Abstract
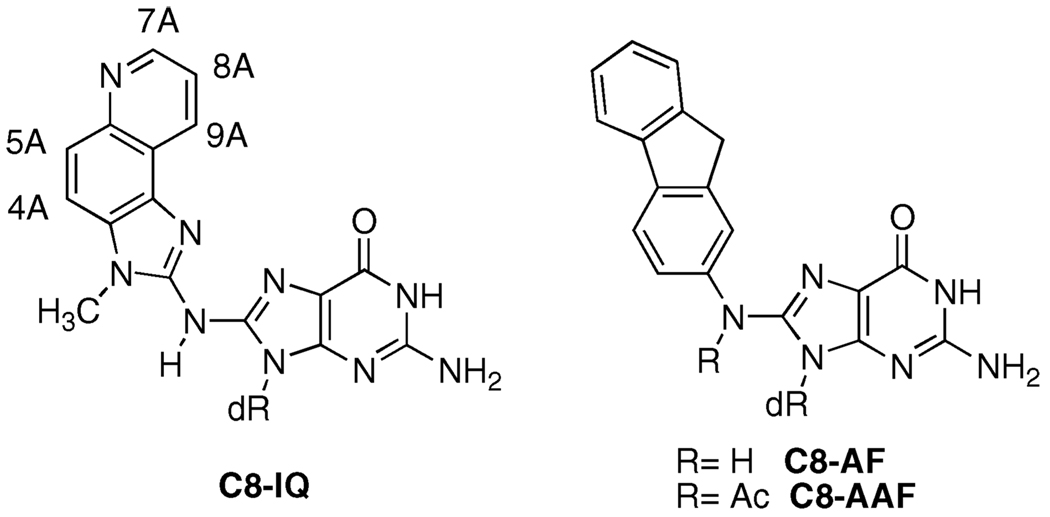
2-Amino-3-methylimidazo[4,5-f]quinoline (IQ) is a highly mutagenic heterocyclic amine found in cooked meats. The major DNA adduct of IQ is at the C8-position of dGuo. We have previously reported the incorporation of the C8-IQ adduct into oligonucleotides, namely, the G1-position of codon 12 of the N-ras oncogene sequence (G1G2T) and the G3-position of the NarI recognition sequence (G1G2CG3CC) (Elmquist et al. (2004) J. Am. Chem. Soc. 126, 11189–11201). Ultraviolet spectroscopy and circular dichroism studies indicated that the conformation of the adduct in the two oligonucleotides was different, and they were assigned as groove-bound and base-displaced intercalated, respectively. The conformation of the latter was subsequently confirmed through NMR and restrained molecular dynamics studies (Wang et al. (2006) J. Am. Chem. Soc. 128, 10085–10095). We report here the incorporation of the C8-IQ adduct into the G1- and G2-positions of the NarI sequence. A complete analysis of the UV, CD, and NMR chemical shift data for the IQ protons are consistent with the IQ adduct adopting a minor groove-bound conformation at the G1- and G2-positions of the NarI sequence. To further correlate the spectroscopic data with the adduct conformation, the C8-aminofluorene (AF) adduct of dGuo was also incorporated into the NarI sequence; previous NMR studies demonstrated that the AF-modified oligonucleotides were in a sequence-dependent conformational exchange between major groove-bound and base-displaced intercalated conformations. The spectroscopic data for the IQ- and AF-modified oligonucleotides are compared. The sequence-dependent conformational preferences are likely to play a key role in the repair and mutagenicity of C8-arylamine adducts.
Introduction
2-Amino-3-methylimidazo[4,5-f]quinoline (IQ1) is part of a family of heterocyclic aromatic amines (HCAs) that are formed through the pyrolysis of carbohydrates and amino acids; the conditions for HCA formation are achieved during the cooking of proteinaceous food, such as red meats, poultry, and fish (1–5). Some of these compounds have been shown to be highly mutagenic in Ames assays and carcinogenic in laboratory animals (4). 2-Amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) is the most abundantly formed HCA, whereas IQ and 2-amino-3,4-dimethylimidazo[4,5-f]quinoline (MeIQ) are the most mutagenic in the Ames assay. Human exposure to HCAs is a daily occurrence, making these compounds likely contributors to the onset of certain cancers, such as colorectal, prostate, and breast cancer (6–12).
Aromatic amines require a two-stage metabolic activation before they can react with DNA (1, 13, 14). A cytochrome P450 oxidizes the aromatic amine to a hydroxylamine, which is subsequently esterified by N-acetyltransferase or sulfotrans-ferase. Solvolysis of the hydroxylamine ester produces a highly reactive arylnitrenium ion, which is the DNA modifying species. Most aromatic amines, including IQ, react predominantly at the C8-position of dGuo (Figure 1). A minor N2-adduct has also been identified in some cases (15, 16). Mutations caused by the IQ-adduct include −1 and −2 frameshifts and base substitutions (17, 18).
Figure 1.
C8-dGuo adducts of IQ, AF, and AAF.
Perhaps the most well studied of the aromatic amines are 2-aminofluorene (AF) and N-acetyl-2-aminofluorene (AAF) (19). Although structurally similar, these compounds elicit significantly different biological responses. AAF induces SOS-dependent frameshift mutations in bacteria, whereas AF produces base-pair substitution mutations by an SOS-independent mechanism. There is good evidence that the local sequence context can play a significant role in the bypass of the C8-dGuo adducts of arylamines. Site-specific mutagenesis of the C8-dGuo adducts of AAF and AF at all three dGuo positions of the NarI recognition sequence (5′-G1G2CG3CC-3′) has been reported (20, 21). Replication in SOS-induced bacteria resulted in −2 deletions (91% mutational frequency) when the C8-AAF adduct was incorporated at the G3-position, which is part of a CpG dinucleotide repeat. However, the mutation frequency for frameshifts was only 8.4% and 2.8% when incorporated at G2 and G1, respectively (21). The C8-AF adduct produced only base-pair substitution mutations, primarily G → T transversions. Interestingly, both the AF and AAF adducts produce base-pair substitutions when replicated in mammalian cells (COS-7) (21). Crystallographic studies of AAF- and AF-adducted oligonucle-otides bound to replicative DNA polymerases have been reported and provide insight into the mechanism of bypass (22, 23). Bypass of the C8-AAF and AF adducts in vitro have also been reported and support the in vivo results (20, 24–32).
We have previously reported the site-specific incorporation of the C8-IQ adduct into oligonucleotides containing codon-12 of the N-ras oncogene (5′-GGCAG1G2,TGGTG-3′) and at the G3-position of the NarI recognition sequence (CTCG1G2CG3CC-ATC-3′) (33). Preliminary studies based on circular dichorism and UV absorption spectroscopy indicated that the C8-IQ adduct adopts a groove-bound conformation at the G1-position of the N-ras-12 sequence and a base-displaced intercalated conformation at the G3-position of the NarI sequence (1a and 4a, Table 1). We subsequently confirmed the base-displaced intercalated conformation of 4a by NMR (34). We report here the synthesis and characterization of oligonucleotides containing the C8-dGuo adduct of IQ at the G1- and G2-positions of the NarI restriction sequence. In contrast to the reiterated G3-position, UV analysis of the IQ-adduct at the non-iterated G1- and G2-positions is consistent with a largely groove-bound adduct. Preliminary NMR analysis provided further support for these assignments.
Table 1.
IQ- and AF-modified Oligonucleotides Used in This Study and Their Mass Spectral Characterization
|
m/z (Da) |
|||
|---|---|---|---|
| oligonucleotide | a. G = C8-IQa | b. G = C8-AFb | |
| 1 | 5′-GGCA-GGT-GGT–3′ | 3647.76 | |
| 2 | 5′-CTC-G1GCGCC-ATC–3′ | 3776.07 | 3759.33 |
| 3 | 5′-CTC-GG2CGCC-ATC–3′ | 3777.33 | 3758.98 |
| 4 | 5′-CTC-GGCG3CC-ATC–3′ | 3776.38 | 3760.35 |
The calculated masses for oligonucleotides 1a and 2a–4a (M–H) are 3647.22 and 3776.58 Da, respectively.
The calculated mass for oligo-nucleotides 2b–4b is 3759.58 Da.
Experimental Procedures
Oligonucleotide Synthesis
Unmodified oligonucleotides were purchased from Midland Certified Reagents (Midland, TX). The adducted oligonucleotides were synthesized on an Expedite 8909 DNA synthesizer (PerSeptive Biosystems, Inc., Framingham, MA) on a 1 µmol scale using the UltraMild line of phosphoramidites (phenoxyacetyl-protected dA, 4-isopropyl-phenoxyacetyl-protected dG, acetyl-protected dC, and T phosphoramidites) and solid supports from Glen Research Corporation (Sterling, VA). The manufacturer’s standard synthesis protocol was followed except for the incorporation of the modified phosphoramidites, which was accomplished off-line by a manual coupling protocol (33). At this point, the column was removed from the instrument and sealed with two 1 mL syringes, one contained 250–300 µL of the manufacturer’s 1H-tetrazole-activator solution (1.9–4.0% in acetonitrile) and the other contained 250 µL of the phosphoramidite (25 mg, 0.098 M in anhydrous methylene chloride). The 1H-tetrazole and the phosphoramidite solutions were sequentially drawn through the column (1H-tetrazole first), and this procedure was repeated periodically over 30 min. After this time, the column was washed with anhydrous manufacturer’s grade acetonitrile and returned to the instrument for the capping, oxidation, and detritylation steps. The remainder of the synthesis was carried out as normal. The oligonucleotides were initially purified by HPLC using a 0.1 M ammonium formate buffer (solvent 1) and methanol (solvent 2) on a C-18 reversed-phase column with UV detection (Method A). The solvent gradient was as follows: initially 99% solvent 1, then a 40 min linear gradient to 50% solvent 1; 5 min isocratic at 50% solvent 1, then a 5 min linear gradient to the initial conditions. Subsequent optimization of the purification method (Method B) included purification by HPLC using a 20 mM sodium phosphate buffer (pH 7.0) (solvent 1) and methanol (solvent 2). The solvent gradient was as follows: initially 99% solvent 1, then a 27.5 min linear gradient to 35% solvent 1; 2.5 min linear gradient to 50% solvent 1 followed by 5 min isocratic at 50% solvent 1, then a 5 min linear gradient to initial conditions.
Melting Temperature Determination
Thermal melting analyses of modified and unmodified duplex oligonucleotides were performed as previously reported (33).
Circular Dichroism Measurements
CD measurements of modified and unmodified oligonucleotides were performed at 23 °C as previously reported (33).
NMR Measurements
One- and two-dimensional NMR spectra for unmodified d(CTCGGCGCCATC)2 and IQ-modified duplexes (1a–4a) were carried out at a 1H NMR frequency of 800 MHz; all spectra were acquired at 15 °C. 1H NOESY experiments in D2O were recorded at mixing times of 250, 200, 150, and 90 ms, with 1024 points in the t1 dimension and 2048 points in the t2 dimension. The relaxation delay was 2 s. The data in the t1 dimension were zero-filled to give a matrix of 2K × 2K real points. Magnitude COSY spectra in D2O were obtained with 16 scans. The data matrix was 256 (t1) × 1024 (t2) complex points. The data in the t1 dimension were zero-filled to give a matrix size of 512 (D1) × 1024 (D2) real points. The NMR data was processed using FELIX (v 97.0, Accelyris, Inc., San Diego, CA) on Silicon Graphics (Mountain View, CA) Octane workstations. Chemical shifts of proton resonances were referenced to the water resonance. Sample preparation for the oligonucleotides was as follows: the unmodified and modified (2a–4a) oligonucleotides were annealed with the complementary d(GATGGCGCCGAG) 12-mer strand at 70 °C with stoichiometry followed by monitoring single-proton resonances in both strands. The resulting 12-mer duplexes were dissolved in 0.250 mL of 10 mM phosphate buffer (pH 7.0) containing 100 mM NaCl and 0.05 mM EDTA in 99.996% D2O. The oligonucleotide concentrations were determined to be in the region of 0.35–0.7 mM using an extinction coefficient of 1.10 × 105 M−1 cm−1 at 260 nm.
Mass Spectrometry
MALDI-TOF mass spectrometry of oli-gonucleotides was performed at the Vanderbilt University Mass Spectrometry Resource Center using a 3-hydroxypicolinic acid (HPA) matrix containing ammonium hydrogen citrate (7 mg/mL) to suppress sodium and potassium adducts. MALDI-TOF mass spectrometric sequencing of modified oligonucleotides was performed by treatment of purified oligonucleotides (0.03 optical density units, OD) in 24 µL of ammonium hydrogen citrate buffer (pH 9.4) containing 20 mM MgSO4 with 2 milliunits of phosphodi-esterase I (PI). Aliquots (4 µL) were taken before enzyme addition and at 1, 8, 18, 28, and 38 min time points and added sequentially to the same vial, which was kept frozen on dry ice. The digest mixture was then desalted using Millipore (Billerica, MA) C18 Ziptips and eluted directly onto a MALDI plate in 3-hydroxypi-colinic acid (HPA) containing ammonium hydrogen citrate (7 mg/ mL). MALDI-TOF analysis gave a mass ladder corresponding to the sequential loss of nucleotides from the 3′-end of the oligo-nucleotide. An identical analysis was performed using phosphodi-esterase II (PII), which gave a mass ladder corresponding to the sequential lose of nucleotides from the 5′-end of the oligonucleotide (35).
Capillary Gel Electrophoresis
Analyses were carried out using a Beckman (Fullerton, CA) P/ACE Instrument System 5500 Series monitored at 260 nm on a 27 cm × 100 mm column packed with the manufacturer’s 100-R gel (for ss DNA) using a Tris-borate buffer system containing 7.0 M urea and a Beckman P/ACE MDQ Capillary Electrophoresis System monitored at 254 nm on a 31.2 cm column with the same manufacturer’s packing.
Results
Site-Specific Synthesis of Oligonucleotides Containing C8-dGuo Adducts of IQ and AF
We previously described the synthesis of C8-IQ-modified oligonucleotides 1a and 4a (33, 36). Using this chemistry, we prepared the C8-IQ-modified oligonucleotides at the G2- and G3-positions of the NarI recognition sequence (2a and 3a). Conformational studies of an identical sequence containing the C8-AF adduct at all three dGuo positions have been reported (37–39). We synthesized the AF-modified oligonucleotides 2b–4b (Table 1) using the corresponding phosphoramidite (see Supporting Information for synthetic schemes and procedures) in order to compare their properties with those of the IQ-modified oligonucleotides. The modified oligonucleotides were purified by HPLC, and their purity is estimated to be greater than 95% by capillary gel electrophoresis. Oligonucleotides were characterized by MALDI-TOF mass spectrometry (Table 1) and sequenced by a MALDI-TOF analysis of controlled exonuclease digestions with nu-cleases PI and PII (35). In all cases, the exonuclease was unable to digest past the C8-adduct (see Supporting Information for sequencing mass ladders).
Thermal Melting (Tm) Studies of IQ-Adducted Oligo-nucleotides
Oligonucleotides 2a, 3a, and 2b–4b were hybridized to the appropriate full-length complementary strands, and the influence of the C8-IQ and AF adducts on the thermal stability of the oligonucleotide was examined by UV melting temperature analysis (Tm). This data is summarized in Table 2. Fuchs and Romano independently reported Tm data for AAF-modified oligonucleotides in the NarI sequence (40, 41). The results from the Fuchs study (entries e–g) as well as our previously reported data on 1a and 4a are also listed in Table 2 for comparison. It should be noted that the oligonucleotide sequence flanking the NarI recognition sequence and Tm buffer conditions employed by Fuchs are different from ours.
Table 2.
Thermal Melting Temperatures (Tm) of IQ-, AF-, and AAF-Modified Oligonucleotides
| oligonucleotide a |
Tm (Tm)b |
|||
|---|---|---|---|---|
| IQc | AFc | AAFd | ||
| a | 5 ′ -GGC AGG TGG TG-3 ′ (1) | 51° (−9°)e | ||
| 3 ′ -CCG TCC ACC AC-5 ′ | ||||
| b | 5 ′ -CTC GGC GCC ATC-3 ′ (2) | 58° (−7°)f | 57° (−8°)f | |
| 3 ′ -GAG CCG CGG TAG-5 ′ | ||||
| c | 5 ′ -CTC GGC GCC ATC-3 ′ (3) | 60° (−5°)f | 56° (−9°)f | |
| 3 ′ -GAG CCG CGG TAG-5 ′ | ||||
| d | 5 ′ -CTC GGC GCC ATC-3 ′ (4) | 61°(−4°)e | 52°(−13°)f | |
| 3 ′ -GAG CCG CGG TAG-5 ′ | ||||
| e | 5 ′ -ACC GGC GCC ACA-3 ′ | 51° (−10°)g | ||
| 3 ′ -TGG CCG CGG TGT-5 ′ | ||||
| f | 5 ′ -ACC GGC GCC ACA-3 ′ | 48° (−13°)g | ||
| 3 ′ -TGG CCG CGG TGT-5 ′ | ||||
| g | 5 ′ -ACC GGC GCC ACA-3 ′ | 48° (−13°)g | ||
| 3 ′ -TGG CCG CGG TGT-5 ′ | ||||
| h | 5 ′ -CTC GGC GCC ATC-3 ′ (4) | 48°(+10°)e | 44° (+6°)f | |
| 3 ′ -GAG CC- -GG TAG-5 ′ | ||||
| i | 5 ′ -ACC GGC GCC ACA-3 ′ (4) | 49° (+15°)g | ||
| 3 ′ -TGG CC- -GG TGT-5 ′ | ||||
| j | 5 ′ -CTC GGC GCC ATC-3 ′ (4) | 55° (+4°)f | ||
| 3 ′ -GAG CCG -GG TAG-5 ′ | ||||
G is the C8-modified dGuo.
ΔTm= Tm (modified) — Tm (unmodified). The Tm values for the unmodified Ras-12 (entry a, 11-mer), our NarI (entries b—d, 12-mer), and Fuchs’ NarI (entries e—g, 12-mer) oligonucleotides were 60, 65, and 61 °C, respectively. The Tm values for our unmodified NarI (entry h) and Fuch’s unmodified NarI (entry i) oligonucleotide opposite a two-base deletion were 38 and 34 °C, respectively. The Tm values for our unmodified NarI (entry j) oligonucleotide opposite a one-base deletion was 51 °C.
Conditions: 10 mM phosphate buffer (pH 7.0) containing 100 mM NaCl, 0.05 mM EDTA, and 0.5 A260/mL of each oligonucleotide. The temperature was raised 1 °C min−1.
Conditions: 10 mM Tris, 50 mM NaCl, and 1 mM EDTA; DNA concentration was 40 µg/mL.
Ref 33.
This work.
Ref 40.
The C8-IQ adduct had the least overall destabilizing effect on the oligonucleotides, lowering the Tm between 4 and 9 °C relative to the unmodified duplex. Of these, the least destabilized was 4a in which the IQ group is intercalated (4a < 3a < 2a < 1a), although the differences are small (33, 34). The opposite trend is observed for the AF-adducted oligonucleotides. Previous NMR analyses of these oligonucleotides showed that the AF-adduct was 70 and 90% groove-bound when at the G1- (2b) and G2-positions (3b), respectively, whereas the G3-adducted oligonucleotide (4b) had an equal mixture of the groove-bound and base-displaced intercalated conformers (37–39). We observed an increase in the thermal destabilization (8–13 °C) as the proportion of the intercalated conformation increases. The AAF-modified oligonucleotides showed the largest overall destabilization, lowering the Tm by 10–13 °C, although they were the least sensitive to the sequence (40, 41). Complete structural information for the AAF-modified oligonucleotides in the NarI sequence is not currently available. The C8-AAF adduct was shown to exist predominantly (∼70%) in the base-displaced intercalated conformation in a sequence in which the modified dGuo was flanked by dCyd, as is the case for the G3-position of the NarI sequence (42). Cho and Zhou utilized a C7-fluorinated AAF-derivative and 19F NMR spectroscopy to examine the conformational heterogeneity of the C8-dGuo adduct. These studies showed that the C8-(7-fluoro-AAF) adduct adopted two distinct base-displaced intercalated conformations in a local -GGA- sequence that differed by the torsional angle of the N-acetyl group (43).
Fuchs and co-workers showed that incorporation of the C8-AAF adduct at the G3-position of the NarI sequence resulted in one- and two-base deletions during the in vivo and in vitro replication by E. coli pol II (27, 44); we observed a similar outcome for the in vitro replication of the C8-IQ adduct in the same local sequence with pol II (45). To simulate the two-base slippage product, Fuchs and Romano independently examined the thermal stability of an oligonucleotide duplex in which the GpC nucleotides opposite the C8-AAF adduct were deleted (41, 46). The C8-AAF adduct at the G3-position was found to stabilize the duplex relative to the unmodified oligonucleotide by 15 °C. Interestingly, the Tm of the C8-AAF-modified oligonucleotide paired opposite a two-base deletion was the same as that for a full-length complement, leading to the speculation that these two duplexes had similar conformations (41). We examined the thermal stability of oligonucleotides containing the C8-IQ and AF adducts at the G3-positions of the NarI sequence when hybridized to a 10-mer complement missing the CpG nucleotides opposite the adduct. Both adducts stabilized the slippage product relative to the unmodified duplex (10 and 6 °C, respectively), although to a lesser degree than was reported for the AAF adduct. In contrast to the C8-AAF adduct, the IQ- and AF-modified duplexes were significantly less stable opposite a two-base deletion than when hybridized to a full-length oligonucleotide complement (13 and 8 °C, respectively). The Tm of the C8-IQ adduct opposite a one-base deletion was 4 °C more stable than the unmodified duplex and 7 °C higher than that of the two-base deletion product but was still 6 °C lower than when paired with the full-length complement.
UV Absorption, and Circular Dichroism Studies of Ad-ducted Oligonucleotides
We previously reported that the circular dichroism and UV spectra of oligonucleotides containing the C8 IQ-adduct at the non-iterated G1-position of the N-ras-12 sequence (1a) and the reiterated G3-position of the NarI recognition sequence (4a) suggested dramatically different adduct conformations. On the basis of the UV and CD spectra, we assigned 1a and 4a as possessing groove-bound and based-displaced intercalated conformations, respectively.
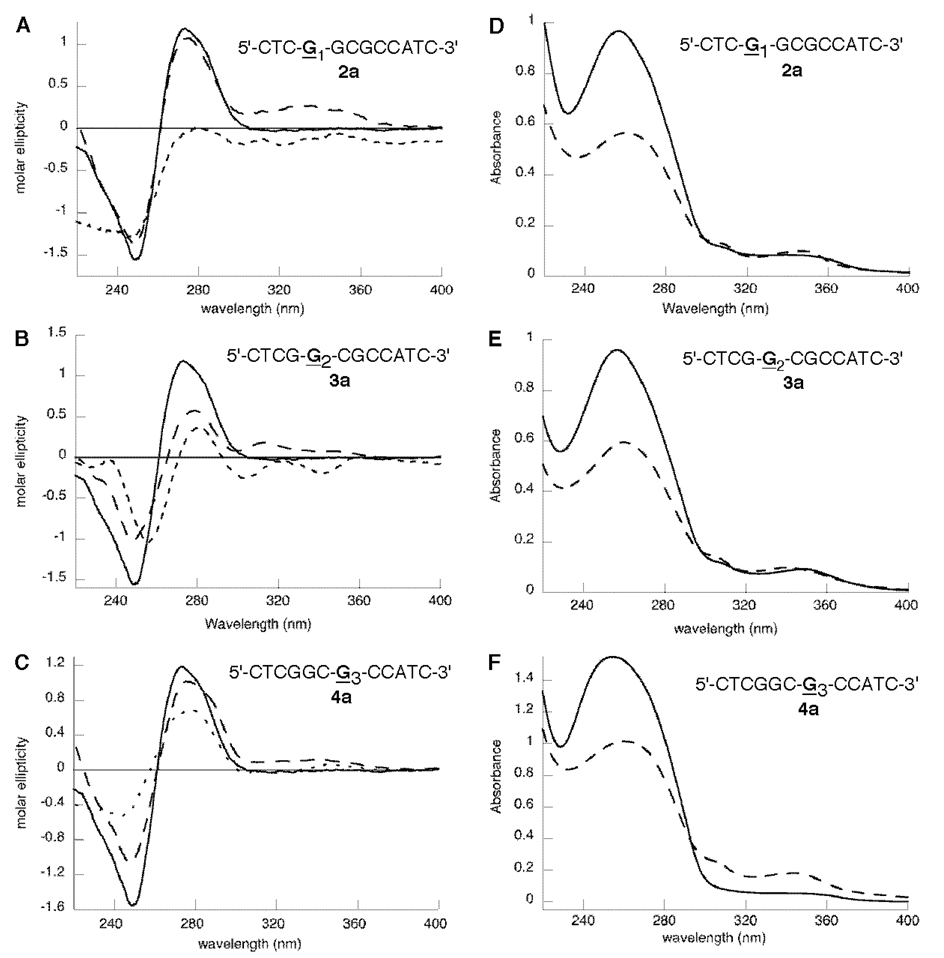
We examined the CD and UV spectra of 2a and 3a in which the C8-IQ adduct was incorporated at the G1- and G2-positions of the NarI sequence, respectively (Figure 2). The CD spectra of the single-stranded oligonucleotides 2a and 3a more closely resemble that of 1a. The positive ellipticty at ∼270 nm was greatly reduced for 2a and 3a compared to that for the unmodified oligonucleotide, and in all cases, the induced CD of the IQ chromophore is weak. The spectral properties changed significantly for the double-stranded oligonucleotides. The induced CD of the IQ group is positive in all cases but differs in intensity. The duplex oligonucleotide 2a (Figure 2A) more closely resembles that of 4a (Figure 2C) with the main difference being that the induced CD of the IQ chromophore is significantly stronger. The CD spectrum of duplex 3a is similar to that of 1a but with a slightly less intense induced CD of the IQ group. In contrast to oligonucleotide 4a (Figure 2F), the intensity of the IQ absorbance in the UV spectra of 2a and 3a (Figure 2D and E) were essentially unchanged upon hybridization to a complementary strand, which is more consistent with the groove-bound conformation of the IQ adduct.
Figure 2.
(A–C) CD spectra of the unmodified duplex (—), the C8-IQ-modified single strand (---), and the C8-IQ-modified duplex (---) oligonucleotides at positions G1 (2a, panel A), G2 (3a, panel B), and G3 (4a, panel C) of the NarI restriction sequence. (D–F) UV spectra of the C8-modified single strand (---) and double strand (—) oligonucleotides at positions G1 (2a, panel D), G2 (3a, panel E), and G3 (4a, panel F) of the NarI restriction sequence.
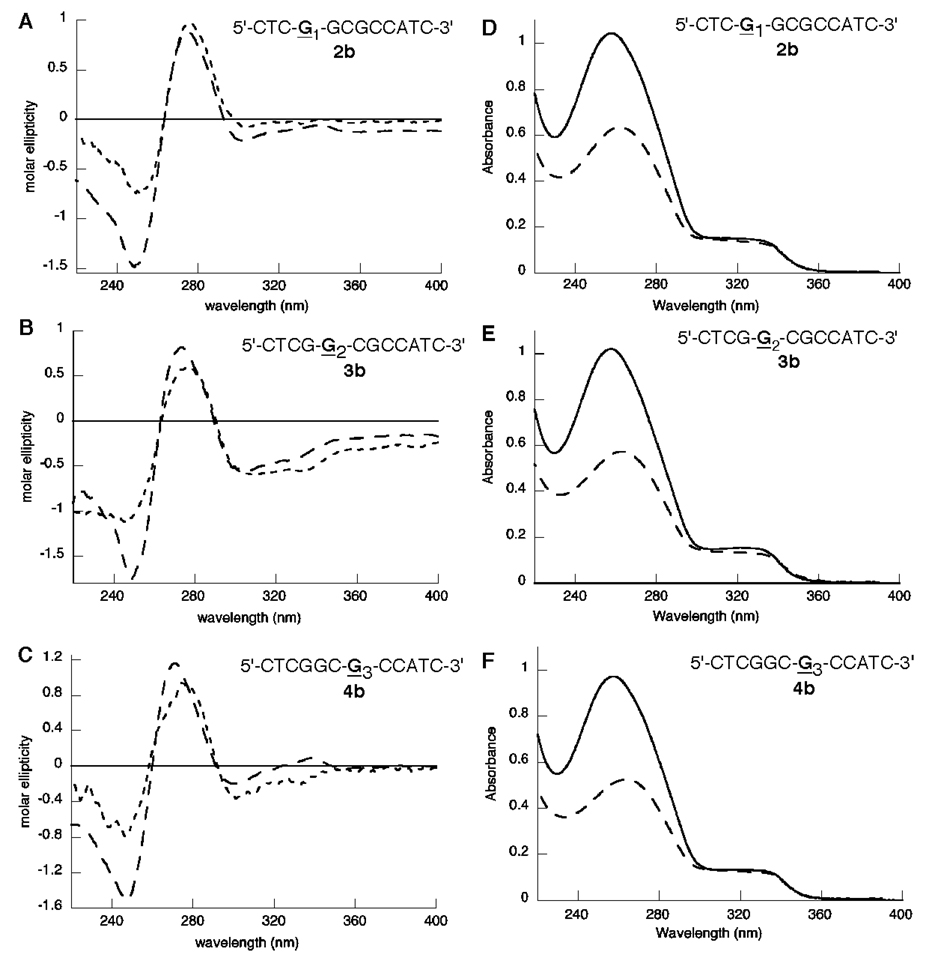
Previous NMR studies of the AF adduct at all three positions of the NarI sequence (2b–4b) demonstrated that AF-modified dGuo was in conformational exchange; the relative proportions of the groove-bound and base-displaced intercalated conformations were estimated to be 70:30, 90:10, and 50:50 for the AF adduct at the G1-, G2-, and G3-positions (2b–4b, respectively) (37–39). We examined the CD and UV spectra of these oligonucleotides (2b–4b) in order to further correlate the conformation with the CD and UV spectral properties (Figure 3). The UV spectra were nearly identical for all three oligo-nucleotides; the intensity of the AF absorbance was virtually unchanged upon hybridization to the appropriate complementary strand. The CD spectra of 2b–4b were similar, differing only in the intensity of the induced CD of the AF group (Figure 3A–C). The negative absorbance at ∼250 nm and positive absor-bance at ∼270 nm assigned to the DNA helix for oligonucle-otides 2b–4b were similar to those of the corresponding unmodified duplexes. For all three duplexes, the induced CD of the AF group was negative; this signal was significantly more intense for 3b than for 2b or 4b.
Figure 3.
(A–C) CD spectra of the C8-AF-modified single strand (---) and duplex (---) oligonucleotides at positions G1 (2b, panel A), G2 (3b, panel B), and G3 (4b, panel C) of the NarI restriction sequence. (D–F) UV spectra of the C8-AF-modified single strand (–––) and double strand (—) oligonucleotides at positions G1 (2b, panel D), G2 (3b, panel E), and G3 (4b, panel F) of the NarI restriction sequence.
NMR Spectroscopy
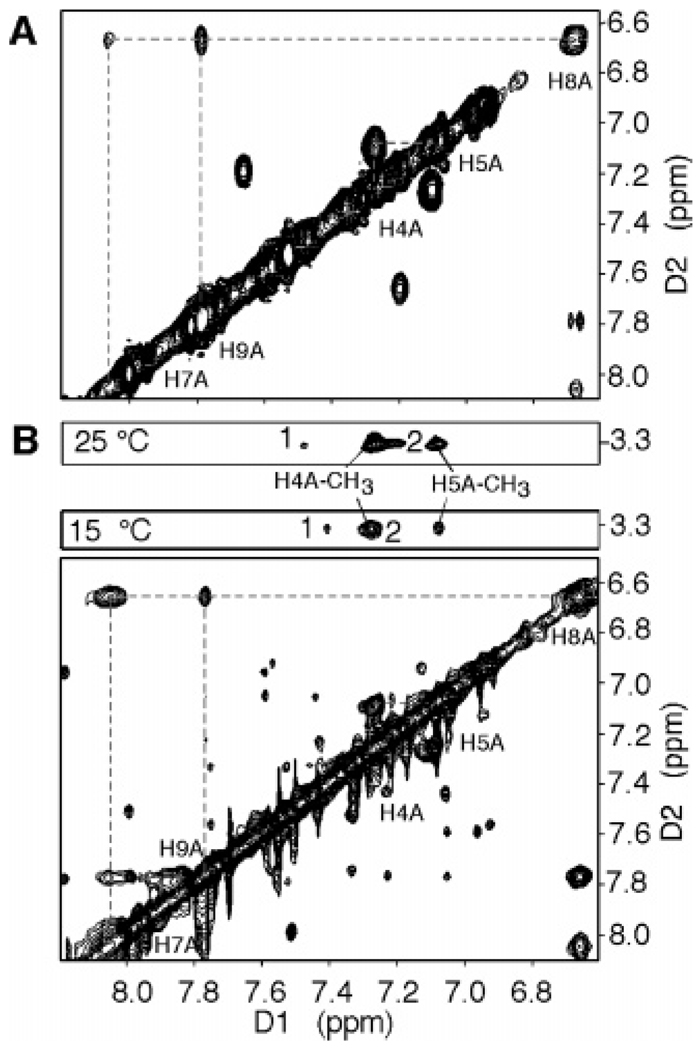
We reasoned that the NMR chemical shifts of the IQ protons of oligonucleotides 1a–4a would provide further insight into the adduct conformation. We have established that duplex 4a prefers a base-displaced intercalated conformation (34). We anticipated that the IQ protons of the base-displaced intercalated conformation of oligonucleotide 4a would be more shielded by the neighboring base-pairs relative to the groove-bound conformation of 1a, 2a, and 3a. We used the standard numbering for IQ, followed by an A to distinguish it from the numbering of the guanine (C8-IQ, Figure 1). The NMR spectra of the IQ-modified duplexes 1a–4a were recorded at 15 °C, where well-resolved spectra were obtained, and one major conformation was observed for each duplex. The adduct protons were assigned from a combination of 1D NOESY and COSY spectra. Proton H4A was readily assigned by the observation of a cross-peak to the N3A-methyl resonance in the NOESY spectrum as shown in Figure 4B for duplex 4a. Proton H5A was then readily assigned by COSY (Figure 4A) and NOESY cross-peaks to H4A. Protons H7A, H8A, and H9A were assigned on the basis of the comparison of their relative chemical shift and JHH couplings to those of quinoline and the adducted nucleoside (C8-IQ). Strong cross-peaks in the NOESY spectrum were observed for H8A and H9A and for H8A and H7A; the latter cross-peak was broadened because of the presence of the neighboring ring nitrogen. No exchanged cross-peaks were observed.
Figure 4.
Expanded plots of the COSY (A) and NOESY (B) spectra showing the IQ aromatic region of duplex 4a. The NOESY spectrum was recorded at 15 °C with the mixing time at 90 ms or 25 °C with a mixing time of 200 ms. 1 and 2 are the NOESY cross-peaks between the IQ N-CH3 group and the 3′-flanking Cyt H5 and 3′-flanking Gua H8 of the complementary strand, respectively, and are consistent with a base-displaced intercalated conformation.
The chemical shifts of the IQ protons H4A–H9A are listed in Table 3. The chemical shifts for protons H4A and H5A were consistent between the three oligonucleotides, indicating that they shared a similar chemical environment. However, the chemical shifts for protons H7A, H8A, and H9A were significantly different in 1a–4a and provided insight into the conformational preferences of these duplexes. These protons were most upfield in oligonucleotide 4a, which was predicted to be in the base-displaced intercalated conformation by UV spectroscopy, and the NMR chemical shift data is consistent with this assignment. In addition, NOEs were observed between the aromatic IQ protons and the aromatic protons of the flanking bases (35); two cross-peaks were observed between the IQ-N3A-methyl resonance and the H6 proton of the flanking 5′-Cyt bases and the H8 proton of the flanking Gua on the complementary strand (cross-peaks 1 and 2 in Figure 4B). Cross-peak 2 was overlapped with the H4A-CH3 cross-peak at 15 °C but was clearly visible at 25 °C (200 ms mixing time). The strong cross-peaks at ∼7.6 and 7.1 ppm (Figure 4) are currently unassigned.
Table 3.
Comparison of the Chemical Shifts of the IQ Aromatic Protons for Oligonucleotides 1a–4a and the C8-IQ-Modified Nucleoside
| chemical shift of the IQ protons |
|||||||
|---|---|---|---|---|---|---|---|
| H4A | H5A | H7A | H8A | H9A | N-CH3 | conformation | |
| laa | 7.34 | 7.26 | 8.57 | 7.24 | 8.30 | 3.06 | groove-bound |
| 2aa | 7.22 | 7.14 | 8.60 | 7.10 | 8.12 | 3.22 | groove-bound |
| 3aa | 7.30 | 7.05 | 8.32 | 6.96 | 8.00 | 3.22 | groove-bound |
| 4aa | 7.27 | 7.11 | 8.07 | 6.70 | 7.80 | 3.37 | base-displaced |
| intercalated | |||||||
| C8-Qb | 7.39 | 7.28 | 8.34 | 7.10 | 8.20 | 3.74 | |
NMR spectra recorded at 800 MHz is at 15 °C in 10 mM phosphate buffer at pH 7.0, containing 100 mM NaCl and 0.05 mM EDTA in 99.996% D2O.
NMR was recorded at 25 °C in DMSO-d6.
Oligonucleotide 1a had the most downfield chemical shift for protons H7A–H9A, which is consistent with a groove-bound conformation in accord with the assignment based on its UV absorbance spectrum. The chemical shifts for the H7A–H9A protons of 2a were similar to those of 1a, although these values for oligonucleotide 3a were intermediate between 2a and 4a. It is interesting to note that the chemical shift of the N3A-methyl group, which occurs over a 0.31 ppm range, shows a trend opposite that of the H7A–H9A protons. The N3A-methyl group for groove-bound adduct 1a was the most shielded and was observed at 3.06 ppm, whereas this resonance was observed at 3.37 ppm for the intercalated structure 4a. An intermediate value of 3.22 ppm was observed for duplexes 2a and 3a. In contrast to 4a, no NOEs were observed between the IQ N3A-methyl group and the flanking DNA bases for duplexes 1a–3a, consistent with a groove-bound structure (Figures S21–S23 of the Supporting Information).
Discussion
The Buchwald–Hartwig palladium-catalyzed N-arylation reaction has emerged as a valuable and versatile method for the preparation of carcinogen-modified nucleosides (47, 48). Our lab and others have utilized this reaction for the synthesis of C8-arylamine adducts of dGuo (33, 36, 49–58). The C8-dGuo adducts of IQ and AF were synthesized using this chemistry and incorporated into oligonucleotides via their phosphoramidite. Oligonucleotides containing the C8-IQ adduct at all three dGuo positions of the NarI recognition sequences and in the N-ras-12 sequence were prepared. The C8-AF adduct was also incorporated into the NarI sequence.
Fuchs and co-workers observed that the treatment of bacteria with N-acetoxy-AAF resulted in a large number of two-base deletions in the NarI recognition sequence (59). The NarI sequence is regarded as a hotspot for arylamine modification in bacteria and has been used extensively for studying frameshift mutations (60). We utilized CD, UV, and NMR spectroscopy to examine the conformation of the C8-IQ and C8-AF adducts at all three dGuo positions of the NarI sequence and codon-12 of the N-ras oncogene (1–4). Geacintov and co-workers utilized absorbance and fluorescence emission spectroscopy to characterize the conformation of oligonucleotides containing N2-PAH adducts (61, 62). It was observed that base-displaced intercalated conformations (termed site I) resulted in an ∼10 nm red shift, a measurable reduction in the intensity of the UV absorbance, and a significantly quenched fluorescence emission. The groove-bound conformations (site II), however, exhibited an ∼2 nm red shift in the absorbance maximum with no loss of intensity and a modest quenching of the fluorescence emission. This analysis was also applied to an oligonucleotide containing a C8-aminopyrene adduct (AP) (63). Unfortunately, the C8-modified dGuo is not fluorescent, and the broad absorbance bands assigned to the IQ chromophore make distinguishing relatively small shifts in wavelength tenuous. We found that the intensity of the UV absorbance of the IQ chromophore at ∼340 nm was greatly reduced when 4a was hybridized to a complementary strand, but relatively unchanged for oligonucle-otides 1a, 2a, and 3a, suggesting that 4a adopts a predominantly base-displaced intercalated conformation while the others were groove-bound. When intercalated, the IQ ring system is likely to be within van der Waals contact with the neighboring base pairs. The π–π stacking interactions between the IQ ring system and neighboring base pairs in the intercalated conformation as well as the significant change in the solvation of the adduct are likely contributors to the perturbation of the IQ chromophore. Such a perturbation would not be expected in the groove-bound conformation where the IQ group and the stacked DNA bases pairs are not in the optimal spatial arrangement for π-interactions.
To further expand the relationship between the UV absor-bance spectra and the conformation of C8-arylamine adducts, we examined the C8-AF adduct at all three positions of the NarI recognitions sequence (2b–4b). Previous NMR analyses demonstrated that these oligonucleotides are in conformational exchange between the base-displaced intercalated and groove-bound conformations (37, 38). The UV spectra of all three AF-modified oligonucleotides were nearly identical, and little or no change in the absorbance intensity of the AF chromophore was observed upon hybridization to a complementary strand. This behavior is peculiar, particularly for oligonucleotide 4b, which was shown to exist as an equal mixture of the base-displaced intercalated and groove-bound conformers (37, 38). Because electronic transitions occur on a much faster time scale than the conformational exchange of the C8-AF group, each conformation should contribute proportionally to the overall absorbance spectrum. Thus, we would expect the intensity of the AF absorbance for oligonucleotide 4b to be reduced relative to that of the spectra of 2b and 3b, which were shown to exist largely in the groove-bound conformation (70 and 90%, respectively). The smaller aromatic framework of the AF group may make it less sensitive to perturbations caused by π–π interactions or solvation effects as seen with the IQ, PAHs, and AP adducts. Unfortunately, this indicates that simply comparing the intensity of the adduct chromophore in the single strand and the duplex is not a general indication of a groove-bound versus base-displaced intercalated adduct conformation. Although a perturbation of the adduct’s UV absorbance is likely to indicate a base-displaced intercalated conformation, the absence of such a perturbation does not preclude the presence of this conformation.
Cho and co-workers have related the induced CD of the C8-AF adduct to specific conformations of the modified oligonucleotide (64). They observed that the induced CD of the C8-AF moiety is positive when in a base-displaced intercalated (stacked) conformation and negative when the adduct is bound in the major groove (B-type). The induced CD assigned to the AF group was of low intensity when the AF adduct was located in a local 5′-CGC-3′ sequence, as for 4b (Figure 3C). This observation correlates well to the NMR analysis that showed the adduct to be in conformational exchange in this sequence with an approximately equal population of the groove-bound and base-displaced intercalated conformations (37–39). The negative induced CD displayed for 2b and 3b reflects that the AF adducts of these duplexes are largely in the groove-bound conformation. The more intense negative ellipticity for 3b is indicative of the adduct being ∼90% groove-bound in this particular local sequence, versus 70% for 2b.
We believe that these observations can be extended to the IQ-modified oligonucleotides as well. The induced CD of the IQ chromophore for duplex oligonucleotides 1a–4a are positive, although the intensity of 4a is very weak. The weak, positive induced CD for 4a is a reflection of the perturbed UV absorbance because the intensity of the induced CD is, in part, also dependent upon the intensity of the UV absorbance. The positive induced CD suggests that an intercalated conformation exists for all four duplexes (1a–4a). However, Cho noted that the minor groove-bound adduct (termed a wedged conformation) also gave a positive induced CD for the C8-AF adducts. This conformation was observed when the C8-AF-modified dGuo was opposite mismatched purine bases (65, 66). The positive induced CD for 1a–3a, taken together with the observation that the UV absorbance of the IQ chromophore was not significantly perturbed upon hybridization, is consistent with a minor groove-bound adduct. It is interesting to note that molecular modeling studies of the C8-IQ adduct at the G3-position of the NarI sequence found a minor groove-bound (wedged) conformation to be favored (67).
Preliminary NMR analysis of the IQ-modified duplexes (1a–4a) further supports these assignments. The aromatic IQ protons H7A–H9A exhibited chemical shift differences up to 0.54 ppm depending on the sequence context. This is compared to the 0.8–1.2 ppm chemical shifts changes for the base-displaced intercalated and major groove-bound C8-AF adduct in the same sequence (38).2 However, the chemical shifts of the AF protons of the base-displaced intercalated conformation was between 0.32–0.63 ppm upfield of the minor groove-bound adduct (66). These chemical shift differences are much more in line with the values we observed for the IQ adduct.
The most shielded resonances were for duplex 4a, which is consistent with a base-displaced intercalated conformation. We initially proposed that a hydrogen-bonding interaction between the quinoline nitrogen of the IQ moiety and the complementary Cyt in duplex 4a contributed to the preference for the base-displaced intercalated conformation (33). We have recently determined the structure of the C8-IQ adduct at the G3-position (4a), and although the base-displaced intercalated conformation for this duplex was confirmed, the structure did not support the proposed hydrogen-bonding interactions involving the quinoline nitrogen (34). As part of our earlier study, we examined the UV and CD spectra of 4a when the C8-IQ adduct is opposite a thymine (33, 64). We observed that the IQ chromophore had an intense UV absorbance and an intense positive induced CD signal. These results suggest minor groove-bound (wedged) conformations analogous to the observations of Cho and co-workers for the mismatched C8-AF adduct (33, 64).
NMR studies of a duplex containing the C8-PhIP adduct were also reported to adopt a largely base-displaced intercalated conformation (68, 69). Although the overall sequence of the PhIP-containing oligonucleotide was significantly different from 4a, the adduct was similarly flanked by dCyd’s. The conformation of a C8-AP-containing duplex in which the flanking bases were dCyd’s was also determined to exist exclusively in a base-displaced intercalated conformation (70); the larger hydrophobic surface of AP certainly contributes to this preference. These structural studies suggest that favorable π-stacking arrangements between the C8-arylamine group and the flanking Gua’s of the complementary strand significantly contribute to the intercalated conformation.
The most downfield chemical shifts for IQ protons H7A–H9A were observed for oligonucleotides 1a and 2a and is consistent with a groove-bound IQ adduct; the conclusions from the NMR chemical shift data correlate well with the very small changes in the UV spectra between the single-stranded and duplex oligonucleotides, and the CD spectra further suggest that the adduct is bound in the minor groove with the modified dGuo adopting a syn conformation about the glycosidic bond. The chemical shift data of oligonucleotide 3a is intermediate between the groove-bound (1a and 2a) and base-displaced intercalated (4a) structures. The UV and CD spectra of 3a were similar to those assigned as a minor groove-bound conformation (1a and 2a). Collectively, our data leads to the tentative assignment of the G2-modified duplex 3a possessing a minor groove-bound conformation; however, the possibility that 3a is in rapid conformational exchange must also be considered. NMR studies to distinguish these possibilities are currently underway.
The outcome of in vivo and in vitro replication bypass of the C8-AAF adduct is strongly influenced by its specific location within the NarI recognition sequence. In bacterial systems, two-base deletions were observed when the G3-position was modified, whereas base-pair substitutions occurred when the adduct was at the G1- and G2-positions. We have recently reported the in vitro bypass of the C8-IQ adduct at the G1- and G3-positions of the NarI recognition sequences with prokaryotic and human DNA polymerases. Replication with E. coli pol I Klenow fragment (exo−), pol II (exo−), and Dpo4 resulted in two-base deletions when the C8-IQ adduct was located at the reiterated G3-position, whereas error-free bypass and extension was observed when the G1-position was modified (45). The correlation of these results with the present conformational studies suggests a possible structural basis to the sequence-dependent mutations generated by IQ and in particular the susceptibility of reiterated CpG sequences toward arylamine-induced two-base deletion. It should be noted that in vitro replication of the C8-IQ adduct with human pol η resulted in error-free bypass and extension (71), indicating that specific interactions between the polymerase active site and the adduct are highly influential in the outcome of lesion bypass. It should also be noted that Fuchs reported that the induced CD spectra of the C8-AAF adduct at all three dGuo-positions of the NarI sequence were negative (40), suggesting a significant contribution of a major groove-bound conformation. Thus, although the results of trans-lesion bypass of the C8-IQ and C8-AAF adducts are very similar, the conformation of the C8-AAF adduct may be more similar to that of the C8-AF adduct than that of the C8-IQ adduct.
Conclusions
Oligonucleotides containing the C8-IQ adduct were prepared in a variety of sequence contexts. The UV absorbance and induced CD of the adduct in single strand and duplex were used to determine the preference for a groove-bound or base-displaced intercalated conformation. The intensity of the adduct chro-mophore was significantly perturbed when incorporated at the G3-position of the NarI recognition sequence (4a), consistent with a base-displaced intercalated conformation. The intensity of this absorbance was relatively unchanged when positioned in non-iterated sequences, indicative of a groove-bound conformation (1a–3a). This simple UV analysis could not be definitively extended to the conformation of the related C8-AF adduct (2b–4b). The CD spectra are consistent with the IQ group bound in the minor groove of duplexes 1a–3a. An examination of the NMR chemical shifts of the IQ protons in the different sequences further supports these assignments. The relationship between the local sequence and the conformational preferences for the C8-IQ adduct is intriguing and may play a significant role in the carcinogenicity of these dietary genotoxins. For example, the specific local distortion of DNA is likely to influence the recognition of the lesion by repair proteins. The specific conformations could also lead to specific interactions with DNA polymerase and thereby lead to differential rates and selectivity during trans-lesion synthesis.
Supplementary Material
Synthetic procedures for the preparation of the C8-AF dGuo phosphoramidite reagent and related 1H, 13C, and 31P NMR spectra; MALDI-TOF spectra and MALDI-TOF sequencing of adducted oligonucleotides (2–4); and expanded NMR COSY and NOSEY spectra for duplexes 1a–3a used for the conformational assignments. This material is available free of charge via the Internet at http://pubs.acs.org.
Acknowledgment
This work was supported by NIH through research grant CA-55678 (M.P.S.) and center grant ES-00267 (C.J.R and M.P.S). J.S.S. was supported by an NIH predoctoral traineeship (ES-07028). Funding for the NMR spectrometers and CD instrument was supplied by NIH Grants RR-05805, RR-006381, ES-00267, and Vanderbilt University.
Footnotes
This article is dedicated to the memory of Christopher J. Michejda.
Abbreviations: AF, 2-aminofluorene; AAF, 2-acetylaminofluorene; IQ, 2-amino-3-methylimidazo[4,5-f]quinoline; PhIP, 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine; HCA, heterocyclic arylamine; C8-IQ, 8-[(3-methyl-3H-imidazo[4,5-f]quinolin-2-yl)amino]-2′-deoxy-guanosine; C8-AF, 8-(9H-fluoren-2-ylamino)-2′-deoxyguanosine; C8-AAF, 8-(acetyl-9H-fluoren-2-ylamino)-2′-deoxyguanosine; C8-PhIP, 8-[(1-methyl-6-phenyl-1Himidazo[4,5-b]pyridin-2-yl)amino]-2′-deoxyguanosine; C8-AP, 8-(1-pyrenylamino)-2′-deoxyguanosine; PAH, polycyclic aromatic hydrocarbon; AP, aminopyrene; DMSO, dimethylsulfoxide; Tm, thermal melting temperature; CD, circular dichroism; MALDI-TOF, matrix-assisted laser desorption/time-of-flight (MS).
The chemical shift differences of the IQ protons between the intercalated and groove-bound conformations (up to 0.54 ppm) is smaller than those for the C8-AF adduct. As a result, a reviewer suggested that the NMR chemical shift differences observed for the IQ protons may be due to differing ring current effects of the flanking base pairs rather than different conformations. Mao et al. have assigned the chemical shifts of the AF-protons for the intercalated conformations at the three dGuo positions of the NarI sequence; in general, they were found to be within 0.1 ppm of each other (38). The only protons that showed significant differences in chemical shift were H1 and H3, which are adjacent to the point of attachment to Gua. It is likely that these protons showed different chemical shifts because of subtle difference in the intercalated structure. Therefore, differing ring current effects are probably not the source of the chemical shift difference we observed for the IQ protons of 1a–4a.
References
- 1.Turesky RJ. Heterocyclic aromatic amine metabolism, DNA adduct formation, mutagenesis, and carcinogenesis. Drug Metab. Rev. 2002;34:625–650. doi: 10.1081/dmr-120005665. [DOI] [PubMed] [Google Scholar]
- 2.Schut HAJ, Snyderwine EG. DNA adducts of heterocyclic amine food mutagens: Implications for mutagenesis and carcinogenesis. Carcinogenesis. 1999;20:353–368. doi: 10.1093/carcin/20.3.353. [DOI] [PubMed] [Google Scholar]
- 3.Skog KI, Johansson MAE, Jagerstad MI. Carcinogenic heterocyclic amines in model systems and cooked foods: A review on formation, occurrence and intake. Food Chem. Toxicol. 1998;36:879–896. doi: 10.1016/s0278-6915(98)00061-1. [DOI] [PubMed] [Google Scholar]
- 4.Sugimura T. Overview of carcinogenic heterocyclic amines. Mutat. Res. 1997;376:211–219. doi: 10.1016/s0027-5107(97)00045-6. [DOI] [PubMed] [Google Scholar]
- 5.Felton JS, Knize MG, Dolbeare FA, Wu R. Mutagenic activity of heterocyclic amines in cooked foods. Environ. Health Perspect. 1994;102:201–204. doi: 10.1289/ehp.94102s6201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Potter JD. Nutrition and colorectal cancer. Cancer, Causes Control. 1996;7:127–146. doi: 10.1007/BF00115644. [DOI] [PubMed] [Google Scholar]
- 7.Willet WC. Diet, nutrition, and avoidable cancer. Environ. Health Perspect. 1995;103:165–170. doi: 10.1289/ehp.95103s8165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lang NP, Butler MA, Massengill JP, Lawson M, Stotts RC, Hauer-Jensen M, Kadlubar FF. Rapid metabolic phenotypes for acetyltransferase and cytochrome P4501A2 and putative exposure to food-borne heterocyclic amines increase the risk for colorectal cancer for polyps. Cancer Epidemiol., Biomarkers Prev. 1994;3:675–682. [PubMed] [Google Scholar]
- 9.Shirai T, Sano M, Tamano S, Takahashi S, Hirose M, Futakuchi M, Hasegawa R, Imaida K, Matsumoto K, Wakabayashi K, Sugimura T, Ito N. The prostate: a target for carcino-genicity of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) derived from cooked foods. Cancer Res. 1997;57:195–198. [PubMed] [Google Scholar]
- 10.Nagao M, Sugimura T. Carcinogenic factors in food with relevance to colon cancer development. Mutat. Res. 1993;290:43–51. doi: 10.1016/0027-5107(93)90031-a. [DOI] [PubMed] [Google Scholar]
- 11.Shirai T, Asamoto M, Takahashi S, Imaida K. Diet and prostate cancer. Toxicology. 2002;181–182:89–94. doi: 10.1016/s0300-483x(02)00260-3. [DOI] [PubMed] [Google Scholar]
- 12.Snyderwine EG. Diet and mammary gland carcinogenesis. Recent Results Cancer Res. 1998;152:3–10. doi: 10.1007/978-3-642-45769-2_1. [DOI] [PubMed] [Google Scholar]
- 13.Kim D, Guengerich FP. Cytochrome P450 activation of arylamines and heterocyclic amines. Annu. Rev. Pharmacol. Toxicol. 2005;45:27–49. doi: 10.1146/annurev.pharmtox.45.120403.100010. [DOI] [PubMed] [Google Scholar]
- 14.Turesky RJ. Interspecies metabolism of heterocyclic aromatic amines and the uncertainties in extrapolation of animal toxicity data for human risk assessment. Mol. Nutr. Food Res. 2005;49:101–117. doi: 10.1002/mnfr.200400076. [DOI] [PubMed] [Google Scholar]
- 15.Turesky RJ, Markovic J. DNA adduct formation of the food carcinogen 2-amino-3-methylimidazo[4,5-f]quinoline at the C-8 and N2 atoms of guanine. Chem. Res. Toxicol. 1994;7:752–761. doi: 10.1021/tx00042a007. [DOI] [PubMed] [Google Scholar]
- 16.Turesky RJ, Rossi SC, Welti DH, Lay JO, Kadlubar FF. Characterization of DNA adducts formed in vitro by reaction of N-hydroxy-2-amino-3-methylimidazo[4,5-f]quinoline and N-hy-droxy-2-amino-3,8-dimethylimidazo[4,5-f]quinoxaline at the C-8 and N2 atoms of guanine. Chem. Res. Toxicol. 1992;5:479–490. doi: 10.1021/tx00028a005. [DOI] [PubMed] [Google Scholar]
- 17.Burnouf D, Bichara M, Dhalluin C, Garcia A, Janel-Bintz R, Koffel-Schwartz N, Lambert I, Lefevre JF, Lindsley JE, Maenhaut-Michel G, Milhe C, Lobo-Napolitano R, Valladier-Belguise P, Fuchs RP. Induction of frameshift mutations at hotspot sequences by carcinogen adducts. Recent Results Cancer Res. 1997;143:1–20. doi: 10.1007/978-3-642-60393-8_1. [DOI] [PubMed] [Google Scholar]
- 18.Leong-Morgenthaler PM, Op Het Velt C, Jaccaud E, Turesky RJ. Mutagenicity of 2-amino-3-methylimidazo[4,5-f]quinoline in human lymphoblastoid cells. Carcinogenesis. 1998;19:1749–1754. doi: 10.1093/carcin/19.10.1749. [DOI] [PubMed] [Google Scholar]
- 19.Heflich RH, Neft RE. Genetic toxicity of 1-acetylaminofluorene, 2-aminofluorene and some of their metabolites and model metabolites. Mutat. Res. 1994;318:73–114. doi: 10.1016/0165-1110(94)90025-6. [DOI] [PubMed] [Google Scholar]
- 20.Belguise-Valladier P, Fuchs RPP. N-2-Aminofluorene and N-2-acetylaminofluorene adducts: The local sequence context of the adduct and its chemical structure determines its replication properties. J. Mol. Biol. 1995;249:903–913. doi: 10.1006/jmbi.1995.0347. [DOI] [PubMed] [Google Scholar]
- 21.Tan X, Suzuki N, Grollman AP, Shibutani S. Mutagenic events in Escherichia coli and mammalian cells generated in response to acetylaminofluorene-derived DNA adducts positioned in the Nar I restriction enzyme site. Biochemistry. 2002;41:14255–14262. doi: 10.1021/bi0202878. [DOI] [PubMed] [Google Scholar]
- 22.Hsu GW, Kiefer JR, Burnouf D, Becherel OJ, Fuchs RPP, Beese LS. Observing translesion synthesis of an aromatic amine DNA adduct by a high-fidelity DNA polymerase. J. Biol. Chem. 2004;279:50280–50285. doi: 10.1074/jbc.M409224200. [DOI] [PubMed] [Google Scholar]
- 23.Dutta S, Li Y, Johnson D, Dzantiev L, Richardson CC, Romano LJ, Ellenberger T. Crystal structures of 2-acetylami-nofluorene and 2-aminofluorene in complex with T7 DNA polymerase reveal mechanisms of mutagenesis. Proc. Natl. Acad. Sci. U.S.A. 2004;101:16186–16191. doi: 10.1073/pnas.0406516101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gill JP, Romano LJ. Mechanism for N-acetyl-2-aminofluorene-induced frameshift mutagenesis by Escherichia coli DNA polymerase I (Klenow fragment) Biochemistry. 2005;44:15387–15395. doi: 10.1021/bi051437s. [DOI] [PubMed] [Google Scholar]
- 25.Shibutani S, Suzuki N, Grollman AP. Mechanism of frameshift (deletion) generated by acetylaminofluorene-derived DNA adducts in vitro. Biochemistry. 2004;43:15929–15935. doi: 10.1021/bi048087e. [DOI] [PubMed] [Google Scholar]
- 26.Lone S, Romano LJ. Mechanistic insights into replication across from bulky DNA adducts: a mutant polymerase I allows an N-acetyl-2-aminofluorene adduct to be accommodated during DNA synthesis. Biochemistry. 2003;42:3826–3834. doi: 10.1021/bi027297t. [DOI] [PubMed] [Google Scholar]
- 27.Becherel OJ, Fuchs RPP. Mechanism of DNA polymerase II-mediated frameshift mutagenesis. Proc. Natl. Acad. Sci. U.S.A. 2001;98:8566–8571. doi: 10.1073/pnas.141113398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suzuki N, Ohashi E, Hayashi K, Ohmori H, Grollman AP, Shibutani S. Translesional synthesis past acetylaminofluorene-derived DNA adducts catalyzed by human DNA polymerase κ and Escherichia coli DNA polymerase IV. Biochemistry. 2001;40:15176–15183. doi: 10.1021/bi010702g. [DOI] [PubMed] [Google Scholar]
- 29.Ohashi E, Ogi T, Kusumoto R, Iwai S, Masutani C, Hanaoka F, Ohmori H. Error-prone bypass of certain DNA lesions by the human DNA polymerase κ. Genes Dev. 2000;14:1589–1594. [PMC free article] [PubMed] [Google Scholar]
- 30.Burnouf DY, Miturski R, Fuchs RPP. Sequence context modulation of translesion synthesis at a single N-2-acetylaminofluorene adduct located within a mutation hot spot. Chem. Res. Toxicol. 1999;12:144–150. doi: 10.1021/tx9801920. [DOI] [PubMed] [Google Scholar]
- 31.Doisy R, Tang MS. Effect of aminofluorene and (acetylamino)fluorene adducts on the DNA replication mediated by Escherichia coli polymerases I (Klenow fragment) and III. Biochemistry. 1995;34:4358–4368. doi: 10.1021/bi00013a027. [DOI] [PubMed] [Google Scholar]
- 32.Lindsley JE, Fuchs RPP. Use of single-turnover kinetics to study bulky adduct bypass by T7 DNA polymerase. Biochemistry. 1994;33:764–772. doi: 10.1021/bi00169a018. [DOI] [PubMed] [Google Scholar]
- 33.Elmquist CE, Stover JS, Wang Z, Rizzo CJ. Site-specific synthesis and properties of oligonucleotides containing C8-deoxyguanosine adducts of the dietary mutagen IQ. J. Am. Chem. Soc. 2004;126:11189–11201. doi: 10.1021/ja0487022. [DOI] [PubMed] [Google Scholar]
- 34.Wang F, DeMuro NE, Elmquist CE, Stover JS, Rizzo CJ, Stone MP. Base-displaced intercalated structure of the food mutagen 2-amino-3-methylimidazo[4,5-f]quinoline (IQ) in the recognition sequence of the NarI restriction enzyme, a hotspot for-2 bp deletions. J. Am. Chem. Soc. 2006;128:10085–10095. doi: 10.1021/ja062004v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tretyakova N, Matter B, Ogdie A, Wishnok JS, Tannen-baum SR. Locating nucleobase lesions within DNA sequences by MALDI-TOF mass spectral analysis of exonuclease ladders. Chem. Res. Toxicol. 2001;14:1058–1070. doi: 10.1021/tx010062i. [DOI] [PubMed] [Google Scholar]
- 36.Wang Z, Rizzo CJ. Synthesis of the C8-deoxyguanosine adduct of the food mutagen IQ. Org. Lett. 2001;3:565–568. doi: 10.1021/ol006968h. [DOI] [PubMed] [Google Scholar]
- 37.Mao B, Hingerty BE, Broyde S, Patel DJ. Solution structure of the aminofluorene [AF]-intercalated conformer of the syn-[AF]-C8-dG adduct opposite dC in a DNA duplex. Biochemistry. 1998;37:81–94. doi: 10.1021/bi972257o. [DOI] [PubMed] [Google Scholar]
- 38.Mao B, Hingerty BE, Broyde S, Patel DJ. Solution structure of the aminofluorene [AF]-external conformer of the anti-[AF]-C8-dG adduct opposite dC in a DNA duplex. Biochemistry. 1998;37:95–106. doi: 10.1021/bi972258g. [DOI] [PubMed] [Google Scholar]
- 39.Patel DJ, Mao B, Gu Z, Hingerty BE, Gorin A, Basu AK, Broyde S. Nuclear magnetic resonance solution structures of covalent aromatic amine-DNA adducts and their mutagenic relevance. Chem. Res. Toxicol. 1998;11:391–407. doi: 10.1021/tx9702143. [DOI] [PubMed] [Google Scholar]
- 40.Koehl P, Valladier P, Lefevre JF, Fuchs RPP. Strong structural effect of the position of a single acetylaminofluorene within a mutation hot spot. Nucleic Acids Res. 1989;17:9531–9541. doi: 10.1093/nar/17.23.9531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou Y, Romano LJ. Solid-phase synthesis of oligonucleotides containing site-specific N-(2′-doxyguanosin-8-yl)-2-(acetylamino)fluorene adducts using 9-fluorenylmethoxycarbonyl as the base-protecting group. Biochemistry. 1993;32:14043–14052. doi: 10.1021/bi00213a038. [DOI] [PubMed] [Google Scholar]
- 42.O’Handley SF, Sanford DG, Xu R, Lester CC, Hingerty BE, Broyde S, Krugh TR. Structural characterization of an N-acetyl-2-aminofluorene (AAF) modified DNA oligomer by NMR, energy minimization, and molecular dynamics. Biochemistry. 1993;32:2481–2497. doi: 10.1021/bi00061a005. [DOI] [PubMed] [Google Scholar]
- 43.Cho BP, Zhou L. Probing the conformational heterogeneity of the acetylaminofluorene-modified 2′-deoxyguanosine and DNA by 19F NMR spectroscopy. Biochemistry. 1999;38:7572–7583. doi: 10.1021/bi990182d. [DOI] [PubMed] [Google Scholar]
- 44.Fuchs RPP, Koffel-Schwartz N, Pelet S, Janel-Bintz R, Napolitano R, Becherel OJ, Broschard TH, Burnouf DY, Wagner J. DNA polymerases II and V mediate respectively mutagenic(-2 frameshift) anderror-free bypass of a single N-2-acetylaminofluorene adduct. Biochem. Soc. Trans. 2001;29:191–195. doi: 10.1042/0300-5127:0290191. [DOI] [PubMed] [Google Scholar]
- 45.Stover JS, Chowdhury G, Zang H, Guengerich FP, Rizzo CJ. Translesion synthesis past the C8-and N2-deoxyguanosine adducts of the dietary mutagen 2-amino-3-methylimidazo[4,5-f]-quinoline in the NarI recognition sequence by prokaryotic DNA polymerases. Chem. Res. Toxicol. 2006;19:1506–1517. doi: 10.1021/tx0601455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Milhe C, Fuchs RPP, Lefevre JF. NMR data show that the carcinogen N-2-acetylaminoflurorene stabilizes an intermediate of -2 frameshift mutagenesis in a region of high mutation frequency. Eur. J. Biochem. 1996;235:120–127. doi: 10.1111/j.1432-1033.1996.00120.x. [DOI] [PubMed] [Google Scholar]
- 47.Lakshman MK. Palladium-catalyzed C-N and C-C cross-coupling as versatile, new avenues for modifications of purine 2′-deoxynucleosides. J. Organomet. Chem. 2002;653:234–251. [Google Scholar]
- 48.Lakshman MK. Synthesis of biologically important nucleoside analogs by palladium-catalyzed C-N bond-formation. Curr. Org. Synth. 2005;2:83–112. [Google Scholar]
- 49.Schoffers E, Olsen PD, Means JC. Synthesis of C8-adenosine adducts of arylamines using palladium catalysis. Org. Lett. 2001;3:4221–4223. doi: 10.1021/ol016900h. [DOI] [PubMed] [Google Scholar]
- 50.Meier C, Grasl S, Detmer I, Marx A. Synthesis of oligonucleotides bearing an arylamine modification in the C8-position of 2′-deoxyguanosine. Nucleosides, Nucleotides Nucleic Acids. 2005;24:691–694. doi: 10.1081/ncn-200060267. [DOI] [PubMed] [Google Scholar]
- 51.Meier C, Grasl S. Highly efficient synthesis of a phosphoramidite building block of C8-deoxyguanosine adducts of arylamines. Synlett. 2002:802–804. [Google Scholar]
- 52.Grasl S, Meier C. Synthesis of oligonucleotide building blocks of 2′-deoxyguanosine bearing C8-arylamine modification. Nucleosides, Nucleotides Nucleic Acids. 2003;22:1119–1121. doi: 10.1081/NCN-120022751. [DOI] [PubMed] [Google Scholar]
- 53.Gillet LCJ, Scharer OD. Preparation of C8-amine and acetylamine adducts of 2′-deoxyguanosine suitably protected for DNA synthesis. Org. Lett. 2002;4:4205–4208. doi: 10.1021/ol026474f. [DOI] [PubMed] [Google Scholar]
- 54.Gillet LCJ, Alzeer J, Scharer OD. Site-specific incorporation of N-(deoxyguanosin-8-yl)-2-acetylaminofluorene (dG-AAF) into oligonucleotides using modified ‘ultra-mild’ DNA synthesis. Nucleic Acids Res. 2005;33:1961–1969. doi: 10.1093/nar/gki335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Takamura-Enya T, Ishikawa S, Mochizuki M, Wakabayashi K. A practical approach for the chemical synthesis of 2′-deoxyguanosine-C8 adducts with mutagenic/carcinogenic aminoor nitro-arenes. Tetrahedron Lett. 2003;44:5969–5973. [Google Scholar]
- 56.Takamura-Enya T, Enomoto S, Wakabayashi K. Palladium-catalyzed direct N-arylation of nucleosides, nucleotides, and oligonucleotides for efficient preparation of dG-N2 adducts with carcinogenic amino-/nitroarenes. J. Org. Chem. 2006;71:5599–5606. doi: 10.1021/jo0605243. [DOI] [PubMed] [Google Scholar]
- 57.Takamura-Enya T, Ishikawa S, Mochizuki M, Wakabayashi K. Chemical synthesis of 2′-deoxyguanosine-C8 adducts with heterocyclic amines: An application to synthesis of oligonucleotides site-specifically adducted with 2-amino-1-methyl-6-phenylimidazo-[4,5-b]pyridine. Chem. Res. Toxicol. 2006;19:770–778. doi: 10.1021/tx050296s. [DOI] [PubMed] [Google Scholar]
- 58.Bonala R, Torres MC, Iden CR, Johnson F. Synthesis of the PhIP adduct of 2′-deoxyguanosine and its incorporation into oligomeric DNA. Chem. Res. Toxicol. 2006;19:734–738. doi: 10.1021/tx0600191. [DOI] [PubMed] [Google Scholar]
- 59.Fuchs RPP, Schwartz N, Daune MP. Hot spots of frameshift mutations induced by the ultimate carcinogen N-acetoxy-N-2-acetylaminofluorene. Nature. 1981;294:657–659. doi: 10.1038/294657a0. [DOI] [PubMed] [Google Scholar]
- 60.Hoffmann GR, Fuchs RPP. Mechanisms of frameshift mutations: Insight from aromatic amines. Chem. Res. Toxicol. 1997;10:347–359. doi: 10.1021/tx960128n. [DOI] [PubMed] [Google Scholar]
- 61.Geacintov NE, Cosman M, Mao B, Alfano A, Ibanez V, Havvey RG. Spectroscopic characterization and site I/site II classification of cis and trans benzo[a]pyrene diolepoxide enantiomersguanosine adducts in oligonucleotides and polynucleotides. Carcinogenesis. 1991;12:2099–2108. doi: 10.1093/carcin/12.11.2099. [DOI] [PubMed] [Google Scholar]
- 62.Huang W, Amin S, Geacintov NE. Fluorescence characteristics of site-specific and stereochemically distinct benzo[a]-pyrene diol epoxide-DNA adducts as probes of adduct conformation. Chem. Res. Toxicol. 2002;15:118–126. doi: 10.1021/tx010135y. [DOI] [PubMed] [Google Scholar]
- 63.Nolan SJ, Vyas RR, Hingerty BE, Ellis S, Broyde S, Shapiro R, Basu AK. Solution properties and computational analysis of an oligodeoxynucleotide containing N-(deoxyguanosin-8-yl)-1-aminopyrene. Carcinogenesis. 1996;17:133–144. doi: 10.1093/carcin/17.1.133. [DOI] [PubMed] [Google Scholar]
- 64.Liang F, Meneni S, Cho BP. Induced circular dichroism characteristics as conformational probes for carcinogenic aminofluo-rene-DNA adducts. Chem. Res. Toxicol. 2006;19:1040–1043. doi: 10.1021/tx0601253. [DOI] [PubMed] [Google Scholar]
- 65.Norman D, Abuaf P, Hingerty BE, Live D, Grunberger D, Broyde S, Patel DJ. NMR and computational characterization of the N-(deoxyguanosin-8-yl)aminofluorene adduct [(AF)G] opposite adenosine in DNA: (AF)G[syn]•A[anti] pair formation and its pH dependence. Biochemistry. 1989;28:7462–7476. doi: 10.1021/bi00444a046. [DOI] [PubMed] [Google Scholar]
- 66.Abuaf P, Hingerty BE, Broyde S, Grunberger D. Solution conformation of the N-(deoxyguanosin-8-yl)aminofluorene adduct opposite deoxyinosine and deoxyguanosine in DNA by NMR and computational characterization. Chem. Res. Toxicol. 1995;8:369–378. doi: 10.1021/tx00045a008. [DOI] [PubMed] [Google Scholar]
- 67.Wu X, Shapiro R, Broyde S. Conformational analysis of the major DNA adduct derived from the food mutagen 2-amino-3-methylimidazo[4,5-f]quinoline. Chem. Res. Toxicol. 1999;12:895–905. doi: 10.1021/tx990108w. [DOI] [PubMed] [Google Scholar]
- 68.Brown K, Guenther EA, Dingley KH, Cosman M, Harvey CA, Shields SJ, Turteltaub KW. Synthesis and spectroscopic characterization of site-specific 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine oligodeoxyribonucleotide adducts. Nucleic Acids Res. 2001;29:1951–1959. doi: 10.1093/nar/29.9.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Brown K, Hingerty BE, Guenther EA, Krishnan VV, Broyde S, Turteltaub KW, Cosman M. Solution structure of the 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine C8-deoxyguanosine adduct in duplex DNA. Proc. Natl. Acad. Sci. U.S.A. 2001;98:8507–8512. doi: 10.1073/pnas.151251898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mao B, Vyas RR, Hingerty BE, Broyde S, Basu AK, Patel DJ. Solution conformation of the N-(deoxyguanosin-8-yl)-1-aminopyrene ([AP]dG) adduct opposite dC in a DNA duplex. Biochemistry. 1996;35:12659–12670. doi: 10.1021/bi961078o. [DOI] [PubMed] [Google Scholar]
- 71.Choi J-Y, Stover JS, Angel KC, Chowdhury G, Rizzo CJ, Guengerich FP. Biochemical basis of genotoxicity of heterocyclic arylamine food mutagens: Human DNA polymerase η selectively produces a two-base deletion in copying the N2-guanyl adduct of 2-amino-3-methylimidazo[4,5-f]quinoline but not the C8-adduct at the NarI G3 site. J. Biol. Chem. 2006;281:25297–25306. doi: 10.1074/jbc.M605699200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Synthetic procedures for the preparation of the C8-AF dGuo phosphoramidite reagent and related 1H, 13C, and 31P NMR spectra; MALDI-TOF spectra and MALDI-TOF sequencing of adducted oligonucleotides (2–4); and expanded NMR COSY and NOSEY spectra for duplexes 1a–3a used for the conformational assignments. This material is available free of charge via the Internet at http://pubs.acs.org.