Abstract
This evidence-based review addresses the question of whether testing for UGT1A1 mutations in patients with metastatic colorectal cancer treated with irinotecan leads to improvement in outcomes (e.g., irinotecan toxicity, response to treatment, morbidity, and mortality), when compared with no testing. No studies were identified that addressed this question directly. The quality of evidence on the analytic validity of current UGT1A1 genetic testing methods is adequate (scale: convincing, adequate, inadequate), with available data indicating that both analytic sensitivity and specificity for the common genotypes are high. For clinical validity, the quality of evidence is adequate for studies reporting concentration of the active form of irinotecan (SN-38), presence of severe diarrhea, and presence of severe neutropenia stratified by UGT1A1 common genotypes. The strongest association for a clinical endpoint is for severe neutropenia. Patients homozygous for the *28 allele are 3.5 times more likely to develop severe neutropenia compared with individuals with the wild genotype (risk ratio 3.51; 95% confidence interval 2.03–6.07). The proposed clinical utility of UGT1A1 genotyping would be derived from a reduction in drug-related adverse reactions (benefits) while at the same time avoiding declines in tumor response rate and increases in morbidity/mortality (harms). At least three treatment options for reducing this increased risk have been suggested: modification of the irinotecan regime (e.g., reduce initial dose), use of other drugs, and/or pretreatment with colony-stimulating factors. However, we found no prospective studies that examined these options, particularly whether a reduced dose of irinotecan results in a reduced rate of adverse drug events. This is a major gap in knowledge. Although the quality of evidence on clinical utility is inadequate, two of three reviewed studies (and one published since our initial selection of studies for review) found that individuals homozygous for the *28 allele had improved survival. Three reviewed studies found statistically significant higher tumor response rates among individuals homozygous for the *28 allele. We found little or no direct evidence to assess the benefits and harms of modifying irinotecan regimens for patients with colorectal cancer based on their UGT1A1 genotype; however, results of our preliminary modeling of prevalence, acceptance, and effectiveness indicate that reducing the dose would need to be highly effective to have benefits outweigh harms. An alternative is to increase irinotecan dose among wild-type individuals to improve tumor response with minimal increases in adverse drug events. Given the large number of colorectal cancer cases diagnosed each year, a randomized controlled trial of the effects of irinotecan dose modifications in patients with colorectal cancer based on their UGT1A1 genotype is feasible and could clarify the tradeoffs between possible reductions in severe neutropenia and improved tumor response and/or survival in patients with various UGT1A1 genotypes.
Keywords: colorectal cancer, UGT1A1, pharmacogenetics, systematic review
INTRODUCTION
Medical disorder and treatment
Colorectal cancer (CRC) is the third leading cause of new cancer in the United States, with about 150,000 new cases per year. More than 55,000 deaths from CRC were expected in 2006. At least 15% of individuals with new CRC cancers (20,000 –25,000) might be candidates for irinotecan therapy.1–3 For the 70 – 80% of patients who present with “apparently resectable localized disease,” optimal treatment is usually considered to be surgery followed by adjuvant therapy for high-risk cases.3– 6 CRC patients with advanced disease at diagnosis may receive first-line systemic chemotherapy or chemotherapy and radiation therapy, either followed by surgery or used palliatively if surgery is not indicated.5 Fluorouracil continues to be the first choice of drugs for use in chemotherapy and may be used in combination with leucovorin.4,6,7 However, other combination chemotherapy regimens involving the use of irinotecan and other drugs seem to improve the median survival over fluorouracil and leucovorin and are increasingly being prescribed for first line and sequential therapy for patients with metastatic CRC.4,8,9
UGT enzymes and metabolism of irinotecan
Irinotecan is a topoisomerase I inhibitor that interrupts DNA replication in cancer cells, resulting in cell death.10 –12 The irinotecan prodrug is activated by the enzyme carboxylesterase to the active metabolite SN-38, which is 100–1000 times more cytotoxic than the parent drug.10 SN-38 is further catalyzed into an inactive glucuronide derivative, SN-38G by several hepatic and extrahepatic UGT enzymes. The major isozyme involved in this catalyzation is UGT1A1, but others (UGT1A 6, 7, 9, and 10) also have some role.13 A decrease in the level of functional UGT1A1 enzyme reduces a person’s ability to metabolize SN-38 to an inactive form, and low UGT1A1 enzyme levels have been associated with a higher risk for adverse reactions caused by relatively high levels of and/or prolonged exposure to the active form of the drug.6,14 Based on available tests and the proposed clinical scenario, the UGT1A1 enzyme was the focus of the evidence review with the associated key questions contained in Table 1.
Table 1.
Key questions relating to the analytic framework
|
Testing for UGT1A1 variants
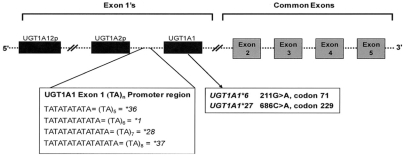
The UGT1A gene family includes nine protein coding genes and four pseudogenes, and encodes 13 different isoforms of the UGT1A enzyme (UGT1A1 through UGT1A13p). The isoforms result from alternative splicing of promoters and regions encoding substrate binding domains (multiple exon 1 sites) to common exons 2–5 (Fig. 1).12,15–20 At least 63 UGT1A1 variants have been described, including single base pair changes, frame shift mutations, insertions, and deletions in the promoter region, five exons and two introns of the gene.21 Most are associated with absent, reduced, or inactive enzyme; one is associated with an increased enzyme level, and the effects of some are unknown.
Fig. 1.
Schematic of the partial UGT1A1 gene showing locations of the polymorphisms of interest for this review in the exon 1 promotor region and in exon 1. First exons are alternatively spliced to common exons to produce UGT isoforms. Adapted from Clin Pharmacol Ther. 2004;75:495–500 and Oncology (Williston Park). 2003;17:52–55.17,20
This review focuses on the more commonly tested mutations (Table 2). The first is a two-base pair insertion (TA) in the TATA box in the promoter region of the gene.22 The result of this mutation is that the (TA)6TAA sequence, found in the promoter of the wild genotype UGT1A1*1 allele, becomes (TA)7TAA; this variant is designated as UGT1A1*28. The (TA)5TAA (UGT1A1*36) and (TA)8TAA (UGT1A1*37) variants are also described (Table 2), but they are less common and less routinely tested. Other mutations include polymorphisms in exon 1, c.211G>A (UGT1A1*6), and g.686C>A (UGT1A1*27) (Table 2).
Table 2.
UGT1A1 allele naming conventions, locations, and associated phenotypes
| UGT1A1 allelea | Variant | Location | Enzyme activity | Associated phenotype |
|---|---|---|---|---|
| UGT1A1*1 | (TA)6TAA | Promoter | Normal | Wild type |
| UGT1A1*28 | (TA)7TAA | Promoter | Reduced | Gilbert syndrome |
| UGT1A1*36 | (TA)5TAA | Promoter | Increased | |
| UGT1A1*37 | (TA)8TAA | Promoter | Reduced | Crigler-Najjar, type II |
| UGT1A1*6 | c.211G>A; G71R | Exon 1 | Reduced | Gilbert syndrome |
| UGT1A1*27 | g.686C>A; P229Q | Exon 1 | Reduced | Gilbert syndrome |
Allele frequencies, stratified by race, are shown in Table 4.
Because information on additional functional polymorphisms in the promoter (e.g., −3279T>G; UGT1A1*60) and coding regions (e.g., 1456T>G; UGT1A1*7) of the enzyme were limited at the time of the initial review, we did not include studies of these polymorphisms in the analysis.23–26 However, these studies and others published since the initial review,26 –28 have shown that some polymorphisms are relatively common in specific racial/ethnic groups (e.g., Asians) and may influence metabolism of irinotecan.
Clinical scenario
As noted, the UGT1A1 *28 allele is associated with reduced levels of enzyme. Therefore, individuals with the wild genotype sequence (*1/*1) who have average levels of the enzyme will metabolize SN-38 more quickly than those who are either heterozygous (*1/*28) or homozygous (*28/*28) for this allele. Higher or more prolonged exposure to the active form of irinotecan is thought to explain many of the adverse drug events associated with irinotecan use, including severe neutropenia and severe diarrhea. Thus, if irinotecan dosage can be modified on the basis of patients’ UGT1A1 genotype, some proportion of these adverse events might be avoided. However, a reduction in dosage might also be associated with reduced tumor response and/or increased morbidity.
In 2004, a change to the prescribing information29,30 in the Camptosar (irinotecan) Injection Package Insert was announced through an Food and Drug Administration (FDA) Center for Drug Evaluation and Research email alert (NDA 20-571/S-024/S-027/S-028), which stated that:
“. . . a reduction in the starting dose by at least one level should be considered for patients known to be homozygous for the UGT1A1*28 allele . . . . However, the precise dose reduction in this patient population is not known and subsequent dose modifications should be considered based on individual patient tolerance to treatment.”
“Individuals who are homozygous for the UGT1A1*28 allele are at increased risk for neutropenia following initiation of Camptosar treatment. A reduced initial dose should be considered . . . . Heterozygous patients . . . may be at increased risk for neutropenia; however, clinical results have been variable and such patients have been shown to tolerate normal starting doses.”
Subsequently, in August 2005, the Invader® UGT1A1 Molecular Assay (Third Wave Technologies, Inc., Madison, WI) was cleared by the US FDA Center for Devices and Radiologic Health under 510(k) rules for Drug Metabolizing Enzyme Genotyping Systems.31,32 The Invader test and other laboratory developed UGT1A1 tests are currently available from multiple laboratories in the United States and are being marketed to oncologists and pathologists as an aid to clinical decision making.33,34 In its package insert,35 Third Wave Technologies, Inc. describes the assay as follows:
“. . . . . an in vitro diagnostic test for the detection and genotyping of the *1 (TA6) and *28 (TA7) alleles of the UDP glucuronosyltransferase 1A1 (UGT1A1) gene in genomic DNA from whole peripheral blood as an aid in the identification of patients with greater risk for decreased UDP-glucuronosyltransferase activity.”
METHODS
The purpose of this article is to provide a summary and extend the findings of a more formal evidence report (see Resources section). These evidence reports are to be used by the Evaluation of Genomic Applications in Practice and Prevention (EGAPP) Working Group (EWG) to inform the development of formal recommendations for clinical practice.36 The methods used to identify, review, evaluate, analyze, and summarize the evidence are detailed in Appendix C of the original evidence report (see Resources section). Investigators at Research Triangle Institute (RTI) International were contracted by the National Office of Public Health Genomics at the Centers for Disease Control and Prevention to conduct the initial stage of the review of this evidence, focusing on clinical validity and utility. RTI staff conducted a MEDLINE literature search (through May 2006) for studies addressing the clinical validity and utility of UGT1A1 genotyping in metastatic CRC patients treated with irinotecan. Based on key questions and discussion with the UGT1A1 Technical Expert Panel, a list of article inclusion and exclusion criteria was generated. Articles were also identified by search of the references included in selected articles. RTI reviewed abstracts and selected articles found in the search, abstracted data into evidence tables, assessed the quality of individual articles, and prepared a preliminary report. When the RTI report was submitted, members of the Centers for Disease Control and Prevention-sponsored EGAPP initiative team and EGAPP consultants performed searches for and reviewed articles on the analytic validity of UGT1A1 genotyping and on UGT1A1 allele/genotype frequencies, and updated the clinical validity and utility searches through December 2006. They also performed additional summarization and statistical analyses, integrated the component sections, and produced a draft evidence report for consideration by the EWG.
With a focus on the application of study data to specific key questions, EGAPP reviewers assessed the quality of evidence for the evaluation components (i.e., analytic and clinical validity, clinical utility) based on standard criteria, including study design and conduct, consistency and generalizability of data, and appropriateness of statistical analyses. Short summaries were written for all individual studies included for assessment of analytic validity, clinical validity, and clinical utility, and included EGAPP and RTI quality ratings (see Resources section). Feedback was sought throughout the review from the Technical Expert Panel, other technical consultants, and the EWG.36 In addition, a draft of the evidence report was sent to nine expert peer reviewers. The report was revised in response to comments from the reviewers and resubmitted to the EWG along with a summary of comments and their disposition.
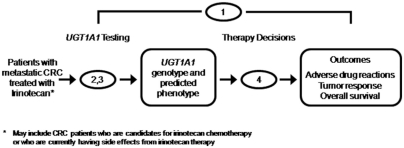
The focus of this report is on patients with metastatic CRC treated with irinotecan. The analytic framework is shown in Figure 2, with the numbers indicating the key questions shown in Table 1. These key questions were developed by the EWG and further refined in discussions with a Technical Expert Panel. Key question 1 is the overarching question: “Does testing for UGT1A1 mutations in patients with metastatic CRC treated with irinotecan lead to improvement in outcomes (e.g., irinotecan toxicity, response to treatment, morbidity and mortality) compared to no testing?” If direct evidence is insufficient to answer key question 1, key questions 2 through 4 are used to elicit intermediate information to address the overarching question through a “chain of evidence.” In reviewing the available evidence, we often used questions from the ACCE (Analytic validity, Clinical validity, Clinical utility, and Ethical, Legal and Social implication) review framework37 to identify and organize the specific information needed to address the key questions.
Fig. 2.
The analytic framework: testing for UGT1A1 mutations in patients with metastatic colorectal cancer (CRC) treated with irinotecan (Camptosar). This schematic shows the analytic framework underlying the current review. The numbers indicate the four key questions contained in Table 1.
RESULTS
Analytic validity
Identification of relevant literature
Through MEDLINE searches, we identified 17 articles that were included in analyses (Appendix C of the original evidence report, see Resources section). Searches of the gray literature (e.g., unpublished reports, web sites, government documents) identified laboratories offering UGT1A1 testing, as well as a US FDA 510(k) summary and relevant committee reports (see Resources section).
Analytic sensitivity and specificity
Table 3 summarizes the results of four method comparison studies on testing for *28 (TA7) and *1 (TA6) alleles, using sequencing as the referent method.18,31,38,39 In these studies, genotypes for all of 190 samples homozygous or heterozygous for *28 were correctly identified (estimated analytic sensitivity 100%; 95% confidence interval [CI] 98 –100%). The homozygous wild genotype (*1/*1) was correctly identified in all of 131 samples (estimated analytic specificity 100%; 95% CI 97–100%). A 2007 study (not included in the Table 3 analysis) was very consistent, reporting 100% concordance in 88 samples tested by sequencing and PCR/capillary electrophoresis and the Invader assay (*28/*28, N = 13; *28/*1, N = 46; *1/*1, N = 29).40 Very little data are available to support estimates of analytic sensitivity for other promoter (*36, *37) and exon 1 variants (*6, *27).38
Table 3.
Analytic validity of UGT1A1 testing for genotypes involving the *28 variant
| Analytic
|
||||||
|---|---|---|---|---|---|---|
| Sensitivitya |
Specificitya |
|||||
| Source | N | Test method | Referent method | *28/*28 | *28/*1 | *1/*1 |
| Monaghan19 | 12 | Radioactive PCR | Sequencing | 4/4 | 5/5 | 3/3 |
| Pirulli40 | 40 | DHPLC | Sequencing | 19/19 | 8/8 | 13/13 |
| Hasegawa39 | 60 | Invader (RUO) | Sequencing | 4/4 | 11/11 | 42/42b |
| Invader32 | 212 | Invader (IVD) | Sequencing | 30/30 | 109/109 | 73/73 |
| All | 324 | 57/57 | 133/133 | 131/131 | ||
| Overall estimate (95% confidence interval) | 100% (98–100%) | 100% (97–100%) | ||||
Test result/referent result.
Does not include three sample failures.
Test reproducibility and failure rates
Data on the reproducibility of Invader UGT1A1 Molecular Assay results and assay failure rates were reported as part of the FDA 510(k) approval process.31 Twenty blood samples (six *1/*1 wild genotypes, five *28/*1 heterozygotes, four *28/*28 homozygotes, and five undisclosed genotypes) were each tested five times at three different sites (300 test results). Of the 49 initial “invalid calls” or sample failures, 40 were due to invalid positive or negative control results, and nine to low signal intensity. Failure rates on the first run were 9.3%, 0%, and 7.0% for the three sites. Six samples failed again when retested (6 of 600; 1%, 95% CI 0.4 –2.2%). Incorrect results were reported for 11 samples, all from one site, for an overall correct call rate of 98.8% (883/894; 95% CI 97.8 –99.4%). Nine of these 11 incorrect results may have been sample mix ups, an example of preanalytic errors that are also expected to occur in clinical practice. If the erroneous results caused by sample mix ups are excluded, the overall correct analytic call rate for the Invader assay was 99.8% (95% CI 99.2–99.9%). In a report of Invader testing for UGT1A1*1, *28, *6, and *27, Hasegawa et al.38 observed failure rates of 6.7% (3 of 45) in tests of*1/*1 homozygotes, 10% (6 of 60) in UGT1A1*6 testing, and 21.7% (13 of 60) in UGT1A1*27 testing, but these relatively high-failure rates may have been related to low DNA concentrations in the samples tested.
Since we completed this review, a 2007 study compared the failure rates for sequencing, PCR/capillary electrophoresis, and the Invader assay in tests of 119 samples containing *1, *28, *36, and *37 variants.40 The study’s authors reported first-run failure rates of 5.0% for sequencing and 1.7% for the PCR/ capillary electrophoresis method, with all failures resolved by repeat analysis. The Invader assay failed on the first and second runs in 7.6% of samples (9 of 119; three *1/*1, two *28/*1, one *28/*28, and three other).
Limitations
Although most data were collected using the Invader technology, other technologies are being used. In addition, these data mainly focus on the analytic phase of testing and do not include errors in the preanalytic (e.g., sample handling or labeling errors) and postanalytic (e.g., data entry or interpretive/ reporting errors) phases.41,42 A large proportion of the data were reported by early Invader investigators38,43 or by the manufacturer of the Invader kit as part of its FDA submission.31 Only two of four studies31,39 reported that the samples were blinded to those performing the assays, to rule out retesting to get the “right” answer.
Clinical Validity
Identification of relevant literature
Because of the limited literature, the analysis was restricted to nine studies, including several that allowed entry to individuals with tumors other than CRC.14,44 –51 UGT1A1*1 and *28 account for 98 –99% of the UGT1A1 polymorphisms in the white population and were the focus of most studies identified. Studies on polymorphisms that are more commonly found in other racial/ethnic groups (e.g., UGT1A1*6 and *27 in Asians) were more limited. In addition, homozygosity for *28 is specified as the primary risk factor in the Camptosar (irinotecan) package insert.29 Consequently, we chose to limit the review of clinical validity to these common alleles for which testing is broadly available.
UGT1A1 genotypes and SN-38 levels
The metabolism of the prodrug irinotecan and its relationship to UGT1A1 genotypes has been described earlier. One way to assess the “exposure” to SN-38 is a ratio of the area under the curve (AUC) for SN-38G (the inactive form of irinotecan) to the AUC for SN-38 (the active form). Essentially, this compares the integrated time dose exposure for the inactive form (SN-38G) to that of the active form (SN-38). High values indicate that most exposure is to the inactive form; low values indicate increased exposure to the active form. The results of six published studies26,45,50,52–54 showed that the AUC ratios were lowest among individuals homozygous for *28 (*28/*28), intermediate among those heterozygous for *28 (*1/*28), and highest among those with the wild genotype (*1/*1). This indicates that the highest relative exposure to the active form of irinotecan, SN-38, occurs among the individuals homozygous for *28 (*28/*28). The SN-38 to SN-38G AUC ratios should be viewed as an intermediate measure of irinotecan exposure.
A more appropriate measure of exposures would include the irinotecan dose. The biliary index (BI) is the irinotecan AUC times the ratio of the SN-38 to SN38G AUCs. Two studies provided the BI for cancer patients stratified by UGT1A1 genotype. One studied 71 CRC patients,50 the other reported on 20 patients with solid tumors, four of whom had CRC.45,55 Both found a significant and consistent dose response in the BI from the wild type, through the heterozygotes and homozygotes. These data strongly indicate the highest time-weighted exposure to the active form of irinotecan occurs in individuals homozygous for *28 (*28/*28). There were no apparent differences in these findings between studies in whites and Asians (see Table KQ3.1 in full evidence review, Resources section).
Irinotecan treatment regimens
Studies selected for evaluating clinical validity (and clinical utility) did not use standardized treatment regimens. Table 5 provides a brief description of the treatment regimens used in the studies we evaluated.14,44 –51 In several studies, multiple treatment protocols were evaluated. Because we could not account for the effect of variations in treatment regimens in subsequent analyses, we examined the homogeneity of results to determine whether treatment variations had a significant effect on study results. In other words, if the analysis of a clinical validity measure (e.g., severe neutropenia) was found to be homogeneous within a comparison group (e.g., *28 homozygotes compared with *1/*1 wild genotype), it was assumed that a given treatment regimen did not have a significant impact on that measure.
Table 5.
Chemotherapy treatment regimens used in studies selected for analysis
Carlini et al.51
|
UGT1A1 genotypes and severe diarrhea
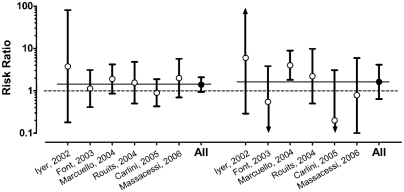
The severity of diarrhea is graded on a subjective scale from 1 (mild) to 4 (severe or life threatening).56 The overall observed rates of severe diarrhea (Grades 3 and 4) among participants in the six studies selected for analysis44 – 48,51 was 24% (95% CI 19 –30%). When stratified by UGT1A1 genotypes, the rates of severe diarrhea were 18% (95% CI 11–28%) among those with the wild genotype, 27% (95% CI 20 –36%) among those heterozygous for *28, and 27% (95% CI 12– 48%) among those homozygous for *28. The analysis showed the severe diarrhea rates between studies to be homogeneous among participants with the same genotype (Q values of 10.4, 7.1, and 9.5; P values of 0.1, 0.3, and 0.2). Figure 3 shows the corresponding summary risk ratios (RRs) from these studies (with the risk among 154 study participants with the wild genotype serving as the referent category) of 1.40 (95% CI 0.94 –2.08) for 155 participants heterozygous for *28, and 1.63 (95% CI 0.64 – 4.14) for 41 participants homozygous for *28. The results were homogeneous within groupings (Q values of 3.0 and 8.5; P values of 0.7 and 0.1, respectively).
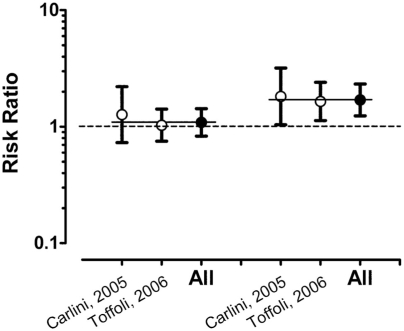
Fig. 3.
Risk ratios for severe diarrhea among cancer patients treated with Irinotecan by UGT1A1 genotype from six published studies.44–48,51 The studies are listed on the x-axis, sorted by the risk ratio comparing rates in heterozygotes (*1/*28) to wild-type individuals (*1/*1) on the left-hand side. The risk ratios for homozygotes (*28/*28) versus wild type is the right-hand side. Bars indicate the 95% confidence interval (CI) with the consensus estimate (All) for the two comparison groups. The dotted line indicates a risk ratio of 1.00 (no difference). The two thin solid lines indicate the consensus estimates for the two groups of 1.40 (95% CI 0.94–2.08) and 1.63 (95% CI 0.64–4.14), respectively.
UGT1A1 genotypes and severe neutropenia
Neutropenia is a decrease in the number of circulating neutrophils (a type of white blood cell that usually accounts for 50 –70% of circulating white blood cells) and can be caused by bone marrow suppression associated with the use of antineo-plastic chemotherapy drugs. Because neutrophils help defend against bacterial infections, chronic neutropenia can be life threatening. Neutropenia is graded based on the absolute neutrophil count, or number of neutrophil cells per mm3 (Grade 1, 1500 –1999; Grade 2, 1000 –1499; Grade 3, 500 –999; and Grade 4, <500 cells/mm3).56 The overall observed rate of severe neutropenia (Grades 3 and 4 combined) among participants from the eight studies14,45–51 selected for analysis was 16% (95% CI 13–19%). When stratified by UGT1A1 genotypes, the rates were 9.8% (6.8 –14%) among those with the wild genotype, 18% (14 –23%) among those heterozygous for *28, and 38% (22–57%) among those homozygous for *28. The results were homogeneous within genotype (Q values of 4.5, 7.1, and 7.9, and P values of 0.7, 0.4, and 0.2, respectively).
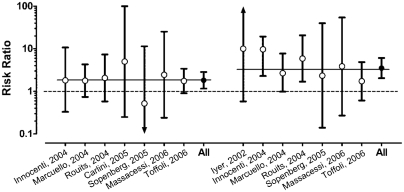
Figure 4 shows the corresponding severe neutropenia RRs from these studies, with the risk among participants with the wild genotype serving as the referent category. The summary RRs were computed using original data and a random effects model. Summary RRs were 1.82 (95% CI 1.16 –2.85) for participants heterozygous for *28 (based on samples from 276 heterozygotes and 282 participants with the wild genotype) and 3.51 (95% CI 2.03– 6.07) for those homozygous for *28 (based on samples from 57 homozygotes and 263 participants with the wild genotype). The RRs were homogeneous within comparison groups (Q values of 1.2 and 5.2, and P values of 0.9 and 0.5, respectively). Overall, these data provide clear evidence that rates of severe neutropenia differ significantly based on the three major UGT1A1 genotypes, and that there is a dose response relationship between the number of mutant alleles and rate of severe neutropenia.
Fig. 4.
Risk ratios for severe neutropenia by UGT1A1 genotype from eight published studies.14,45–51 The studies are listed on the x-axis, stratified by heterozygote individuals (*1/*28) versus wild type (*1/*1) on the left-hand side and homozygote individuals (*28/*28) versus wild type on the right-hand side. Two results (Iyer 2002 for heterozygotes, Carlini 2005 for homozygotes) are not shown as the risk ratio could not be computed due to no observations in one or more groups. The bars indicate the 95% confidence interval (CI) with the consensus estimate (All) for the two comparison groups. The dotted line indicates a risk ratio of 1.00 (no difference). The two thin solid lines indicate the consensus estimates for the two groups of 1.82 (95% CI 1.16–2.85) and 3.51 (95% CI 2.03–6.07), respectively.
Clinical sensitivity and specificity of UGT1A1 genotypes as an indicator of risk for severe neutropenia
Table 6 shows the clinical sensitivity and specificity for severe neutropenia of UGT1A1 genotyping among participants from the eight studies included in the analysis.14,45,47–51 A positive UGT1A1 test is defined as an individual homozygous for the *28 allele (*28/*28), and the outcome of interest is severe (Grade 3 or 4) neutropenia. We defined clinical sensitivity as the proportion of individuals with severe neutropenia who were homozygous for *28, and clinical specificity as the proportion of individuals without severe neutropenia who were not homozygous for *28. We estimated (based on a random effects model) that the tests in these studies had an overall clinical sensitivity of 23% (95% CI 15–34%) and an overall clinical specificity of 92% (95% CI 90 –94%).
Table 6.
Clinical sensitivity and specificity of UGT1A1 genotyping for severe neutropenia
| Study | True positive | False negative | True negative | False positive | Sensitivity (%) | Specificity (%) |
|---|---|---|---|---|---|---|
| Carlini 200551 | 0 | 2 | 59 | 5 | 0 | 92 |
| Innocenti 200414 | 4 | 5 | 48 | 2 | 44 | 96 |
| Iyer 200245 | 2 | 0 | 18 | 2 | 100 | 90 |
| Marcuello 200446 | 4 | 18 | 73 | 6 | 18 | 92 |
| Massacesi 200647 | 1 | 3 | 52 | 6 | 25 | 90 |
| Rouits 200448 | 4 | 10 | 59 | 3 | 29 | 95 |
| Soepenberg 200549 | 0 | 1 | 21 | 1 | 0 | 95 |
| Toffoli 200650 | 4 | 33 | 195 | 18 | 11 | 92 |
| Alla (95% confidence interval) | 23 (15–34) | 92 (90–94) |
Using a random effects model.
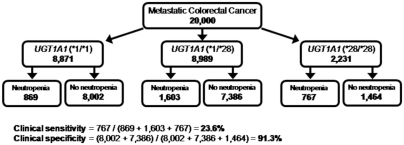
It is also possible to compute the expected clinical sensitivity and specificity from parameters obtained earlier in this review as shown by the flowchart in Figure 5. The chart shows how a theoretical population of 20,000 white metastatic CRC patients initiating irinotecan therapy is first stratified by UGT1A1 genotypes on the basis of the consensus estimate of the *28 allele frequency (0.334 from Table 4) and the Hardy-Weinberg principle. Among the 8871 patients with the wild genotype, the baseline rate of severe neutropenia (9.8%) would result in 869 of them experiencing this adverse drug reaction. Using the RRs of 1.82 and 3.51, the number of adverse reactions in the individuals heterozygous and homozygous for *28 can also be computed. The expected clinical sensitivity and specificity of 24% and 91%, respectively, agree closely with the consensus rates of 23% and 92% computed from published observations (Table 6). The corresponding positive predictive value derived from Figure 5 is 52% (767/1464) and the negative predictive value is 86% (1 − (869 + 1603)/(8871 + 8898)).
Fig. 5.
Flow diagram showing the derivation of clinical sensitivity and specificity of UGT1A1 genotyping to identify severe neutropenia in a hypothetical cohort of 20,000 white individuals with metastatic colorectal cancer. The clinical sensitivity and specificity are derived using previously reported parameters (e.g., allele frequency, risk ratios), stratified by UGT1A1 genotype. Overall, the clinical sensitivity is 24% with a specificity of 91% (false positive rate of 9%).
Table 4.
Consensus UGT1A1 allele frequencies stratified by race
| Common allele frequencies (95% confidence interval)
|
Other allele frequencies (95% confidence interval)
|
|||||
|---|---|---|---|---|---|---|
| Race | Studies (patients) | *28 (TA7) | *36 (TA5) | *37 (TA8) | *6 (211G>A) | *27 (686C>A) |
| White | 11 (2517)19,20,58–66,72 | 0.334 (0.309–0.361) | 0.003 (0.001–0.008)58,64,65 | 0.002 (0.001–0.009)58,64 | 0.005 (0.001–0.03)60 | 0.00067 |
| Asian/Asian Americana | 4 (454)58,64,68,69 | 0.139 (0.112–0.171) | 0.000 (0.00–0.09)58,64 | 0.000 (0.00–0.09)58,64 | 0.13b (0.10–0.17)67–69 | 0.023 (0.014–0.035)67,69 |
| African/African American | 3 (411)58,70,71 | 0.404 (0.358–0.452)58,70,71 | 0.058 (0.039–0.085)58,70,71 | 0.043 (0.026–0.070)58,70,71 | 0.0067 | 0.0067 |
Limitations
Nearly all of the clinical validity information was collected from populations consisting of non-Hispanic whites. When studies did include people of other racial/ethnic groups, the results were not stratified by race/ethnicity. Treatment protocols (including irinotecan dosage, method of delivery, and frequency of treatment) varied widely both within- and between-studies, and these variations could affect both the overall rate of adverse events and the RRs for specific adverse events. Although we did not find strong evidence of such an impact, most studies contained too few subjects to be confident. Some studies reported adverse events after the first cycle of treatment, others after the completion of treatment, and still others provided both. Although one study showed that adverse events among patients homozygous for the *28 allele most often occurred in the first cycle of treatment,50 not enough studies provided clinical outcomes at both times to allow for a meaningful subanalysis by treatment time. Several studies included patients who had cancer at sites other than the colon. However, we could not determine the impact of including these studies, because none of the studies stratified their results by cancer site. Lastly, several studies included patients with less common genotypes. However, because these patients were always included in larger groupings, it was not possible to combine results for patients with these genotypes across studies.
UGT1A1 allele frequencies by racial group
The pharmacokinetics of irinotecan does not seem to differ based on gender or race. However, clinically relevant UGT1A1 alleles and genotype frequencies do differ by race/ethnicity. Table 4 shows estimated UGT1A1 *28, *36, *37, *6, and *27 allele frequencies among people of white, Asian, and African descent. Estimates of allele frequencies for the *28 allele are based on 11 studies in white populations,18,19,57– 66 four studies in mixed Asian and Asian American populations,58,63,67,68 and three studies in African and African American populations.57,69,70 Fewer studies reported on allele frequencies of the less common alleles (*36 and *37 promoter alleles; *6 and *27 polymorphisms) in whites,57,59,63,64,66 Asians28,57,63,66 – 68,71 and people of African descent.57,66,69,70 Differences between allele frequencies by racial groups are clear, including statistically higher frequencies of *36 (TA5) in African/African American populations,57,69,70 and of *6 in Asian populations.28,66 – 68
UGT1A1 genotypes and tumor response
Three studies provided information on tumor response, stratified by UGT1A1 genotype.44,50,51 One study44 found a higher rate of stable or partially responsive tumors among *28 heterozygotes and homozygotes combined than among patients with the wild genotype (RR of 1.6; 95% CI 0.8 –3.0). The two other studies50,51 defined a responsive tumor as “partial or complete response” and provided sufficient data to examine response rates by UGT1A1 genotype. Summary results from the two other studies50,51 showed tumor response rates of 41% (95% CI 33– 40%) among patients with the wild genotype, 47% (95% CI 33– 63%) among *28 heterozygotes, and 70% (95% CI 40 – 84%) among *28 homozygotes. The results were homogeneous within genotype (Q values of 0.2, 2.2, and 0.6, and P values of 0.6, 0.1, and 0.4, respectively). Figure 6 shows an analysis of the tumor response rate (as defined in the studies) versus UGT1A1 genotype, with individuals having the wild genotype used as the referent category.50,51 Overall, the *28 heterozygotes had a nonsignificantly higher response rate (RR, 1.09; 95% CI 0.83–1.43), and *28 homozygotes had a significantly higher response rate (RR, 1.70; 95% CI 1.24 –2.33; P = < 0.001). The studies were homogeneous within genotype (Q values of 0.4 and 0.8, and P values of 0.6 and 0.8, respectively).
Fig. 6.
Risk ratios for tumor response by UGT1A1 genotype from two published studies.50,51 The studies are listed on the x-axis, with the risk ratios for heterozygote individuals (*1/*28) versus wild type (*1/*1) on the left hand side, and the risk ratios for homozygote individuals (*28/*28) versus wild type on the right hand side. The bars indicate the 95% confidence interval (CI) with the consensus estimate (All) for the two comparison groups. The dotted line indicates a risk ratio of 1.00 (no difference). The two thin solid lines indicate the consensus estimates for the two groups of 1.09 (95% CI 0.83–1.43) and 1.70 (95% CI 1.24–2.33), respectively.
UGT1A1 genotypes and mortality
Two of the three studies providing information on tumor response among patients treated with irinotecan also provided some information about mortality.44,50 The other study46 provided information only on survival. It was not possible to combine the information from these three studies in a formal analysis. Instead, the findings are summarized in Table 7. The data from Font et al.44 are for combined heterozygotes and homozygotes, and all patients had lung cancer. The data from Toffoli et al.50 compare homozygotes (and heterozygotes) with the wild type, and all patients had CRC. The data from Marcuello et al.46 include 95 patients with CRC and represent combined heterozygotes and homozygotes. None of the differences were statistically significant. Findings from two studies44,50 were in the direction of improved survival for *28 homozygotes versus nonwild genotype patients, whereas the third reported a survival advantage for the wild genotype individuals.46
Table 7.
Mortality-related outcome data from studies of cancer patients treated with irinotecan, stratified by UGT1A1 genotype
| Study | Outcome measure | Finding |
|---|---|---|
| Font et al.44 | Time to progression | 3 mo (*1/*1) vs. 4 mo (othera) |
| Median survival | 8 mo (*1/*1) vs. 11 mo (other) | |
| 1 yr survival | 21% (*1/*1) vs. 41% (other) | |
| 2 yr survival | 14% (*1/*1) vs. 31% (other) | |
| Marcuello et al.46 | Median survival | 32 mo (*1/*1) vs. 24 mo (other1) |
| Toffoli et al.56 | Hazard ratio | 0.81 (95% CI 0.45–1.44) (*28/*28 vs. *1/*1) |
| Hazard ratio | 0.84 (95% CI 0.58–1.21) (*1/*28 vs. *1/*1) | |
| Median survival | 613 days (*1/*1) vs. 686 days (*28/*28) | |
| Median survival | 613 days (*1/*1) vs. 669 days (*1/*28) |
Other includes both heterozygotes (*1/*28) and homozygotes (*28/*28).
Clinical Utility
Options for modifying patient care
There is insufficient information for the less common genotypes to provide clear options for patient management. For CRC patients who have the common *28 polymorphism, the three main options for modifying patient care have been summarized and discussed.72
Modify the irinotecan regimen: The Camptosar (irinotecan) package insert provides suggested modified (reduced) dose levels (mg/m2) for two single-drug regimens of Camptosar (125 mg/m2 weekly and 350 mg/m2 every 3 weeks).29 It states that a reduction by one dose level may be considered for patients 65 years or older, those having low performance status, or those with increased bilirubin levels; reduction in starting dose by at least one level “should be considered for patients known to be homozygous for the UGT1A1*28 allele.” However, the package insert also notes that “the appropriate dose reduction in this patient population is not known.”
Use other drugs: Newer drugs (e.g., cetuximab, bevaci-zumab) can be substituted in a variety of regimens that vary the combination of drugs, as well as the doses, schedules, and duration of infusion for each drug.
Treat patients with colony-stimulating factors before the first cycle of chemotherapy to prevent the occurrence of febrile neutropenia: Such treatments, which cost 2–3000 dollars per dose, are currently recommended by the National Comprehensive Cancer Network for patients with a 20% or greater risk of febrile neutropenia.73 Although *28 homozygous patients have a 36% risk of severe neutropenia, the proportion associated with fever is unknown. This pretreatment and monitoring of white cell counts might be an acceptable alternate indicator of acceptable dosing.
Additionally, treatment options need to be placed in the context of overall care. The choice of treatment for CRC patients should also reflect the level of risk for various adverse effects that they consider to be acceptable. Thus, the UGT1A1 test may be useful to identify *28 homozygous patients who may prefer a treatment with low risk of toxicity even if it may not be as effective in fighting their cancer, whereas the testing may not be as useful for those seeking aggressive therapy and willing to accept risk of higher toxicity. Decisions about testing may also be based on the specific planned regimen and dosing. McLeod74 has proposed that, unless patients receive irinotecan at a dose >150 mg/m2 (either alone or in combination with a myelotoxic drug) or irinotecan >100 mg/m2 in combination with another marrow-toxic agent (e.g., oxaliplatin), their increased risk for toxicity is “neither statistically nor clinically significant” and testing may not be warranted.
Will reduction in the irinotecan dose reduce patients’ risk of having a severe drug-related adverse event?
Based on the clinical validity and additional information on the pharmacokinetics of irinotecan, it is biologically plausible that a reduced initial dose in *28 homozygous patients could result in a reduction in severe neutropenia. However, no studies (with or without randomization) have genotyped patients before their first treatment, modified starting dosages, and then compared the clinical outcomes (e.g., severe neutropenia, tumor response) based on these modified dosage with outcomes among patients receiving a standard dose. Currently, reducing the irinotecan dosage in subsequent cycles is the standard method of avoiding additional instances of neutropenia. For example, Toffoli et al.50 have shown that reducing dosage from 180 mg/m2 to between 90 and 150 mg/m2 in all individuals having neutropenia reduced the rate of neutropenia in *28 homozygotes in subsequent cycles. They reported that the odds ratio (OR) for neutropenia among *28 homozygotes patients relative to the wild genotype dropped from 8.6 (95% CI 1.3–57) after the first cycle to 2.0 (95% CI 0.6 –7) after the end of therapy on lower doses (2– 6 cycles). However, they also found that the point estimates for tumor-related morbidity and mortality were lower among *28 homozygous patients (and to a lesser extent heterozygous patients), possibly due to the effects of “over-dosing.” Thus, the reduced drug metabolism rate among these patients in these two groups (i.e., slower inactivation of SN-38) that may cause the increased rate of severe adverse drug events (harm) is possibly also responsible for the apparent increase in tumor response and improved survival (benefit).
Comparing the benefits and harms
The proposed benefit of testing metastatic CRC patients for UGT1A1 genotype is that the risk for adverse drug-related side effects (e.g., severe neutropenia) among patients found to be homozygous for the *28 genotype (and to a lesser extent for those found to be heterozygous) can be reduced by lowering their initial and/or subsequent doses of irinotecan. The concomitant harm is that reduction in irinotecan dosage many also reduce the effectiveness of chemotherapy in tumor suppression and long-term survival.
To compare these competing effects, we used a model (shown in Table 8) that incorporates estimates of the effect of reducing the initial irinotecan dosage given to *28 homozygous CRC patients on the number of severe neutropenia episodes avoided and on the number of additional CRC tumors nonresponsive to treatment. The numbers are based on the hypothetical population shown in Figure 5, with results shown for projections of the effectiveness of an irinotecan dose reduction from 20% to 100%. Effectiveness of 100% means that the rate of severe neutropenia among *28 homozygous patients receiving the reduced dose will be equivalent to that among patients with the wild genotype receiving the full dose. From the literature, that rate is expected to be about 9.8%. The number needed to test shown in Table 8 indicates the total number of cancer patients that would need to be genotyped (and have reduced dose in all found to be *28 homozygotes) to avoid one case of severe neutropenia in a homozygous patient. Our calculations assume that the reduced dose will cause homozygotes to have the same tumor response rate as individuals with the wild genotype. This may be an oversimplification of the model, as response rates seem to also be dose dependent.
Table 8.
Preliminary estimates of the clinical utility of testing metastatic CRC patients for UGT1A1 polymorphisms: Benefits and harms among UGT1A1*28 homozygotes
| Effectiveness of irinotecan dose reduction in preventing neutropenia (%) | Projected Total number of neutropenia cases | Cases of neutropenia avoided | Number needed to test to avoid one case of neutropenia | Additional nonresponsive CRC tumors: case of neutropenia avoided |
|---|---|---|---|---|
| 100 | 219 | 548 | 36 | 1.2:1 |
| 90 | 274 | 493 | 41 | 1.3:1 |
| 80 | 328 | 439 | 46 | 1.5:1 |
| 70 | 383 | 384 | 52 | 1.7:1 |
| 60 | 438 | 329 | 61 | 2.0:1 |
| 50 | 493 | 274 | 73 | 2.4:1 |
| 40 | 548 | 219 | 91 | 2.9:1 |
| 30 | 603 | 164 | 122 | 3.9:1 |
| 20 | 658 | 109 | 183 | 5.9:1 |
| 10 | 713 | 54 | 370 | 12:1 |
As an example, consider that reducing the irinotecan dose among *28 homozygous individuals is 100% effective in reducing excess neutropenia (Table 8, row 1). How many excess events might be avoided among a hypothetical population of 20,000 whites. A total of 2231 homozygotes (Fig. 5) would occur and with a background rate of 9.8% for severe neutropenia, 219 events would occur if homozygotes had the same rate as wild-type individuals. Because the RR for severe neutropenia among homozygotes is 3.51 (Fig. 4), the actual number would be expected to be 767 or 548 more than the baseline of 218 events. The number needed to test to avoid one individual with severe neutropenia would be 20,000 tests divided by 548 avoided severe neutropenia events or 36 (Table 8, Column 4). The number of nonresponsive CRC tumors among homozygous individuals (*28/*28) receiving a reduced dose is considered a constant (647) and is computed as follows. The baseline response rate in wild-type individuals is 41%, and the observed response rate for homozygotes is 1.70 times higher, or 69% (Fig. 6). Thus, there were originally 1539 responsive tumors among the homozygotes (0.69 * 2231), but only 892 might still be responsive with a reduced dose (0.40 * 2231). This is a drop of 647 responsive tumors. These 647 additional nonresponsive tumors (harms) can be compared with the avoidance of 548 cases of neutropenia (benefits) and result in an OR of 647:548 or 1.2:1 (Table 8, Column 5).
An alternative approach to comparing the benefits and harms would be to compare number of nonresponsive CRC tumors to the number of deaths resulting from severe neutropenia. An estimated 1 in 110 cases of severe neutropenia might result in death in individuals receiving irinotecan as a first-line treatment,75–77 and the ratio of nonresponsive tumors to avoided events of severe neutropenia (last column in Table 8) could be converted to this measure by dividing the right-hand side by 110. For example, using the numbers in Row 1, the OR of nonresponsive tumor versus death resulting from severe neutropenia change from 647:494 to 647:(494/110) or about 140:1. According to the very preliminary analysis reported in Table 8, it seems that at high rates of effectiveness (70 –100%), each avoided case of neutropenia is associated with one nonresponsive tumor. At lower rates of effectiveness (20 –50%), there are likely to be 2–5 times as many nonresponsive tumors as avoided cases of severe neutropenia.
Might individuals with the wild genotype be underdosed?
Given some limited evidence that individuals homozygous for *28 have improved survival72 (Table 7), it is possible that individuals with the wild genotype are underdosed. Original Phase I studies did not stratify patients by UGT1A1 genotype and, therefore, higher doses may be well tolerated by wild genotype individuals (*1/*1). Preliminary data from new Phase I dose-escalation trials with patients stratified by genotype have been recently published (see Resources section).78 – 80
Limitations
In general, the same problems with studies of clinical validity are applicable to clinical utility. The study populations were mainly non-Hispanic whites, treatment regimens varied widely, patients with rare genotypes were grouped with patients with common genotypes, and patients with cancers other than CRC were included in some studies. As a result, the modeling of the benefits (reduction in risk for severe neutropenia) and harms (reduction in treatment effectiveness that may occur among patients homozygous for *28 whose irinotecan treatment dose is reduced) is based on weak evidence that is, in the case of harms, nonsignificant. These limitations underscore the need for caution in interpreting the results and indicate the need for further study.
Recent information
Since the formal literature search, one additional trial81 has been reported that would have been included in the analysis of clinical validity and clinical utility. Specifically, that study found a higher rate of severe neutropenia among patients homozygous for *28 (RR, 5.4; 95% CI 2.4 –12) but no difference among *28 heterozygous patients (RR, 0.8; 95% CI 0.2–2.8). The study reported little or no relationship between patients’ UGT1A1 genotype and risk for severe diarrhea. This study found an improved rate of survival among patients homozygous for *28 (P = 0.06).
In another study published since the initial review, Hoskins et al.82 conducted a meta-analysis that provided more evidence for an association between irinotecan dose and risk of irinotecan-related Grades 3– 4 neutropenia. On the basis of commonly used treatment regimens, irinotecan dose levels were stratified into three groups low (<150 mg/m2), medium (150 –250 mg/m2), and high (>250 mg/m2). They reported a statistically significant association between genotype and hematologic toxicity at medium and high doses of irinotecan, with ORs of 3.22 for *28 homozygotes versus *28 heterozygotes (95% CI 1.52– 6.81, P = 0.008), and 27.8 for *28 homozygotes versus wild genotype (95% CI 4.0 –195, P = 0.005). They did not find these associations at low doses (OR = 1.80; 95% CI 0.37– 8.84; P = 0.41).
All of these findings are consistent with those reported in the main body of the review and strengthen the findings of the existing evidence review.
DISCUSSION
Quality of evidence
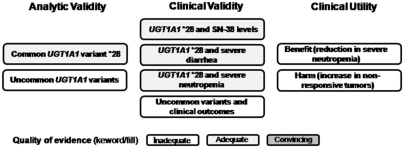
Figure 7 summarizes the quality of evidence for the key questions (Table 1). We rated the quality of evidence as adequate for the analytic validity of the common UGT1A1 variant *28, as there are two or more relatively high quality studies providing consistent results. However, the number of challenges do not allow for a confident estimates of the analytic sensitivity and specificity of the tests, even though the point estimates were high. Lastly, the data are restricted mainly to the analytic phase of testing. There are little or no data to estimate the analytic validity of tests for the less common UGT1A1 variants.
Fig. 7.
Graphic display of the quality of evidence for selected components of the current evidence review. For analytic validity, clinical validity, and clinical utility, each of the main components of the evidence review is represented by the text within a box. The quality of evidence is shown by the shading (no shading, inadequate; light shading, adequate; dark shading, convincing).
We also rated the quality of evidence for the association of the *28 variant with the active form of irinotecan (SN-38), severe diarrhea and severe neutropenia as adequate, based on a systematic review of lower quality studies for these three outcome measures. Little or no data are available to examine these three outcomes with respect to the less common UGT1A1 variants. Although it remains plausible, the evidence was inadequate to prospectively examine whether an initial reduction in irinotecan dosage in CRC patients homozygous for *28 was associated with a reduced risk for severe neutropenia. Little or no data are available to allow a direct, prospective comparison of the possible benefits and harms association with dosage reduction, but the previous modeling suggests further studies are warranted.
Important gaps in knowledge are as follows:
There seems to be a clear relationship between UGT1A1 genotype and severe neutropenia (and some evidence of a relationship with severe diarrhea), but there is no direct or indirect (chain of evidence) evidence to support the clinical utility of modifying an initial and/or subsequent dose of irinotecan in patients with metastatic CRC as a way to change the rate of adverse drug events (e.g., severe neutropenia).
Even if adverse drug-related events were reduced, this risk reduction may come at the expense of a reduction in tumor responsiveness in *28 homozygotes, leading to an overall net harm.
The data on the clinical validity of tests for UGT1A1 variants other than *28 are limited.
The analytic validity of UGT1A1 testing in clinical practice is unknown. Laboratories offering such testing may include variants in addition to *28 for which little evidence is available.
Pre- and postanalytic errors in UGT1A1 testing have not been reported, but the rate of such errors is likely to be similar to that reported for other genetic tests done in high-complexity laboratories.41,42 A new external proficiency testing program jointly offered by the American College of Medical Genetics and the College of American Pathologists is likely to provide important evidence about the analytic validity of UGT1A1 testing in clinical settings.
There are limited data on UGT1A1 variants in Hispanic and African American populations.
If UGT1A1 testing were recommended for routine use in clinical practice, additional studies would be needed to understand the potential effects of alleles that are rare in whites but more common in other racial/ethnic groups (e.g., *6 in Asians), and testing panels would need to include all variants of clinical significance in the population to be tested.
Research agenda
Analysis of data from the American College of Medical Genetics and College of American Pathologists proficiency testing program will provide needed information about the analytic validity of UGT1A1 tests offered for clinical use. However, additional studies concerning the clinical validity of tests for the less common UGT1A1 variants are needed. Given the rarity of these genotypes, these studies will need to include large numbers of subjects receiving treatment. This is feasible, however, because metastatic CRC is relatively common, as is chemotherapy with irinotecan. The most appropriate way to collect the evidence needed to document whether, or how, to modify irinotecan dosage for patients with particular UGT1A1 genotypes is to mount prospective studies (preferably including randomized trials) comparing outcomes among patients who receive targeted doses of irinotecan versus outcomes among those who receive doses currently recommended for all CRC patients. Such a study should be considered ethical, as it is not known whether the supposed benefits outweigh the possible harms. There are sufficient numbers of subjects for recruitment to be completed in a relatively short period of time.
Acknowledgments
Funding for this report was provided by National Office of Public Health Genomics, Centers for Disease Control and Prevention. The RTI preliminary evidence review for clinical validity was developed under project number 0208234.036.
The authors acknowledge Nedra Whitehead, PhD; Meera Viswanathan, PhD; and Eric Gillis, MS of RTI International, Research Triangle Park, NC, for their contributions to the full evidence report.
Members of the UGT1A1 Technical Evaluation Panel (TEP). From the EGAPP Working Group: Kathryn A. Phillips, PhD; Joan A. Scott, MS; Steven Teutsch, MD, MPH. Others included EGAPP Consultant Glenn Palomaki, BS; CDC representative Linda A. Bradley, PhD; RTI International representatives Meera Viswanathan, PhD; Nedra Whitehead, PhD; Kathleen N. Lohr, PhD; and RTI consultant Bert O’Neil, MD.
Reviewers of the evidence report for EGAPP included: EGAPP Working Group, D Joe Boone, PhD; Ralph Coates, PhD; Scott Grosse, PhD; Steve Gutman, MD, MBA; Howard McLeod, PharmD; Janelle Hoskins, PhD; Bert O’Neil, MD; Giuseppe Toffoli, MD; Marc Williams, MD; Jim Gudgeon, MS, MBA.
Reviewers of this manuscript for EGAPP included: EGAPP Working Group; Amy Brower, PhD; Janelle Hoskins, PhD; Giuseppe Toffoli, MD.
Footnotes
Disclaimer: The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
Disclosure: The authors declare no conflicts of interest.
Resources
Bradley LA, Palomaki GE, Dotson WD, et al. Evidence review: can UGT1A1 genotyping reduce morbidity and mortality in patients with metastatic colorectal cancer treated with irinotecan? 2008. Available at: http://www.egappreviews.org.
REFERENCES
- 1.Centers for Disease Control and Prevention. Colorectal cancer: the importance of prevention and early detection (fact sheet) [Accessed July 18, 2008]. Available at: http://www.cdc.gov/cancer/colorectal/pdf/about2004.pdf.
- 2.Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2006. CA Cancer J Clin. 2006;56:106–130. doi: 10.3322/canjclin.56.2.106. [DOI] [PubMed] [Google Scholar]
- 3.Wu X, Cokkinides V, Chen VW, et al. Associations of subsite-specific colorectal cancer incidence rates and stage of disease at diagnosis with county-level poverty, by race and sex. Cancer. 2006;107:1121–1127. doi: 10.1002/cncr.22009. [DOI] [PubMed] [Google Scholar]
- 4.Pessino A, Sobrero A. Optimal treatment of metastatic colorectal cancer. Expert Rev Anticancer Ther. 2006;6:801–812. doi: 10.1586/14737140.6.5.801. [DOI] [PubMed] [Google Scholar]
- 5.Saunders M, Iveson T. Management of advanced colorectal cancer: state of the art. Br J Cancer. 2006;95:131–138. doi: 10.1038/sj.bjc.6603233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van Cutsem E, Verslype C, Demedts I. The treatment of advanced colorectal cancer: where are we now and where do we go? Best Pract Res Clin Gastroenterol. 2002;16:319–330. doi: 10.1053/bega.2002.0288. [DOI] [PubMed] [Google Scholar]
- 7.Board RE, Valle JW. Metastatic colorectal cancer: current systemic treatment options. Drugs. 2007;67:1851–1867. doi: 10.2165/00003495-200767130-00004. [DOI] [PubMed] [Google Scholar]
- 8.Kelly H, Goldberg RM. Systemic therapy for metastatic colorectal cancer: current options, current evidence. J Clin Oncol. 2005;23:4553–4560. doi: 10.1200/JCO.2005.17.749. [DOI] [PubMed] [Google Scholar]
- 9.Wolpin BM, Meyerhardt JA, Mamon HJ, Mayer RJ. Adjuvant treatment of colorectal cancer. CA Cancer J Clin. 2007;57:168–185. doi: 10.3322/canjclin.57.3.168. [DOI] [PubMed] [Google Scholar]
- 10.Mathijssen RH, van Alphen RJ, Verweij J, et al. Clinical pharmacokinetics and metabolism of irinotecan (CPT-11) Clin Cancer Res. 2001;7:2182–2194. [PubMed] [Google Scholar]
- 11.Miners JO, McKinnon RA, Mackenzie PI. Genetic polymorphisms of UDP-glucuronosyltransferases and their functional significance. Toxicology. 2002:181–182. 453–456. doi: 10.1016/s0300-483x(02)00449-3. [DOI] [PubMed] [Google Scholar]
- 12.Toffoli G, Cecchin E, Corona G, Boiocchi M. Pharmacogenetics of irinotecan. Curr Med Chem Anticancer Agents. 2003;3:225–237. doi: 10.2174/1568011033482477. [DOI] [PubMed] [Google Scholar]
- 13.Nagar S, Blanchard RL. Pharmacogenetics of uridine diphosphoglucurono-syltransferase (UGT) 1A family members and its role in patient response to irinotecan. Drug Metab Rev. 2006;38:393–409. doi: 10.1080/03602530600739835. [DOI] [PubMed] [Google Scholar]
- 14.Innocenti F, Undevia SD, Iyer L, et al. Genetic variants in the UDP-glucuronosyltransferase 1A1 gene predict the risk of severe neutropenia of irinotecan. J Clin Oncol. 2004;22:1382–1388. doi: 10.1200/JCO.2004.07.173. [DOI] [PubMed] [Google Scholar]
- 15.Bosma PJ. Inherited disorders of bilirubin metabolism. J Hepatol. 2003;38:107–117. doi: 10.1016/s0168-8278(02)00359-8. [DOI] [PubMed] [Google Scholar]
- 16.Gagnon JF, Bernard O, Villeneuve L, Tetu B, Guillemette C. Irinotecan inactivation is modulated by epigenetic silencing of UGT1A1 in colon cancer. Clin Cancer Res. 2006;12:1850–1858. doi: 10.1158/1078-0432.CCR-05-2130. [DOI] [PubMed] [Google Scholar]
- 17.Innocenti F, Ratain MJ. “Irinogenetics” and UGT1A: from genotypes to haplotypes”. Clin Pharmacol Ther. 2004;75:495–500. doi: 10.1016/j.clpt.2004.01.011. [DOI] [PubMed] [Google Scholar]
- 18.Monaghan G, Ryan M, Seddon R, Hume R, Burchell B. Genetic variation in bilirubin UPD-glucuronosyltransferase gene promoter and Gilbert’s syndrome. Lancet. 1996;347:578–581. doi: 10.1016/s0140-6736(96)91273-8. [DOI] [PubMed] [Google Scholar]
- 19.Mercke Odeberg J, Andrade J, Holmberg K, Hoglund P, Malmqvist U, Odeberg J. UGT1A polymorphisms in a Swedish cohort and a human diversity panel, and the relation to bilirubin plasma levels in males and females. Eur J Clin Pharmacol. 2006;62:829–837. doi: 10.1007/s00228-006-0166-3. [DOI] [PubMed] [Google Scholar]
- 20.Innocenti F, Ratain MJ. Irinotecan treatment in cancer patients with UGT1A1 polymorphisms. Oncology (Williston Park) 2003;17:52–55. [PubMed] [Google Scholar]
- 21.Human UGT Allele Tables. [Accessed July 18, 2008]. Available at: http://som.flinders.edu.au/FUSA/ClinPharm/UGT/allele_table.html.
- 22.von Ahsen N, Oellerich M, Schutz E. DNA base bulge vs unmatched end formation in probe-based diagnostic insertion/deletion genotyping: genotyping the UGT1A1 (TA)(n) polymorphism by real-time fluorescence PCR. Clin Chem. 2000;46:1939–1945. [PubMed] [Google Scholar]
- 23.Ando Y, Fujita K, Sasaki Y, Hasegawa Y. UGT1AI*6 and UGT1A1*27 for individualized irinotecan chemotherapy. Curr Opin Mol Ther. 2007;9:258–262. [PubMed] [Google Scholar]
- 24.Minami H, Sai K, Saeki M, et al. Irinotecan pharmacokinetics/pharmacodynamics and UGT1A genetic polymorphisms in Japanese: roles of UGT1A1*6 and *28. Pharmacogenet Genomics. 2007;17:497–504. doi: 10.1097/FPC.0b013e328014341f. [DOI] [PubMed] [Google Scholar]
- 25.Saeki M, Saito Y, Jinno H, et al. Comprehensive UGT1A1 genotyping in a Japanese population by pyrosequencing. Clin Chem. 2003;49:1182–1185. doi: 10.1373/49.7.1182. [DOI] [PubMed] [Google Scholar]
- 26.Sai K, Saeki M, Saito Y, et al. UGT1A1 haplotypes associated with reduced glucuronidation and increased serum bilirubin in irinotecan-administered Japanese patients with cancer. Clin Pharmacol Ther. 2004;75:501–515. doi: 10.1016/j.clpt.2004.01.010. [DOI] [PubMed] [Google Scholar]
- 27.Sai K, Saito Y, Sakamoto H, et al. Importance of UDP-glucuronosyltransferase 1A1*6 for irinotecan toxicities in Japanese cancer patients. Cancer Lett. 2008;261:165–171. doi: 10.1016/j.canlet.2007.11.009. [DOI] [PubMed] [Google Scholar]
- 28.Sandanaraj E, Jada SR, Shu X, et al. Influence of UGT1A9 intronic I399C>T polymorphism on SN-38 glucuronidation in Asian cancer patients. Pharmacogenomics J. 2008;8:174–185. doi: 10.1038/sj.tpj.6500473. [DOI] [PubMed] [Google Scholar]
- 29.Camptosar (irinotecan) package insert. [Accessed July 17, 2008]. Available at: http://www.fda.gov/medwatch/SAFETY/2002/camptosar[lowem]label_highlighted.pdf#search=%22camptosar%20package%20insert%22.
- 30.FDA Clinical Pharmacology Subcommittee Proceedings. Pharmacogenetics of irinotecan: scientific and clinical impact of UGT polymorphisms. Nov 3, 2004. [Accessed July 17, 2008]. Available at: http://www.fda.gov/ohrms/dockets/AC/04/briefing/2004-4079B1_03_Topic1-TabA.pdf.
- 31.Invader UGT1A1 Molecular Assay 510K Summary. [Accessed July 17, 2008]. Available at: http://www.fda.gov/cdrh/pdf5/K051824.pdf#search=%22Invader%C2%AE%20UGT1A1%20Molecular%20Assay%20%20510k%20summary%22.
- 32.Invader UGT1A1 molecular assay for irinotecan toxicity. A genetic test for an increased risk of toxicity from the cancer chemotherapy drug irinotecan (Camptosar) Med Lett Drugs Ther. 2006;48:39–40. [PubMed] [Google Scholar]
- 33.Press Release: Third Wave and Genzyme Genetics Announce Preferred Marketing Relationship for Colorectal Cancer Personalized Medicine Test. [Accessed July 17, 2008]. Available at: http://www.genzymegenetics.com/about/news/gene_p_news_thirdwave.asp.
- 34.US Food and Drug Administration. FDA news. FDA clears genetic test that advances personalized medicine: test helps determine safety of drug therapy. [Accessed July 17, 2008]. Available at: http://www.fda.gov/bbs/topics/NEWS/2005/NEW01220.html.
- 35.Third Wave, Inc. Invader UGT1A1 Molecular Assay Package Insert. [Accessed July 17, 2008]. Available at: http://www.ons.org/fda/documents/FDA93005insert.pdf.
- 36.Teutsch SM, Bradley LA, Palomaki GE, et al. The Evaluation of Genomic Applications in Practice and Prevention (EGAPP) initiative: methods of the EGAPP working group. Genet Med. 2009;11:3–14. doi: 10.1097/GIM.0b013e318184137c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haddow JE, Palomaki GE. ACCE: a model process for evaluating data on emerging genetic tests. In: Khoury M, Little J, Burke W, editors. Human Genome Epidemiology: A Scientific Foundation for Using Genetic Information to Improve Health and Prevent Disease. New York: Oxford University Press; 2003. pp. 217–233. [Google Scholar]
- 38.Hasegawa Y, Sarashina T, Ando M, et al. Rapid detection of UGT1A1 gene polymorphisms by newly developed Invader assay. Clin Chem. 2004;50:1479–1480. doi: 10.1373/clinchem.2004.034694. [DOI] [PubMed] [Google Scholar]
- 39.Pirulli D, Giordano M, Puzzer D, et al. Rapid method for detection of extra (TA) in the promoter of the bilirubin-UDP-glucuronosyl transferase 1 gene associated with Gilbert syndrome. Clin Chem. 2000;46:129–131. [PubMed] [Google Scholar]
- 40.Baudhuin LM, Highsmith WE, Skierka J, Holtegaard L, Moore BE, O’Kane DJ. Comparison of three methods for genotyping the UGT1A1 (TA)n repeat polymorphism. Clin Biochem. 2007;40:710–717. doi: 10.1016/j.clinbiochem.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 41.Palomaki GE, Bradley LA, Richards CS, Haddow JE. Analytic validity of cystic fibrosis testing: a preliminary estimate. Genet Med. 2003;5:15–20. doi: 10.1097/00125817-200301000-00003. [DOI] [PubMed] [Google Scholar]
- 42.Palomaki GE, Haddow JE, Bradley LA, Richards CS, Stenzel TT, Grody WW. Estimated analytic validity of HFE C282Y mutation testing in population screening: the potential value of confirmatory testing. Genet Med. 2003;5:440–443. doi: 10.1097/01.gim.0000096500.66084.85. [DOI] [PubMed] [Google Scholar]
- 43.Hasegawa Y, Ando Y, Shimokata K. Screening for adverse reactions to irinotecan treatment using the Invader UGT1A1 Molecular Assay. Expert Rev Mol Diagn. 2006;6:527–533. doi: 10.1586/14737159.6.4.527. [DOI] [PubMed] [Google Scholar]
- 44.Font A, Sanchez JM, Taron M, et al. Weekly regimen of irinotecan/docetaxel in previously treated non-small cell lung cancer patients and correlation with uridine diphosphate glucuronosyltransferase 1A1 (UGT1A1) polymorphism. Invest New Drugs. 2003;21:435–443. doi: 10.1023/a:1026251202137. [DOI] [PubMed] [Google Scholar]
- 45.Iyer L, Das S, Janisch L, et al. UGT1A1*28 polymorphism as a determinant of irinotecan disposition and toxicity. Pharmacogenomics J. 2002;2:43–47. doi: 10.1038/sj.tpj.6500072. [DOI] [PubMed] [Google Scholar]
- 46.Marcuello E, Altes A, Menoyo A, Del Rio E, Gómez-Pardo M, Baiget M. UGT1A1 gene variations and irinotecan treatment in patients with metastatic colorectal cancer. Br J Cancer. 2004;91:678–682. doi: 10.1038/sj.bjc.6602042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Massacesi C, Terrazzino S, Marcucci F, et al. Uridine diphosphate glucuronosyl transferase 1A1 promoter polymorphism predicts the risk of gastrointestinal toxicity and fatigue induced by irinotecan-based chemotherapy. Cancer. 2006;106:1007–1016. doi: 10.1002/cncr.21722. [DOI] [PubMed] [Google Scholar]
- 48.Rouits E, Boisdron-Celle M, Dumont A, Guérin O, Morel A, Gamelin E. Relevance of different UGT1A1 polymorphisms in irinotecan-induced toxicity: a molecular and clinical study of 75 patients. Clin Cancer Res. 2004;10:5151–5159. doi: 10.1158/1078-0432.CCR-03-0548. [DOI] [PubMed] [Google Scholar]
- 49.Soepenberg O, Dumez H, Verweij J, et al. Phase I pharmacokinetic, food effect, and pharmacogenetic study of oral irinotecan given as semisolid matrix capsules in patients with solid tumors. Clin Cancer Res. 2005;11:1504–1511. doi: 10.1158/1078-0432.CCR-04-1758. [DOI] [PubMed] [Google Scholar]
- 50.Toffoli G, Cecchin E, Corona G, et al. The role of UGT1A1*28 polymorphism in the pharmacodynamics and pharmacokinetics of irinotecan in patients with metastatic colorectal cancer. J Clin Oncol. 2006;24:3061–3068. doi: 10.1200/JCO.2005.05.5400. [DOI] [PubMed] [Google Scholar]
- 51.Carlini LE, Meropol NJ, Bever J, et al. UGT1A7 and UGT1A9 polymorphisms predict response and toxicity in colorectal cancer patients treated with capecitabine/irinotecan. Clin Cancer Res. 2005;11:1226–1236. [PubMed] [Google Scholar]
- 52.Mathijssen RH, Marsh S, Karlsson MO, et al. Irinotecan pathway genotype analysis to predict pharmacokinetics. Clin Cancer Res. 2003;9:3246–3253. [PubMed] [Google Scholar]
- 53.Mathijssen RH, de Jong FA, van Schaik RH, et al. Prediction of irinotecan pharmacokinetics by use of cytochrome P450 3A4 phenotyping probes. J Natl Cancer Inst. 2004;96:1585–1592. doi: 10.1093/jnci/djh298. [DOI] [PubMed] [Google Scholar]
- 54.Paoluzzi L, Singh AS, Price DK, et al. Influence of genetic variants in UGT1A1 and UGT1A9 on the in vivo glucuronidation of SN-38. J Clin Pharmacol. 2004;44:854–860. doi: 10.1177/0091270004267159. [DOI] [PubMed] [Google Scholar]
- 55.Gupta E, Lestingi TM, Mick R, Ramirez J, Vokes EE, Ratain MJ. Metabolic fate of irinotecan in humans: correlation of glucuronidation with diarrhea. Cancer Res. 1994;54:3723–3725. [PubMed] [Google Scholar]
- 56.National Cancer Institute. Common Toxicity Criteria for Grading the Severity of Diarrhea. [Accessed July 17, 2008]. Available at: http://www.cancer.gov/cancertopics/pdq/supportivecare/gastrointestinalcomplications/HealthProfessional/Table2.
- 57.Beutler E, Gelbart T, Demina A. Racial variability in the UDP-glucurono-syltransferase 1 (UGT1A1) promoter: a balanced polymorphism for regulation of bilirubin metabolism? Proc Natl Acad Sci USA. 1998;95:8170–8174. doi: 10.1073/pnas.95.14.8170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Borlak J, Thum T, Landt O, Erb K, Hermann R. Molecular diagnosis of a familial nonhemolytic hyperbilirubinemia (Gilbert’s syndrome) in healthy subjects. Hepatology. 2000;32:792–795. doi: 10.1053/jhep.2000.18193. [DOI] [PubMed] [Google Scholar]
- 59.Bosch TM, Doodeman VD, Smits PH, Meijerman I, Schellens JH, Beijnen JH. Pharmacogenetic screening for polymorphisms in drug-metabolizing enzymes and drug transporters in a Dutch population. Mol Diagn Ther. 2006;10:175–185. doi: 10.1007/BF03256456. [DOI] [PubMed] [Google Scholar]
- 60.Cecchin E, Russo A, Corona G, et al. UGT1A1*28 polymorphism in ovarian cancer patients. Oncol Rep. 2004;12:457–462. [PubMed] [Google Scholar]
- 61.Danoff TM, Campbell DA, McCarthy LC, et al. A Gilbert’s syndrome UGT1A1 variant confers susceptibility to tranilast-induced hyperbiliru-binemia. Pharmacogenomics J. 2004;4:49–53. doi: 10.1038/sj.tpj.6500221. [DOI] [PubMed] [Google Scholar]
- 62.Kohle C, Mohrle B, Munzel PA, et al. Frequent co-occurrence of the TATA box mutation associated with Gilbert’s syndrome (UGT1A1*28) with other polymorphisms of the UDP-glucuronosyltransferase-1 locus (UGT1A6*2 and UGT1A7*3) in Caucasians and Egyptians. Biochem Pharmacol. 2003;65:1521–1527. doi: 10.1016/s0006-2952(03)00074-1. [DOI] [PubMed] [Google Scholar]
- 63.Lampe JW, Bigler J, Horner NK, Potter JD. UDP-glucuronosyltransferase (UGT1A1*28 and UGT1A6*2) polymorphisms in Caucasians and Asians: relationships to serum bilirubin concentrations. Pharmacogenetics. 1999;9:341–349. doi: 10.1097/00008571-199906000-00009. [DOI] [PubMed] [Google Scholar]
- 64.Rauchschwalbe SK, Zuhlsdorf MT, Schuhly U, Kuhlmann J. Predicting the risk of sporadic elevated bilirubin levels and diagnosing Gilbert’s syndrome by genotyping UGT1A1*28 promoter polymorphism. Int J Clin Pharmacol Ther. 2002;40:233–240. doi: 10.5414/cpp40233. [DOI] [PubMed] [Google Scholar]
- 65.Sampietro M, Lupica L, Perrero L, Romano R, Molteni V, Fiorelli G. TATA-box mutant in the promoter of the uridine diphosphate glucuronosyl-transferase gene in Italian patients with Gilbert’s syndrome. Ital J Gastroenterol Hepatol. 1998;30:194–198. [PubMed] [Google Scholar]
- 66.Kaniwa N, Kurose K, Jinno H, et al. Racial variability in haplotype frequencies of UGT1A1 and glucuronidation activity of a novel single nucleotide polymorphism 686C> T (P229L) found in an African-American. Drug Metab Dispos. 2005;33:458–465. doi: 10.1124/dmd.104.001800. [DOI] [PubMed] [Google Scholar]
- 67.Sugatani J, Yamakawa K, Yoshinari K, et al. Identification of a defect in the UGT1A1 gene promoter and its association with hyperbilirubinemia. Biochem Biophys Res Commun. 2002;292:492–497. doi: 10.1006/bbrc.2002.6683. [DOI] [PubMed] [Google Scholar]
- 68.Tang KS, Chiu HF, Chen HH, et al. Link between colorectal cancer and polymorphisms in the uridine-diphosphoglucuronosyltransferase 1A7 and 1A1 genes. World J Gastroenterol. 2005;11:3250–3254. doi: 10.3748/wjg.v11.i21.3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Guillemette C, Millikan RC, Newman B, Housman DE. Genetic polymorphisms in uridine diphospho-glucuronosyltransferase 1A1 and association with breast cancer among African Americans. Cancer Res. 2000;60:950–956. [PubMed] [Google Scholar]
- 70.Goldberg RM, McLeod HL, Sargent DJ, et al. Genetic polymorphisms, toxicity, and response rate in African Americans with metastatic colorectal cancer compared to Caucasians when treated with IFL, FOLFOX, or IROX in Intergroup N9741. J Clin Oncol. 2006;24:3503. [Google Scholar]
- 71.Liu JY, Qu K, Sferruzza AD, Bender RA. Distribution of the UGT1A1*28 polymorphism in Caucasian and Asian populations in the US: a genomic analysis of 138 healthy individuals. Anticancer Drugs. 2007;18:693–696. doi: 10.1097/CAD.0b013e32803a46fe. [DOI] [PubMed] [Google Scholar]
- 72.Innocenti F, Ratain MJ. Pharmacogenetics of irinotecan: clinical perspectives on the utility of genotyping. Pharmacogenomics. 2006;7:1211–1221. doi: 10.2217/14622416.7.8.1211. [DOI] [PubMed] [Google Scholar]
- 73.NCCN Myeloid Growth Factors Panel Members. Myeloid growth factors V. 1.2008-NCCN clinical practice guidelines in oncology. [Accessed July 18, 2008]. Available at: http://www.nccn.org/professionals/physician_gls/PDF/myeloid_growth.pdf.
- 74.McLeod HL. To test or not to test: an update on UGT1A1 testing. Oncology Issues. 2006 Nov–Dec;:20–22. [Google Scholar]
- 75.Goldberg RM, Sargent DJ, Morton RF, et al. Randomized controlled trial of reduced-dose bolus fluorouracil plus leucovorin and irinotecan or infused fluorouracil plus leucovorin and oxaliplatin in patients with previously untreated metastatic colorectal cancer: a North American Intergroup Trial. J Clin Oncol. 2006;24:3347–3353. doi: 10.1200/JCO.2006.06.1317. [DOI] [PubMed] [Google Scholar]
- 76.Cunningham D, Pyrhonen S, James RD, et al. Randomised trial of irinotecan plus supportive care versus supportive care alone after fluorouracil failure for patients with metastatic colorectal cancer. Lancet. 1998;352:1413–1418. doi: 10.1016/S0140-6736(98)02309-5. [DOI] [PubMed] [Google Scholar]
- 77.Saltz LB, Cox JV, Blanke C, et al. Irinotecan plus fluorouracil and leucovorin for metastatic colorectal cancer. Irinotecan study group. N Engl J Med. 2000;343:905–914. doi: 10.1056/NEJM200009283431302. [DOI] [PubMed] [Google Scholar]
- 78.Goetz MP, Reid JM, Safgren SL, et al. UGT1A1*28 genotype determines the maximum tolerated dose (MTD) and pharmacokinetics (PK) of irinotecan-based chemotherapy: a phase 1 dose-escalation trial. Proceedings of American Society of Clinical Oncology, Gastrointestinal Cancers Symposium; Orlando, Florida. 2007; Abstract 235. [Google Scholar]
- 79.Hazama S, Koudo H, Yoshida S, et al. UGT1As polymorphisms predict toxicity in colorectal cancer patients treated with different recommended doses of irinotecan oriented by UGT1A1*28 polymorphism based on previous phase I study. J Clin Oncol. 2007;25:14511. [Google Scholar]
- 80.Innocenti F, Janisch L, Das S, et al. A genotype-directed phase I study of irinotecan in advanced cancer patients. J Clin Oncol, 2007 ASCO Annu Meet Proc Part I. 2007;25:2502. [Google Scholar]
- 81.Cote JF, Kirzin S, Kramar A, et al. UGT1A1 polymorphism can predict hematologic toxicity in patients treated with irinotecan. Clin Cancer Res. 2007;13:3269–3275. doi: 10.1158/1078-0432.CCR-06-2290. [DOI] [PubMed] [Google Scholar]
- 82.Hoskins JM, Goldberg RM, Qu P, Ibrahim JG, McLeod HL. UGT1A1*28 genotype and irinotecan-induced neutropenia: dose matters. J Natl Cancer Inst. 2007;99:1290–1295. doi: 10.1093/jnci/djm115. [DOI] [PubMed] [Google Scholar]