Abstract
The nuclear envelope of higher eukaryotic cells reforms at the exit from mitosis, in concert with the assembly of nuclear pore complexes (NPCs). The first step in postmitotic NPC assembly involves the “seeding” of chromatin with ELYS and the Nup107-160 complex. Subsequent steps in the assembly process are poorly understood and different mechanistic models have been proposed to explain the formation of the full supramolecular structure. Here, we show that the initial step of chromatin seeding is negatively regulated by importin β. Direct imaging of the chromatin attachment sites reveals single sites situated predominantly on the highest substructures of chromatin surface and lacking any sign of annular structures or oligomerized pre-NPCs. Surprisingly, the inhibition by importin β is only partially reversed by RanGTP. Importin β forms a high-molecular-weight complex with both ELYS and the Nup107-160 complex in cytosol. We suggest that initiation sites for NPC assembly contain single copies of chromatin-bound ELYS/Nup107-160 and that the lateral oligomerization of these subunits depends on the recruitment of membrane components. We predict that additional regulators, besides importin β and Ran, may be involved in coordinating the initial seeding of chromatin with subsequent steps in the NPC assembly pathway.
INTRODUCTION
In multicellular eukaryotes that undergo an open mitosis, the nuclear envelope reforms during every cell cycle. Postmitotic nuclear assembly involves the coordinated formation of the double nuclear membranes and the massive proteinaceous assemblies of nuclear pore complexes (NPCs; Gerace and Burke, 1988; Burke and Ellenberg, 2002; Hetzer et al., 2005; Antonin et al., 2008). NPCs, embedded at the junction of the two nuclear membranes, mediate bidirectional transport of macromolecules across the nuclear envelope and are composed of multiple copies of ∼30 different nucleoporins (Vasu et al., 2001; Weis, 2003; Fahrenkrog et al., 2004; Tran and Wente, 2006; Alber et al., 2007). NPC assembly is thought to be a tightly regulated, stepwise process, which is coordinated with mitotic exit events, such as chromosome decondensation (Burke and Ellenberg, 2002; Wozniak and Clarke, 2003; Anderson and Hetzer, 2008). NPCs also form during interphase, in metazoan cells, by insertion into the intact nuclear envelope (Maul et al., 1972; D'Angelo et al., 2006). This second type of NPC assembly more closely resembles the situation in many fungi, which undergo a closed mitosis, preserving an intact nuclear envelope throughout the cell cycle (Ryan et al., 2007).
Two major mechanistic models for NPC assembly have been proposed (Macaulay and Forbes, 1996; Harel et al., 2003a; Walther et al., 2003a; reviewed in Wozniak and Clarke, 2003). These models differ in the role and time-of-entry ascribed to membranes and in the mechanistic link drawn between postmitotic and interphase NPC assembly. The unified model of assembly suggests that a similar sequence of events occurs on the surface of decondensing chromosomes at telophase and during NPC assembly in interphase. In this model, NPCs form within patches of double nuclear membranes after a critical fusion event between the inner and outer membranes. Most of the soluble subunits are integrated into the forming structure after the formation of an initial “pore,” or aqueous channel, by this fusion event (Macaulay and Forbes, 1996; Harel et al., 2003a). By contrast, the “prepore” model suggests a different sequence of events, which is unique to postmitotic NPC assembly. In this second model, soluble nucleoporin subunits are recruited to the surface of chromatin, where they are able to oligomerize into an annular prepore (or pre-NPCs) structure, in the absence of membranes. Additional soluble subunits are sequentially recruited to this structure, with the membrane components added at a late stage and sealing around the mature NPC (Walther et al., 2003a; Hetzer et al., 2005). Much of the difference between these two mechanistic models boils down to the definition of what constitutes a “pre-NPC” and at which point is the contribution of membrane components required (Wozniak and Clarke, 2003; Antonin et al., 2008). Because mature NPCs have an eightfold rotational symmetry, it is thought that individual nucleoporin subcomplexes are present within the structure in multiples of eight copies (Cronshaw et al., 2002; Beck et al., 2004; Schwartz, 2005). The oligomerization of individual subunits to form the structural scaffold of the NPC has been suggested to be a self assembly process (Hetzer et al., 2005; Hsia et al., 2007; Brohawn et al., 2008). The complex interactions between scaffold and peripheral nucleoporins are still poorly understood, although some attempts to decipher and model these interactions have been made for the yeast NPC (Allen et al., 2002; Shulga and Goldfarb, 2003; Alber et al., 2007).
Much of the mechanistic evidence for NPC assembly models has been derived from cell-free reconstitution assays, such as the Xenopus egg extract system. These in vitro assays mimic the early events after fertilization and facilitate the biochemical dissection of the complex nuclear assembly process (Forbes et al., 1983; Lohka and Masui, 1983; Newport, 1987; Harel et al., 2003a; Walther et al., 2003b). The sequential recruitment of individual nucleoporins to the nuclear periphery has been extensively documented in cultured mammalian cells (Bodoor et al., 1999; Rabut et al., 2004; Dultz et al., 2008) and has been interpreted as support for the prepore model of assembly. A few scattered electron microscopy reports have indicated ring-like structures on the surface of chromatin, which were claimed to have been formed in the absence of membranes (Maul, 1977; Sheehan et al., 1988; Drummond et al., 2006). However, in the most recent report of this kind, the use of detergent (which would have removed membranes during the preparation protocol) prevents any mechanistic conclusions from being drawn (Drummond et al., 2006). A finding that is consistent with both models of NPC assembly is that the large Nup107-160 nucleoporin subcomplex is recruited at a very early stage to the surface of chromatin (Belgareh et al., 2001; Vasu et al., 2001; Harel et al., 2003b; Walther et al., 2003a). This nine-member protein complex is a critical subunit, which also needs to be incorporated from the outer side of the nuclear envelope during interphase NPC assembly (D'Angelo et al., 2006). Thus, the essential role of the Nup107-160 complex can be interpreted as being the first building block of a prepore on chromatin or as an essential component needed on both sides of double membrane patches to initiate NPC assembly.
A new player in the early stages of NPC assembly has recently been identified as the large vertebrate protein ELYS, or the homologous MEL-28 in Caenorhabditis elegans (Rasala et al., 2006; Franz et al., 2007; Gillespie et al., 2007). ELYS contains an AT-hook DNA-binding motif and was initially thought to be a transcription factor (Kimura et al., 2002). Subsequent work demonstrated that ELYS acts as an essential adaptor between the Nup107-160 complex and chromatin and targets NPC assembly to the nuclear periphery at the exit from mitosis. The results of RNAi-mediated depletion or genetic mutations of ELYS/MEL-28 in mammalian cells and in C. elegans are both consistent with this essential role at an early stage of NPC assembly (Fernandez and Piano, 2006; Rasala et al., 2006; Franz et al., 2007). A recessive lethal mutation in the zebrafish ortholog of ELYS results in the flotte lotte (flo) phenotype. Early flo embryos survive thanks to a maternal pool of the protein, but subsequently die because of cell cycle arrest and apoptosis in proliferative tissues (Davuluri et al., 2008). The most recent mechanistic insight on NPC assembly has been obtained from the Xenopus reconstitution system. A comprehensive analysis of nucleoporin subcomplexes and their role in nuclear reconstitution demonstrated that only ELYS and the Nup107-160 complex are recruited to chromatin in the absence of membranes (Rasala et al., 2008). These results provide strong support for the unified model of assembly and for an early, essential role of membranes immediately after the “seeding” of chromatin with ELYS and Nup107-160.
The prototypic nuclear import receptor importin β is a master regulator of mitotic and interphase events. Importin β has been shown to negatively regulate multiple steps in postmitotic nuclear envelope and NPC assembly, as well as NPC assembly during interphase (reviewed in Harel and Forbes, 2004; Ryan et al., 2007; D'Angelo and Hetzer, 2008). The inhibitory effect of importin β has been suggested to arise from its binding to multiple nucleoporins and the suppression of their interactions (Harel et al., 2003a; Walther et al., 2003b; Hetzer et al., 2005; Delmar et al., 2008). A recent review has hypothesized that importin β may also sequester ELYS in a separate inactive form (D'Angelo and Hetzer, 2008).
Here, we investigate the nature of the seeding sites on the surface of chromatin and redefine the requirements for creating a prepore or initiation site for NPC assembly. We find a negative regulatory role for importin β in this early step in the assembly process. We show that rather than dissociating ELYS from the Nup107-160 complex, importin β forms a high-molecular-weight complex with both of these components in cytosol.
MATERIALS AND METHODS
Antibodies
Commercially obtained antibodies included monoclonal anti-importin β (Sigma-Aldrich, St. Louis, MO), rabbit anti-histone H3 (#9715, Cell Signaling, Beverly, MA) and rabbit IgG (Calbiochem, La Jolla, CA). Affinity purification of polyclonal rabbit antibodies was performed as described by Shah et al. (1998). Previously described antibodies included anti-hNup133, anti-hNup160, anti-mNup85 (Harel et al., 2003b); anti-XNup43 (Orjalo et al., 2006); and anti-Transportin and anti-CRM 1 (Shah et al., 1998). Anti-Nup107 polyclonal antibodies were a gift from Ulrike Kutay (ETH Zurich, Zurich, Switzerland), generated against aa 3–73 of Xenopus laevis Nup107 and affinity-purified on the antigen coupled to Affi-Gel 10 (Bio-Rad, Richmond, CA). Affinity-purified anti-Exportin-t was generated against aa 1–411 of the human protein and cross-reacts with the Xenopus homolog (Harel and Forbes, unpublished data). Polyclonal antibodies against Xenopus laevis ELYS aa 1820–1864 were generated for this study, affinity-purified, and used for immunoprecipitation, immunofluorescence, immunoelectron microscopy, and immunoblot analysis. A second anti-XELYS antibody, directed against the extreme C-terminus of the protein (Rasala et al., 2008) was used to confirm the immunoblot results shown for ELYS.
Recombinant Protein Expression and Purification
The coding sequence for Xenopus ELYS aa 1820–1864 (LOC397707) was inserted into pET28A and expressed as a soluble hexahistidine-T7–tagged protein in the Escherichia coli strain BL21 (DE3) Rosetta. The purified protein was used to immunize two rabbits, and antisera were first passed over a 6xHis-T7-GFP column to deplete antibodies against the tags, before affinity purification on the immobilized protein (Shah et al., 1998). Histidine-tagged proteins were purified on Ni-NTA resin (Qiagen, Cahtsworth, CA) according to standard procedures. The use of histidine-tagged human importin β and RanQ69L has been described (Harel et al., 2003a). To purify untagged Xenopus importin β, the pGEX6P-Xbfl clone (a gift from Rene Chan and Douglass Forbes, University of California, San Diego, La Jolla, CA) was expressed, purified, and cleaved by PreScission protease (GE Healthcare, Waukesha, WI) as previously described (Delmar et al., 2008). Expression, purification, and loading of RanQ69L with GTP by the EDTA method were performed as in Kutay et al. (1997) and Orjalo et al. (2006). The efficiency of the nucleotide-exchange reaction was monitored by reverse-phase HPLC on a C-18 column (Supelco, Bellefonte, PA), run isocratically in 100 mM KH2PO4/K2HPO4, pH 6.5, 10 mM tetrabutylammonium bromide, and 8.5% acetonitrile (Smith and Rittinger, 2002). Protein preparations were extensively dialyzed and concentrated to a similar extent on Amicon Ultra-4 microconcentrators (Millipore, Bedford, MA), and samples of the last filtrates were run against blank samples by HPLC, to control for any loosely bound nucleotides released into solution. Samples containing 1–2 nmol of protein were withdrawn for analysis, proteins were denatured and removed by centrifugation, and the supernatant was loaded on the HPLC column. Calibration was with known nucleotide standards, and absorbance was measured at 254 nm. The zzRanQ69L clone was a gift from Dirk Görlich (Max Planck Institute for Biophysical Chemistry, Göttingen, Germany) and was expressed and immobilized on IgG Sepharose as previously described (Kutay et al., 1997).
Anchored Chromatin Assay for Immunofluorescence
Xenopus egg cytosol, crude nucleoplasmin, and demembranated sperm chromatin were prepared as previously described (Macaulay and Forbes, 1996; Harel et al., 2003b; Rasala et al., 2008). Chromatin was decondensed and allowed to settle onto poly-lysine–coated coverslips essentially as described (Rasala et al., 2008), at a final concentration of 1500 sperm units/μl in 1× ELB (10 mM HEPES, pH 7.6, 50 mM KCl, and 2.5 mM MgCl2). Each chromatin unit is derived from one sperm head. Tethered chromatin templates were washed once in 1× ELB and blocked in 5% BSA-ELB for 20 min. Egg cytosol was diluted in an equal volume of 1× ELBS (10 mM HEPES, pH 7.6, 250 mM sucrose, 50 mM KCl, and 2.5 mM MgCl2) and centrifuged at 14,000 × g for 20 min to remove residual membranes. The resulting cytosol was designated as membrane-free by probing for ribophorin, as in Rasala et al. (2008). Recombinant proteins (total volume of addition not exceeding 20% of the reaction) were preincubated for 15 min in membrane-free cytosol supplemented with an ATP-regenerating system and 5 μg/ml nocodazole. Reaction mixtures (30 μl) were added to the chromatin-coated coverslips and incubated for 30 min at room temperature in a humidified chamber. The coverslips were washed three times with 1× ELBK (10 mM HEPES, pH 7.6, 100 mM KCl, and 2.5 mM MgCl2), fixed in 4% formaldehyde (Electron Microscopy Sciences, Fort Washington, PA) in 1× ELB, and processed for immunofluorescence microscopy. Chromatin was stained with Hoechst 33258 (Sigma-Aldrich). Affinity-purified anti-ELYS and anti-Nup107 were each used at a dilution of 1:300 (final concentration: 6–7 μg/ml), and TRITC goat anti-rabbit (Jackson ImmunoResearch, West Grove, PA) was used at 1:200. Images were acquired on an Olympus BX61TRF motorized microscope (Melville, NY), equipped with a DP70 digital camera. Quantitative analysis of anti-ELYS immunofluorescent staining was performed with custom written software (OpenView; Tsuriel et al., 2006) on images captured under identical settings from 12 randomly chosen, nonoverlapping chromatin templates in each category. The chromatin surface area was first delineated from Hoechst 33258 staining of the same fields of view. To that end, a threshold was set resulting in images composed of only suprathreshold pixels representing bright Hoechst 33258 staining. These images were then used as masks to define the relevant pixels (i.e., pixels located within chromatin regions) in images of antibody-labeled chromatin. Fluorescence intensities of all such pixels were averaged for each chromatin template and for each condition. Nonspecific staining was measured on coverslips in which the primary antibody was omitted. Normalized fluorescence intensity was calculated after the subtraction of nonspecific staining (∼6% on average) and compiled from different experiments.
Anchored Chromatin Assay for Immunoblotting
Chromatin templates were prepared as detailed above and allowed to settle onto coverslips at a final concentration of 2500 sperm units/μl. Chromatin-coated coverslips were washed, blocked, and incubated with reaction mixtures (70 μl). Recombinant proteins were preincubated for 15 min in membrane-free cytosol, except for reactions in which the order of addition was changed. Chromatin binding was carried out at room temperature for 30 min, followed by three washes in 1× ELBK to remove unbound proteins. Chromatin-bound proteins were solubilized in SDS-PAGE sample loading buffer and subjected to immunoblot analysis.
Field Emission Scanning Electron Microscopy
To facilitate the visualization of chromatin binding reactions by field emission scanning electron microscopy (FESEM), chromatin templates were attached to silicon chips (Ted Pella, Irvine, CA). Silicon chips were pretreated with 0.2 mg/ml poly-lysine for 15 min and washed with H2O. Decondensed chromatin was allowed to settle by gravity on the silicon chips, washed, blocked, and prepared for binding as detailed above for the immunofluorescence assay. Tethered chromatin templates were incubated with membrane-free cytosol for 30 min in a humidified chamber at room temperature. The silicon chips were then moved to 24-well plates, washed three times in 1× ELBK, and fixed in 3.7% formaldehyde, 0.2% glutaraldehyde (Electron Microscopy Sciences) in buffer E (80 mM PIPES, pH 6.8, 150 mM sucrose, and 1 mM MgCl2) for 30 min at room temperature. Subsequent preparation steps were essentially as in Allen et al. (2007), with postfixation in aqueous 0.5% osmium tetroxide, critical-point drying performed on a CPD030 apparatus (Bal-Tec AG, Liechtenstein) and sputter coating with 2 nm of chromium (Emitech, London, England; K575X). For immunogold labeling, the optimal dilution for each primary antibody was determined based on initial trials by indirect immunofluorescence and subsequent adjustment for the FESEM protocol. Dilution of the secondary gold-conjugated probes was optimized for minimal background staining by comparison to reactions in which the primary antibody was omitted. The number of gold particles counted on identical chromatin surface areas of such control reactions was <0.5% for protein A-gold (15 nm; Department of Cell Biology, Utrecht School of Medicine; used with anti-Nup107) and ∼3% for 12-nm gold-conjugated goat anti-rabbit (Jackson ImmunoResearch; used with anti-ELYS, anti-Nup133, and anti-H3). Samples were examined using a Zeiss Ultra 55 or Ultra plus field emission scanning electron microscope (Thornwood, NY).
Immunodepletion, Solution Binding, and Functional Assays
For the immunodepletion of ELYS from membrane-free cytosol, 150 μg of anti-ELYS or preimmune IgG were bound to 40 μl of protein A-Sepharose beads (GE Healthcare) and used for two rounds of depletion as previously described (Harel et al., 2003b). To follow chromatin seeding in solution, untethered chromatin templates were incubated for 30 min in membrane-free cytosol and probed by direct immunofluorescence, essentially as in Rasala et al. (2008), using anti-ELYS labeled by Oregon Green-488 succinimidyl ester (Molecular Probes, Eugene, OR). To verify that RanQ69L-GTP was functional, NLS-BSA import and GST-IBB (glutathione S-transferase–importin β–binding domain of importin α) pulldown assays were performed. Briefly, nuclei were reconstituted from Xenopus egg extract and the import of TRITC-NLS-BSA was assayed as in Harel et al. (2003b), with RanQ69L-GTP added into the reaction 10 min before the import substrate. GST-IBB (a gift from Matt Michael, Harvard University, Cambridge, MA) was expressed in bacteria, bound to glutathione-Sepharose 4B beads (GE Healthcare), and incubated in egg extract with or without the addition of 5 μM RanQ69L-GTP. The beads were washed once, and bound proteins were eluted directly in SDS-PAGE sample loading buffer.
Subfractionation of Xenopus Egg Cytosol
The crude soluble fraction of Xenopus egg extract (typically 10–25 ml starting material) was diluted in two volumes of buffer A (25 mM Tris, pH 7.4, 10 mM MgCl2, 10% glycerol, and 1 mM DTT) and centrifuged for 1 h at 270,000 × g to remove membranes and large aggregates. The supernatant was filtered through a 0.45-μm filter (Schleicher & Schuell, Keene, NH), loaded on a 28-ml DEAE Affi-Gel Blue column (Bio-Rad), and eluted with 150 mM NaCl in buffer A. The resulting material was diluted in buffer A and loaded on a 16-ml Q-Sepharose FF anion-exchange column (GE Healthcare). Proteins were resolved on a 300-ml linear gradient of 0.1–1 M NaCl in buffer A. For functional analysis in the chromatin-binding assay or for further purification by gel filtration chromatography, specific fractions were pooled, desalted, and concentrated by use of Amicon Ultra-4 microconcentrators (10,000 MWCO, Millipore). Gel filtration chromatography was performed on a Superose-6 column (GE Healthcare), and 0.5-ml fractions were collected and precipitated with TCA for immunoblot analysis or pooled for immunoprecipitation. Immunoprecipitation out of specific enriched fractions was performed essentially as described by Shah et al. (1998), including coupling of the antibodies to protein A-Sepharose (GE Healthcare) by dimethylpimelimidate (Sigma-Aldrich). Affinity-purified antibodies or microgram equivalent amounts of control rabbit IgG were used, and all dilutions and washes were in PBS. Immunoprecipitated proteins were eluted from the beads by the addition of 100 mM glycine, pH 2.5, and analyzed by immunoblotting. Affinity chromatography of endogenous ELYS/Nup107-160 on immobilized zzRanQ69L-GTP was performed on an IgG Sepharose 6 FF column (GE Healthcare), prepared as described by Kutay et al. (1997). ELYS-enriched fractions from the Q-Sepharose column were pooled, concentrated, and exchanged into buffer B (10 mM HEPES, pH 7.6, 60 mM sucrose, 50 mM KCl, and 2.5 mM MgCl2), before being loaded on the affinity column. The column was washed with four column volumes of buffer B and then sequentially eluted in buffer B containing 250 mM, 450 mM, and 1 M KCl.
RESULTS
Importin β Negatively Regulates the Binding of the Nup107-160 Complex to Chromatin
The binding of the Nup107-160 complex to chromatin, via the essential adaptor protein ELYS, has been shown to be the earliest detectible step in postmitotic NPC assembly (Rasala et al., 2006; Franz et al., 2007; Gillespie et al., 2007; Rasala et al., 2008). To test for a potential regulatory effect of importin β on this process, we utilized an in vitro nuclear reconstitution system derived from Xenopus egg extracts (Forbes et al., 1983; Lohka and Masui, 1983; Macaulay and Forbes, 1996). To focus on the earliest stages of assembly, we used a simplified assay consisting of anchored chromatin and cytosol, in the absence of membranes. ELYS and the Nup107-160 complex have been recently shown to be the only nucleoporins recruited to chromatin under these conditions (Rasala et al., 2008). Decondensed Xenopus sperm chromatin was attached to poly-lysine–coated coverslips, washed, blocked, and incubated with egg cytosol. Chromatin binding was visualized by indirect immunofluorescence using anti-ELYS and anti-Nup107 antibodies (Figure 1A, top row). Importin β is normally present in egg cytosol at a concentration of ∼3 μM (Kutay et al., 1997). The addition of 20 μM recombinant importin β resulted in a dramatic reduction in the signal of ELYS and Nup107 on the surface of chromatin. The inhibitory effect of importin β was observed both with a histidine-tagged human protein (Figure 1A, bottom row) and with an untagged Xenopus protein (see Materials and Methods and Figure 4). To probe for the presence of additional complex members, we performed Western blot analysis on samples of the chromatin-bound proteins. Antibodies directed against ELYS, Nup160, Nup133, Nup107, Nup85, and Nup43 all gave a clear signal in the chromatin-bound fraction, but were absent in reactions containing an excess of recombinant importin β (Figure 1B). Histone H3 served as a loading control (Figure 1B, bottom row), ruling out a global effect of importin β on chromatin organization. Both histidine-tagged human importin β and untagged Xenopus importin β showed this negative regulatory effect in the Western blot assay. A partial effect on the chromatin binding of ELYS was observed in both the immunofluorescence and immunoblot assays, when recombinant importin β was added at a concentration of 7–15 μM (data not shown). To ask if the negative effect of importin β was primarily due to an interaction with soluble or chromatin-bound components, we changed the order of addition in the reaction. When excess importin β was preincubated with chromatin or when it was added only after the incubation of cytosol with chromatin templates, it did not show an inhibitory effect in the binding assay (Figure 1C). Thus, importin β needs prior access to cytosol in order to block the binding of ELYS and the Nup107-160 complex to chromatin.
Figure 1.
Importin β inhibits the binding of ELYS and Nup107-160 to chromatin. Anchored chromatin-binding assays were conducted with membrane-free Xenopus egg cytosol and processed for indirect immunofluorescence (A) or immunoblot analysis (B and C), as described in Materials and Methods. (A) Cytosol was preincubated with 20 μM ovalbumin (control), or 20 μM human importin β (hImp β), before addition on to coverslips with poly-lysine–tethered chromatin templates. Chromatin binding was carried out for 30 min at room temperature. DNA was stained with Hoechst 33258, and binding was visualized by staining with affinity-purified antibodies directed against ELYS and Nup107. Scale bar, 10 μm. (B) Immunoblot analysis of the chromatin-bound fraction from reactions performed on chromatin-coated coverslips. Membrane-free cytosol was preincubated with different amounts of histidine-tagged full-length human importin β (hImp β) or untagged full-length Xenopus importin β (XImp β) and then added on to chromatin-coated coverslips. The blot was probed for the endogenous proteins: ELYS, Nup160, Nup133, Nup107, Nup85, Nup43, and histone H3. ELYS and Nup107-160 complex members were absent from reactions containing an excess of recombinant importin β. (C) The inhibitory effect of importin β occurs through interactions in cytosol. Immunoblot analysis was performed as in B, with 20 μM human importin β added to the reactions in lanes 1–3 and 20 μM ovalbumin added to the control in lane 4. In lanes 1 and 4, the recombinant proteins were preincubated with cytosol before the addition onto chromatin-coated coverslips. In lane 2, importin β was preincubated with chromatin and washed once, and cytosol was subsequently added. In lane 3, cytosol was added to the chromatin-coated coverslip, followed by a wash and a subsequent incubation with importin β. Only the preincubation of importin β in cytosol (lane 1) resulted in the inhibition of binding.
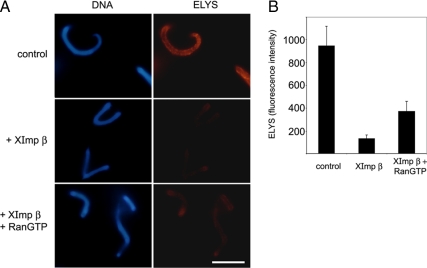
Figure 4.
RanGTP is not sufficient to counteract the negative effect of importin β. Anchored chromatin-binding assays were analyzed by indirect immunofluorescence with anti-ELYS antibody as in Figure 1. Recombinant proteins were preincubated with cytosol before the addition onto chromatin-coated coverslips. (A) Representative images from binding reactions containing cytosol supplemented with 20 μM ovalbumin (control), 20 μM Xenopus importin β (XImp β), or 20 μM Xenopus importin β + 40 μM RanQ69L-GTP (XImp β + RanGTP). Scale bar, 10 μm. (B) Quantitative analysis summarizing three separate experiments of the type shown in A. Anti-ELYS immunofluorescent signal intensity was measured exclusively from the chromatin surface on 12 randomly chosen, nonoverlapping templates in each category. Normalized fluorescence intensity is shown after the subtraction of nonspecific staining, measured on identical coverslips for which the primary antibody was omitted from the procedure. Error bar, SD.
Scanning Electron Microscopy Reveals the Distribution of Chromatin Attachment Sites
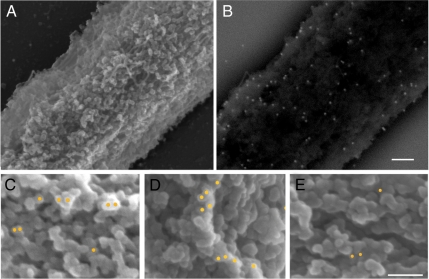
To directly visualize the attachment sites of ELYS and the Nup107-160 complex on the surface of chromatin, we used field emission scanning electron microscopy (FESEM). Chromatin templates were attached to silicon chips, incubated in egg cytosol and then fixed and processed for FESEM. Immunogold labeling was performed with antibodies directed against ELYS, Nup107, and Nup133, followed by protein A-gold or gold-conjugated secondary antibodies. Because membranes were omitted from this assembly reaction, immunolocalization can be expected to reveal the “seeding” or initiation sites for nuclear pore assembly (Franz et al., 2007; Rasala et al., 2008). The in-lens image shown in Figure 2A depicts the complex surface topology of a relatively large area of chromatin. The same area is imaged in Figure 2B through a backscatter electron detector, revealing the exact position of gold particles. The staining patterns of ELYS, Nup107, and Nup133 were identical, with gold particles quite evenly distributed over the surface of chromatin. This is the first time that the chromatin-seeding sites are imaged at high resolution. Higher magnifications (Figure 2, C and D) show that the attachment sites of ELYS/Nup107-160 occur predominantly on the highest ridges, or elevated structures, of the three-dimensional chromatin landscape. This is particularly evident from a comparison to the staining pattern of unmodified histone H3, which is prevalent in the deeper recesses and lower structures of chromatin (Figure 2E). Thus, the primary and gold-conjugated antibodies used in this assay have access to the whole surface of chromatin, but the ELYS/Nup107-160 epitopes appear to be preferentially located at the protruding or elevated structures. Quantitative analysis of the relative positions of gold particles on the chromatin topography for all four antibodies described above, is shown in Supplemental Figure S1.
Figure 2.
Direct visualization of ELYS/Nup107-160 attachment sites on chromatin by FESEM. Chromatin-coated silicon chips were incubated in egg cytosol, fixed, and processed for immunogold labeling. (A) In-lens image showing the surface topology of chromatin, and (B) the corresponding backscatter detector image revealing the position of gold particles after labeling with anti-Nup107. (C) Anti-ELYS and (D) anti-Nup133, immunolabeling with pseudocolored gold particle positions superimposed from the backscatter images. Note that the ELYS/Nup107-160 attachment sites occur predominantly on the elevated substructures of chromatin. (E) Immunogold labeling with anti-histone H3 is preferentially localized to lower substructures. Scale bars, 100 nm.
It should be noted that we saw no evidence for structures with eightfold rotational symmetry, rings, or oligomerized immunostained structures. The most closely situated gold particles appear in groups of two or three, which can be interpreted as closely positioned copies of ELYS/Nup107-160. Because we use affinity-purified polyclonal antibodies, these can also be interpreted as two antibodies binding to the same target protein. Thus, we see no indication that ELYS or the Nup107-160 complex can oligomerize on the surface of chromatin in the absence of membranes. Supplemental Figure S1C demonstrates that the inclusion of membranes in the anchored assembly reaction results in the formation of abundant NPCs, with clear rotational symmetry, embedded within a nuclear envelope. Therefore, our inability to detect ring-like structures in the membrane-free reactions is not a consequence of the attachment of chromatin to silicon chips or the fixation and visualization methods for FESEM.
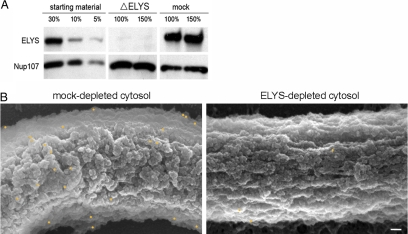
To confirm the specificity of the anti-ELYS antibody and the chromatin surface staining pattern observed by FESEM, we immunodepleted ELYS from Xenopus egg cytosol (essentially as described by Franz et al., 2007). As shown in Figure 3A, this resulted in the removal of >97% of the endogenous ELYS protein from cytosol (compare 5% starting material to 150% ΔELYS). Mock-depleted and ELYS-depleted cytosol were then incubated with anchored chromatin templates and processed for immunogold labeling and FESEM. Figure 3B demonstrates that the chromatin surface staining was drastically reduced in the reaction containing ELYS-depleted cytosol, confirming the specificity of the staining pattern observed in Figure 2, A–D, and reaffirming the essential role of ELYS as the adaptor for chromatin seeding.
Figure 3.
Immunodepletion of ELYS inhibits chromatin seeding. (A) Membrane-free cytosol was treated by two rounds of incubation with immobilized preimmune (mock depletion) or anti-ELYS antibodies. The amount of ELYS in mock-depleted (mock) and ELYS-depleted (ΔELYS) cytosol were compared by immunoblotting. Varying amounts of untreated cytosol were loaded for comparison. Nup107 was not codepleted with ELYS. (B) Mock-depleted and ELYS-depleted cytosol were incubated with chromatin-coated silicon chips and processed for FESEM with anti-ELYS and gold-conjugated secondary antibodies as in Figure 2. Pseudocolored gold particle positions were superimposed from the backscatter images. Scale bar, 100 nm.
The Inhibitory Effect of Importin β Is Partially Reversed by RanGTP
Importin β has been shown to regulate several distinct steps during mitotic progression and postmitotic nuclear assembly. In most of these regulatory circuits importin β is counteracted by RanGTP (reviewed in Harel and Forbes, 2004; Clarke and Zhang, 2008). To ask whether the inhibition of ELYS/Nup107-160 binding to chromatin could be similarly reversed, we used the RanQ69L mutant (Kutay et al., 1997). This mutant form of the Ran GTPase can be preloaded with nucleotide and remains locked in the GTP-bound form. The anchored chromatin immunofluorescence assay was used to follow ELYS/Nup107-160 binding (Figure 4A, top row) and excess recombinant importin β was added at a concentration of 20 μM to inhibit this binding (Figure 4A, middle row). When RanQ69L-GTP was added together with excess importin β, only a partial reversal of the inhibitory effect was observed (Figure 4A, bottom row). RanQ69L-GTP was unable to relieve the negative regulation by importin β, when added at equimolar concentration or up to 40 μM. Quantitative analysis of the immunofluorescence signals demonstrated that ELYS binding was reduced to ∼14% in the presence of 20 μM importin β. A twofold molar excess of RanQ69L-GTP incubated together with 20 μM importin β in cytosol was only able to restore ELYS binding to ∼39% of the control (Figure 4B).
The insensitivity of some importin β functions to reversal by RanGTP has been suggested to arise from an N-terminal histidine-tag present on the human recombinant construct used in many studies (Delmar et al., 2008). We therefore used untagged Xenopus importin β in our assay. The extent of GTP loading and the functional effect of the RanQ69L-GTP preparation used in this experiment were verified in three separate assays, described in Supplemental Figure S2. We note that both histidine-tagged human importin β and Xenopus importin β consistently produced similar inhibitory effects in all the assays presented in Figures 1 and 4. To control for possible adverse effects of chromatin tethering to a solid support, the chromatin-binding assays were repeated in solution, confirming the strong inhibition of ELYS binding by excess importin β and partial reversal by RanQ69L-GTP (Supplemental Figure S3). In conclusion, Ran in its GTP-bound form only partially reversed the negative regulation by importin β.
Different Molecular Species of the Nup107-160 Complex Exist in Egg Cytosol
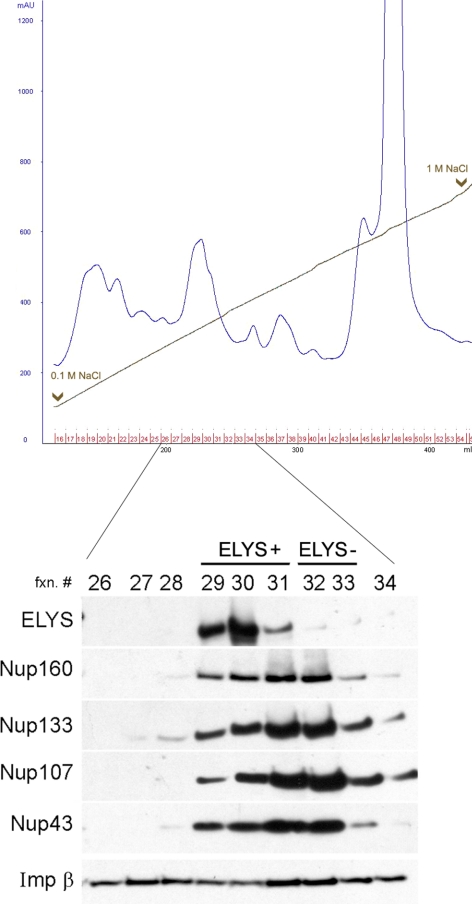
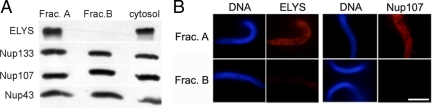
To gain a deeper understanding of the binding of Nup107-160 to chromatin and its regulation by importin β, we set out to purify the endogenous complex from Xenopus egg extract. The Xenopus Nup107-160 complex has been previously analyzed by specific pulldowns on fragments of Nup153 and Nup98, as well as immunoprecipitations (Vasu et al., 2001; Harel et al., 2003b; Walther et al., 2003a; D'Angelo et al., 2006). Here, we have attempted to subfractionate egg cytosol and follow the endogenous complex through several purification steps, in order to determine if it represents one or more molecular species. Multiple step column chromatography, starting from complete cytosol, led to a considerable enrichment and a partial purification of the nucleoporin subcomplex. Surprisingly, a simple two-column procedure resulted in a clear separation of two distinct subpopulations containing known complex members. The general elution profile of a Q-Sepharose ion exchange column is shown in Figure 5. ELYS and the Nup107-160 complex appeared together in one region of this elution profile. Fractions eluted from the column at a salt concentration of 200–230 mM NaCl were enriched in ELYS and Nup107-160 members, whereas fractions eluted between 230 and 260 mM NaCl lacked ELYS, but still contained all the complex members that we probed for (Figure 5). This elution profile is very reproducible and enabled us to test two separate forms of the complex (fraction A from the “ELYS+” region and fraction B from the “ELYS−” region) in the immunofluorescence chromatin-binding assay. In agreement with the suggested role of ELYS as an adaptor, only fraction A was able to significantly bind to chromatin, as probed by anti-ELYS and anti-Nup107 antibodies (Figure 6). Contrary to previous suggestions (Franz et al., 2007; D'Angelo and Hetzer, 2008; Rasala et al., 2008), we did not find clear evidence for a separate free pool of ELYS that might be capable of binding chromatin on its own. Our results suggest that a particular subpopulation of the endogenous Nup107-160 complex in egg extract is preassociated with ELYS and that this high-molecular-weight species is the unit that “seeds” chromatin for NPC assembly.
Figure 5.
Two distinct subpopulations of the Nup107-160 complex in egg cytosol differ in ELYS content. Xenopus egg cytosol was subfractionated on a DEAE Affi-Gel Blue column followed by a Q-Sepharose column. The second column was eluted with a linear 0.1–1 M NaCl gradient. The column elution profile is shown by OD280 absorbance (in blue). Fractions 26–34, from the region containing ELYS and Nup107-160 complex members, were analyzed by immunoblotting as shown in the bottom panel. The blot was probed with antibodies to ELYS and Nup107-160 complex members. Only fractions 29–31, eluted at 200–230 mM NaCl, contained significant amounts of ELYS, whereas Nup107-160 complex members peaked over the whole 200–260 mM NaCl range (fractions 29–34).
Figure 6.
Only a subfraction of the endogenous Nup107-160 complex in egg extract is associated with ELYS and capable of seeding chromatin. (A) Fractions of equal volume, eluted from the Q-Sepharose column at 225 mM NaCl (fraction A, enriched in ELYS) and 250 mM NaCl (fraction B, lacking ELYS) were desalted, concentrated, and analyzed by immunoblotting. The two fractions contain comparable amounts of Nup133, Nup107, and Nup43, but only fraction A contains ELYS. (B) Fractions A and B were tested in the immunofluorescence binding assay, as in Figure 1. Chromatin binding was visualized by staining with antibodies directed against ELYS and Nup107. Only fraction A was able to bind to chromatin. Scale bar, 10 μm.
The results of our chromatin-binding assays (Figure 1) indicate that the importin β exerts its inhibitory effect on the chromatin “seeding” capacity of ELYS and the Nup107-160 complex through interactions in cytosol. We asked whether endogenous importin β in cytosol could be associated with either of the two subpopulations of Nup107-160 that were detected by ion exchange chromatography. Fraction A (the ELYS+ form) and fraction B (the ELYS− form) were subjected to immunoprecipitation with affinity-purified anti-Nup107 antibodies. As shown in Figure 7, multiple Nup107-160 complex members were efficiently pulled out from both fractions. However, endogenous importin β was only found in the immunoprecipitation from the ELYS+ subpopulation. Importantly, the anti-Nup107 antibodies pulled out both ELYS and importin β from this fraction, suggesting that they may be present in a common complex.
Figure 7.
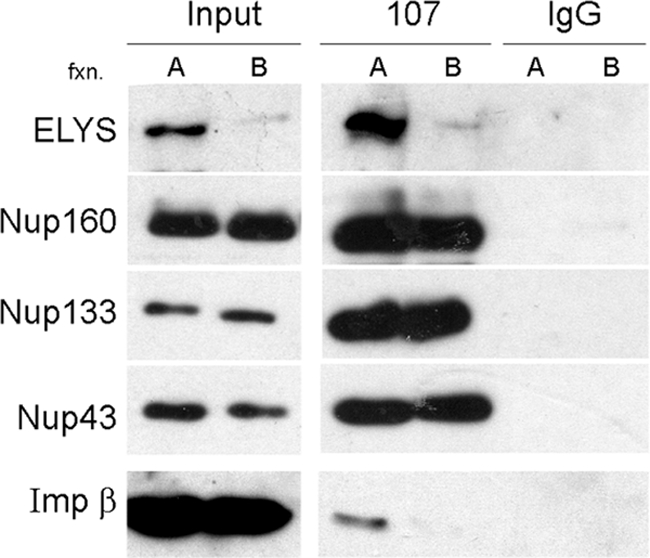
Importin β is associated with the ELYS+ subpopulation of the Nup107-160 complex. Fraction A (enriched in ELYS) and fraction B (lacking ELYS) eluted from a Q-Sepharose column as in Figure 6 were desalted and used for immunoprecipitation with affinity-purified anti-Nup107 or control rabbit IgG. Nup160, Nup133, and Nup43 were coimmunoprecipitated by anti-Nup107 from both fractions, whereas ELYS and importin β were only significantly pulled out from fraction A. Note that importin β is a very abundant protein in egg cytosol and is present throughout the entire elution profile of the Q-Sepharose column.
Importin β Forms a High-Molecular-Weight Complex with ELYS and Nup107-160
To further investigate the potential interactions between the Nup107-160 complex, ELYS and importin β, the ELYS+ fractions eluted from the Q-Sepharose column were subjected to gel filtration chromatography on a Superose-6 column. An identical sample of the ELYS+ fractions was first incubated with 10 μM recombinant untagged Xenopus importin β and then subjected to gel filtration. A comparison of these two gel filtration runs was carried out by immunoblot analysis of the resulting fractions, shown in Figure 8A. For each of the three antibodies used (anti-ELYS, anti-Nup133, and anti-Nup107), a slight shift to higher molecular weight fractions is seen in the gel filtration column run after incubation with excess importin β (Figure 8A, +XImp β rows). No other significant changes were detected, and in particular, there was no indication that the addition of importin β had caused the release of ELYS from the Nup107-160 complex (monomeric ELYS or a complex with importin β would be expected to shift below the 440-kDa marker). Thus, rather than dissociating ELYS from the Nup107-160 complex, importin β appears to be added on to these components, forming a higher molecular weight complex.
Figure 8.
Importin β forms a high-molecular-weight complex with both ELYS and Nup107-160. (A) ELYS-enriched fractions eluted from a Q-Sepharose column, as in Figure 5, were pooled and loaded on a Superose-6 column. One-half of the sample was preincubated with 10 μM Xenopus importin β before gel filtration (+XImp β rows). Samples eluted from these two gel filtration runs (−/+β) were analyzed by immunoblotting. Note that the addition of excess importin β does not shift ELYS to lower molecular weight fractions. (B) Two Superose-6 gel filtration runs (−/+ importin β) were performed as in A. Fractions 5–8 eluted from each column were pooled together and subjected to immunoprecipitation with anti-ELYS or anti-Nup107 antibodies. Immunoblotting confirms the existence of a high-molecular-weight complex containing ELYS, Nup107-160, and importin β.
To directly probe the higher molecular weight species of ELYS/Nup107-160 appearing in this gel filtration chromatography, two identical columns were run (−/+ recombinant importin β) and fractions 5–8 of each column were pooled together and subjected to immunoprecipitation with anti-ELYS or anti-Nup107 antibodies (Figure 8B). The results confirm the existence of a complex containing ELYS, Nup107-160, and importin β. The comparison of experiments with and without added recombinant importin β shows that only a fraction of the excess recombinant protein remained associated with ELYS/Nup107-160 throughout the procedure. This implies that the interaction of importin β with ELYS/Nup107-160 is of rather low affinity.
In conclusion, we see no indication that importin β is able to release ELYS from the Nup107-160 complex and sequester it in a separate inactive form. Instead, our results point to the formation of a high-molecular-weight complex containing importin β, ELYS, and the Nup107-160 complex. It is presumably this high-molecular-weight complex that corresponds to the inhibited form, which is unable to bind chromatin.
A RanGTP Affinity Column Pulls Out ELYS and Nup107-160 from Cytosol
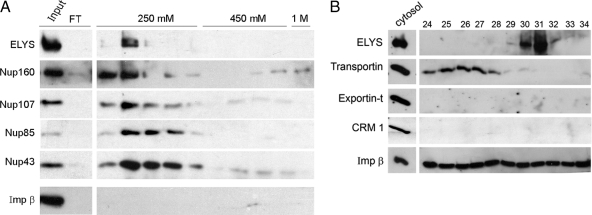
The results of our chromatin-binding assay performed in complete cytosol (Figure 4) imply that RanGTP cannot easily displace importin β from ELYS and/or the Nup107-160 complex. Gel filtration chromatography and immunoprecipitation demonstrate that at least a fraction of the endogenous Nup107-160 complex in egg cytosol is associated with both importin β and ELYS (Figure 8). We therefore decided to directly test the ability of the complex-bound, endogenous importin β to interact with RanGTP. Fractions eluted from the Q-Sepharose ion exchange column were subjected to an additional affinity chromatography step on immobilized RanQ69L-GTP (Kutay et al., 1997). Most known cargoes and interacting partners of importin β are thought to be released after the binding of RanGTP (Gorlich et al., 1996; Shah et al., 1998; Bayliss et al., 2000; Ben-Efraim and Gerace, 2001; Pemberton and Paschal, 2005). One can also envision a situation in which the binding of importin β to ELYS/Nup107-160 would obscure the Ran binding domain and prevent the interaction with the column. Neither of these scenarios is compatible with our observation that the vast majority of ELYS and Nup107-160 complex members were retained on the affinity column (Figure 9A). Elution with a moderate salt concentration of 250 mM KCl released ELYS/Nup107-160 from the column in a fraction that was virtually free of importin β. Importin β was also absent from elutions with higher salt concentrations (450 mM and 1 M KCl, Figure 9A) and was only quantitatively released by a 100 mM glycine, pH 2.5, wash (data not shown; see also Kutay et al., 1997).
Figure 9.
A RanGTP affinity column pulls out ELYS and the Nup107-160 complex from cytosol. (A) ELYS-enriched fractions eluted from a Q-Sepharose column were pooled and loaded on an affinity column of immobilized RanQ69L-GTP. Samples were analyzed by immunoblotting. Equivalent samples of the starting material and the flowthrough fraction of the RanQ69L-GTP column were loaded in the first two lanes. ELYS, Nup107-160, and importin β were retained on the affinity column. The column was washed with four column volumes and sequentially eluted with buffer containing 250 mM, 450 mM, and 1 M KCl. Samples from consecutive eluted fractions were loaded for each step. Only ELYS and Nup107-160 complex members were eluted in the first step, whereas importin β remained bound to the column. (B) Fractions eluted from a Q-Sepharose column were probed with antibodies directed against four different transport receptors of the importin β superfamily and compared with a sample of complete cytosol (0.3 μl). Importin β was present in all of these fractions, CRM 1 and exportin-t were not detected, whereas transportin peaked in fractions 24–28, showing minimal overlap with ELYS/Nup107-160.
One possible explanation for these observations is that importin β in egg cytosol can simultaneously bind RanGTP and ELYS/Nup107-160. According to this interpretation, importin β may serve as an intermediary, or molecular adaptor, retaining ELYS and the Nup107-160 complex on the RanGTP affinity column. Conceivably, other nuclear transport receptors present within this cytosolic subfraction could also play this role. A set of fractions from the relevant area of the Q-Sepharose elution gradient was probed for the presence of importin β and three other transport receptors (Figure 9B). Importin β was present throughout the elution profile and overlapped the peak of the ELYS+ subpopulation of the Nup107-160 complex. Two major export receptors, CRM 1 and exportin-t, were absent from all of these fractions, whereas the import receptor transportin was present in peak fractions that largely preceded ELYS and the Nup107-160 complex. Thus, importin β remains our best candidate for providing the physical link to the RanGTP column, although we cannot exclude the possible involvement of additional factors that were not probed in this experiment.
In the practical sense, the immobilized RanGTP affinity column is a very efficient means of enrichment for the purification of the endogenous Nup107-160 complex, which also provides relatively mild conditions for eluting the complex in an importin β-free form. These findings can now be used to improve the biochemical purification of endogenous forms of the Nup107-160 complex toward a structural analysis by electron microscopy.
DISCUSSION
The exit from mitosis in higher eukaryotic cells is marked by the reformation of the nuclear envelope, together with the rapid assembly of NPCs. Different mechanistic models have been proposed for postmitotic NPC assembly, but a recent emerging consensus is that the first essential step of the process involves the “seeding” of chromatin with ELYS and the Nup107-160 complex (Rasala et al., 2006; Franz et al., 2007; Antonin et al., 2008; D'Angelo and Hetzer, 2008). In this study, we show that this initial step of chromatin seeding is negatively regulated by importin β. High-resolution scanning electron microscopy reveals the attachment sites on the surface of chromatin as single sites, lacking any annular structures or oligomerized “prepores.” The inhibitory effect of importin β on the chromatin-binding step is only partially reversed by RanGTP. Moreover, the analysis of endogenous proteins in Xenopus egg extract suggests that importin β may be able to simultaneously bind ELYS/Nup107-160 and RanGTP. These findings have direct implications for the mechanism of NPC assembly.
Importin β negatively regulates nuclear membrane fusion and NPC assembly, as we have previously shown (Harel et al., 2003a; see also D'Angelo et al., 2006; Ryan et al., 2007). Walther et al. proposed a mechanism in which importin β is dissociated from multiple nucleoporins (Nup107-160, Nup153, and Nup358) to facilitate the formation of “prepores” or “pre-NPCs” on the surface of chromatin (Walther et al., 2003a; Hetzer et al., 2005). It has also been suggested that nuclear transport receptors can act as molecular chaperones, suppressing interactions between FG-Nups during their transit to NPC assembly sites (Frey and Gorlich, 2007). Importin β should therefore be viewed, not only as the prototypic nuclear transport receptor, but also as a key regulator of multiple steps in the NPC assembly pathway (Harel and Forbes, 2004). We now add to this view negative regulation at the earliest known step of postmitotic NPC assembly. The addition of excess importin β to the Xenopus nuclear reconstitution system caused a dramatic reduction in the binding of ELYS and Nup107-160 complex members to chromatin. The simplest model based on this observation would have importin β bound to ELYS or to the complex of ELYS/Nup107-160 in cytosol and displaced (under normal conditions) by the high concentration of RanGTP generated close to the surface of chromatin. Indeed, in the scenario proposed by Walther et al., RanGTP displaces importin β from multiple, soluble nucleoporins enabling their assembly into prepore structures on chromatin. Our FESEM analysis reveals no sign of such putative structures, hypothesized to be annular multicopy assemblies of a subset of nucleoporins that form before membrane recruitment (Walther et al., 2003a; Wozniak and Clarke, 2003; Hetzer et al., 2005; Drummond et al., 2006). Immunogold labeling of ELYS, Nup107, and Nup133 reveals single sites dispersed over the topmost ridges of chromatin structure and lacking any clear sign of lateral oligomerization. Although pairs of closely situated gold particles are often observed, we find no evidence that the underlying protein epitopes can further multimerize. There are also no detectable architectural elements resembling any part of mature NPCs in these specimens.
These results, together with the recent demonstration that other nucleoporins cannot be recruited to chromatin in the absence of membranes (Rasala et al., 2008), suggest a different view of the first stages in NPC assembly. We propose that the initial attachment sites, or prepores, on the surface of chromatin are composed of single copies of ELYS and the Nup107-160 complex. No other components are needed for seeding the chromatin landscape for NPC assembly. Further oligomerization and the formation of the basic structural scaffold of the NPC appear to depend on the recruitment of specific membrane components. This suggests that repeated interactions between copies of the Nup107-160 complex, such as head-to-tail concentric rings of eight copies (Hsia et al., 2007), may not be sufficient for the first stages in the assembly of the central scaffold. Lateral oligomerization may depend on interdigitation of a specific “linker,” such as a membrane-nucleoporin domain. Alternatively, soluble copies of Nup107-160 could be prevented from oligomerizing (for instance by importin β), and this inhibition could be released by an interaction with a membrane component. One implication of this model is that only the first attachment site on chromatin needs to contain ELYS. Subsequently recruited copies of the Nup107-160 complex may be free of ELYS. Indeed, we find evidence for the existence of a large pool of ELYS-free Nup107-160 complex in egg cytosol (Figure 5).
Our FESEM analysis indicates that the attachment sites of ELYS and the Nup107-160 complex appear to be preferentially located on the highest ridges and elevated structures of chromatin. Although the formal possibility exists that all the epitopes recognized by our antibodies are only accessible when localized to the highest structures of chromatin, we note that this staining pattern was observed with antibodies to three different proteins (Supplemental Figure S1). It is therefore more likely that the immunogold staining pattern reflects a real difference in the positioning of the seeding nucleoporins. This could be the result of a predetermined, uneven distribution of ELYS binding sites on the three-dimensional surface of chromatin or a consequence of on-going chromatin decondensation. The chromatin-binding elements of ELYS have been minimally mapped to the conserved AT-hook motif and an adjacent site, close to the C-terminus of the protein (Rasala et al., 2008). It is still unclear whether this reflects binding to specific sequences in DNA and if there is a distinct spatial relationship between the binding sites of ELYS/Nup107-160 and replication-licensing complexes (Gillespie et al., 2007; Rasala et al., 2008). In the future, it will be interesting to see if the elevated structures of chromatin correspond to some type of AT-rich sequences in DNA. It also remains to be determined whether the connection to chromatin, via ELYS, is maintained after NPC assembly is completed.
How is the negative regulation by importin β released, to allow NPC assembly to initiate at the correct sites? As mentioned above, a simple model would have importin β displaced by the high concentration of RanGTP close to the surface of chromatin, whereas the binding sites for ELYS could be exposed by chromatin decondensation at anaphase/telophase (Hetzer et al., 2005; Antonin et al., 2008; D'Angelo and Hetzer, 2008). Our results do not support such a mechanism, because RanQ69L-GTP did not fully reverse importin β's inhibitory effect on chromatin binding by ELYS/Nup107-160. Our assay was conducted with untagged Xenopus importin β, which has been shown to be free to interact with RanGTP (Delmar et al., 2008). Importantly, these conclusions are reinforced by the chromatographic separation on immobilized RanQ69L-GTP. As stated above, the simplest interpretation of our data is that endogenous importin β remains bound to ELYS/Nup107-160 while being “fished out” of egg cytosol through the interaction with RanGTP. This would suggest that the GTPase ON-state is insufficient to trigger the release of importin β from Nup107-160 and/or ELYS. Alternative explanations for the RanGTP affinity column results might involve a different exportin (other than CRM 1 and exportin-t that were tested in this study) or a hitherto unrecognized Ran-binding domain in ELYS or one of the Nup107-160 complex members. None of these interpretations is compatible with a simple on-off switch model based on Ran and importin β. RanGTP does counteract importin β in other aspects of nuclear assembly, most notably at a later stage in NPC assembly (Delmar et al., 2008; see also Ryan et al., 2007) and in the formation of chromatin free annulate lamellae pores (Walther et al., 2003b). However, we predict that an additional regulator may be involved in the first critical step of chromatin seeding. This may be analogous to the complex relationship between importin β and Ran functions during mitotic progression, in which an initial on-off switch model has been replaced with more elaborate mechanistic schemes. Current mitotic models emphasize the importance of complex spatial gradients, posttranslational modifications and combined effects of additional mitotic regulators, such as Aurora kinases (Di Fiore et al., 2004; Bastiaens et al., 2006; Clarke and Zhang, 2008; Kalab and Heald, 2008). Intriguingly, a recent report suggests a new role for Cdc48/p97 in promoting postmitotic nuclear assembly by extracting the Aurora B kinase from chromatin (Ramadan et al., 2007). The p97 ATPase complex is suggested to promote nuclear assembly by removing an inhibitor: the chromatin associated kinase, which is marked for inactivation by polyubiquitination. Similar mechanisms may be at work for the removal of importin β from ELYS/Nup107-160.
Using a classical chromatography approach, we find evidence for the existence of multiple high-molecular-weight forms of the Nup107-160 complex in Xenopus egg cytosol. This is consistent with the notion that most of the individual complex members are tightly held together (Belgareh et al., 2001; Vasu et al., 2001; Orjalo et al., 2006; Boehmer et al., 2008; Brohawn et al., 2008; Chakraborty et al., 2008), but interactions with ELYS and importin β may vary. Immunoprecipitation experiments indicate that at least some of the subpopulation of the Nup107-160 complex, which is preassociated with ELYS, is also bound by importin β. This may provide a means of regulating the number of initiation sites for NPC assembly in relation to chromatin decondensation or at different developmental stages. We find no evidence that importin β releases ELYS from the Nup107-160 complex or sequesters ELYS in an inactive form. We propose that the high-molecular-weight complex of importin β, ELYS and Nup107-160 represents the inhibited form of the chromatin-seeding module in NPC assembly. Importin β could be acting by direct binding and blocking of the C-terminus of ELYS, while ELYS remains bound to Nup107-160. Alternatively, importin β may bind to other regions of the large ELYS/Nup107-160 complex and cause a conformational change affecting the chromatin binding sites at the C-terminus of ELYS.
In conclusion, we propose that the initiation sites for NPC assembly consist of single copies of the Nup107-160 complex, bound to elevated sites on chromatin through the adaptor protein ELYS. Importin β negatively regulates the seeding of chromatin at these sites, and subsequent assembly steps are strictly dependent on the recruitment of membrane components. Very little is known about any of the subsequent steps in the complex assembly process. An important question for the future will be to determine if individual copies of the Nup107-160 complex can directly interact with each other or whether other nucleoporins serve as mediators for lateral oligomerization.
Supplementary Material
ACKNOWLEDGMENTS
We thank Douglass Forbes, Ulrike Kutay, Dirk Görlich, Matt Michael, and Beate Sodeik (Hannover Medical School, Hannover, Germany) for the kind gift of reagents. We also thank Michael Elbaum (Weizmann Institute of Science, Rehovot, Israel) for stimulating discussions and continuous help with FESEM and Noam Ziv (Technion, Haifa, Israel) for instruction in the use of the OpenView software. This work was supported by grants from the Israel Science Foundation (813/05) and the European Commission FP6 Marie Curie IRG (031161) to A.H. FESEM work by A.R. was supported by training and research grants from the Russell Berrie Nanotechnology Institute– Technion.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-02-0150) on July 22, 2009.
REFERENCES
- Alber F., et al. The molecular architecture of the nuclear pore complex. Nature. 2007;450:695–701. doi: 10.1038/nature06405. [DOI] [PubMed] [Google Scholar]
- Allen N. P., Patel S. S., Huang L., Chalkley R. J., Burlingame A., Lutzmann M., Hurt E. C., Rexach M. Deciphering networks of protein interactions at the nuclear pore complex. Mol. Cell Proteom. 2002;1:930–946. doi: 10.1074/mcp.t200012-mcp200. [DOI] [PubMed] [Google Scholar]
- Allen T. D., Rutherford S. A., Murray S., Sanderson H. S., Gardiner F., Kiseleva E., Goldberg M. W., Drummond S. P. A protocol for isolating Xenopus oocyte nuclear envelope for visualization and characterization by scanning electron microscopy (SEM) or transmission electron microscopy (TEM) Nat. Protoc. 2007;2:1166–1172. doi: 10.1038/nprot.2007.137. [DOI] [PubMed] [Google Scholar]
- Anderson D. J., Hetzer M. W. The life cycle of the metazoan nuclear envelope. Curr. Opin. Cell Biol. 2008;20:386–392. doi: 10.1016/j.ceb.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonin W., Ellenberg J., Dultz E. Nuclear pore complex assembly through the cell cycle: regulation and membrane organization. FEBS Lett. 2008;582:2004–2016. doi: 10.1016/j.febslet.2008.02.067. [DOI] [PubMed] [Google Scholar]
- Bastiaens P., Caudron M., Niethammer P., Karsenti E. Gradients in the self-organization of the mitotic spindle. Trends Cell Biol. 2006;16:125–134. doi: 10.1016/j.tcb.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Bayliss R., Littlewood T., Stewart M. Structural basis for the interaction between FxFG nucleoporin repeats and importin-beta in nuclear trafficking. Cell. 2000;102:99–108. doi: 10.1016/s0092-8674(00)00014-3. [DOI] [PubMed] [Google Scholar]
- Beck M., Forster F., Ecke M., Plitzko J. M., Melchior F., Gerisch G., Baumeister W., Medalia O. Nuclear pore complex structure and dynamics revealed by cryoelectron tomography. Science. 2004;306:1387–1390. doi: 10.1126/science.1104808. [DOI] [PubMed] [Google Scholar]
- Belgareh N., et al. An evolutionarily conserved NPC subcomplex, which redistributes in part to kinetochores in mammalian cells. J. Cell Biol. 2001;154:1147–1160. doi: 10.1083/jcb.200101081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Efraim I., Gerace L. Gradient of increasing affinity of importin beta for nucleoporins along the pathway of nuclear import. J. Cell Biol. 2001;152:411–417. doi: 10.1083/jcb.152.2.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodoor K., Shaikh S., Salina D., Raharjo W. H., Bastos R., Lohka M., Burke B. Sequential recruitment of NPC proteins to the nuclear periphery at the end of mitosis. J. Cell Sci. 1999;112:2253–2264. doi: 10.1242/jcs.112.13.2253. [DOI] [PubMed] [Google Scholar]
- Boehmer T., Jeudy S., Berke I. C., Schwartz T. U. Structural and functional studies of Nup107/Nup133 interaction and its implications for the architecture of the nuclear pore complex. Mol. Cell. 2008;30:721–731. doi: 10.1016/j.molcel.2008.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brohawn S. G., Leksa N. C., Spear E. D., Rajashankar K. R., Schwartz T. U. Structural evidence for common ancestry of the nuclear pore complex and vesicle coats. Science. 2008;322:1369–1373. doi: 10.1126/science.1165886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke B., Ellenberg J. Remodelling the walls of the nucleus. Nat. Rev. Mol. Cell Biol. 2002;3:487–497. doi: 10.1038/nrm860. [DOI] [PubMed] [Google Scholar]
- Chakraborty P., et al. Nucleoporin levels regulate cell cycle progression and phase-specific gene expression. Dev. Cell. 2008;15:657–667. doi: 10.1016/j.devcel.2008.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke P. R., Zhang C. Spatial and temporal coordination of mitosis by Ran GTPase. Nat. Rev. Mol. Cell Biol. 2008;9:464–477. doi: 10.1038/nrm2410. [DOI] [PubMed] [Google Scholar]
- Cronshaw J. M., Krutchinsky A. N., Zhang W., Chait B. T., Matunis M. J. Proteomic analysis of the mammalian nuclear pore complex. J. Cell Biol. 2002;158:915–927. doi: 10.1083/jcb.200206106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Angelo M. A., Anderson D. J., Richard E., Hetzer M. W. Nuclear pores form de novo from both sides of the nuclear envelope. Science. 2006;312:440–443. doi: 10.1126/science.1124196. [DOI] [PubMed] [Google Scholar]
- D'Angelo M. A., Hetzer M. W. Structure, dynamics and function of nuclear pore complexes. Trends Cell Biol. 2008;18:456–466. doi: 10.1016/j.tcb.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davuluri G., Gong W., Yusuff S., Lorent K., Muthumani M., Dolan A. C., Pack M. Mutation of the zebrafish nucleoporin elys sensitizes tissue progenitors to replication stress. PLoS Genet. 2008;4:e1000240. doi: 10.1371/journal.pgen.1000240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmar V. A., Chan R. C., Forbes D. J. Xenopus importin beta validates human importin beta as a cell cycle negative regulator. BMC Cell Biol. 2008;9:14. doi: 10.1186/1471-2121-9-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Fiore B., Ciciarello M., Lavia P. Mitotic functions of the Ran GTPase network: the importance of being in the right place at the right time. Cell Cycle. 2004;3:305–313. [PubMed] [Google Scholar]
- Drummond S. P., Rutherford S. A., Sanderson H. S., Allen T. D. High resolution analysis of mammalian nuclear structure throughout the cell cycle: implications for nuclear pore complex assembly during interphase and mitosis. Can. J. Physiol. Pharmacol. 2006;84:423–430. doi: 10.1139/y05-148. [DOI] [PubMed] [Google Scholar]
- Dultz E., Zanin E., Wurzenberger C., Braun M., Rabut G., Sironi L., Ellenberg J. Systematic kinetic analysis of mitotic dis- and reassembly of the nuclear pore in living cells. J. Cell Biol. 2008;180:857–865. doi: 10.1083/jcb.200707026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahrenkrog B., Koser J., Aebi U. The nuclear pore complex: a jack of all trades? Trends Biochem. Sci. 2004;29:175–182. doi: 10.1016/j.tibs.2004.02.006. [DOI] [PubMed] [Google Scholar]
- Fernandez A. G., Piano F. MEL-28 is downstream of the Ran cycle and is required for nuclear-envelope function and chromatin maintenance. Curr. Biol. 2006;16:1757–1763. doi: 10.1016/j.cub.2006.07.071. [DOI] [PubMed] [Google Scholar]
- Forbes D. J., Kirschner M. W., Newport J. W. Spontaneous formation of nucleus-like structures around bacteriophage DNA microinjected into Xenopus eggs. Cell. 1983;34:13–23. doi: 10.1016/0092-8674(83)90132-0. [DOI] [PubMed] [Google Scholar]
- Franz C., Walczak R., Yavuz S., Santarella R., Gentzel M., Askjaer P., Galy V., Hetzer M., Mattaj I. W., Antonin W. MEL-28/ELYS is required for the recruitment of nucleoporins to chromatin and postmitotic nuclear pore complex assembly. EMBO Rep. 2007;8:165–172. doi: 10.1038/sj.embor.7400889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey S., Gorlich D. A saturated FG-repeat hydrogel can reproduce the permeability properties of nuclear pore complexes. Cell. 2007;130:512–523. doi: 10.1016/j.cell.2007.06.024. [DOI] [PubMed] [Google Scholar]
- Gerace L., Burke B. Functional organization of the nuclear envelope. Annu. Rev. Cell Biol. 1988;4:335–374. doi: 10.1146/annurev.cb.04.110188.002003. [DOI] [PubMed] [Google Scholar]
- Gillespie P. J., Khoudoli G. A., Stewart G., Swedlow J. R., Blow J. J. ELYS/MEL-28 chromatin association coordinates nuclear pore complex assembly and replication licensing. Curr. Biol. 2007;17:1657–1662. doi: 10.1016/j.cub.2007.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorlich D., Pante N., Kutay U., Aebi U., Bischoff F. R. Identification of different roles for RanGDP and RanGTP in nuclear protein import. EMBO J. 1996;15:5584–5594. [PMC free article] [PubMed] [Google Scholar]
- Harel A., Chan R. C., Lachish-Zalait A., Zimmerman E., Elbaum M., Forbes D. J. Importin beta negatively regulates nuclear membrane fusion and nuclear pore complex assembly. Mol. Biol. Cell. 2003a;14:4387–4396. doi: 10.1091/mbc.E03-05-0275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harel A., Forbes D. J. Importin beta: conducting a much larger cellular symphony. Mol. Cell. 2004;16:319–330. doi: 10.1016/j.molcel.2004.10.026. [DOI] [PubMed] [Google Scholar]
- Harel A., Orjalo A. V., Vincent T., Lachish-Zalait A., Vasu S., Shah S., Zimmerman E., Elbaum M., Forbes D. J. Removal of a single pore subcomplex results in vertebrate nuclei devoid of nuclear pores. Mol. Cell. 2003b;11:853–864. doi: 10.1016/s1097-2765(03)00116-3. [DOI] [PubMed] [Google Scholar]
- Hetzer M. W., Walther T. C., Mattaj I. W. Pushing the envelope: structure, function, and dynamics of the nuclear periphery. Annu. Rev. Cell Dev. Biol. 2005;21:347–380. doi: 10.1146/annurev.cellbio.21.090704.151152. [DOI] [PubMed] [Google Scholar]
- Hsia K. C., Stavropoulos P., Blobel G., Hoelz A. Architecture of a coat for the nuclear pore membrane. Cell. 2007;131:1313–1326. doi: 10.1016/j.cell.2007.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalab P., Heald R. The RanGTP gradient—a GPS for the mitotic spindle. J. Cell Sci. 2008;121:1577–1586. doi: 10.1242/jcs.005959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura N., Takizawa M., Okita K., Natori O., Igarashi K., Ueno M., Nakashima K., Nobuhisa I., Taga T. Identification of a novel transcription factor, ELYS, expressed predominantly in mouse foetal haematopoietic tissues. Genes Cells. 2002;7:435–446. doi: 10.1046/j.1365-2443.2002.00529.x. [DOI] [PubMed] [Google Scholar]
- Kutay U., Bischoff F. R., Kostka S., Kraft R., Gorlich D. Export of importin alpha from the nucleus is mediated by a specific nuclear transport factor. Cell. 1997;90:1061–1071. doi: 10.1016/s0092-8674(00)80372-4. [DOI] [PubMed] [Google Scholar]
- Lohka M. J., Masui Y. Formation in vitro of sperm pronuclei and mitotic chromosomes induced by amphibian ooplasmic components. Science. 1983;220:719–721. doi: 10.1126/science.6601299. [DOI] [PubMed] [Google Scholar]
- Macaulay C., Forbes D. J. Assembly of the nuclear pore: biochemically distinct steps revealed with NEM, GTP gamma S, and BAPTA. J. Cell Biol. 1996;132:5–20. doi: 10.1083/jcb.132.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maul G. G. The nuclear and the cytoplasmic pore complex: structure, dynamics, distribution, and evolution. Int. Rev. Cytol. Suppl. 1977:75–186. [PubMed] [Google Scholar]
- Maul G. G., Maul H. M., Scogna J. E., Lieberman M. W., Stein G. S., Hsu B. Y., Borun T. W. Time sequence of nuclear pore formation in phytohemagglutinin-stimulated lymphocytes and in HeLa cells during the cell cycle. J. Cell Biol. 1972;55:433–447. doi: 10.1083/jcb.55.2.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newport J. Nuclear reconstitution in vitro: stages of assembly around protein-free DNA. Cell. 1987;48:205–217. doi: 10.1016/0092-8674(87)90424-7. [DOI] [PubMed] [Google Scholar]
- Orjalo A. V., Arnaoutov A., Shen Z., Boyarchuk Y., Zeitlin S. G., Fontoura B., Briggs S., Dasso M., Forbes D. J. The Nup107-160 nucleoporin complex is required for correct bipolar spindle assembly. Mol. Biol. Cell. 2006;17:3806–3818. doi: 10.1091/mbc.E05-11-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pemberton L. F., Paschal B. M. Mechanisms of receptor-mediated nuclear import and nuclear export. Traffic. 2005;6:187–198. doi: 10.1111/j.1600-0854.2005.00270.x. [DOI] [PubMed] [Google Scholar]
- Rabut G., Doye V., Ellenberg J. Mapping the dynamic organization of the nuclear pore complex inside single living cells. Nat. Cell Biol. 2004;6:1114–1121. doi: 10.1038/ncb1184. [DOI] [PubMed] [Google Scholar]
- Ramadan K., Bruderer R., Spiga F. M., Popp O., Baur T., Gotta M., Meyer H. H. Cdc48/p97 promotes reformation of the nucleus by extracting the kinase Aurora B from chromatin. Nature. 2007;450:1258–1262. doi: 10.1038/nature06388. [DOI] [PubMed] [Google Scholar]
- Rasala B. A., Orjalo A. V., Shen Z., Briggs S., Forbes D. J. ELYS is a dual nucleoporin/kinetochore protein required for nuclear pore assembly and proper cell division. Proc. Natl. Acad. Sci. USA. 2006;103:17801–17806. doi: 10.1073/pnas.0608484103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasala B. A., Ramos C., Harel A., Forbes D. J. Capture of AT-rich chromatin by ELYS recruits POM121 and NDC1 to initiate nuclear pore assembly. Mol. Biol. Cell. 2008;19:3982–3996. doi: 10.1091/mbc.E08-01-0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan K. J., Zhou Y., Wente S. R. The karyopherin Kap95 regulates nuclear pore complex assembly into intact nuclear envelopes in vivo. Mol. Biol. Cell. 2007;18:886–898. doi: 10.1091/mbc.E06-06-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz T. U. Modularity within the architecture of the nuclear pore complex. Curr. Opin. Struct. Biol. 2005;15:221–226. doi: 10.1016/j.sbi.2005.03.003. [DOI] [PubMed] [Google Scholar]
- Shah S., Tugendreich S., Forbes D. Major binding sites for the nuclear import receptor are the internal nucleoporin Nup153 and the adjacent nuclear filament protein Tpr. J. Cell Biol. 1998;141:31–49. doi: 10.1083/jcb.141.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehan M. A., Mills A. D., Sleeman A. M., Laskey R. A., Blow J. J. Steps in the assembly of replication-competent nuclei in a cell-free system from Xenopus eggs. J. Cell Biol. 1988;106:1–12. doi: 10.1083/jcb.106.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulga N., Goldfarb D. S. Binding dynamics of structural nucleoporins govern nuclear pore complex permeability and may mediate channel gating. Mol. Cell. Biol. 2003;23:534–542. doi: 10.1128/MCB.23.2.534-542.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S. J., Rittinger K. Preparation of GTPases for structural and biophysical analysis. Methods Mol. Biol. 2002;189:13–24. doi: 10.1385/1-59259-281-3:013. [DOI] [PubMed] [Google Scholar]
- Tran E. J., Wente S. R. Dynamic nuclear pore complexes: life on the edge. Cell. 2006;125:1041–1053. doi: 10.1016/j.cell.2006.05.027. [DOI] [PubMed] [Google Scholar]
- Tsuriel S., Geva R., Zamorano P., Dresbach T., Boeckers T., Gundelfinger E. D., Garner C. C., Ziv N. E. Local sharing as a predominant determinant of synaptic matrix molecular dynamics. PLoS Biol. 2006;4:e271. doi: 10.1371/journal.pbio.0040271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasu S., Shah S., Orjalo A., Park M., Fischer W. H., Forbes D. J. Novel vertebrate nucleoporins Nup133 and Nup160 play a role in mRNA export. J. Cell Biol. 2001;155:339–354. doi: 10.1083/jcb.200108007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther T. C., et al. The conserved Nup107-160 complex is critical for nuclear pore complex assembly. Cell. 2003a;113:195–206. doi: 10.1016/s0092-8674(03)00235-6. [DOI] [PubMed] [Google Scholar]
- Walther T. C., Askjaer P., Gentzel M., Habermann A., Griffiths G., Wilm M., Mattaj I. W., Hetzer M. RanGTP mediates nuclear pore complex assembly. Nature. 2003b;424:689–694. doi: 10.1038/nature01898. [DOI] [PubMed] [Google Scholar]
- Weis K. Regulating access to the genome: nucleocytoplasmic transport throughout the cell cycle. Cell. 2003;112:441–451. doi: 10.1016/s0092-8674(03)00082-5. [DOI] [PubMed] [Google Scholar]
- Wozniak R., Clarke P. R. Nuclear pores: sowing the seeds of assembly on the chromatin landscape. Curr. Biol. 2003;13:R970–R972. doi: 10.1016/j.cub.2003.11.046. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.