EA controls using a series of 2X2 contingency tables (Tables 5 and 6). Among EAs, two haplotypes comprised >90% of chromosomes for the GABRG1 haplotype block and >95% of chromosomes for GABRA2. AA subjects had a third common haplotype for both haplotype blocks. Haplotype frequencies differed significantly between EA and AA controls. For GABRG1, there was a significantly greater frequency of the ATCC haplotype in AD than controls in both EA groups (Δ=0.078 in CT EA cases, p<0.001; 0.064 in MATCH EA cases, p=0.002), but this did not reach significance in AA cases (Δ=0.059). For the 6-SNP GABRA2 3′-region haplotype, there was a non-significantly greater frequency of the AGACTC minor haplotype in EA alcoholics than controls (Δ=0.046 in CT cases, p=0.052 and 0.028 in MATCH cases, p=0.20). The frequency of the GABRA2 AGACTC minor haplotype did not differ by diagnosis among AAs.
Table 5.
GABRG1 4-SNP haplotype (SNPs 3–6; rs1391166, 7654165, 10033451, 4280776) frequencies for self-identified EA and AA subjects.
| EA -Connecticut sample | EA - Project Match | AA Subjects | EA c/w AA controls | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Haplotype* | Controls(n=535) | AD (n=372) | aχ2; p-value | AD (n=727) | bχ2; p-value | Controls(n=100) | AD (n=148) | cχ2; p-value | dχ2; p-value |
| TCTT | .490 | .394 | 15.74; < .001 | .426 | 10.43; .001 | .291 | .261 | 0.57; .45 | 26.13; <.001 |
| ATCC | .432 | .510 | 11.47; < .001 | .496 | 9.77; .002 | .261 | .320 | 1.93; .16 | 19.23; <.001 |
| ATTT | .026 | .024 | 0.07; .79 | .025 | 0.01; .92 | .359 | .359 | 0.00; .96 | 255.4; <.001 |
Only those haplotypes with frequency >0.05 in at least one subgroup are shown.
2X2 χ2 table (haplotype (y) vs. all others) EA Controls compared with EA Connecticut Alcoholics
EA Controls compared with EA MATCH Alcoholics
AA Controls compared with AA Alcoholics (Connecticut and MATCH combined)
EA Controls compared with AA Controls
Table 6.
GABRA2 6-SNP haplotype (snps 7, A-E; rs1440133, 567926, 534459, 529826, 279869, 279858) frequencies for self-identified EA and AA subjects.
| EA -Connecticut sample | EA - Project Match | AA Subjects | EA c/w AA controls | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Haplotype* | Controls (n=535) | AD (n=372) | aχ2; p-value | AD (n=727) | bχ2; p-value | Controls (n=100) | AD (n=148) | cχ2; p-value | dχ2; p-value |
| GAGTGT | .585 | .524 | 6.18; .013 | .561 | 1.94; .16 | .281 | .278 | 0.00; .95 | 60.50; <.001 |
| AGACTC | .386 | .432 | 3.78; .052 | .414 | 1.63; .20 | .229 | .228 | 0.00; .98 | 17.13; <.001 |
| GAACTT | .005 | .001 | N/A | .001 | N/A | .453 | .440 | 0.16; .69 | 469.8; <.001 |
Only those haplotypes with frequency >0.05 in at least one subgroup are shown.
2X2 χ2 table (haplotype (y) vs. all others) EA Controls compared with EA Connecticut Alcoholics
EA Controls compared with EA MATCH Alcoholics
AA Controls compared with AA Alcoholics (Connecticut and MATCH combined)
EA Controls compared with AA Controls
N/A–χ2 table cell expected counts <5
There was a moderate degree of LD between markers in the two haplotype blocks (r = 0.51–0.59 for EA subjects and r=0.00–0.53 for AA subjects). To examine whether variation in one or the other of the two adjacent genes accounted for the association to AD, we examined the phase and pattern of linkage of the adjacent GABRG1 and GABRA2 haplotype blocks by estimating the frequency of extended haplotypes defined by the 10 SNPs from these two blocks. Table 7 lists the frequency of the most common 10-SNP haplotypes covering the larger 208,000 bp interval. The most common extended haplotype among EAs (TCTT-GAGTGT) was under represented in both of the EA alcoholic samples (Δ=0.098 in CT cases and Δ=0.059 in MATCH cases; p<0.001 and 0.002, respectively). Chromosomes with the GABRG1 risk haplotype (ATCC) were over-represented in EA alcoholics, irrespective of the GABRA2 haplotype in both EA samples. The distortion was greatest when the less common GABRG1 risk haplotype was paired with the major GABRA2 haplotype (GAGTGT) (Δ=0.042 in CT cases, and Δ=0.044 in MATCH cases; p=0.04 and p=0.005, respectively). In contrast, the GABRA2 haplotype defined by the minor allele at each marker (AGACTC), which we previously found to be associated with AD (Covault et al. 2004), when paired with the GABRG1 non-risk haplotype (TCTT), did not differ in frequency between EA alcoholics and controls in the present analysis. A more formal restatement of this analysis is to examine the ratio in cases vs. controls of the two putative risk haplotypes with each conditional on the presence of the other. Both haplotype ratios, f(AB)/f(Ab) and f(aB)/f(ab), are expected not to differ in cases and controls if the “A vs. a” component of the haplotype identifies a disease-associated marker and the “B vs. b” site represents a neutral marker, irrespective of genetic mode of inheritance (Valdes&Thomson 1997). The χ2 test of the null hypothesis that these ratios are equivalent is rejected (p < .01) in EA AD samples, if the GABRA2 AD-associated haplotype is considered the risk marker and the GABRG1 haplotype is considered the neutral marker. In this context, markers in the GABRA2 haplotype block do not capture all of the disease risk associated with this chromosomal region. In contrast, the null hypothesis that the ratios are equivalent is sustained (p > 0.2) when the GABRG1 AD-associated haplotype block is treated as the conditional risk marker. Equivalent results were obtained examining 2-SNP haplotypes using a single representative SNP for the GABRG1 and GABRA2 blocks (e.g., SNPs 4 and A). These observations suggest that the allelic and haplotypic association of GABRA2 SNPs with AD in our CT sample in both this and our prior study (Covault et al. 2004), may in part be secondary to LD of these markers with risk-related variants in the adjacent GABRG1 5′-region. In AAs, although not statistically significant, the extended 10-SNP haplotype pattern was qualitatively similar to the pattern in EAs, in that there was a greater frequency of chromosomes with an extended haplotype containing the GABRG1 ATCC motif.
Table 7.
GABRG1-A2 10-SNP extended haplotype (SNPs 3–7 & A-E; rs1391166, 7654165, 10033451, 4280776, 1440133, 567926, 534459, 529826, 279869, 279858) frequencies for self-identified EA and AA subjects.
| EA - Connecticut sample | EA - Project Match | AA Subjects | EA c/w AA controls |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Haplotype* g1/a2 risk |
Controls (n=535) |
AD (n=372) |
aχ2; p-value |
AD (n=727) |
bχ2; p-value |
Controls (n=100) |
AD (n=148) |
cχ2; p-value |
dχ2; p-value |
| −/− TCTT-GAGTGT | .425 | .327 | 14.46; <.001 | .366 | 9.29; .002 | .148 | .146 | 0.07; .79 | 48.52; <.001 |
| +/+ ATCC-AGACTC | .309 | .346 | 3.26; .07 | .334 | 1.69; .19 | .135 | .165 | 0.41; .52 | 17.86; <.001 |
| +/− ATCC-GAGTGT | .109 | .141 | 4.06; .044 | .153 | 8.02; .005 | .081 | .095 | 0.41; .52 | 1.56; .21 |
| −/− ATTT-GAACTT | .003 | .000 | N/A | .001 | N/A | .294 | .280 | 0.09; .76 | 315.5; <.001 |
| −/− TCTT-GAACTT | .003 | .003 | N/A | .001 | N/A | .114 | .073 | 1.43; .23 | 103.8; <.001 |
| −/+ TCTT-AGACTC | .055 | .052 | 0.21; .64 | .047 | 0.91; .34 | .026 | .024 | 0.08; .78 | 2.66; .10 |
Only those extended haplotypes with frequency >0.05 in at least one subgroup are shown. Risk haplotype blocks over-represented in alcoholics are indicated by + (ATCC for GABRG1 and AGACTC for GABRA2), all other haplotype blocks are noted with a–sign.
2X2 χ2 table (haplotype (y) vs. all others) EA Controls compared with EA Connecticut Alcoholics
EA Controls compared with EA MATCH Alcoholics
AA Controls compared with AA Alcoholics (Connecticut and MATCH combined)
EA Controls compared with AA Controls
N/A–χ2 table cell expected counts <5
Significant racial population differences were observed in the allele frequencies for 13 of the 15 SNPs, as well as for the most common haplotypes for the GABRG1and GABRA2 blocks (Tables 3–6). To evaluate population genetic stratification, we used the program STRUCTURE v2.1 (Pritchard et al. 2000; Falush et al. 2003) to generate estimates of the proportion of African vs. European genetic ancestry using genotype results from a panel of 34 ancestry informative markers (Stein et al. 2004; Luo et al. 2005; Yang et al. 2005). We found no differences in the degree of EA genetic ancestry in the three EA samples [controls = 0.980 ± 0.043, CT alcoholics = 0.980 ± 0.043, and MATCH alcoholics = 0.983 ± 0.029; F(2,1631)=1.56, p=0.21]. Consequently, population stratification is unlikely to be an explanation for the observed allele frequency differences between EA alcoholics and controls. Similarly, for AAs, there was no significant difference in estimated European genetic admixture for alcoholics and controls [0.084 ± 0.149 and 0.054 ± 0.143, respectively; F(1,246)=1.61, p=0.11].
Quantitative estimates of genetic ancestry proportion from STRUCTURE were also used as a covariate in binary logistic regression analysis (together with age and sex, which differed by diagnosis). This yielded corrected odds ratios (ORs) for AD as a function of GABRG1 or GABRA2 AD-associated haplotype copy number as determined using PHASE. Dominant, recessive, and additive models were compared using dummy coding: 0, 1, 1; 0, 0, 1 and 0, 0.5, 1, respectively, for 0, 1 or 2 copies of the GABRG1 or GABRA2 AD-associated haplotype (i.e., ATCC or AGACTC, respectively). For EAs (combined MATCH and CT alcoholics), an additive genetic model provided the best fit for the GABRG1 ATCC haplotype (Table 8), with an OR=1.26 (95% CI=1.06–1.50) for one copy (i.e. ATCC/x) and OR=1.60 (95% CI=1.13–2.25) for two copies of the AD-associated haplotype (i.e. ATCC/ATCC). Qualitatively similar results were seen when age, sex and genetic ancestry were omitted from the model. For the GABRA2 AGACTC haplotype, a dominant genetic model provided the best fit and an OR=1.39 (95% CI=1.08–1.80) for carriers. These models did not show interactive effects of haplotype with either gender or genetic ancestry proportion. Due to the co-linearity of the two risk haplotypes, we were unable simultaneously to examine the independent effects on AD risk of the GABRG1 and GABRA2 haplotype blocks. There was no evidence that the association of AD with GABRG1 or GABRA2 markers in the CT sample was due to co-morbid drug dependence (relevant data were not available for Project MATCH). Considering the impact of drug dependence, ORs for the AD risk haplotypes were marginally greater (GABRG1) or unchanged (GABRA2) for subjects in the CT EA sample with no co-morbid lifetime diagnosis of cocaine, opioid or cannabis dependence compared with all CT EA alcoholics [GABRG1 additive model OR=1.35 (95% CI=1.05–1.74) vs. 1.25 (95% CI=0.99–1.58) for one copy and OR=1.82 (95% CI=1.09–3.04) vs. 1.56 (95% CI=0.98–2.48) for two copies of the GABRG1 risk haplotype; GABRA2 dominant model OR=1.45 (95% CI=0.99–2.14) vs. 1.40 (95% CI=0.98–1.99) for carriers].
Table 8.
Binary logistic regression analysis of GABRG1 and GABRA2 AD-risk haplotypes as a function of the genetic model (including age, sex and % EA genetic heritage as covariates).
| Risk haplotype | Genetic model | B | S.E. | Wald | p-value |
|---|---|---|---|---|---|
| GABRG1 | additive | 0.47 | 0.18 | 7.1 | 0.008 |
| ATCC haplotype | dominant | 0.27 | 0.14 | 4.0 | 0.45 |
| recessive | 0.37 | 0.15 | 5.7 | 0.016 | |
| GABRA2 | additive | 0.32 | 0.18 | 3.1 | 0.079 |
| AGACTC haplotype | dominant | 0.33 | 0.13 | 6.3 | 0.012 |
| recessive | 0.00 | 0.17 | 0.0 | 0.99 |
Finally, based on the apparent difference in genetic mode of action of risk elements potentially represented in the two haplotype blocks, we used binary logistic regression analysis to derive corrected measures for association of individual SNPs with AD in the combined EA sample using variable coding for both additive and dominant genetic models. Figure 2 illustrates the log10 (p-value) from binary logistic regression analyses in which age, sex, and genetic ancestry were used as covariates. The strongest association with AD was seen for SNPs 4 and 5 (rs7654165 and rs10033451) in the 5′-upstream region of GABRG1 assuming an additive genetic model followed by SNPs 7, A, and B (rs1440133, rs567926, and rs534459) in the 3′ region of GABRA2 assuming a dominant genetic model. This difference in the best-fit genetic model for markers in the two haplotype block regions suggests that there may be separate contributions to risk for AD by GABRG1 and GABRA2.
Figure 2.
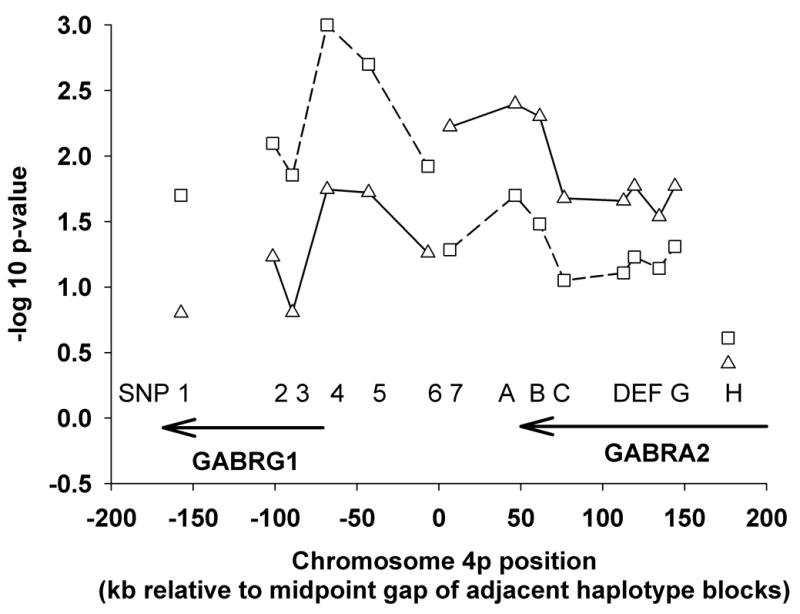
Negative log10 of binary logistic regression significance (p-value) for allelic association with AD after correction for age, sex, and proportion of EA genetic ancestry among all EA subjects (1099 AD and 535 controls) as a function of GABRG1 and GABRA2 SNP location assuming additive (--□--) or dominant (–Δ–) genetic models. SNPs within each of the two haplotype blocks identified in Figure 1A for EA subjects are joined by a line. Marker positions are shown relative to the mid-point between these two haplotype blocks. Consistent with results from haplotype analysis regarding potential mode of risk transmission, individual markers in the GABRG1 haplotype block show a larger effect under an additive model while those in the GABRA2 block are more consistent with a dominant genetic effect model.
DISCUSSION
The findings reported here suggest that the association of GABRA2 with AD, as reported in four independent studies (Covault et al. 2004; Edenberg et al. 2004; Lappalainen et al. 2005; Fehr et al. 2006), may be complicated by a moderate degree of LD between markers in the 3′ region of GABRA2 with those in the 5′ region of GABRG1. Although prior studies showed consistent evidence for association of AD to markers and haplotypes in the middle and 3′ region of GABRA2, the extent of the AD-associated GABRA2 haplotype block and the potential association of AD to markers in the adjacent haplotype block were not well described. Edenberg et al. (2004) examined six SNPs in the adjacent GABRG1 gene (0.4 kb upstream to IVS8) and found nominal evidence for association (p=0.05) to AD for one of the markers within the 5′-region haplotype block of GABRG1 that was examined in this report. Seeking to define the extent of the GABRA2 3′-region haplotype block, we genotyped markers extending into the 5′-region of GABRG1, where we found evidence among EAs of association to AD that exceeded the evidence for allelic and haplotypic association for GABRA2. Indeed, in the Project MATCH EA sample, the distortion of allele and haplotype frequencies related to AD for GABRA2 markers was not statistically significant, while allele and haplotype frequencies in the haplotype block in the GABRG1 5′ region showed significant evidence of association in both EA samples. These findings were unaffected when corrections for age, sex, and ancestry proportion were included in the analysis. These results suggest that our prior reported findings of association of AD with the GABRA2 gene (Covault et al. 2004) were partly due to LD of GABRA2 markers with functional genetic variation in the adjacent GABRG1 gene.
It is of interest to note that in the three published case-control studies (Covault et al. 2004; Lappalainen et al. 2005; Fehr et al. 2006), AD was associated with the minor allele and haplotype for markers in the 3′-region of GABRA2, consistent with the findings reported here for the EA alcoholic sample from CT. In contrast, in the COGA multiplex family sample, the more common haplotype was over-represented among alcoholics (Edenberg et al. 2004). Re-analysis of the COGA dataset revealed that the association of the major allele at markers in the 3′ region of GABRA2 with AD was observed only among subjects dependent on both alcohol and drugs (Agrawal et al. 2006).
We observed large differences in allele frequencies for AA vs. EA subjects at 13 of the 15 SNPs examined. Population differences in marker frequencies have been observed in other candidate genes related to substance use disorders (Gelernter et al. 1997; Gelernter et al. 1999; Covault et al. 2001; Luo et al. 2003; Herman et al. 2006). Such population differences highlight the need to consider population genetic stratification as a potential source of artifact in genetic association studies. Further, results from a recent gene expression microarray study noted that differences in frequency of cis-acting SNPs among ethnic groups may also be associated with significant differences in RNA expression levels of the associated genes (Spielman et al. 2007).
Limitations of our study include the lack of a sample of controls collected at each of the Project MATCH treatment sites, which required comparison of allele and haplotype frequencies for both samples of AD subjects with a common control sample recruited exclusively from Connecticut. However, we found no evidence of differences in genetic admixture between the CT and MATCH alcoholic samples. Although we observed a similar magnitude of increase in frequency of the GABRG1 risk haplotype ATCC in AA and EA subjects, our small sample of AA subjects yielded limited statistical power. The sample of female alcoholics also limited our capacity to examine with high statistical power interactive effects of sex × genotype. Finally, the lack of data on lifetime drug dependence diagnoses in the Project MATCH sample limited our ability to examine whether in the CT AD sample the association of AD to GABRG1 markers was independent of co-morbid drug dependence.
The findings from this study provide important new information regarding the likely physical location of functional genetic variants responsible for the reported association of AD to the GABAA receptor subunit gene cluster on chromosome 4p. The region of strongest association to AD that we observed includes potential regulatory regions immediately upstream of the GABRG1 gene. The markers with the strongest association, rs7654165 and rs10033451, are located 1 and 26 kb upstream of the 5′ end of the GABRG1 transcript, respectively, suggesting that functional variants related to AD in this interval might alter the patterns of regional, cellular, or temporal expression of GABRG1. The γ1 subunit is notable, in that unlike most GABAA subunits, its expression is limited to very few brain areas, including the pallidum, septum, bed nucleus of the stria terminalis and the central and medial amygdaloid nuclei (Ymer et al. 1990; Araki et al. 1992; Persohn et al. 1992; Wisden et al. 1992; Pirker et al. 2000). Other brain regions that show selective expression of γ1 subunits (e.g., hypothalamic medial preoptic area) vs. γ2 (e.g., ventromedial nucleus of the hypothalamus) (Herbison&Fenelon 1995; Nett et al. 1999) differ in GABAA pharmacologic properties and show opposite effects with respect to receptor modulation by anabolic steroids (Jorge-Rivera et al. 2000). Additional differences in the pharmacology of γ1-containing receptors include an insensitivity to the benzodiazepine antagonist flumazenil (Ymer et al. 1990; Khom et al. 2006) and an increased sensitivity of γ1-containing receptors to the neuroactive steroid allopregnanolone in transfected human embryonic kidney (HEK293) cells compared with γ2-containing receptors (Puia et al. 1993). Pharmacologic agents that are selective for γ1-containing GABAA receptors may be of clinical interest given the limited CNS distribution of this receptor subunit.
Acknowledgments
The assistance of Linda Burian, Dawn Perez, and Tracy Drzyzga in the conduct of this work and statistical consultation by Hongyu Zhao, Ph.D. are greatly appreciated. We are also grateful to Project MATCH investigators and NIAAA for providing access to clinical data and blood samples from that multi-center study. This study was supported by NIH grants P50 AA03510, M01 RR06192, R01 AA11330, R01 AA015606, K24 AA13736, and K24 DA15105.
Footnotes
Conflicts of interest statement. None of the authors have conflicts of interest related to this work and have not had any conflict related to the work within the past 3 years.
References
- Agrawal A, Edenberg HJ, Foroud T, Bierut LJ, Dunne G, Hinrichs AL, Nurnberger JI, Crowe R, Kuperman S, Schuckit MA, Begleiter H, Porjesz B, Dick DM. Association of GABRA2 with Drug Dependence in the Collaborative Study of the Genetics of Alcoholism Sample. Behav Genet. 2006;36(5):640–50. doi: 10.1007/s10519-006-9069-4. [DOI] [PubMed] [Google Scholar]
- Araki T, Kiyama H, Tohyama M. The GABAA receptor gamma 1 subunit is expressed by distinct neuronal populations. Brain Res Mol Brain Res. 1992;15(1–2):121–32. doi: 10.1016/0169-328x(92)90159-9. [DOI] [PubMed] [Google Scholar]
- American Psychiatric Association. Diagnostic and Statistical Manual of Mental Disorders. 4. Washington, D.C: American Psychiatric Press; 1994. [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Blouin AG, Perez EL, Blouin JH. Computerized administration of the Diagnostic Interview Schedule. Psychiatry Res. 1988;23(3):335–44. doi: 10.1016/0165-1781(88)90024-8. [DOI] [PubMed] [Google Scholar]
- Caetano R, Cunradi C. Alcohol dependence: a public health perspective. Addiction. 2002;97(6):633–45. doi: 10.1046/j.1360-0443.2002.00184.x. [DOI] [PubMed] [Google Scholar]
- Covault J, Gelernter J, Kranzler H. Association study of cannabinoid receptor gene (CNR1) alleles and drug dependence. Mol Psychiatry. 2001;6(5):501–2. doi: 10.1038/sj.mp.4000925. [DOI] [PubMed] [Google Scholar]
- Covault J, Gelernter J, Nellissery M, Kranzler HR. Allelic and Haplotypic Association of GABRA2 with Alcohol Dependence. Am J Med Genetics. 2004;129B:104–9. doi: 10.1002/ajmg.b.30091. [DOI] [PubMed] [Google Scholar]
- Dick DM, Edenberg HJ, Xuei X, Goate A, Hesselbrock V, Schuckit M, Crowe R, Foroud T. No association of the GABAA receptor genes on chromosome 5 with alcoholism in the collaborative study on the genetics of alcoholism sample. Am J Med Genet B Neuropsychiatr Genet. 2005;132(1):24–8. doi: 10.1002/ajmg.b.30058. [DOI] [PubMed] [Google Scholar]
- Dick DM, Edenberg HJ, Xuei X, Goate A, Kuperman S, Schuckit M, Crowe R, Smith TL, Porjesz B, Begleiter H, Foroud T. Association of GABRG3 with alcohol dependence. Alcohol Clin Exp Res. 2004;28(1):4–9. doi: 10.1097/01.ALC.0000108645.54345.98. [DOI] [PubMed] [Google Scholar]
- Edenberg HJ, Dick DM, Xuei X, Tian H, Almasy L, Bauer LO, Crowe RR, Goate A, Hesselbrock V, Jones K, Kwon J, Li TK, Nurnberger JI, Jr, O’Connor SJ, Reich T, Rice J, Schuckit MA, Porjesz B, Foroud T, Begleiter H. Variations in GABRA2, Encoding the alpha 2 Subunit of the GABAA Receptor, Are Associated with Alcohol Dependence and with Brain Oscillations. Am J Hum Genet. 2004;74(4):705–14. doi: 10.1086/383283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164(4):1567–87. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr C, Sander T, Tadic A, Lenzen KP, Anghelescu I, Klawe C, Dahmen N, Schmidt LG, Szegedi A. Confirmation of association of the GABRA2 gene with alcohol dependence by subtype-specific analysis. Psychiatr Genet. 2006;16(1):9–17. doi: 10.1097/01.ypg.0000185027.89816.d9. [DOI] [PubMed] [Google Scholar]
- First MB, Spitzer RL, Gibbon M, Williams JBW. Strutured Clinical Interview for DSM-IV Axis I Disorders. Washington, D.C: APA Press; 1997. [Google Scholar]
- Gelernter J, Kranzler H, Cubells J. Genetics of two mu opioid receptor gene (OPRM1) exon I polymorphisms: population studies, and allele frequencies in alcohol- and drug-dependent subjects. Mol Psychiatry. 1999;4(5):476–83. doi: 10.1038/sj.mp.4000556. [DOI] [PubMed] [Google Scholar]
- Gelernter J, Kranzler H, Cubells JF. Serotonin transporter protein (SLC6A4) allele and haplotype frequencies and linkage disequilibria in African- and European-American and Japanese populations and in alcohol-dependent subjects. Hum Genet. 1997;101(2):243–6. doi: 10.1007/s004390050624. [DOI] [PubMed] [Google Scholar]
- Grant BF, Dawson DA, Stinson FS, Chou SP, Dufour MC, Pickering RP. The 12-month prevalence and trends in DSM-IV alcohol abuse and dependence: United States, 1991–1992 and 2001–2002. Drug Alcohol Depend. 2004;74(3):223–34. doi: 10.1016/j.drugalcdep.2004.02.004. [DOI] [PubMed] [Google Scholar]
- Herbison AE, Fenelon VS. Estrogen regulation of GABAA receptor subunit mRNA expression in preoptic area and bed nucleus of the stria terminalis of female rat brain. J Neurosci. 1995;15(3 Pt 2):2328–37. doi: 10.1523/JNEUROSCI.15-03-02328.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman AI, Kranzler HR, Cubells JF, Gelernter J, Covault J. Association study of the CNR1 gene exon 3 alternative promoter region polymorphisms and substance dependence. Am J Med Genet B Neuropsychiatr Genet. 2006;141B(5):499–503. doi: 10.1002/ajmg.b.30325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorge-Rivera JC, McIntyre KL, Henderson LP. Anabolic steroids induce region- and subunit-specific rapid modulation of GABA(A) receptor-mediated currents in the rat forebrain. J Neurophysiol. 2000;83(6):3299–309. doi: 10.1152/jn.2000.83.6.3299. [DOI] [PubMed] [Google Scholar]
- Kendler KS. Twin studies of psychiatric illness: an update. Arch Gen Psychiatry. 2001;58(11):1005–14. doi: 10.1001/archpsyc.58.11.1005. [DOI] [PubMed] [Google Scholar]
- Khom S, Baburin I, Timin EN, Hohaus A, Sieghart W, Hering S. Pharmacological properties of GABAA receptors containing gamma1 subunits. Mol Pharmacol. 2006;69(2):640–9. doi: 10.1124/mol.105.017236. [DOI] [PubMed] [Google Scholar]
- Koob GF. A role for GABA mechanisms in the motivational effects of alcohol. Biochem Pharmacol. 2004;68(8):1515–25. doi: 10.1016/j.bcp.2004.07.031. [DOI] [PubMed] [Google Scholar]
- Krystal JH, Staley J, Mason G, Petrakis IL, Kaufman J, Harris RA, Gelernter J, Lappalainen J. Gamma-aminobutyric acid type A receptors and alcoholism: intoxication, dependence, vulnerability, and treatment. Arch Gen Psychiatry. 2006;63(9):957–68. doi: 10.1001/archpsyc.63.9.957. [DOI] [PubMed] [Google Scholar]
- Lappalainen J, Krupitsky E, Remizov M, Pchelina S, Taraskina A, Zvartau E, Somberg LK, Covault J, Kranzler HR, Krystal JH, Gelernter J. Association between alcoholism and gamma-amino butyric acid alpha2 receptor subtype in a Russian population. Alcohol Clin Exp Res. 2005;29(4):493–8. doi: 10.1097/01.alc.0000158938.97464.90. [DOI] [PubMed] [Google Scholar]
- Loh EW, Higuchi S, Matsushita S, Murray R, Chen CK, Ball D. Association analysis of the GABA(A) receptor subunit genes cluster on 5q33–34 and alcohol dependence in a Japanese population. Mol Psychiatry. 2000;5(3):301–7. doi: 10.1038/sj.mp.4000719. [DOI] [PubMed] [Google Scholar]
- Long JC, Knowler WC, Hanson RL, Robin RW, Urbanek M, Moore E, Bennett PH, Goldman D. Evidence for genetic linkage to alcohol dependence on chromosomes 4 and 11 from an autosome-wide scan in an American Indian population. Am J Med Genet. 1998;81(3):216–21. doi: 10.1002/(sici)1096-8628(19980508)81:3<216::aid-ajmg2>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- Luo X, Kranzler HR, Zhao H, Gelernter J. Haplotypes at the OPRM1 locus are associated with susceptibility to substance dependence in European-Americans. Am J Med Genet B Neuropsychiatr Genet. 2003;120(1):97–108. doi: 10.1002/ajmg.b.20034. [DOI] [PubMed] [Google Scholar]
- Luo X, Kranzler HR, Zuo L, Yang BZ, Lappalainen J, Gelernter J. ADH4 gene variation is associated with alcohol and drug dependence: results from family controlled and population-structured association studies. Pharmacogenet Genomics. 2005;15(11):755–68. doi: 10.1097/01.fpc.0000180141.77036.dc. [DOI] [PubMed] [Google Scholar]
- Nett ST, Jorge-Rivera JC, Myers M, Clark AS, Henderson LP. Properties and sex-specific differences of GABAA receptors in neurons expressing gamma1 subunit mRNA in the preoptic area of the rat. J Neurophysiol. 1999;81(1):192–203. doi: 10.1152/jn.1999.81.1.192. [DOI] [PubMed] [Google Scholar]
- Persohn E, Malherbe P, Richards JG. Comparative molecular neuroanatomy of cloned GABAA receptor subunits in the rat CNS. J Comp Neurol. 1992;326(2):193–216. doi: 10.1002/cne.903260204. [DOI] [PubMed] [Google Scholar]
- Pierucci-Lagha A, Gelernter J, Feinn R, Cubells JF, Pearson D, Pollastri A, Farrer L, Kranzler HR. Diagnostic reliability of the Semi-structured Assessment for Drug Dependence and Alcoholism (SSADDA) Drug Alcohol Depend. 2005;80(3):303–12. doi: 10.1016/j.drugalcdep.2005.04.005. [DOI] [PubMed] [Google Scholar]
- Pirker S, Schwarzer C, Wieselthaler A, Sieghart W, Sperk G. GABA(A) receptors: immunocytochemical distribution of 13 subunits in the adult rat brain. Neuroscience. 2000;101(4):815–50. doi: 10.1016/s0306-4522(00)00442-5. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155(2):945–59. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Project MATCH Research Group. Matching alcoholism treatments to client heterogeneity: treatment main effects and matching effects on drinking during treatment. J Stud Alcohol. 1998;59:631–639. doi: 10.15288/jsa.1998.59.631. [DOI] [PubMed] [Google Scholar]
- Puia G, Ducic I, Vicini S, Costa E. Does neurosteroid modulatory efficacy depend on GABAA receptor subunit composition? Receptors Channels. 1993;1(2):135–42. [PubMed] [Google Scholar]
- Radel M, Vallejo RL, Iwata N, Aragon R, Long JC, Virkkunen M, Goldman D. Haplotype-based localization of an alcohol dependence gene to the 5q34 {gamma}-aminobutyric acid type A gene cluster. Arch Gen Psychiatry. 2005;62(1):47–55. doi: 10.1001/archpsyc.62.1.47. [DOI] [PubMed] [Google Scholar]
- Reich T, Edenberg HJ, Goate A, Williams JT, Rice JP, Van Eerdewegh P, Foroud T, Hesselbrock V, Schuckit MA, Bucholz K, Porjesz B, Li TK, Conneally PM, Nurnberger JI, Jr, Tischfield JA, Crowe RR, Cloninger CR, Wu W, Shears S, Carr K, Crose C, Willig C, Begleiter H. Genome-wide search for genes affecting the risk for alcohol dependence. Am J Med Genet. 1998;81(3):207–15. [PubMed] [Google Scholar]
- Sander T, Ball D, Murray R, Patel J, Samochowiec J, Winterer G, Rommelspacher H, Schmidt LG, Loh EW. Association analysis of sequence variants of GABA(A) alpha6, beta2, and gamma2 gene cluster and alcohol dependence. Alcohol Clin Exp Res. 1999;23(3):427–31. [PubMed] [Google Scholar]
- Spielman RS, Bastone LA, Burdick JT, Morley M, Ewens WJ, Cheung VG. Common genetic variants account for differences in gene expression among ethnic groups. Nat Genet. 2007;39(2):226–31. doi: 10.1038/ng1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein MB, Schork NJ, Gelernter J. A polymorphism of the beta1-adrenergic receptor is associated with low extraversion. Biol Psychiatry. 2004;56(4):217–24. doi: 10.1016/j.biopsych.2004.05.020. [DOI] [PubMed] [Google Scholar]
- Stephens M, Donnelly P. A comparison of bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet. 2003;73(6):1162–9. doi: 10.1086/379378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68(4):978–89. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdes AM, Thomson G. Detecting disease-predisposing variants: the haplotype method. Am J Hum Genet. 1997;60(3):703–16. [PMC free article] [PubMed] [Google Scholar]
- Wisden W, Laurie DJ, Monyer H, Seeburg PH. The distribution of 13 GABAA receptor subunit mRNAs in the rat brain. I. Telencephalon, diencephalon, mesencephalon. J Neurosci. 1992;12 (3):1040–62. doi: 10.1523/JNEUROSCI.12-03-01040.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang BZ, Zhao H, Kranzler HR, Gelernter J. Practical population group assignment with selected informative markers: characteristics and properties of Bayesian clustering via STRUCTURE. Genet Epidemiol. 2005;28(4):302–12. doi: 10.1002/gepi.20070. [DOI] [PubMed] [Google Scholar]
- Ymer S, Draguhn A, Wisden W, Werner P, Keinanen K, Schofield PR, Sprengel R, Pritchett DB, Seeburg PH. Structural and functional characterization of the gamma 1 subunit of GABAA/benzodiazepine receptors. Embo J. 1990;9(10):3261–7. doi: 10.1002/j.1460-2075.1990.tb07525.x. [DOI] [PMC free article] [PubMed] [Google Scholar]