Abstract
Centromere identity is thought to be determined by epigenetic mechanisms. The centromere-specific histone H3 variant CENP-A plays a central role in specifying the locus where the centromere is constructed. However, the precise mechanisms that target CENP-A to centromeric chromatin are poorly understood. Here, we show that facilitates chromatin transcription (FACT) localizes to centromeres in a CENP-H–containing complex-dependent manner. In conditional mutant cell lines for SSRP1, a subunit of FACT, centromere targeting of newly synthesized CENP-A is severely inhibited. The chromatin remodeling factor CHD1 binds to SSRP1 both in vivo and in vitro and associates with centromeres. The centromeric localization of CHD1 is lost in SSRP1-depleted cells. RNA interference knockdown of CHD1 leads to a decrease in the amount of centromere localized CENP-A. These findings indicate that the CENP-H–containing complex facilitates deposition of newly synthesized CENP-A into centromeric chromatin in cooperation with FACT and CHD1.
INTRODUCTION
The centromere is a specialized chromatin region that incorporates the centromere-specific histone H3 variant CENP-A. Although the chromatin that contains CENP-A is normally constructed on repetitive α-satellite DNA in human cells, the DNA sequences themselves are thought to be neither necessary nor sufficient for determination of centromere position (Cheeseman and Desai, 2008). Studies of neocentromeres have shown that functional centromeres can be ectopically constructed and stably maintained at loci that do not possess α-satellite sequences (Marshall et al., 2008). This indicates that the centromeric region is specified by epigenetic mechanisms rather than by cis-acting elements. One candidate for an epigenetic marker that specifies centromeres is CENP-A (Black and Bassett, 2008). CENP-A is localized at functional centromeres, including neocentromeres and is required for the centromeric localization of other centromere proteins (Warburton et al., 1997; Howman et al., 2000; Oegema et al., 2001; Goshima et al., 2003; Regnier et al., 2005; Liu et al., 2006). Ectopic mislocalization of human CENP-A, or its Drosophila counterpart CID, induces the colocalization of other centromere proteins to the noncentromeric loci (Van Hooser et al., 2001; Heun et al., 2006). Overall, the available information indicates that CENP-A is the key player for targeting centromere proteins to the correct sites by acting as an epigenetic marker. Thus, to understand how the centromere is specified in the correct chromosomal region, it is important to elucidate the mechanisms that target CENP-A to centromeric chromatin.
Recent studies identified various proteins involved in the centromeric localization of CENP-A. RbAp46/48, hMis18α/β, and M18BP1/KNL2 are required for centromeric localization of CENP-A in vertebrates and nematodes (Hayashi et al., 2004; Fujita et al., 2007; Maddox et al., 2007). HJURP binds to CENP-A and is required for centromeric localization of CENP-A in vertebrates (Foltz et al., 2009; Dunleavy et al., 2009). CENP-A mislocalization in hMis18 depleted cells is rescued by trichostatin A, suggesting that the hMis18 complex might acetylate histones or centromere localized proteins to enhance CENP-A deposition.
The constitutive centromere associated network (CCAN) proteins have been found to localize to centromeres throughout the cell cycle and have important roles in the construction of functional kinetochores (Cheeseman and Desai, 2008). We previously identified the CENP-H-I complex as a member of the CCAN in chicken and human cells. The CENP-H-I complex can be subdivided into several groups. The CENP-H–containing subcomplex, including CENP-H, -I, -K, and -M, is required for the efficient targeting of newly synthesized CENP-A into centromeres (Okada et al., 2006), but it does not include typical chromatin remodeling factors. This suggests that the CENP-H–containing complex might act in concert with other proteins that function to remodel chromatin structures.
Recent proteomic analyzes revealed that several chromatin remodeling factors and histone chaperones are associated with CENP-A–containing chromatin (Obuse et al., 2004; Foltz et al., 2006). On the basis of published data, we hypothesized that the chromatin remodeling complex named facilitates chromatin transcription (FACT) might be involved in CENP-A incorporation into centromeric chromatin. FACT was first reported as a factor that enhances transcription elongation from chromatin templates in vitro (Orphanides et al., 1998), and in budding yeast, FACT was shown to alter chromatin structure without using ATP hydrolysis (Ruone et al., 2003; Rhoades et al., 2004). In fission yeast, FACT contributes to centromeric heterochromatin formation (Lejeune et al., 2007). In addition, Drosophila FACT enhances the deposition of histone H3.3 on the nucleosomes adjacent to FACT binding sites (Nakayama et al., 2007), suggesting that FACT alters chromatin structure to make the chromatin competent for replacement of canonical histone H3 by histone H3 variants. These findings raise the possibility that the centromere localized FACT plays an important role in deposition of the centromere-specific histone H3 variant CENP-A into the centromeric chromatin.
To elucidate the possible role of the CENP-H–containing complex in deposition of newly synthesized CENP-A into the centromeric chromatin, we investigated the functional interaction of the CENP-H–containing complex with FACT. Here, we report that FACT localizes to centromeres in the CENP-H–containing complex dependent manner and that FACT is required for the efficient incorporation of newly synthesized CENP-A into centromeres. In addition, the ATP-dependent chromatin remodeling factor CHD1 is localized to centromeres through a physical interaction with FACT and is required for the maintenance of the amount of CENP-A at centromeres. Therefore, the CENP-H–containing complex seems to facilitate CENP-A incorporation into centromeric chromatin in cooperation with FACT and CHD1.
MATERIALS AND METHODS
Molecular Biology, Cell Culture, and Transfection
Chicken target disruption constructs for SSRP1 was generated such that genomic fragments encoding 13 exons were replaced with a histidinol- (hisD) or puromycin- (puro) resistance cassette under control of the β-actin promoter. Target constructs were transfected with a Gene Pulser II electroporator (Bio-Rad Laboratories, Hercules, CA). DT40 cells were cultured and transfected as described previously (Nishihashi et al., 2002). All DT40 cells were cultured at 38.5°C in Dulbecco's modified medium supplemented with 10% fetal calf serum, 1% chicken serum, penicillin, and streptomycin. To repress the expression of the tet-responsive transgenes, tetracycline (Sigma-Aldrich, St. Louis, MO) was added to the culture medium to a final concentration of 2 μg/ml. Fluorescence-activated cell sorting (FACS) analysis was performed as described previously (Fukagawa et al., 2001).
For the CENP-A-GFP incorporation assay, cells were resuspended in Nucleofector T reagent containing 2 μg of cytomegalovirus (CMV)-chicken CENP-A-green fluorescent protein (GFP) plasmid and 1 μg of CMV-monomeric red fluorescent protein (mRFP) plasmid and electroporation was performed using a Nucleofector instrument (Amaxa Biosystems, Gaithersburg, MD). Cells were cultured for 16 h and fixed with 3% paraformaldehyde. In each experiment, only the cells expressing mRFP signals were counted for scoring the efficiency of CENP-A incorporation.
Human HeLa S3 cells were cultured on a seesaw shaker placed in a CO2 incubator for suspension culture. Control small interfering RNAs (siRNAs) or siRNAs against CHD1 (Invitrogen, Carlsbad, CA) were transfected using Lipofectamine RNAi MAX (Invitrogen). The predesigned siRNAs against CHD1 and control siRNAs can be ordered as Stealth Select RNAi reagent HSS101841 and Stealth RNAi Negative Control LO GC (12935-200), respectively.
Immunocytochemistry
Cells were fixed with 3% paraformaldehyde and incubated with antibodies in 0.5% bovine serum albumin (BSA) for 1 h. DNA was counterstained with 4,6-diamidino-2-phenylindole (DAPI). For staining with anti-SSRP1, anti-SPT16, and anti-CHD1 antibodies, cells were treated with nocodazole (1 μg/ml) for 2 h before fixation to accumulate mitotic cells. Cells were then extracted with 0.1% Triton X-100 in phosphate-buffered saline (PBS) for 2 min followed by 15-min fixation with 3% paraformaldehyde. Antibody incubations were performed for 1 h by using CanGetSignal Immunostaining Solution A (Toyobo Engineering, Osaka, Japan). CHD1 was detected using N-16 polyclonal antibody (Santa Cruz Biotechnology, Santa Cruz, CA) and human CENP-A by using D115–3 monoclonal antibody (MBL International, Woburn, MA). All images were collected with a cooled charge-coupled device camera (CoolSNAP HQ, Photometrics Image Point; Photometrics, Tucson, AZ) mounted on an IX71 inverted microscope (Olympus, Tokyo, Japan) with a 60× objective lens (PlanApo 60×, numerical aperture 1.40) together with a filter wheel. Images were analyzed with an IPLab software (Signal Analytics, Vienna, VA). All images for a given condition were acquired, processed, and scaled identically.
Glutathione Transferase (GST)-Pull Down Assay
Bacterially expressed GST-tagged chicken SSRP1 polypeptides were conjugated with glutathione-Sepharose beads (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom) and incubated with in vitro translated proteins for 2 h at 4°C in buffer A (20 mM HEPES-KOH, pH 7.4, 150 mM KCl, 10 mM MgCl2, 10% glycerol, 0.1% NP-40, 1% BSA, and 50 μg/ml ethidium bromide) and then washed with buffer B (20 mM HEPES-KOH, pH 7.4, 150 mM KCl, 10 mM MgCl2, 10% glycerol, and 0.1% NP40). Benzonase nuclease (100 U) (Novagen, Madison, WI) was added separately to the in vitro-translated proteins and purified GST-tagged SSRP1 and incubated for 1 h at 25°C before the binding reaction when described.
Microarray
Total RNA was prepared from SSRP1 conditional mutant cells cultured in the absence or presence of tet for 42 h and subjected to biotin labeling according to the manufacturer's protocols. Gene expression profiles were analyzed using GeneChip Chicken Genome Array (Affymetrix, Santa Clara, CA).
DNA Data Bank of Japan/European Molecular Biology Laboratory/GenBank Accession Numbers
The nucleotide sequences of cDNA for chicken SSRP1 and CHD1 are listed under accession AB465209 and AB465210, respectively.
RESULTS
FACT Localizes to Centromeres
SSRP1 and SPT16, which constitute FACT, copurified with endogenous CENP-A or CENP-A-TAP in the partially digested chromatin fraction of HeLa cells (Obuse et al., 2004; Foltz et al., 2006). We also found that both SSRP1 and SPT16 coimmunoprecipitated with CENP-A-GFP from chicken DT40 cells by a similar method to others (Supplemental Figure S1). However, the precise cellular localization of SSRP1 and SPT16 had not yet been determined. To investigate whether FACT localizes to centromeres, we analyzed the localization of endogenous SSRP1 and SPT16 by using specific antibodies. The chicken DT40 cell line that stably expresses CENP-H-RFP was used to enable visualization of centromere positions. As reported previously (Kelley et al., 1999), in cells fixed with paraformaldehyde, we obtained strong signals for both SSRP1 and SPT16 throughout the nuclei (Figure 1A). In contrast, when cells were treated with 0.1% Triton X-100 to eliminate soluble proteins before fixation, we found that both SSRP1 and SPT16 colocalized with CENP-H-RFP (Figure 1B). In interphase nuclei, immunostaining for SSRP1 and SPT16 proteins showed strong signals at centromeres, but some signals were also present at noncentromeric positions. At mitosis, SSRP1 and SPT16 were mainly localized at the centromeres. These results demonstrate that FACT associates with centromeres throughout the cell cycle.
Figure 1.
FACT localizes to centromeres. Localization of endogenous SSRP1 and SPT16 in CENP-H-RFP–expressing cells was visualized using anti-SSRP1 and anti-SPT16 antibodies. (A) Cells fixed with paraformaldehyde. (B) Cells treated with 0.1% Triton X-100 in PBS for 2 min before paraformaldehyde fixation. SSRP1 and SPT16 (green), CENP-H-RFP (red), and DAPI (blue) staining are shown.
The CENP-H–containing Complex Is Required for the Centromeric Localization of FACT
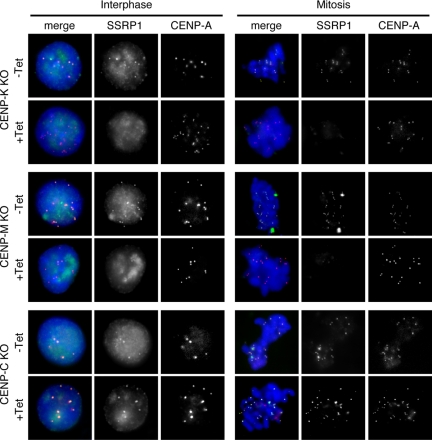
Because FACT is recruited to specific sites via protein–protein interactions with various factors, such as a DNA-binding protein (Shimojima et al., 2003) or DNA polymerase α (Zhou and Wang, 2004), we postulated that one or more members of the CCAN might be involved in the centromeric localization of FACT. To test this possibility, we immunostained the conditional mutant cell lines for CENP-K, CENP-M, and CENP-C with anti-SSRP1 or anti-SPT16 antibodies. In these conditional mutant cell lines, expression of the corresponding CENPs is blocked by the addition of tet to the medium. CENP-A-RFP expression was used to visualize centromere loci in these cells. We found that the centromeric localization of both SSRP1 and SPT16 was abolished at 48 h after the addition of tet in CENP-K mutant cells and at 72 h in CENP-M mutant cells. In contrast, in CENP-C mutant cells, both SSRP1 and SPT16 were present in centromeres after the addition of tet (Figure 2 and Supplemental Figure S2). We reported previously that the centromeric localization of the CENP-H–containing complex was not dependent on CENP-C (Kwon et al., 2007; Hori et al., 2008). These results suggest that the CENP-H–containing complex but not CENP-C is required for centromeric localization of FACT.
Figure 2.
CENP-H–containing complex is required for centromeric localization of FACT. Immunostaining using an anti-SSRP1 antibody in CENP-K, CENP-M, and CENP-C conditional mutant cells that stably expressed CENP-A-RFP to visualize centromeres. CENP-K and CENP-C conditional mutant cells were cultured for 48 h in the absence (−Tet) or presence (+Tet) of tet. CENP-M conditional mutant cells were cultured for 72 h in the presence of tet (+Tet). SSRP1 (green), CENP-A-RFP (red), and DAPI (blue) staining are shown.
Construction of SSRP1 Conditional Mutant Cell Lines
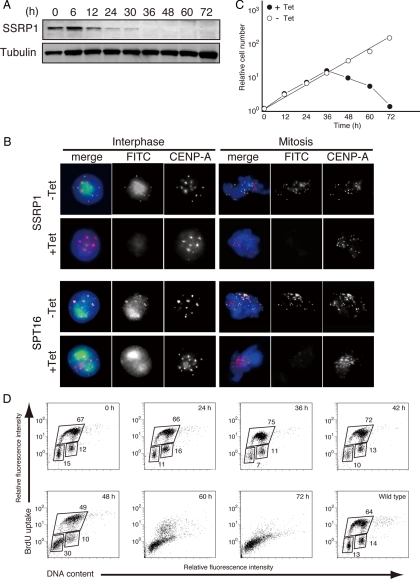
The CENP-H–containing complex but not CENP-C is required for the centromeric targeting of newly synthesized CENP-A (Okada et al., 2006). As is shown in Figure 2, centromeric localization of FACT is dependent on the CENP-H–containing complex but not CENP-C. Therefore, we hypothesized that centromere localized FACT plays an important role in centromeric targeting of nascent CENP-A. To test whether FACT is required for CENP-A incorporation into centromeres, we generated SSRP1 conditional mutants in which the viability of homozygous mutant cells was maintained by expression of SSRP1 cDNA under the control of a tet-repressible promoter (Supplemental Figure S3). We found that the SSRP1 protein decreased to an undetectable level by 36 h after the addition of tet in the SSRP1 conditional mutant cells (Figure 3A).
Figure 3.
Construction of SSRP1 conditional mutant cell lines. (A) Western blot analysis of total cell lysates from SSRP1 conditional mutant cells by using an anti-SSRP1 antibody at the indicated times after addition of tet to the cultures. Anti-tubulin antibody was used as the loading control. (B) Immunostaining using anti-SSRP1 and anti-SPT16 antibodies of SSRP1 conditional mutant cells that stably expressed CENP-A-RFP to visualize centromeres. Cells were cultured for 45 h in the presence of tet (+Tet). SSRP1 or SPT16 (green), CENP-A-RFP (red), and DAPI (blue) staining are shown. (C) Representative growth curves for the SSRP1 conditional mutant cells in the presence or absence of tet. Tet was added at time 0 to the SSRP1 conditional mutant cells (+Tet), and the number of living cells was counted. (D) BrdU incorporation was detected using fluorescein isothiocyanate (FITC)-anti-BrdU, and DNA contents were measured after propidium iodide staining. The bottom-left box, top box, and bottom right box represent to G1, S, and G2/M phase cells, respectively. The numbers in the boxes indicate the percentages of gated events.
To further confirm the centromeric localization of SSRP1, SSRP1 conditional mutant cell lines that stably express SSRP1-GFP were established and cultured in the presence of tet. Cells were fixed with methanol, and CENP-A was visualized with specific antibodies. We observed SSRP1-GFP signals at centromeres without Triton extraction before fixation (Supplemental Figure S4).
Because SSRP1 and SPT16 form a heterodimer, we tested whether SSRP1 was required for the centromeric localization of SPT16. CENP-A-RFP was used to visualize the centromere loci in the SSRP1 conditional mutant cells. We found that both SSRP1 and SPT16 had disappeared from the centromeres at 45 h after the addition of tet (Figure 3B).
SSRP1 conditional mutant cells stopped proliferating ∼48 h after the addition of tet (Figure 3C). We examined the cell cycle profile by measuring both cellular DNA content and DNA synthesis by using FACS after pulse labeling with 5-bromo-2′-deoxyuridine (BrdU). SSRP1-deficient cells started dying at 48 h after the addition of tet but did not show arrest at any particular cell cycle stage (Figure 3D). FACT is known to be involved in diverse cellular functions (Reinberg and Sims, 2006), such as transcription regulation (Shimojima et al., 2003; Nakayama et al., 2007), replication (Zhou and Wang, 2004), and repair (Heo et al., 2008). Thus, SSRP1 seems to be required for cell viability, and SSRP1-deficient cells may have died because of defects in these vital functions.
Newly synthesized CENP-A Is Not Efficiently Incorporated into Centromeres in SSRP1-deficient Cells
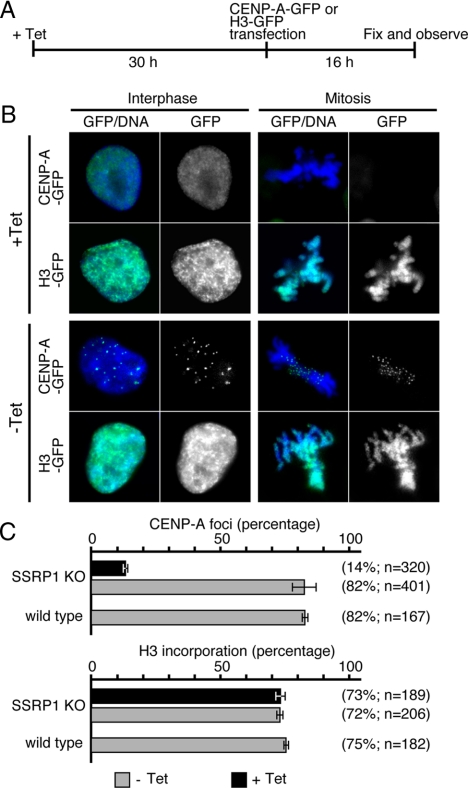
To investigate whether FACT is required for centromeric targeting of newly synthesized CENP-A, a CENP-A-GFP–expressing plasmid was transiently transfected into SSRP1 conditional mutant cells that had been cultured for 30 h in the presence of tet; the localization of CENP-A-GFP was analyzed at 16 h after transfection (Figure 4A). In the absence of tet, 82% of the cells exhibited centromeric localization of transiently expressed CENP-A-GFP. In contrast, the efficiency of centromeric targeting of nascent CENP-A-GFP was significantly reduced (14%) in SSRP1-depleted cells (Figure 4C). To determine whether this effect was specific to CENP-A, the efficiency of incorporation of transiently expressed histone H3-GFP was examined. In contrast to CENP-A incorporation, histone H3-GFP was efficiently assembled into the chromatin both in the presence or absence of tet (Figure 4C). These data show that FACT is required for efficient incorporation of newly synthesized CENP-A into centromeres.
Figure 4.
Newly synthesized CENP-A is not efficiently incorporated into centromeres in SSRP1 conditional mutant cells. (A) Schematic of the CENP-A-GFP incorporation assay. SSRP1 conditional mutant cells were cultured for 30 h in the presence of tet before transfection with CENP-A-GFP– or H3-GFP–expressing plasmids by using a Nucleofector instrument. Cells were then fixed 16 h after transfection and evaluated for the incorporation of CENP-A-GFP or H3-GFP. (B) Representative images of CENP-A-GFP foci in interphase and mitotic cells. (C) Quantification of cells with punctate CENP-A-GFP foci.
Because CENP-A is known to associate stably to centromeres (Okada et al., 2006; Jansen et al., 2007), multiple times of cell cycle progression in the SSRP1-depleted condition should be required to detect the reduction of pre-existing CENP-A signals. However, as is shown in Figure 3, SSRP1 conditional mutant cells immediately stopped proliferating when SSRP1 was depleted, we could not detect decrease in the signal intensities of pre-existing CENP-A at centromeres (Figure 3B).
To further confirm the requirement of SSRP1 for CENP-A incorporation into centromeres, we analyzed the domain of SSRP1 that is required for CENP-A incorporation. The GST-fused polypeptides of SSRP1 containing N-terminal region (1-193 aa), pleckstrin homology (PH) domain (194-544 aa), high-mobility group (HMG) domain (545-613 aa), and C-terminal region (614-706 aa) (Supplemental Figure S5A) were bacterially expressed and incubated with in vitro translated chicken CENP-A. We found that the C-terminal region of SSRP1 binds to CENP-A in vitro (Supplemental Figure S5B). We then tested whether the mutant SSRP1 that lacks the C-terminal region is able to rescue the CENP-A incorporation defect observed in SSRP1-depleted cells. The SSRP1 conditional mutant cells that stably express wild-type or C-terminal truncated SSRP1 were established, and the efficiency for centromeric targeting of newly synthesized CENP-A was determined. We found that CENP-A-GFP was not efficiently targeted to centromeres in the cells expressing SSRP1 mutant that lacks C-terminal region (Supplemental Figure S5C). These data suggest that C-terminal region of SSRP1 plays an important role for targeting of CENP-A to centromeres.
SSRP1 Is Not Required for Centromeric Localization of the CENP-H–containing Complex
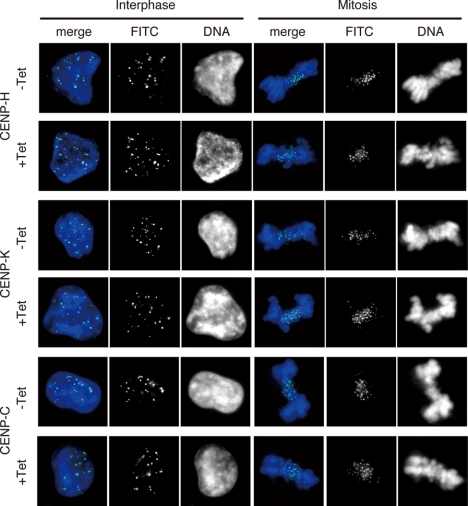
To confirm that FACT is required for incorporation of nascent CENP-A, we examined whether SSRP1 depletion affected the centromeric localization of the CENP-H-containing complex. We performed immunostaining with antibodies raised against CENP-H, CENP-K and CENP-C in SSRP1 conditional mutant cells cultured in the absence or presence of tet. We found that the centromeric localization of CENP-H, CENP-K and CENP-C was not altered at interphase or mitosis at 45 h after the addition of tet (Figure 5). We conclude that depletion of FACT does not affect the centromeric localization of the CENP-H–containing complex.
Figure 5.
Depletion of FACT does not affect localization of centromere proteins. Immunostaining of SSRP1 conditional mutant cells with anti-CENP-H, anti-CENP-K and anti-CENP-C antibodies. Cells were cultured for 45 h in the presence of tet (+Tet). Immunostaining signals were detected with fluorescein isothiocyanate (FITC)-conjugated secondary antibodies (green). DNA was counterstained with DAPI (blue).
Because FACT is known to be involved in transcription regulation, it is possible that the expression of genes with important functions in CENP-A incorporation into centromeres might be altered in SSRP1 conditional mutant cells. To examine this possibility, we analyzed the changes in the global gene expression profile of SSRP1-deficient cells by using a cDNA microarray. cDNA samples were prepared from SSRP1 conditional mutant cells cultured in the absence or presence of tet for 42 h. We found that the expression of genes for centromere proteins, such as CENP-H, CENP-I, CENP-K, and CENP-M, that are required for the centromeric localization of CENP-A was not significantly affected by depletion of SSRP1 (Supplemental Figure S6A). We also analyzed the expression of the CENP-A gene by reverse transcription-polymerase chain reaction (RT-PCR) and confirmed that CENP-A expression was not changed in SSRP1-deficient cells (Supplemental Figure S6B). These data suggest that FACT does not influence either the localization or expression of the CENP-H–containing complex subunits and that FACT may be directly involved in the deposition of nascent CENP-A into centromeres.
CHD1 Localizes to Centromeres through Interaction with SSRP1
FACT alters chromatin structure without using ATP hydrolysis (Ruone et al., 2003; Rhoades et al., 2004). This alteration to chromatin structure may require an additional chromatin remodeling factor because FACT can displace a histone H2A/H2B dimer from a nucleosome but does not have remodeling activity against a histone H3/H4 tetramer. The chromatin remodeling factor CHD1 was reported to interact directly with SSRP1 in human, Drosophila, and yeast cells (Kelley et al., 1999; Simic et al., 2003). Hrp1, the Schizosaccharomyces pombe orthologue of CHD1, is involved in the maintenance of the level of SpCENP-A at centromeres (Walfridsson et al., 2005). Thus, it is possible that CHD1 is involved in CENP-A deposition into centromeres in concert with FACT and the CENP-H–containing complex.
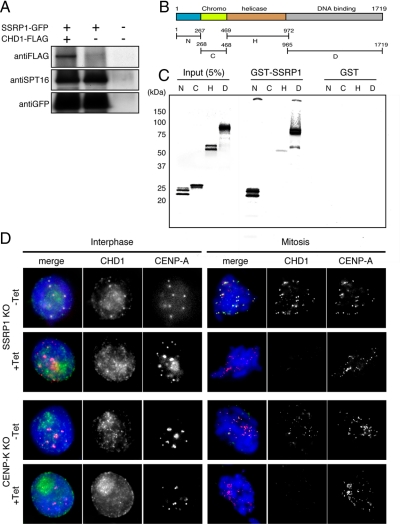
As a first step to examine this possibility, we investigated whether chicken CHD1 physically interacts with FACT. SSRP1 conditional mutant cell lines that stably express SSRP1-GFP and CHD1-FLAG were established and cultured in the presence of tet. These cell lines grew continuously in the presence of tet, indicating that the GFP-tagged SSRP1 was functional. Immunoprecipitation was performed using an anti-GFP antibody, and CHD1-FLAG was detected using an anti-FLAG antibody. We found that CHD1-FLAG coimmunoprecipitated with SSRP1-GFP when both proteins were coexpressed (Figure 6A).
Figure 6.
SSRP1 is required for centromere localization of CHD1. (A) SSRP1 conditional mutant cells that stably express both SSRP1-GFP and CHD1-FLAG were established and cultured in the presence of tet. Total cell lysates were prepared and subjected to coimmunoprecipitation using an anti-GFP antibody. CHD1-FLAG and SSRP1-GFP were detected by Western blot analysis by using anti-FLAG and anti-GFP antibodies, respectively. (B) Conserved domains in chicken CHD1. The polypeptide regions containing the N-terminal region (N; 1-267 aa), the chromodomain (C; 268-468 aa), the ATPase/helicase domain (H; 469-972 aa), and the DNA binding domain (D; 965-1719 aa) were used for the in vitro binding assay shown in C. (C) GST-pull down assay. Each polypeptide was expressed and labeled in vitro and incubated with GST-SSRP1 or GST. (D) Immunostaining using an anti-CHD1 antibody in SSRP1 and CENP-K conditional mutant cells that stably expressed CENP-A-RFP to visualize centromeres. SSRP1 and CENP-K conditional mutant cells were cultured for 45 h and 48 h in the absence (−Tet) or presence (+Tet) of tet, respectively. CHD1 (green), CENP-A (red), and DAPI (blue) staining are shown.
We also performed an in vitro binding assay to assess direct protein–protein interactions between CHD1 and SSRP1. CHD1 contains several conserved domains, such as the chromodomain, the ATPase/helicase domain, and the DNA binding domain (Figure 6B). To test for direct binding to SSRP1, each domain was expressed in vitro and incubated with bacterially expressed GST-SSRP1. We found that the N-terminal region (1-267 aa) and the DNA binding domain (965-1719 aa) bound to GST-SSRP1 (Figure 6C). To eliminate the possibility that DNA may bridge the interaction between CHD1 and SSRP1, in vitro binding assay was repeated in the presence of Benzonase nuclease to degrade DNA present in the reaction. We found that the N-terminal region and the DNA binding domain of CHD1 bound to GST-SSRP1 (Supplemental Figure S7A). Next, we established the SSRP1 conditional mutant cell lines that stably express SSRP1-GFP and the FLAG-tagged CHD1 fragments, and immunoprecipitation was performed using an anti-GFP antibody. We found that the N-terminal region and the DNA binding domain of CHD1 coimmunoprecipitated with SSRP1-GFP (Supplemental Figure S7B). We then determined the region of SSRP1 that is required for binding of CHD1. The N-terminal region and the DNA binding domain of CHD1 were expressed and labeled in vitro, and incubated with the GST-fused polypeptides of SSRP1 containing N-terminal region (1-193 aa), PH domain (194-544 aa), HMG domain (545-613 aa), and C-terminal region (614-706 aa). We found that the N-terminal region of CHD bound to the PH domain-containing region of SSRP1 and that the DNA binding domain of CHD1 bound to the PH domain-containing region and the C-terminal region of SSRP1 (Supplemental Figure S7C). These results indicate that chicken CHD1 can bind directly to SSRP1.
The biochemical association of CHD1 with SSRP1 suggests that CHD1 might function at centromeres together with FACT. Therefore, we analyzed whether CHD1 localizes to centromeres. Immunostaining was performed using anti-CHD1 and anti-CENP-A antibodies in SSRP1 conditional mutant cells. The cells were cultured in the absence or presence of tet for 45 h, treated with 0.1% Triton X-100 to eliminate soluble proteins, and then fixed with paraformaldehyde. We found that CHD1 localized to centromeres at interphase and mitosis in cells cultured in the absence of tet. However, CHD1 disappeared from centromeres under SSRP1 depleted conditions (Figure 6D). These results indicate that FACT is required for the centromeric localization of CHD1.
As is shown in Figure 2, the CENP-H–containing complex is required for the centromeric localization of FACT. To confirm that the CENP-H–containing complex is also required for the centromeric localization of CHD1, we performed immunostaining of CHD1 in CENP-K and CENP-C conditional mutant cells that stably expressed CENP-A-RFP as a centromeric marker. When these cells were cultured in the presence of tet, the centromeric localization of CHD1 was abolished in CENP-K conditional mutant cells (Figure 6D), whereas the depletion of CENP-C did not affect the centromeric localization of CHD1 (Supplemental Figure S8). These results suggest that the CENP-H–containing complex is required for centromeric targeting of both CHD1 and FACT.
CHD1 Is Required for Centromeric Localization of CENP-A
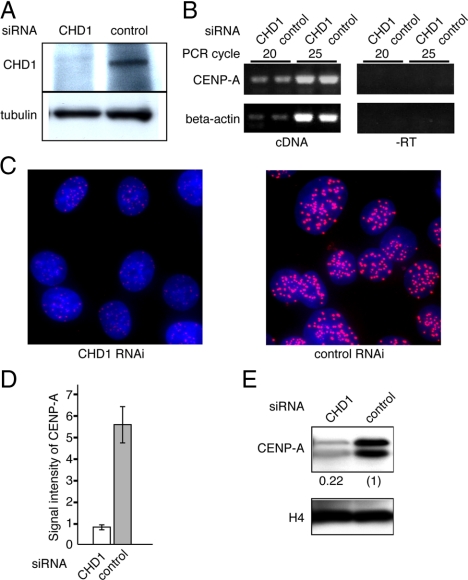
To determine whether CHD1 is involved in the centromeric localization of CENP-A, RNA interference (RNAi) knockdown was performed using HeLa S3 cells. The amount of CHD1 protein was reduced to an undetectable level by 72 h after the transfection of CHD1 specific siRNAs (Figure 7A). We used RT-PCR to confirm that expression of the CENP-A gene was not affected by the CHD1 siRNA treatment (Figure 7B). Next, we examined the effect of CHD1 depletion on the level of endogenous CENP-A localization at centromeres. Cells were cultured for 72 h after the transfection of CHD1-specific siRNAs or control siRNAs, and CENP-A was visualized with a specific antibody. In this time period, cells divided approximately three times by judging from cell number counting (Supplemental Figure S9). We found that the level of endogenous CENP-A localized at centromeres was significantly reduced after the CHD1 specific siRNA treatment (Figure 7C). CENP-A signals at centromere loci (n = 100) were quantified using MetaMorph software (Molecular Devices, Sunnyvale, CA). The intensities of the CENP-A signals in CHD1-depleted cells were reduced to 15% of those in cells treated with the control siRNA (Figure 7D). The reduction in the amounts of CENP-A was further confirmed by Western blot analysis of purified chromatin from CHD1-depleted or control cells. The amount of CENP-A associated with chromatin in CHD1-depleted cells fell to 22% of that in the control cells (Figure 7E). These results suggest that CHD1 is required for incorporation of CENP-A into centromeric chromatin.
Figure 7.
CHD1 is required for centromere localization of CENP-A. (A) The amount of CHD1 at 72 h after siRNA transfection was assessed by Western blot analysis by using an anti-CHD1 antibody. Anti-tubulin antibody was used as a loading control. (B) Expression of the CENP-A gene was analyzed by RT-PCR. cDNA samples were prepared from cells 72 h after transfection with CHD1 siRNA or a control siRNA. Negative control samples without reverse transcriptase are also shown (−RT). (C) Endogenous CENP-A in cells treated with CHD1 or control siRNAs for 72 h was visualized with anti-CENP-A antibody (red). DNA was counterstained with DAPI (blue). (D) Signal intensities of the CENP-A foci (n = 100) of cells prepared as described in C. (E) The amount of chromatin bound CENP-A in cells treated with CHD1 or control siRNAs for 72 h. Chromatin was purified and subjected to Western blot analysis by using anti-CENP-A or anti-histone H4 antibodies. Relative signal intensities of CENP-A are indicated.
DISCUSSION
CENP-A–containing nucleosomes are the fundamental unit to establish centromeric chromatin, because CENP-A is required for centromeric localization of other centromere proteins. Thus, elucidation of the mechanisms by which CENP-A is specifically deposited to centromeric chromatin will help us to understand how the centromere is specified. Recent studies showed that RbAp46/48, hMis18α/β, and M18BP1/KNL2 are involved in loading of CENP-A into centromeres in vertebrates and nematodes (Hayashi et al., 2004; Fujita et al., 2007; Maddox et al., 2007). HJURP interacts directly with CENP-A and is required for centromeric deposition of CENP-A (Foltz et al., 2009; Dunleavy et al., 2009). Remodeling and spacing factor complex mediates stable association of CENP-A to the centromeric chromatin (Perpelescu et al., 2009). However, the additional molecular mechanisms for this process have not yet been determined. We previously identified the CENP-H–containing complex as a member of the CCAN in chicken and human cells and reported that the CENP-H–containing complex is required for the efficient targeting of newly synthesized CENP-A into centromeres (Okada et al., 2006). However, it was unclear how the CENP-H–containing complex is involved in the deposition of nascent CENP-A into centromeres. In this study, we demonstrate that the chromatin remodeling complex FACT and the ATP-dependent chromatin remodeling factor CHD1 are localized at centromeres in the CENP-H–containing complex dependent manner and are required for CENP-A incorporation into centromeres.
How does the CENP-H–containing complex act to incorporate CENP-A into centromeric chromatin? We propose a model for deposition of nascent CENP-A at centromeric chromatin, in which FACT and CHD1 may be directly involved in CENP-A deposition. The CENP-H–containing complex constitutively localizes to centromeres and is required for the centromeric localization of FACT and CHD1. After DNA replication, the level of pre-existing CENP-A–containing nucleosomes is halved in each sister chromatid. CHD1 deposits nascent CENP-A to centromeric chromatin and thereby replenishes the level of CENP-A in centromeric chromatin.
We have shown that newly synthesized CENP-A is not efficiently incorporated to centromeric chromatin in the SSRP1-depleted cells. As is shown in Figure 3, SSRP1 knockout cells stop cell cycle progression without arrest at any particular cell cycle stage shortly after SSRP1 depletion. Thus, we are unable to exclude the possibility that the absence of nascent CENP-A incorporation in the SSRP1-depleted cells may be caused by the arrest in cell cycle before the time window of CENP-A incorporation. However, our data reveal that FACT plays a fundamental role for nascent CENP-A incorporation to centromeric chromatin at least through recruiting CHD1 to centromeres.
We speculate that centromerically localized FACT might participate in the incorporation of CENP-A by changing chromatin structure. FACT binds to histones H2A and H2B (Orphanides et al., 1999) and destabilizes chromatin structure without using ATP hydrolysis (Ruone et al., 2003; Rhoades et al., 2004). Displacement of H2A/H2B dimers is thought to facilitate remodeling of histone H3 molecules, because H3 molecules are physically masked by H2A/H2B dimers in the complete nucleosome structure (Luger et al., 1997). Indeed, we previously showed that FACT facilitates loading of histone H3.3 to pre-existing nucleosomes adjacent to FACT binding sites (Shimojima et al., 2003; Nakayama et al., 2007). It is therefore possible that FACT may change the structure of centromeric chromatin to facilitate the replacement of canonical histone H3 molecules with histone H3 variants; this would help CHD1 to deposit CENP-A.
We believe that an alternative model may also be feasible. Histone H3-containing nucleosomes are interspersed with CENP-A nucleosomes in centromeric chromatin (Blower et al., 2002). We recently reported that the CENP-T and CENP-W proteins of the CCAN associate with H3-containing nucleosomes at the centromeric chromatin (Hori et al., 2008). Centromeric localization of CENP-T/W is upstream of the CENP-H–containing complex, suggesting a close relationship between CENP-T/W and the CENP-H–containing complex. It is therefore possible that the CENP-H–containing complex might associate with the H3-containing nucleosomes at centromeres and enhance CENP-A incorporation through its effect on chromatin structures surrounding the CENP-A–containing nucleosomes. In fission yeast, FACT is involved in the formation of centromeric heterochromatin (Lejeune et al., 2007), and centromeric heterochromatin is required for the incorporation of SpCENP-A to centromeres on naked templates (Folco et al., 2008), suggesting that maintenance of the structural integrity of centromeric chromatin is important for deposition of CENP-A into centromeres.
In vertebrate cells, the CENP-H–containing complex, FACT and CHD1 are localized at centromeres throughout the cell cycle, whereas CENP-A is incorporated into centromeres from telophase to early G1. One possible mechanism for achieving cell cycle dependent CENP-A loading is by the hMis18 complex functioning in concert with the CENP-H–containing complex. hMis18 and M18BP1/KNL2 transiently localize to centromeres in the same time window as CENP-A incorporation. CENP-A mislocalization in hMis18-depleted cells is rescued by trichostatin A, suggesting that the hMis18 complex might alter the chromatin state or modulate the activity of proteins that are involved in CENP-A deposition, such as the CENP-H–containing complex, FACT or CHD1, in a cell cycle-dependent manner.
In conclusion, we show here that the CENP-H–containing complex is required for the centromeric localization of the chromatin remodeling complex FACT and of the ATP-dependent chromatin remodeling factor CHD1. In addition, we show that these chromatin remodeling enzymes are required for the centromeric localization of CENP-A. Our data suggest that the CCAN proteins play the central role in the establishment of centromere-specific chromatin structures through interaction with chromatin remodeling enzymes.
Supplementary Material
ACKNOWLEDGMENTS
We are very grateful to T. Okamoto and K. Kita for technical assistance. We thank S. Hirose for anti-SPT16 antibody and H. Kimura for histone H3-GFP plasmid. This work was supported by grant-in-aid for scientific research (B) and grant-in-aid for young scientists (B) from the Ministry of Education, Science, Sports and Culture of Japan. This work was partially supported by the Asahi Glass Foundation.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-01-0065) on July 22, 2009.
REFERENCES
- Black B. E., Bassett E. A. The histone variant CENP-A and centromere specification. Curr. Opin. Cell Biol. 2008;20:91–100. doi: 10.1016/j.ceb.2007.11.007. [DOI] [PubMed] [Google Scholar]
- Blower M. D., Sullivan B. A., Karpen G. H. Conserved organization of centromeric chromatin in flies and humans. Dev. Cell. 2002;2:319–330. doi: 10.1016/s1534-5807(02)00135-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I. M., Desai A. Molecular architecture of the kinetochore-microtubule interface. Nat. Rev. Mol. Cell Biol. 2008;9:33–46. doi: 10.1038/nrm2310. [DOI] [PubMed] [Google Scholar]
- Dunleavy E. M., Roche D., Tagami H., Lacoste N., Ray-Gallet D., Nakamura Y., Daigo Y., Nakatani Y., Almouzni-Pettinitti G. HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell. 2009;137:485–497. doi: 10.1016/j.cell.2009.02.040. [DOI] [PubMed] [Google Scholar]
- Folco H. D., Pidoux A. L., Urano T., Allshire R. C. Heterochromatin and RNAi are required to establish CENP-A chromatin at centromeres. Science. 2008;319:94–97. doi: 10.1126/science.1150944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltz D. R., Jansen L. E., Black B. E., Bailey A. O., Yates J. R., 3rd, Cleveland D. W. The human CENP-A centromeric nucleosome-associated complex. Nat. Cell Biol. 2006;8:458–469. doi: 10.1038/ncb1397. [DOI] [PubMed] [Google Scholar]
- Foltz D. R., Jansen L.E.T., Bailey A. O., Yates J. R., 3rd, Bassett E. A., Wood S., Black B. E., Cleveland D. W. Centromere-specific assembly of CENP-A nucleosomes is mediated by HJURP. Cell. 2009;137:472–484. doi: 10.1016/j.cell.2009.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita Y., Hayashi T., Kiyomitsu T., Toyoda Y., Kokubu A., Obuse C., Yanagida M. Priming of centromere for CENP-A recruitment by human hMis18alpha, hMis18beta, and M18BP1. Dev. Cell. 2007;12:17–30. doi: 10.1016/j.devcel.2006.11.002. [DOI] [PubMed] [Google Scholar]
- Fukagawa T., Mikami Y., Nishihashi A., Regnier V., Haraguchi T., Hiraoka Y., Sugata N., Todokoro K., Brown W., Ikemura T. CENP-H, a constitutive centromere component, is required for centromere targeting of CENP-C in vertebrate cells. EMBO J. 2001;20:4603–4617. doi: 10.1093/emboj/20.16.4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G., Kiyomitsu T., Yoda K., Yanagida M. Human centromere chromatin protein hMis12, essential for equal segregation, is independent of CENP-A loading pathway. J. Cell Biol. 2003;160:25–39. doi: 10.1083/jcb.200210005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T., Fujita Y., Iwasaki O., Adachi Y., Takahashi K., Yanagida M. Mis16 and Mis18 are required for CENP-A loading and histone deacetylation at centromeres. Cell. 2004;118:715–729. doi: 10.1016/j.cell.2004.09.002. [DOI] [PubMed] [Google Scholar]
- Heo K., Kim H., Choi S. H., Choi J., Kim K., Gu J., Lieber M. R., Yang A. S., An W. FACT-mediated exchange of histone variant H2AX regulated by phosphorylation of H2AX and ADP-ribosylation of Spt16. Mol. Cell. 2008;30:86–97. doi: 10.1016/j.molcel.2008.02.029. [DOI] [PubMed] [Google Scholar]
- Heun P., Erhardt S., Blower M. D., Weiss S., Skora A. D., Karpen G. H. Mislocalization of the Drosophila centromere-specific histone CID promotes formation of functional ectopic kinetochores. Dev. Cell. 2006;10:303–315. doi: 10.1016/j.devcel.2006.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori T., et al. CCAN makes multiple contacts with centromeric DNA to provide distinct pathways to the outer kinetochore. Cell. 2008;135:1039–1052. doi: 10.1016/j.cell.2008.10.019. [DOI] [PubMed] [Google Scholar]
- Howman E. V., Fowler K. J., Newson A. J., Redward S., MacDonald A. C., Kalitsis P., Choo K. H. Early disruption of centromeric chromatin organization in centromere protein A (Cenpa) null mice. Proc. Natl. Acad. Sci. USA. 2000;97:1148–1153. doi: 10.1073/pnas.97.3.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen L.E.T., Black B. E., Foltz D. R., Cleveland D. W. Propagation of centromeric chromatin requires exit from mitosis. J. Cell Biol. 2007;176:795–805. doi: 10.1083/jcb.200701066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley D. E., Stokes D. G., Perry R. P. CHD1 interacts with SSRP1 and depends on both its chromodomain and its ATPase/helicase-like domain for proper association with chromatin. Chromosoma. 1999;108:10–25. doi: 10.1007/s004120050347. [DOI] [PubMed] [Google Scholar]
- Kwon M. S., Hori T., Okada M., Fukagawa T. CENP-C is involved in chromosome segregation, mitotic checkpoint function, and kinetochore assembly. Mol. Biol. Cell. 2007;18:2155–2168. doi: 10.1091/mbc.E07-01-0045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejeune E., Bortfeld M., White S. A., Pidoux A. L., Ekwall K., Allshire R. C., Ladurner A. G. The chromatin-remodeling factor FACT contributes to centromeric heterochromatin independently of RNAi. Curr. Biol. 2007;17:1219–1224. doi: 10.1016/j.cub.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S. T., Rattner J. B., Jablonski S. A., Yen T. J. Mapping the assembly pathways that specify formation of the trilaminar kinetochore plates in human cells. J. Cell Biol. 2006;175:41–53. doi: 10.1083/jcb.200606020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K., Mader A. W., Richmond R. K., Sargent D. F., Richmond T. J. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Maddox P. S., Hyndman F., Monen J., Oegema K., Desai A. Functional genomics identifies a Myb domain-containing protein family required for assembly of CENP-A chromatin. J. Cell Biol. 2007;176:757–763. doi: 10.1083/jcb.200701065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall O. J., Chueh A. C., Wong L. H., Choo K. H. Neocentromeres: new insights into centromere structure, disease development, and karyotype evolution. Am. J. Hum. Genet. 2008;82:261–282. doi: 10.1016/j.ajhg.2007.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama T., Nishioka K., Dong Y. X., Shimojima T., Hirose S. Drosophila GAGA factor directs histone H3.3 replacement that prevents the heterochromatin spreading. Genes Dev. 2007;21:552–561. doi: 10.1101/gad.1503407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishihashi A., Haraguchi T., Hiraoka Y., Ikemura T., Regnier V., Dodson H., Earnshaw W. C., Fukagawa T. CENP-I is essential for centromere function in vertebrate cells. Dev. Cell. 2002;2:463–476. doi: 10.1016/s1534-5807(02)00144-2. [DOI] [PubMed] [Google Scholar]
- Obuse C., Yang H., Nozaki N., Goto S., Okazaki T., Yoda K. Proteomics analysis of the centromere complex from HeLa interphase cells: UV-damaged DNA binding protein 1 (DDB-1) is a component of the CEN-complex, while BMI-1 is transiently co-localized with the centromeric region in interphase. Genes Cells. 2004;9:105–120. doi: 10.1111/j.1365-2443.2004.00705.x. [DOI] [PubMed] [Google Scholar]
- Oegema K., Desai A., Rybina S., Kirkham M., Hyman A. A. Functional analysis of kinetochore assembly in Caenorhabditis elegans. J. Cell Biol. 2001;153:1209–1226. doi: 10.1083/jcb.153.6.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada M., Cheeseman I. M., Hori T., Okawa K., McLeod I. X., Yates J. R., 3rd, Desai A., Fukagawa T. The CENP-H-I complex is required for the efficient incorporation of newly synthesized CENP-A into centromeres. Nat. Cell Biol. 2006;8:446–457. doi: 10.1038/ncb1396. [DOI] [PubMed] [Google Scholar]
- Orphanides G., LeRoy G., Chang C. H., Luse D. S., Reinberg D. FACT, a factor that facilitates transcript elongation through nucleosomes. Cell. 1998;92:105–116. doi: 10.1016/s0092-8674(00)80903-4. [DOI] [PubMed] [Google Scholar]
- Orphanides G., Wu W. H., Lane W. S., Hampsey M., Reinberg D. The chromatin-specific transcription elongation factor FACT comprises human SPT16 and SSRP1 proteins. Nature. 1999;400:284–288. doi: 10.1038/22350. [DOI] [PubMed] [Google Scholar]
- Perpelescu M., Nozaki N., Obuse C., Yang H., Yoda K. Active establishment of centromeric CENP-A chromatin by RSF complex. J. Cell Biol. 2009;185:397–407. doi: 10.1083/jcb.200903088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regnier V., Vagnarelli P., Fukagawa T., Zerjal T., Burns E., Trouche D., Earnshaw W., Brown W. CENP-A is required for accurate chromosome segregation and sustained kinetochore association of BubR1. Mol. Cell Biol. 2005;25:3967–3981. doi: 10.1128/MCB.25.10.3967-3981.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinberg D., Sims R. J., 3rd de FACTo nucleosome dynamics. J. Biol. Chem. 2006;281:23297–23301. doi: 10.1074/jbc.R600007200. [DOI] [PubMed] [Google Scholar]
- Rhoades A. R., Ruone S., Formosa T. Structural features of nucleosomes reorganized by yeast FACT and its HMG box component, Nhp6. Mol. Cell Biol. 2004;24:3907–3917. doi: 10.1128/MCB.24.9.3907-3917.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruone S., Rhoades A. R., Formosa T. Multiple Nhp6 molecules are required to recruit Spt16-Pob3 to form yFACT complexes and to reorganize nucleosomes. J. Biol. Chem. 2003;278:45288–45295. doi: 10.1074/jbc.M307291200. [DOI] [PubMed] [Google Scholar]
- Shimojima T., Okada M., Nakayama T., Ueda H., Okawa K., Iwamatsu A., Handa H., Hirose S. Drosophila FACT contributes to Hox gene expression through physical and functional interactions with GAGA factor. Genes Dev. 2003;17:1605–1616. doi: 10.1101/gad.1086803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simic R., Lindstrom D. L., Tran H. G., Roinick K. L., Costa P. J., Johnson A. D., Hartzog G. A., Arndt K. M. Chromatin remodeling protein Chd1 interacts with transcription elongation factors and localizes to transcribed genes. EMBO J. 2003;22:1846–1856. doi: 10.1093/emboj/cdg179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hooser A. A., Ouspenski I. I., Gregson H. C., Starr D. A., Yen T. J., Goldberg M. L., Yokomori K., Earnshaw W. C., Sullivan K. F., Brinkley B. R. Specification of kinetochore-forming chromatin by the histone H3 variant CENP-A. J. Cell Sci. 2001;114:3529–3542. doi: 10.1242/jcs.114.19.3529. [DOI] [PubMed] [Google Scholar]
- Walfridsson J., Bjerling P., Thalen M., Yoo E. J., Park S. D., Ekwall K. The CHD remodeling factor Hrp1 stimulates CENP-A loading to centromeres. Nucleic Acids Res. 2005;33:2868–2879. doi: 10.1093/nar/gki579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburton P. E., et al. Immunolocalization of CENP-A suggests a distinct nucleosome structure at the inner kinetochore plate of active centromeres. Curr. Biol. 1997;7:901–904. doi: 10.1016/s0960-9822(06)00382-4. [DOI] [PubMed] [Google Scholar]
- Zhou Y., Wang T. S. A coordinated temporal interplay of nucleosome reorganization factor, sister chromatin cohesion factor, and DNA polymerase alpha facilitates DNA replication. Mol. Cell Biol. 2004;24:9568–9579. doi: 10.1128/MCB.24.21.9568-9579.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.