Abstract
Background
Escherichia coli exhibits diauxic growth in sugar mixtures due to CRP-mediated catabolite repression and inducer exclusion related to phosphotransferase system enzyme activity. Replacement of the native crp gene with a catabolite repression mutant (referred to as crp*) enables co-utilization of glucose and other sugars in E. coli. While previous studies have examined the effects of expressing CRP* mutants on the expression of specific catabolic genes, little is known about the global transcriptional effects of CRP* expression. In this study, we compare the transcriptome of E. coli W3110 (expressing wild-type CRP) to that of mutant strain PC05 (expressing CRP*) in the presence and absence of glucose.
Results
The glucose effect is significantly suppressed in strain PC05 relative to strain W3110. The expression levels of glucose-sensitive genes are generally not altered by glucose to the same extent in strain PCO5 as compared to W3110. Only 23 of the 80 genes showing significant differential expression in the presence of glucose for strain PC05 are present among the 418 genes believed to be directly regulated by CRP. Genes involved in central carbon metabolism (including several TCA cycle genes) and amino acid biosynthesis, as well as genes encoding nutrient transport systems are among those whose transcript levels are most significantly affected by CRP* expression.
We present a detailed transcription analysis and relate these results to phenotypic differences between strains expressing wild-type CRP and CRP*. Notably, CRP* expression in the presence of glucose results in an elevated intracellular NADPH concentration and reduced NADH concentration relative to wild-type CRP. Meanwhile, a more drastic decrease in the NADPH/NADP+ ratio is observed for the case of CRP* expression in strains engineered to reduce xylose to xylitol via a heterologously expressed, NADPH-dependent xylose reductase. Altered expression levels of transhydrogenase and TCA cycle genes, among others, are consistent with these observations.
Conclusion
While the simplest model of CRP*-mediated gene expression assumes insensitivity to glucose (or cAMP), our results show that gene expression in the context of CRP* is very different from that of wild-type in the absence of glucose, and is influenced by the presence of glucose. Most of the transcription changes in response to CRP* expression are difficult to interpret in terms of possible systematic effects on metabolism. Elevated NADPH availability resulting from CRP* expression suggests potential biocatalytic applications of crp* strains that extend beyond relief of catabolite repression.
Background
E. coli growing in a mixture of sugars exhibits diauxic growth characteristics, whereby glucose is preferentially assimilated before other sugars. This is due to CRP-mediated catabolite repression and inducer exclusion related to phosphotransferase system enzyme activity. It is well established that cyclic AMP (cAMP) and its receptor protein (CRP) are involved in transcriptional activation of catabolic genes [1,2], but the details of catabolite repression and inducer exclusion mechanisms and their relation to the levels of cAMP and CRP (also known as CAP) are not clear and have motivated many studies [3-7].
Inducer exclusion is a result of dephosphorylation of enzyme IIAGlc of PTS [8,9] and catabolite repression is associated with altered levels of cAMP [10-12] and CRP [13]. Enzyme IIAGlc, when unphosphorylated, inhibits activity of other transport systems (non-PTS transporter) [14-16]. In its phosphorylated form, enzyme IIAGlc stimulates adenylate cyclase activity, resulting in higher intracellular levels of cAMP [17,18] and the cAMP-CRP complex (global transcription activator).
Efforts to study or alleviate catabolite repression mediated by CRP have resulted in a series of CRP mutants isolated from strains lacking adenylate cyclase and having an apparent reduced dependence on cAMP for activating catabolic genes (called CRP*, CRP-in or CAPc) [1,19,20]. Genetically different crp* strains reported are also phenotypically different, showing different sensitivities to cyclic nucleotides and relieving catabolic repression of select genes examined to different extents [21]. For example, six different crp* mutants isolated after UV treatment and selection for a lactose+ phenotype in an adenylate cyclase-deficient E. coli strain showed a variety of utilization patterns for different sugars (lactose, maltose, arabinose, xylose, ribose, mannose, mannitol) as well as different levels of activation of the lac operon by cAMP or cGMP [22]. Similar examples have been reported by others [3,21].
Ability to co-utilize sugars via relief of catabolite repression during microbial production of value-added chemicals has potential to improve bioproduction process economics [23]. We previously engineered E. coli to produce xylitol from xylose while metabolizing glucose as a source of carbon and energy (xylose metabolism is disabled) [24,25]. Expression of CRP* was an effective approach to promote expression of xylose transporters and enhance xylitol production in the presence of glucose. Although plasmid-based, CRP-independent expression of xylose transporters in wild-type crp strains also enhances xylose uptake and xylitol production in the presence of glucose [25], the favorable effects of CRP* expression were found to go beyond improving xylose transport and to include other beneficial phenotypes such as improved xylitol titer in controlled batch fermentation and reduced acetate production and higher yields on xylose reduced per mole of glucose consumed in resting cell transformations [26].
While CRP*s have been studied at the molecular level and the effects of expressing CRP* mutants on the expression of specific catabolic genes have been reported, the global transcriptional effects and regulatory consequences of CRP* expression is not known. Here, we report the results of comparisons between the transcriptome of E. coli W3110 (expressing wild-type CRP) and that of mutant strain PC05 (expressing CRP*) in the presence and absence of glucose through microarray analysis. Our results show that gene expression in PC05 is drastically different from that of W3110 in both the presence and absence of glucose, and that while expression of the CRP* allele used in this study has the general effect of suppressing transcriptional changes due to glucose, a significant response to glucose nonetheless remains. Results are analyzed in light of the observed differences between wild-type and CRP* strains during xylitol production. We identify many genes showing differential expression that are consistent with the observed elevated levels of glucose oxidation and NADPH-dependent xylose reduction for PC05 compared to W3110. A subsequent intracellular cofactor analysis reveals CRP*-correlated effects on cofactor levels that are consistent with the observed expression changes.
Results
The E. coli W3110-derivative CRP* strains used in our studies are derived from E. coli donor strain ET25 [8], which expresses a CRP* mutant with three amino acid substitutions (I112L, T127I, and A144T) identical to those found in an earlier characterized CRP* strain CA8404 [1]. Amino acid position 127 lies in the cAMP binding pocket, and T127I or T127L mutations occur frequently in CRP* alleles [21,27], presumably serving to reduce the cAMP requirement to form an activating CRP complex [27]. Mutation A144T is also frequently found in different CRP* alleles [3,21,28] and can exhibit the CRP* phenotype to some extent even as the only mutation in the protein [29]. This position lies in the DNA binding domain of CRP and is suggested to improve affinity of the protein for CRP binding sites [29]. A fourth base substitution in the crp* sequence results in a T28K mutation, which is the result of native differences in the crp sequence between W3110 and the donor strain.
Genome-wide transcriptional effects of glucose and CRP*
Table 1 summarizes the genome-wide effects of CRP* expression under the conditions tested, while Table 2 lists the average signal values and expression ratios for specific genes mentioned in this paper. Supplementary Table S1 (see Additional file 1) contains signal values for the complete probe set data for the E. coli K-12 genome. Transcriptome analysis of strain W3110 reveals that 629 genes show significant changes in expression level in response to the presence of glucose (comparison between WT G and WT in Figure 1a). 375 of these genes are upregulated by glucose, as depicted in Figure 1a. The complete list of expression levels of the genes that are differentially expressed between WT G and WT is provided in Supplementary Table S2 (see Additional file 2). Catabolic genes, membrane-related components, and sugar transporters (especially non-glucose PTS related enzymes) represent a large portion of genes repressed by glucose. In a study of CRP-dependent gene expression, Gosset and coworkers reported transcriptome analysis of CRP-dependent genes in another E. coli K-12 strain BW25113 [30]. In Table 3 we compare their result to results from our study for common conditions tested (i.e., WT G/WT). While their study did not examine CRP*, this comparison provides an indication of the consistency of glucose-responsive gene expression among different but similar strains. Our comparison focuses on genes involved in central metabolism and shows that the genes which are subject to glucose repression in BW25113 (such as aceA (isocitrate lyase monomer), aldA (aldehyde dehydrogenase A), sdhA (succinate dehydrogenase) and sucA (oxoglutarate dehydrogenase)) are also downregulated in the presence of glucose for W3110. However, not all the genes which are upregulated in the presence of glucose in BW25113 are upregulated in W3110 under the same conditions (examples are aceE (pyruvate dehydrogenase E1 component), guaB (IMP dehydrogenase), rpsQ (30S ribosomal subunit protein S17)). This is likely to be due to differences between these two strains [26,31-33] as well as the differences in experimental methods.
Table 1.
Summary of genome-wide effects of CRP* expression.
| Conditions compared | Number of genes showing significant differential expression | Up-regulated | Down-regulated | Genes in common with WT G/WT comparison |
| WT G/WT | 629 | 375 | 254 | 629 |
| CRP* G/CRP* | 80 | 29 | 51 | 43 |
| (CRP* G/CRP*)/(WT G/WT) | 238 | ----- | ----- | 198 |
| CRP* G/WT G | 349 | 232 | 117 | 163 |
| CRP*/WT | 553 | 392 | 161 | 205 |
| CRP* G/WT | 481 | 330 | 151 | 218 |
Four different conditions were tested: W3110 in LB+glucose medium (WT G), W3110 in LB medium (WT), PC05 in LB+glucose medium (CRP* G), and PC05 in LB medium (CRP*). Differences in gene expression levels with a P-value of <0.05 were considered significant in each comparison.
Table 2.
Expression levels for the genes discussed in this paper.
| Gene | WT G | WT | CRP* G | CRP* | WT G/WT | CRP* G/CRP* | CRP* G/WT G | CRP* G/WT |
| aceA | 1806 | 3171 | 2584 | 1980 | 0.6 | 1.3 | 1.4 | 0.8 |
| aceE | 7646 | 13291 | 10053 | 9167 | 0.6 | 1.1 | 1.3 | 0.8 |
| acs | 56 | 223 | 203 | 386 | 0.3 | 0.5 | 3.6 | 0.9 |
| aldA | 370 | 2471 | 937 | 1613 | 0.2 | 0.6 | 2.5 | 0.4 |
| argD | 44 | 38 | 73 | 158 | 1.1 | 0.5 | 1.7 | 1.9 |
| asnA | 594 | 1741 | 1409 | 1880 | 0.3 | 0.8 | 2.4 | 0.8 |
| cheR | 1588 | 2315 | 2311 | 1007 | 0.7 | 2.3 | 1.5 | 1.0 |
| cheY | 5365 | 6487 | 5893 | 2646 | 0.8 | 2.2 | 1.1 | 0.9 |
| citF | 50 | 25 | 306 | 269 | 2.0 | 1.1 | 6.1 | 12.2 |
| dhaM | 8790 | 1502 | 10556 | 3530 | 5.9 | 3.0 | 1.2 | 7.0 |
| edd | 4178 | 756 | 3582 | 980 | 5.5 | 3.7 | 0.9 | 4.7 |
| fruA | 3105 | 1289 | 5777 | 3659 | 2.4 | 1.6 | 1.9 | 4.5 |
| fruK | 3722 | 1934 | 6807 | 3866 | 1.9 | 1.8 | 1.8 | 3.5 |
| fumA | 956 | 3284 | 4013 | 3738 | 0.3 | 1.1 | 4.2 | 1.2 |
| gapC | 1316 | 581 | 2345 | 2383 | 2.3 | 1.0 | 1.8 | 4.0 |
| gatD | 1560 | 475 | 6080 | 7981 | 3.3 | 0.8 | 3.9 | 12.8 |
| gatR | 1776 | 786 | 6106 | 5039 | 2.3 | 1.2 | 3.4 | 7.8 |
| gcd | 491 | 467 | 171 | 67 | 1.1 | 2.5 | 0.4 | 0.4 |
| glnP | 2682 | 1037 | 1108 | 1374 | 2.6 | 0.8 | 0.4 | 1.1 |
| glpC | 564 | 8563 | 5595 | 13130 | 0.1 | 0.4 | 9.9 | 0.7 |
| glpF | 738 | 12693 | 2384 | 10944 | 0.1 | 0.2 | 3.2 | 0.2 |
| glpG | 1225 | 895 | 938 | 1426 | 1.4 | 0.7 | 0.8 | 1.1 |
| glpK | 360 | 8374 | 1579 | 8395 | 0.04 | 0.2 | 4.4 | 0.2 |
| glpQ | 159 | 4424 | 1747 | 9315 | 0.04 | 0.2 | 11.0 | 0.4 |
| glpT | 54 | 4285 | 893 | 9145 | 0.01 | 0.1 | 16.5 | 0.2 |
| gltB | 3219 | 2423 | 3388 | 1487 | 1.3 | 2.3 | 1.1 | 1.4 |
| gltD | 1881 | 1282 | 1829 | 595 | 1.5 | 3.1 | 1.0 | 1.4 |
| glyA | 1613 | 3148 | 3353 | 3448 | 0.5 | 1.0 | 2.1 | 1.1 |
| gntK | 631 | 217 | 670 | 259 | 2.9 | 2.6 | 1.1 | 3.1 |
| gntP | 87 | 354 | 321 | 799 | 0.2 | 0.4 | 3.7 | 0.9 |
| gntU | 334 | 98 | 260 | 126 | 3.4 | 2.1 | 0.8 | 2.7 |
| guaB | 4670 | 7698 | 2558 | 1733 | 0.6 | 1.5 | 0.6 | 0.3 |
| ldhA | 1823 | 1281 | 6014 | 3669 | 1.4 | 1.6 | 3.3 | 4.7 |
| maeB | 579 | 1420 | 1509 | 1839 | 0.4 | 0.8 | 2.6 | 1.1 |
| malF | 2381 | 2353 | 7365 | 9358 | 1.0 | 0.8 | 3.1 | 3.1 |
| manZ | 3993 | 2734 | 12806 | 9993 | 1.5 | 1.3 | 3.2 | 4.7 |
| mdh | 5878 | 9397 | 12603 | 15821 | 0.6 | 0.8 | 2.1 | 1.3 |
| mglA | 10 | 592 | 400 | 1205 | 0.02 | 0.3 | 40.0 | 0.7 |
| mglB | 38 | 1376 | 1529 | 4340 | 0.03 | 0.4 | 40.3 | 1.1 |
| nrfE | 783 | 21 | 531 | 754 | 36.5 | 0.7 | 0.7 | 24.7 |
| nrfF | 2460 | 41 | 1177 | 2267 | 59.7 | 0.5 | 0.5 | 28.6 |
| nrfG | 611 | 10 | 216 | 435 | 58.6 | 0.5 | 0.4 | 20.7 |
| pntA | 3946 | 6915 | 7400 | 7086 | 0.6 | 1.0 | 1.9 | 1.1 |
| pntB | 1736 | 2546 | 3057 | 3083 | 0.7 | 1.0 | 1.8 | 1.2 |
| ppsA | 247 | 6026 | 1325 | 3494 | 0.04 | 0.4 | 5.4 | 0.2 |
| proA | 2536 | 2397 | 13 | 12 | 1.1 | 1.1 | 0.01 | 0.01 |
| proB | 3589 | 3092 | 7 | 4 | 1.2 | 1.9 | 0.00 | 0.00 |
| ptsG | 13731 | 3926 | 16556 | 5403 | 3.5 | 3.1 | 1.2 | 4.2 |
| rpsQ | 6405 | 12505 | 9398 | 7652 | 0.5 | 1.2 | 1.5 | 0.8 |
| sdhA | 1033 | 7257 | 3292 | 3154 | 0.1 | 1.0 | 3.2 | 0.5 |
| sdhB | 895 | 5161 | 2407 | 2521 | 0.2 | 1.0 | 2.7 | 0.5 |
| sdhD | 556 | 5466 | 2568 | 1980 | 0.1 | 1.3 | 4.6 | 0.5 |
| serA | 2256 | 956 | 1638 | 793 | 2.4 | 2.1 | 0.7 | 1.7 |
| serC | 3904 | 2365 | 3230 | 1738 | 1.7 | 1.9 | 0.8 | 1.4 |
| srlA | 174 | 4601 | 313 | 3037 | 0.04 | 0.1 | 1.8 | 0.1 |
| sthA | 3236 | 3764 | 1936 | 2741 | 0.9 | 0.7 | 0.6 | 0.5 |
| sucA | 3988 | 8390 | 4822 | 4421 | 0.5 | 1.1 | 1.2 | 0.6 |
| tdcA | 74 | 4064 | 3412 | 8483 | 0.02 | 0.4 | 45.9 | 0.8 |
| tdcB | 147 | 4614 | 7708 | 13552 | 0.03 | 0.6 | 52.4 | 1.7 |
| tdcE | 95 | 397 | 3932 | 11414 | 0.2 | 0.3 | 41.4 | 9.9 |
| tdcG | 126 | 182 | 3018 | 14708 | 0.7 | 0.2 | 23.9 | 16.6 |
| thiC | 163 | 179 | 212 | 455 | 0.9 | 0.5 | 1.3 | 1.2 |
| thiE | 136 | 140 | 183 | 412 | 1.0 | 0.4 | 1.4 | 1.3 |
| thrA | 812 | 2686 | 1997 | 1279 | 0.3 | 1.6 | 2.5 | 0.7 |
| thrB | 849 | 2857 | 2165 | 1353 | 0.3 | 1.6 | 2.6 | 0.8 |
| thrC | 783 | 1722 | 1594 | 1378 | 0.5 | 1.2 | 2.0 | 0.9 |
| tnaA | 688 | 16366 | 10805 | 23343 | 0.04 | 0.5 | 15.7 | 0.7 |
| tnaB | 74 | 3825 | 1957 | 9810 | 0.02 | 0.2 | 26.6 | 0.5 |
| treB | 4465 | 17280 | 4711 | 19044 | 0.3 | 0.3 | 1.1 | 0.3 |
| treC | 3007 | 12164 | 2606 | 15416 | 0.3 | 0.2 | 0.9 | 0.2 |
| Upd | 11584 | 3965 | 4736 | 6483 | 2.9 | 0.7 | 0.4 | 1.2 |
| ymfL | 29 | 56 | 364 | 71 | 0.5 | 5.1 | 12.5 | 6.6 |
| ymfO | 114 | 177 | 961 | 137 | 0.7 | 7.0 | 8.4 | 5.4 |
Four different conditions were tested: W3110 in LB+glucose medium (WT G), W3110 in LB medium (WT), PC05 in LB+glucose medium (CRP* G), and PC05 in LB medium (CRP*)
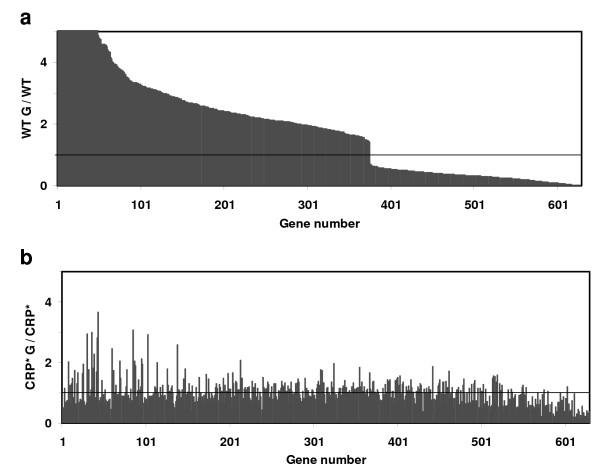
Figure 1.
Genome-wide transcriptional effects of glucose in strain W3110 expressing wild-type CRP, presented as expression ratios for individual genes showing significant differential expression in the presence and absence of glucose (WT G/WT). a) 629 genes show significant changes in expression level in response to the presence of glucose in strain W3110. b) The changes in expression levels of the same genes shown in (a) in CRP* strain PC05, in response to glucose. Gene names and expression levels are given in supplementary Table S2 (see Additional file 2).
Table 3.
Comparison between expression levels (signal values) of the wild-type strain genes in response to the presence of glucose in two different studies.
| Gene symbol | This study | Study by Gosset et al. [30] | ||
| WT G | WT G/WT | WT G | WT G/WT | |
| Upregulated genes in the study by Gosset and coworkers | ||||
| aceE | 7646 | 0.58 | 8177 | 4.4 |
| fis | 3058 | 0.99 | 5726 | 6.9 |
| guaB | 4670 | 0.61 | 2749 | 4.2 |
| ptsG | 13731 | 3.50 | 2387 | 3.2 |
| rplS | 8353 | 0.84 | 5704 | 2.7 |
| rpmE | 13204 | 1.18 | 8773 | 3.9 |
| rpsQ | 6405 | 0.51 | 3311 | 2.7 |
| rpsT | 10344 | 1.8 | 24476 | 2.6 |
| spf | 11308 | 3.40 | 26801 | 11.2 |
| Downregulated genes in the study by Gosset and coworkers | ||||
| aceA | 1806 | 0.57 | 498 | 0.2 |
| aceB | 1466 | 0.65 | 296 | 0.2 |
| aldA | 370 | 0.15 | 305 | 0.1 |
| fumA | 956 | 0.29 | 819 | 0.3 |
| gltA | 126 | 0.27 | 452 | 0.1 |
| mdh | 5878 | 0.62 | 996 | 0.2 |
| pckA | 6733 | 0.56 | 765 | 0.3 |
| sdhA | 1033 | 0.14 | 614 | 0.2 |
| sdhB | 556 | 0.10 | 401 | 0.2 |
| sucA | 3988 | 0.47 | 822 | 0.2 |
| sucB | 6087 | 0.60 | 802 | 0.1 |
| sucC | 5447 | 0.59 | 1250 | 0.2 |
| sucD | 3439 | 0.58 | 526 | 0.1 |
This comparison focuses on genes involved in central metabolism.
Figure 1b depicts the changes in expression levels of the same genes shown in Figure 1a for CRP mutant strain PC05 in response to glucose. The average WT G/WT ratio for the genes which are upregulated in W3110 in the presence of glucose is 4.06 while the average CRP* G/CRP* ratio for the same genes is 1.07. For downregulated genes in W3110 by glucose, WT G/WT and CRP* G/CRP* ratios are 0.32 and 0.81 respectively. These results show that genes whose expression is significantly altered by glucose in strain W3110 are generally not altered to the same extent in strain PCO5 and that CRP* suppresses this effect of glucose.
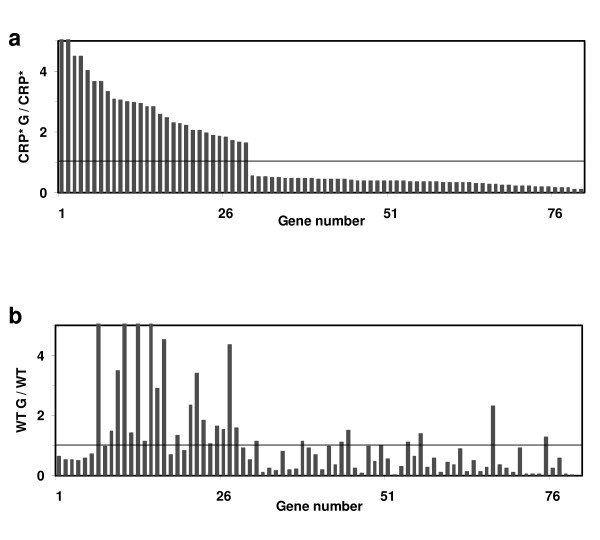
Figure 2 depicts that fewer genes show significant changes in expression level for strain PC05 (80 genes) compared to W3110 (629 genes) when grown in the presence versus absence of glucose. 29 of these genes are upregulated in the presence of glucose. This confirms the expected role of CRP* in the alleviation of glucose repression. Only 43 genes are common between those of Figure 1 and Figure 2. In contrast to W3110, the number of genes that are repressed in the presence of glucose in PC05 is greater than the number of genes that are upregulated. Only 3% of genes that are upregulated in W3110 in the presence of glucose are also upregulated in PC05 in the same condition, while 12% of glucose-repressed genes in W3110 are also repressed by glucose in PC05.
Figure 2.
Genome-wide transcriptional effects of glucose in CRP* strain PC05, presented as expression ratios for individual genes showing significant differential expression in the presence and absence of glucose (CRP* G/CRP*). a) 80 genes show significant changes in expression level for strain PC05. Only 29 are upregulated. b) The changes in expression levels of the same genes shown in (a) for strain W3110, in response to glucose. Gene names and expression levels are given in supplementary Table S3 (see Additional file 3).
The complete list of expression levels of the genes that are differentially expressed between in PC05 with versus without glucose is provided in Supplementary Table S3 (see Additional file 3). Specific examples of genes that are upregulated in PC05 in response to glucose include: the PTS gene ptsG, che genes (involved in regulation of chemotaxis), dhaM (associated with dihydroxyacetone kinase), edd (encoding phosphogluconate dehydratase of the Entner-Doudoroff pathway), gnt genes (gluconate transport and metabolism), genes involved in amino acid metabolism such as glt (glutamate synthase), and ser (serine biosynthesis) genes, and ymf genes of the lambdoid prophage element e14. Genes that are downregulated in PC05 in response to glucose include: argD (involved in lysine and arginine biosynthesis), glp genes (glycerol transport and metabolism), gntP (encoding a gluconate transporter), srlA (glucitol/sorbitol PTS system), thiCE genes (involved in thiamine biosynthesis), and treBC genes (trehalose transport and metabolism). Refer to Table 2 for expression levels and ratios.
418 genes in the E. coli genome are suggested to be regulated in part by CRP, as reported by the most current EcoCyc database [34]. While the modes of regulation of many of these genes are complicated and not well understood (often involving multiple transcription factor binding sites), the CRP-cAMP complex is assigned to be a transcriptional activator for approximately 321 genes (implying upregulation in the absence of glucose) and a repressor for approximately 46 genes (implying downregulation in the absence of glucose) (in some cases the role of CRP is dual or unclear). While 629 genes show significant changes in their expression levels in response to the presence of glucose in W3110 (Figure 1), only 19% of them (118 out of 629) have a CRP binding site close to their start codon (these genes are highlighted in green in Additional file 2 (Table S2)). This result is perhaps not unexpected when considering that most genes under CRP control are also regulated by other transcription factors [34]. Of these 118 genes, 25 are upregulated while 93 show reduced expression in the presence of glucose. 77 out of the 93 genes downregulated in glucose are described in Ecocyc as being activated by CRP-cAMP, showing good agreement with the expected inverse relationship between glucose presence and CRP-cAMP activity. Meanwhile only 23 of the 80 genes showing significant differential expression in the presence of glucose for strain PC05 are present among the list of 418 genes believed to be directly regulated by CRP (highlighted in green in Additional file 3 (Table S3); 7 are upregulated in glucose, and 13 out of the remaining 16 downregulated genes are reported to be activated by CRP-cAMP and therefore expected to show lower expression in the presence of glucose), again demonstrating significant alleviation of catabolite repression. Thus, the majority of expression changes resulting from the presence of glucose or the CRP* mutations are not directly related to altered regulation by CRP at CRP binding sites, but rather due to secondary effects resulting from a smaller number of direct, CRP-mediated expression differences.
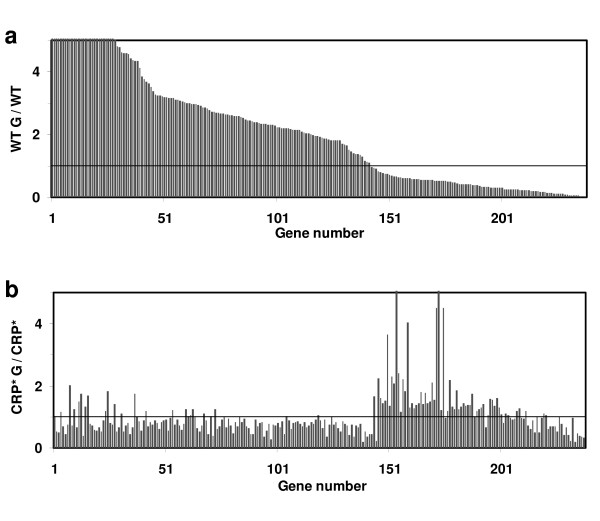
To investigate which genes respond differently to glucose in PC05 compared to W3110, an interaction term ((CRP* G/CRP*)/(WT G/WT)) was examined with the same criteria as pair-wise comparisons. This comparison reports the difference of differences and reveals that 238 genes respond differently to the presence of glucose in W3110 compared to PC05, as illustrated in Figures 3a and 3b (listed in supplementary Table S4 (see Additional file 4)). As shown in Table 2, ppsA (encoding phosphoenolpyruvate synthase), glp genes (involved in glycerol transport and metabolism), mgl genes of the galactose ABC transporter, and TCA cycle genes fumA (fumarate hydratase class I) and sdhA, sdhB, sdhD (succinate dehydrogenase) all show significantly different responses to glucose in W3110 compared to PC05.
Figure 3.
Genes that respond differently to the presence of glucose in W3110 compared to PC05, (CRP* G/CRP*)/(WT G/WT). a) Ratio of expression levels of these 238 genes in strain W3110 in the presence and absence of glucose (WT G/WT). b) Ratio of expression levels of the same genes shown in (a) in strain PC05, in the presence and absence of glucose (CRP* G/CRP*). Gene names and expression levels are given in supplementary Table S4 (see Additional file 4).
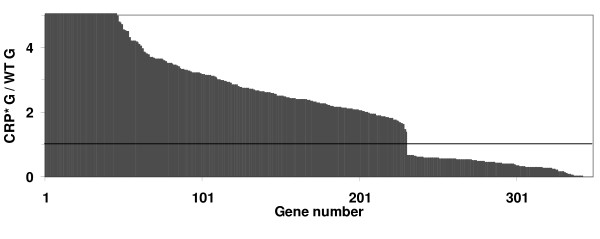
Pair-wise comparison between the CRP* G and WT G conditions identifies the changes in transcriptional levels of genes affected by CRP* in the presence of glucose. In this category, 349 genes show significant changes in their expression levels (Table 1), as depicted in Figure 4 and as listed in supplementary Table S5 (see Additional file 5). Many of the genes upregulated by CRP* (strain PC05) are involved in transport, catabolism, and amino acid metabolism (Table 2), including glp genes (involved in glycerol transport and metabolism), mgl genes (galactose ABC transporter), tdc genes (involved in serine and threonine metabolism), and tnaB (encoding a tryptophan transporter). Genes downregulated by CRP* in this comparison include gcd (encoding glucose dehydrogenase), glnP (glutamine ABC transporter), pro genes (involved in proline biosynthesis), and udp (uridine phosphorylase, involved in pyrimidine ribonucleoside metabolism). Additional differentially expressed genes of relevance to this study are described in the Discussion.
Figure 4.
Significant genome-wide transcription effects of expressing CRP* (strain PC05) instead of wild-type CRP (W3110) in the presence of glucose, presented as individual gene expression ratios (CRP* G/WT G). 349 genes show significant changes in their expression levels between CRP* G and WT G conditions. Gene names and expression levels are given in supplementary Table S5 (see Additional file 5).
Pair-wise comparison of gene expression in PC05 and W3110 grown on LB (without glucose) reveals changes in gene transcription levels as a result of the CRP* mutation. This comparison shows that 553 genes are expressed differently (392 of them are upregulated) between these two strains in the absence of glucose. These results are summarized in Table 1, and the specific genes are listed in supplementary Table S6 (see Additional file 6). gapC (encoding glyceraldehyde 3-phosphate dehydrogenase), nrf genes (involved in anaerobic respiration), and tdc genes (involved in serine and threonine metabolism) are examples of genes upregulated by CRP*.
Finally, to examine the extent to which CRP* reduces the glucose effect, we performed a pair-wise test between two conditions: PC05 in the presence of glucose (CRP* G) and W3110 in the absence of glucose (WT). Our results show that the transcriptional levels of 481 genes are significantly different between these two conditions. These results are summarized in Table 1 and the specific genes and expression values are listed in supplementary Table S7 (see Additional file 7). Examples of genes most significantly upregulated by CRP* in glucose include the nrf genes, citF (encoding citrate lyase), gat genes (involved in hexitol transport and metabolism), edd (encoding phosphogluconate dehydratase), ldhA (lactate dehydrogenase), fru genes (fructose transport and metabolism), manZ (mannose PTS permease), ptsG, and malF (maltose ABC transporter).
Real-Time Reverse Transcription PCR
To confirm the microarray results, the transcript levels of ppsA and pntA, both of which showed significant changes in their transcriptional levels under the various conditions tested, were compared by real-time reverse transcription PCR. The transcript of rpsQ was also analyzed, as this gene was noted to respond quite differently to the presence of glucose with wild-type CRP in our study as compared to the study by Gosset (WT G/WT value of 0.51 compared to 2.7) [30]. Data are presented in supplementary Table S8b as fold-changes (signal ratios) for all conditions tested, and show a good agreement between microarray and real-time RT-PCR results (see Additional file 8).
Cofactor analysis
In order to better understand how CRP* expression may influence NADPH availability for xylose reduction, intracellular cofactor concentrations were quantified for wild-type and CRP* strains engineered to produce xylitol. To prevent xylose metabolism in these strains, xylB encoding xylulokinase was deleted [24]. The wild-type CRP strain (W3110ΔxylB) was transformed with plasmid pPCC207 for inducible co-expression of an NADPH-dependent xylose reductase (CbXR) and the E. coli ATP-dependent xylose transporter system (XylFGH) [25], while the CRP* strain (PC05ΔxylB) was transformed with plasmid pLOI3815 for CbXR expression [24]. The strains were then compared after growth on glucose (no xylitol production) versus growth on glucose plus xylose (resulting in xylose reduction).
Results from the intracellular cofactor concentration measurements are summarized in Table 4. Also listed are the xylitol production results for these strains in batch fermentations and resting cell cultures. Note that the CRP* strain produced considerably more xylitol and with a higher yield. In the absence of xylose, the NADPH concentration is significantly higher in the CRP* strain (0.8 versus 0.5 μmol (g cdw)-1). Meanwhile, given the ability to reduce xylose to xylitol, the NADPH concentration falls to a much lower level in the CRP* strain (from 0.8 to 0.1 μmol (g cdw)-1) compared to wild-type CRP (from 0.5 to 0.3 μmol (g cdw)-1). While the NADPH/NADP+ ratios are nearly identical for both strains in the absence of glucose, the significant consumption of NADPH during xylose reduction coincides with a significantly larger drop in the NADPH/NADP+ ratio for CRP* (from 0.09 to 0.01) compared to wild-type CRP (from 0.10 to 0.05). Also note that the oxidized NADP+ concentrations are significantly elevated in the CRP* strain, while both NADH and NAD+ concentrations are much lower. The net effects are higher total NADP(H) (i.e. NADPH plus NADP+) concentrations and lower NAD(H) concentrations in the CRP* strains compared to wild-type. It is also noteworthy that the NADH/NAD+ ratio is significantly lower in the CRP* strain under both conditions tested.
Table 4.
Culture performance and intracellular cofactor levels and ratios for strains engineered to produce xylitol.
| Batch fermentationa | Resting cellsb | Cofactor concentrations μmol (g cdw)-1 |
Cofactor ratios | |||||||
| Glucose consumed (mM) |
Xylitol produced (mM) |
Acetate produced (mM) | Xylitol yieldc |
NADH | NAD+ | NADPH | NADP+ | NADH/NAD+ | NADPH/NADP+ | |
| W3110ΔxylB + pPCC207, GX | 248 | 223 | 19 | 2.5 | 10.3 | 47.9 | 0.3 ± 0.05 | 5.8 | 0.22 | 0.05 |
| W3110ΔxylB + pPCC207, G | - | - | - | - | 10 ± 1.8 | 47.9 | 0.5 | 5.1 ± 1.0 | 0.21 | 0.10 |
| PC05ΔxylB + pLOI3815, GX | 301 | 680 | 1.3 ± 1.4 | 3.9 | 1.8 ± 0.7 | 33.1 | 0.1 ± 0.04 | 9.9 ± 2.3 | 0.05 | 0.01 |
| PC05ΔxylB + pLOI3815, G | - | - | - | - | 3.8 ± 0.7 | 39.2 | 0.8 | 8.7 ± 2.6 | 0.10 | 0.09 |
W3110ΔxylB harboring plasmid pPCC207 expresses CbXR and XylFGH, while PC05ΔxylB harboring plasmid pPLOI3815 expresses CbXR. Strains were grown in the presence of glucose only (G) or in glucose plus xylose (GX), enabling xylose reduction during glucose metabolism. Standard deviations were less than 15% unless noted.
a Reported in Table 4 of reference [26].
b Resting cells were prepared as described [61]. Values reported were measured after 24 hours of biotransformation with cells suspended to OD600 = 2.0.
c Reported as moles xylitol produced per mole glucose consumed.
Discussion
We previously used a CRP* strain to promote expression of xylose transporters in the presence of glucose to produce xylitol from a glucose+xylose mixture, with xylose metabolism disabled [24]. Plasmid-based, CRP-independent expression of xylose transporters in wild-type crp strains was an alternative strategy we explored to enhance xylose uptake and xylitol production in the presence of glucose [25]. However the favorable effects of CRP* expression were found to go beyond improving xylose transport and to include other beneficial phenotypes such as reduced acetate production and higher yields on xylose reduced per mole of glucose consumed (as shown in Table 4) [26].
Transcription changes associated with CRP* expression are extensive. While many of the genes known to be regulated by CRP show altered expression in the context of CRP*, the majority of differentially expressed genes are not known to be directly under CRP control. The complex network of genes showing altered regulation due to secondary effects of CRP* is not likely to be metabolically systematic, since the mutant CRP used in this study (i.e. CRP*) does not have an evolved physiological role, was isolated under very particular growth conditions, and does not simply serve as a constitutive, cAMP-independent regulator. Therefore the difficulty in identifying clear patterns of differential expression of metabolically related genes is perhaps not surprising (the mutations in CRP* may not uniformly alter the regulator's natural physiological role at different control sites). Rather than attempting to assign physiological meaning to the altered transcriptome, we instead identify gene expression changes that help to explain the beneficial effects of CRP* expression as they relate to xylitol production. Specifically, we focus on genes that may affect NADPH availability.
Improved cofactor availability for xylitol production
In E. coli, complete oxidation of glucose during aerobic growth requires that respiration and anabolic metabolism consume reducing equivalents as they are generated [35]. Elevated glucose flux beyond the capacity of respiration and growth results in incomplete oxidation and acid secretion. Heterologous, NADPH-dependent xylitol production can act as an added electron sink in strains producing xylitol from a mixture of glucose and xylose, and an increased ability to produce xylitol during aerobic growth is expected to increase glucose oxidation and tricarboxylic acid (TCA) cycle flux, provided reducing equivalents from NADH can be converted to NADPH. Alternately, increased expression of genes involved in glucose oxidation would allow for increased xylitol production, provided reducing equivalents can be delivered as NADPH. Our transcription analysis sheds light on the observed ability of CRP* strains expressing xylose reductase to produce more xylitol and secrete less acetate than similar wild-type CRP strains constitutively expressing a xylose transporter.
TCA cycle genes involved in reactions between succinate and oxaloacetate are upregulated in PC05 compared to W3110 in the presence of glucose, as listed in Table 2. These include sdhA, sdhB, sdhD (encoding succinate dehydrogenase), fumA (encoding fumarate hydratase class I), and mdh (encoding malate dehydrogenase). All of these genes are known to have CRP binding sites in their promoter regions, so increased expression in PC05 is likely due to direct regulatory effects of CRP*. A strain with a more active TCA cycle potentially increases glucose oxidation, produces more NADPH and produces less acetate [36,37]. Upregulation of acs (acetyl-CoA synthetase) in CRP* G compared to WT G (3.63-fold) may promote acetate assimilation instead of accumulation. Also shown in Table 2, sthA encoding soluble pyridine nucleotide transhydrogenase is differentially expressed in CRP* G compared to WT G (CRP* G/WT G ratio of 0.6). SthA is believed to primarily oxidize NADPH to regenerate NADH and an increase in NADPH demand corresponds to reduced sthA expression [38,39]. As shown in Table 2, sthA expression is lower with CRP* G compared to WT G. Transcriptional control of sthA is not well understood, but the lack of an apparent CRP binding site suggests that reduced sthA expression may be a result of increased NADPH demand rather than a direct result of CRP*-mediated control. Consistent with this apparent elevated demand for NADPH is the fact that the genes encoding both subunits of the membrane-bound, proton-translocating pyridine nucleotide transhydrogenase (pntA and pntB) are upregulated in CRP* G compared to WT G (1.88 and 1.76-fold respectively). PntAB has been reported to produce 35–45% of the NADPH required for E. coli biosynthesis during aerobic growth [38]. Interestingly, the maeB gene encoding NADP-linked malic enzyme (decarboxylating malate to pyruvate) can serve as another source of NADPH regeneration in E. coli and also shows a higher level of transcription in PC05 compared to W3110 in the presence of glucose (CRP* G/WT G ratio of 2.6). Changes in transcriptional patterns of the above mentioned genes can be in response to increased demands for NADPH in CRP* strains, perhaps as this relates to apparently increased anabolic demands (described below).
To ensure continued growth, E. coli balances intracellular concentrations and ratios of the reduced and oxidized cofactors through a complex interplay between catabolic metabolism, anabolic metabolism, redox-sensitive regulation (both genetic and allosteric) and transhydrogenase activities [35,40-43]. While the mechanisms of maintaining redox balance are not well understood, the expression levels of enzymes and regulators involved in redox metabolism play a critical role. It is thus perhaps not surprising that altering the activity of a global regulator has a significant impact on cofactor concentrations and the range of attainable redox states (as demonstrated in Table 4). NADH regulates the activity of a number of enzymes involved in central metabolism and glucose oxidation (e.g. pyruvate dehydrogenase [44,45], citrate synthase [46,47] and α-ketoglutarate dehydrogenase [48]). CRP* expression results in reduced NADH levels, increased production of NADPH relative to NADH, and increased tolerance to a range of NADPH levels and NADPH/NADP+ ratios, all of which are likely to improve NADPH-dependent xylitol production during glucose metabolism.
Expression of "unnecessary" genes
Most of the genes that are differentially expressed between PC05 and W3110 in the presence of glucose (231 out of 349) are upregulated with CRP* (Figure 4), supporting the generally assumed behavior of CRP* in alleviating glucose-dependent catabolite repression. The otherwise unnecessary upregulation of these genes likely causes a significant increase in demand for carbon and energy, helping to explain the slower growth rate observed for PC05 compared to W3110 [24]. Notable genes that fall into this upregulated category include many involved in amino acids metabolism, such as tnaA (encoding tryptophanase), thrA (aspartokinase I and homoserine dehydrogenase I), glyA (serine hydroxymethyl transferase), tdcB (threonine dehydratase), thrC (threonine synthase), thrB (homoserine kinase), and asnA (asparagine synthetase A) (refer to Table 2). Upregulation of amino acid metabolism pathways may be in response to increased protein synthesis demands caused by upregulation of other genes.
Catabolite repression and inducer exclusion
A relationship between the phosphorylation state of enzyme IIAGlc and the intracellular "phosphoenolpyruvate (PEP)/pyruvate" ratio has been suggested [9]. Decreased levels of phosphorylated enzyme IIAGlc is usually accompanied by decreased PEP/pyruvate ratios. The crucial role of the unphosphorylated form of enzyme IIAGlc in catabolite repression and inducer exclusion is well documented [8,9,14-18]. As shown in Table 2, phosphoenolpyruvate synthase (encoded by ppsA) is expressed to a higher level in PC05 compared to W3110 in the presence of glucose (CRP* G/WT G ratio of 5.35). Upregulation of this enzyme which mediates conversion of pyruvate to PEP may increase the intracellular PEP/pyruvate ratio, resulting in an increase in the phosphorylated form of enzyme IIAGlc. This in turn may increase adenylate cyclase activity [17,18] and further help to alleviate catabolite repression and inducer exclusion in PC05.
Conclusion
We have used microarray analysis to compare the transcriptomes of E. coli W3110 expressing wild-type CRP and mutant strain PC05 expressing CRP* in the presence and absence of glucose. Table 1 summarizes the genome-wide effects of CRP* expression under the conditions tested. Gene expression in the context of CRP* in the presence of glucose is very different from that of wild-type in the absence of glucose. Although fewer genes show expression sensitivity to glucose in PC05 compared to W3110, CRP* does not completely eliminate glucose effects. As expected, CRP* expression causes increased expression of genes involved in nutrient transport and catabolism (among many others). In addition, several genes showing significant differential expression in CRP* versus wild-type CRP help to explain the observed differences in cofactor levels and metabolic behavior of CRP* strains used in xylitol production.
Materials and methods
General
E. coli K-12 strain W3110 (ATCC 27325) and its derivatives were maintained on plates containing Luria-Bertani (LB) medium (10 g tryptone, 5 g yeast extract, 5 g NaCl, and 15 g agar per liter). Methods for construction of strains PC05 (W3110 and crp*), PC07 (W3110ΔxylB), and PC09 (PC05ΔxylB) were described previously [24]. Briefly, the crp* gene and xylB deletion were introduced into W3110 via P1 phage transduction using a lysate from strain ET25 (crp*::Tn10) [8] and PC06 (W3110, ΔxylB::FRT-aac-FRT) [24] followed by selections on tetracycline (for crp*) or apramycin (for ΔxylB) plates. Plasmid pLOI3815 is a medium copy, pBR322-origin vector carrying a kanamycin resistance marker and the xylose reductase gene from Candida boidinii, which is located downstream of tac promoter and upstream of a transcription termination sequence [24]. Xylose transporter genes xylFGH (ATP-dependent xylose transporter system) were cloned downstream of CbXR in pLOI3815 to make plasmid pPCC207.
Amino acid substitutions in the CRP* were confirmed by sequencing. The crp* phenotype was verified in two ways. First, several TetR transductants were grown in LB medium containing glucose (1%) and xylose (1%). Cells were harvested at mid logarithmic growth phase and washed twice in phosphate buffer containing kanamycin (50 μg/mL). After allowing time for residual sugars to be cleared, the cells were resuspended a final time in buffer containing xylose (1%), kanamycin, and 1% triphenyltetrazolium chloride (TTC). Reduction of TTC results in red color formation and indicates constitutive xylose utilization. The crp* phenotype was additionally confirmed using HPLC to verify simultaneous glucose and xylose consumption in batch cultures [24].
Growth conditions
Four different conditions were tested in this study: W3110 in LB medium (WT), W3110 in LB+glucose medium (WT G), PC05 in LB medium (CRP*), and PC05 in LB+glucose medium (CRP* G). All experiments were performed at least in triplicate and all data reported are the average of at least three experiments. Cell culture optical density was measured at 600 nm (OD600) using a SPECTRAMax PLUS384 spectrophotometer (Molecular Devices). Cells grown for harvesting were prepared briefly as follows. Overnight pre-seed cultures were prepared by inoculating 3 ml of LB medium (in 13 × 100 mm tube) with a few colonies from a fresh LB plate. The overnight cultures were used to inoculate, to an OD600 of 0.1, 50 ml LB media (with or without 0.4% glucose supplementation) seed cultures in a 250 ml shake-flask. The seed culture were grown at 37°C to an OD600 of ~2 and then were used to directly inoculate, to an OD600 of 0.02, 100 ml LB media (with or without 0.4% glucose supplementation) cultures in a 500 ml flask. These cultures were grown at 37°C and 250 rpm to an OD600 of 0.5.
Cell harvesting and preparation of RNA
Cells from the 100 ml culture were harvested at an OD600 of 0.5 (early logarithmic growth phase) by immediately placing on ice, transferring to 50 ml falcon tubes and centrifuging at 4°C for 5 minutes before treating with lysozyme. Promega PureYield™RNA Midiprep System kit was used for RNA extraction. As a preliminary check, RNA yield and quality were determined by spectrophotometry according to the manufacturer's protocol and the integrity of the purified RNA was determined by formaldehyde agarose gel electrophoresis.
Labeling, hybridization and scanning
Total RNA concentration and purity were determined using a NanoDrop spectrophotometer and total RNA integrity was examined using an Agilent Bioanalyzer. Total RNA of sufficient concentration, purity, and integrity was labeled and subsequently hybridized to Affymetrix GeneChip microarrays by the Penn State DNA Microarray Facility according to the manufacturer's instructions (Affymetrix Inc, Santa Clara, CA). Briefly, 10 μg of total RNA was converted to cDNA using random primed reverse transcription. cDNA was purified by removing the RNA via hydrolysis with NaOH and then neutralizing the solution. Purified cDNA was fragmented and subsequently end-labeled with biotin. Fragmented, end-labeled cDNA was dissolved in hybridization cocktail and hybridized to Affymetrix GeneChip E. coli Genome 2.0 Arrays (approximately 10000 probe set) for 16 hours at 45°C. The details of GeneChip E. coli Genome 2.0 Arrays are described by Affymetrix [49].
After hybridization, the hybridization cocktail was removed and the arrays were washed to remove unbound and non-specifically bound cDNA. Hybridization was detected by staining the arrays with streptavidin phycoerythrin. All washing and staining was performed using the Affymetrix GeneChip Fluidics Station 450 according to the manufacturer's instructions (Affymetrix Inc, Santa Clara, CA). Stained arrays were scanned using the Affymetrix GCS3000 7G scanner.
Microarray data analysis
A minimum of three data sets was generated for each of the four different conditions tested (based on the combination of the strains W3110 and PC05 in LB and LB+glucose media). Affymetrix Expression Console™software (Version 1.1) was used for background adjustment, normalization and summarization of chip level data in the form of feature intensity (CEL) files in order to generate probe set summarization (CHP) files, using the probe logarithmic intensity error (PLIER) method. Data from CHP files were then exported to a Microsoft Excel spreadsheet for further analysis. Signal values for 10208 probsets from GeneChip E. coli Genome 2.0 Arrays were filtered to extract probe set data for only the E. coli K-12 strain. All calculations and analyses were performed on the 4070 genes remaining after filtration.
Signal values were transformed to the log base for the pair-wise comparisons. A linear model was fitted to each gene using the Bioconductor software package LIMMA [50,51] in the R environment [52]. The linear model coefficients were used to calculate significant differences in expression levels for all pair-wise comparisons. The P-values were adjusted by the Benjamini-Hochberg method [53] and genes with a P-value of <0.05 were considered as those with significantly different expression levels under different conditions tested. Data are reported as expression levels (signal values) or ratios of expression levels. Supplementary Table S1 (Additional file 1) contains signal values for the complete probe set data for the E. coli K-12 genome. Written code in R [52] (file name: sup_code.doc) can be found in supplementary material (see Additional file 9). Gene Annotations were transformed from AFFY probe set ID's to Entrez gene IDs using NETAFFX on the Affymetrix website [49]. The online database for annotation, visualization and integrated discovery (DAVID) [54,55] and Kyoto Encyclopedia of Genes and Genome (KEGG) [56] were used for pathway visualization and gene ontology (GO) classification.
Real-time, Reverse Transcription PCR
Total RNA samples were isolated the same way as for microarray studies. ppsA (phosphoenolpyruvate synthase), pntA (membrane-bound proton-translocating pyridine nucleotide transhydrogenase), and rpsQ (30S ribosomal subunit protein S17) were selected for confirmation by real-time reverse transcription PCR with rrsH (encoding 16S ribosomal RNA) as a control. Primer and probe sequences used for RT-PCR are listed in supplementary Table S8a (Additional file 8) and were designed by Deborah S. Grove of the Penn State Nucleic Acid Facility using Primer Express v2.0 (Applied Biosystems, Foster City, CA). Probes were synthesized by Biosearch (Novato, CA). The Applied Biosystems High Capacity cDNA Reverse Transcription Kit (part number 4368813) was used for reverse transcription according to the manufacturer's instructions for cDNA production. cDNA was amplified in an ABI 7300 real-time machine using TaqMan® Universal PCR Master Mix, No AmpErase® UNG (part number 4324018). Output was analyzed using the 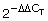 method [57].
method [57].
Cofactor measurements
The cofactor analysis used in this study is based on the methods developed by Bernofsky and Swan [58], and modified by Gibon and Larher [59], and Walton and Stewart [60]. To investigate the effect of xylitol production on intracellular NADP(H) and NAD(H) levels, cofactor concentrations and ratios were measured and compared in PC05ΔxylB strain harboring pLOI3815 and W3110ΔxylB harboring pPCC207. Seed cultures were grown at 37°C to an OD600 of ~2 and then were used to directly inoculate, to an OD600 of 0.02, 100 ml LB medium supplemented with 100 mM glucose, 100 mM xylose (or 200 mM glucose for non-xylitol producing conditions), 50 mM MOPS, kanamycin monosulfate (50 μg/ml) and isopropyl-B-D-thiogalactopyranoside (IPTG, 100 μM) in a 500 ml flask. These cultures were grown at 30°C and 250 rpm to an OD600 of 0.5. Cells were immediately chilled on ice and harvested by pelleting (4°C, 15 min, 3750 rpm) to achieve a final OD600 of 30 in 1 ml. To isolate the oxidized forms, the pellet was resuspended in 0.5 ml of 0.3 M HCl, 50 mM Tricine-NaOH (pH 8.0). To isolate the reduced forms, the pellet was resuspended in 0.5 ml of 0.3 M NaOH. All samples were then heated to 60°C for 7 minutes followed by a neutralization step (0.5 ml 0.3 M NaOH for oxidized forms, 0.3 ml 0.3 M HCl, 0.2 ml 1.0 M Tricine-NaOH (pH 8.0) for reduced forms). The neutralized solutions were then centrifuged (4°C, 60 min, 13000 rpm) and the supernatants were transferred to a new microcentrifuge tube.
Cofactor levels were measured in a 96-well microtiter plate. Either 40 μl of oxidized sample and 40 μl 0.1 M NaCl, or 80 μl of reduced sample was aliquoted to a single well. The 2X stock solution of the reaction mixture consisted of equal volumes of 1.0 M Tricine-NaOH (pH 8.0), 4.2 mM MTT, 40 mM EDTA, 1.67 mM PES, and substrate (either 5 M ethanol or 25 mM glucose-6-phosphate). After addition of the appropriate reaction mixture (ethanol for NAD(H), glucose-6-phosphate for NADP(H)), the plate was incubated at 37°C for 5 minutes. To start the reaction, either 10 units/ml alcohol dehydrogenase (from 100 units/ml stock) or 0.27 units/ml glucose-6-phosphate dehydrogenase (from 2.7 units/ml stock) was added. The formation of reduced MTT was monitored using a SpectraMax384 plate reader, taking readings every 15 seconds for 10 minutes using a wavelength of 570 nm while being incubated at 37°C. The cofactor concentration of the samples was interpolated by comparing the rate of reaction to that observed in a concentration curve run on the same plate, and subtracting the rate from the background of the sample (reaction without enzyme).
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
RK and JWC performed the experiments, DG assisted in experimental design and data analysis, RK and PCC wrote the manuscript. All authors have read and approved the final manuscript.
Note
**NOTE: Average signal values start at column Q in Table S1 and at column D in Tables S2-S7. Expression ratios start at column H in Tables S2-S7.
Supplementary Material
Table S1. Signal values for the complete probe set data for the E. coli K-12 genome.
Table S2. Expression levels of the genes that are differentially expressed between WT G and WT conditions (presented in Figure 1). Genes highlighted in green have a CRP binding site close to their start codon.
Table S3. Expression levels of the genes that are differentially expressed between CRP* G and CRP* conditions (presented in Figure 2). Genes highlighted in green have a CRP binding site close to their start codon.
Table S4. Expression levels of the genes that respond differently to the presence of glucose in W3110 compared to PC05, (CRP* G/CRP*)/(WT G/WT) (presented in Figure 3).
Table S5. Expression levels of the genes that are differentially expressed between CRP* G and WT G conditions (presented in Figure 4).
Table S6. Expression levels of the genes that are differentially expressed between CRP* and WT conditions.
Table S7. Expression levels of the genes that are differentially expressed between CRP* G and WT conditions.
Table S8. a) Primer and probe sequences used for RT-PCR. b) Comparison between microarray and real-time reverse transcription PCR results. Data are presented as fold changes (signal ratio) for all conditions tested.
The file entitled "sup_code.doc" contains the written code in R, used for data analysis.
Acknowledgments
Acknowledgements
We acknowledge W. Boos (University of Konstanz) for providing strain ET25, Dr. Craig Praul at the Penn State DNA Microarray Facility for assistance with all microarray methods and Dr. Deborah S. Grove of the Penn State Nucleic Acid Facility for assistance with RT-PCR. This research was made possible by a grant from the NSF (BES0519516) and support from the Penn State Associate Vice President for Research
Contributor Information
Reza Khankal, Email: khankal@engr.psu.edu.
Jonathan W Chin, Email: jchin@engr.psu.edu.
Debashis Ghosh, Email: ghoshd@psu.edu.
Patrick C Cirino, Email: cirino@engr.psu.edu.
References
- Sabourin D, Beckwith J. Deletion of the Escherichia coli crp gene. J Bacteriol. 1975;122:338–340. doi: 10.1128/jb.122.1.338-340.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmer M, Decrombr B, Pastan I, Perlman R. Cyclic AMP receptor protein of E. coli: its role in synthesis of inducible enzymes. Proc Natl Acad Sci USA. 1970;66:480–487. doi: 10.1073/pnas.66.2.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimova G, Ladant D, Ullmann A. Relief of catabolite repression in a cAMP-independent catabolite gene activator mutant of Escherichia coli. Res Microbiol. 2004;155:76–79. doi: 10.1016/j.resmic.2003.11.002. [DOI] [PubMed] [Google Scholar]
- Tagami H, Aiba H. Role of CRP in transcription activation at Escherichia coli lac promoter: CRP is dispensable after the formation of open complex. Nucleic Acids Res. 1995;23:599–605. doi: 10.1093/nar/23.4.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanchen A, Schicker A, Revelles O, Sauer U. Cyclic AMP-dependent catabolite repression is the dominant control mechanism of metabolic fluxes under glucose limitation in Escherichia coli. J Bacteriol. 2008;190:2323–2330. doi: 10.1128/JB.01353-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettenbrock K, Sauter T, Jahreis K, Kremling A, Lengeler JW, Gilles ED. Correlation between growth rates, EIIACrr phosphorylation, and intracellular cyclic AMP levels in Escherichia coli K-12. J Bacteriol. 2007;189:6891–6900. doi: 10.1128/JB.00819-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saier MH, Reizer J. The bacterial phosphotransferase system: new frontiers 30 years later. Mol Microbiol. 1994;13:755–764. doi: 10.1111/j.1365-2958.1994.tb00468.x. [DOI] [PubMed] [Google Scholar]
- Eppler T, Boos W. Glycerol-3-phosphate-mediated repression of malT in Escherichia coli does not require metabolism, depends on enzyme IIAGlc and is mediated by cAMP levels. Mol Microbiol. 1999;33:1221–1231. doi: 10.1046/j.1365-2958.1999.01570.x. [DOI] [PubMed] [Google Scholar]
- Hogema BM, Arents JC, Bader R, Eijkemans K, Yoshida H, Takahashi H, Alba H, Postma PW. Inducer exclusion in Escherichia coli by non-PTS substrates: The role of the PEP to pyruvate ratio in determining the phosphorylation state of enzyme IIA. Mol Microbiol. 1998;30:487–498. doi: 10.1046/j.1365-2958.1998.01053.x. [DOI] [PubMed] [Google Scholar]
- Ullmann A, Danchin A. Role of cyclic AMP in bacteria. Adv Cyclic Nucleotide Res. 1983;15:1–53. [Google Scholar]
- Kolb A, Busby S, Buc H, Garges S, Adhya S. Transcriptional regulation by cAMP and its receptor protein. Annu Rev Biochem. 1993;62:749–795. doi: 10.1146/annurev.bi.62.070193.003533. [DOI] [PubMed] [Google Scholar]
- Botsford JL, Harman JG. Cyclic AMP in prokaryotes. Microbiol Rev. 1992;56:100–122. doi: 10.1128/mr.56.1.100-122.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagami H, Inada T, Kunimura T, Aiba H. Glucose lowers CRP* levels resulting in repression of the lac operon in cells lacking. Mol Microbiol. 1995;17:251–258. doi: 10.1111/j.1365-2958.1995.mmi_17020251.x. [DOI] [PubMed] [Google Scholar]
- Osumi T, Saier MH. Regulation of lactose permease activity by the phosphoenolpyruvate:sugar phosphotransferase system:Evidence for direct binding of the glucose-specific enzyme III to the lactose permease. Proc Natl Acad Sci USA. 1982;79:1457–1461. doi: 10.1073/pnas.79.5.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson SO, Wright JK, Postma PW. The mechanism of inducer exclusion. Direct interaction between purified IIIGlc of the phosphoenolpyruvate:sugar phosphotransferase system and the lactose carrier of Escherichia coli. EMBO J. 1983;2:715–720. doi: 10.1002/j.1460-2075.1983.tb01490.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saier MH., Jr Protein phosphorylation and allosteric control of inducer exclusion and catabolite repression by the bacterial phosphoenolpyruvate: sugar phosphotransferase system. Microbiol Rev. 1989;53:109–120. doi: 10.1128/mr.53.1.109-120.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JK, Epstein W. Purification and characterization of adenylate cyclase from Escherichia coli K12. J Biol Chem. 1983;258:3750–3758. [PubMed] [Google Scholar]
- Amin N, Peterkofsky A. A dual mechanism for regulating cAMP levels in Escherichia coli. J Biol Chem. 1995;270:11803–11805. doi: 10.1074/jbc.270.20.11803. [DOI] [PubMed] [Google Scholar]
- Guidi-Rontani C, Danchin A, Ullmann A. Isolation and characterization of an Escherichia coli mutant affected in the regulation of adenylate cyclase. J Bacteriol. 1981;148:753–761. doi: 10.1128/jb.148.3.753-761.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botsford JL, Drexler M. The cyclic 3',5'-adenosine monophosphate receptor protein and regulation of cyclic 3',5'-adenosine monophosphate synthesis in Escherichia coli. Molec Gen Genet. 1978;165:47–56. doi: 10.1007/BF00270375. [DOI] [PubMed] [Google Scholar]
- Harman JG, McKenney K, Peterkofsky A. Structure-function analysis of 3 cAMP-independent forms of the cAMP receptor protein. J Biol Chem. 1986;261:6332–6339. [PubMed] [Google Scholar]
- Aiba H, Nakamura T, Mitani H, Mori H. Mutations that alter the allosteric nature of cAMP receptor protein of Escherichia coli. EMBO J. 1985;4:3329–3332. doi: 10.1002/j.1460-2075.1985.tb04084.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aristidou A, Penttila M. Metabolic engineering applications to renewable resource utilization. Curr Opin Biotechnol. 2000;11:187–198. doi: 10.1016/S0958-1669(00)00085-9. [DOI] [PubMed] [Google Scholar]
- Cirino PC, Chin JW, Ingram LO. Engineering Escherichia coli for xylitol production from glucose-xylose mixtures. Biotechnol Bioeng. 2006;95:1167–1176. doi: 10.1002/bit.21082. [DOI] [PubMed] [Google Scholar]
- Khankal R, Chin JW, Cirino PC. Role of xylose transporters in xylitol production from engineered Escherichia coli. J Biotechnol. 2008;134:246–252. doi: 10.1016/j.jbiotec.2008.02.003. [DOI] [PubMed] [Google Scholar]
- Khankal R, Luziatelli F, Chin JW, Frei CS, Cirino PC. Comparison between Escherichia coli K-12 strains W3110 and MG1655 and wild-type E. coli B as platforms for xylitol production. Biotechnol Lett. 2008;30:1645–1653. doi: 10.1007/s10529-008-9720-7. [DOI] [PubMed] [Google Scholar]
- Gorshkova I, Moore JL, McKenney KH, Schwarz FP. Thermodynamics of cyclic nucleotide binding to the cAMP receptor protein and its T127L mutant. J Biol Chem. 1995;270:21679–21683. doi: 10.1074/jbc.270.37.21679. [DOI] [PubMed] [Google Scholar]
- Belduz AO, Lee EJ, Harman JG. Mutagenesis of the cyclic AMP receptor protein of Escherichia coli:targeting positions 72 and 82 of the cyclic nucleotide binding pocket. Nucleic Acids Res. 1993;21:1827–1835. doi: 10.1093/nar/21.8.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garges S, Adhya S. Sites of allosteric shift in the structure of the cyclic-amp receptor protein. Cell. 1985;41:745–751. doi: 10.1016/S0092-8674(85)80055-6. [DOI] [PubMed] [Google Scholar]
- Gosset G, Zhang ZG, Nayyar SN, Cuevas WA, Saier MH. Transcriptome analysis of CRP-dependent catabolite control of gene expression in Escherichia coli. J Bacteriol. 2004;186:3516–3524. doi: 10.1128/JB.186.11.3516-3524.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayendran C, Polen T, Wendisch VF, Friehs K, Niehaus K, Flaschel E. The plasticity of global proteome and genome expression analyzed in closely related W3110 and MG1655 strains of a well-studied model organism, Escherichia coli K-12. J Biotechnol. 2007;128:747–761. doi: 10.1016/j.jbiotec.2006.12.026. [DOI] [PubMed] [Google Scholar]
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006. Article 2006.0008. [DOI] [PMC free article] [PubMed]
- Hayashi K, Morooka N, Yamamoto Y, Fujita K, Isono K, Choi S, Ohtsubo E, Baba T, Wanner BL, Mori H, Horiuchi T. Highly accurate genome sequences of Escherichia coli K-12 strains MG1655 and W3110. Mol Syst Biol. 2006 doi: 10.1038/msb4100049. Article 2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keseler IM, Bonavides-Martinez C, Collado-Vides J, Gama-Castro S, Gunsalus RP, Johnson DA, Krummenacker M, Nolan LM, Paley S, Paulsen IT, Peralta-Gil M, Santos-Zavaleta A, Shearer AG, Karp PD. EcoCyc: A comprehensive view of Escherichia coli biology. Nucleic Acids Res. 2009;37:D464–D470. doi: 10.1093/nar/gkn751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrer T, Sauer U. Different biochemical mechanisms ensure network-wide balancing of reducing equivalents in microbial metabolism. J Bacteriol. 2009;191:2112–2121. doi: 10.1128/JB.01523-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe AJ. The acetate switch. Microbiol Mol Biol Rev. 2005;69:12–50. doi: 10.1128/MMBR.69.1.12-50.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phue JN, Shiloach J. Transcription levels of key metabolic genes are the cause for different glucose utilization pathways in E. coli B (BL21) and E. coli K (JM109) J Biotechnol. 2004;109:21–30. doi: 10.1016/j.jbiotec.2003.10.038. [DOI] [PubMed] [Google Scholar]
- Sauer U, Canonaco F, Heri S, Perrenoud A, Fischer E. The soluble and membrane-bound transhydrogenases UdhA and PntAB have divergent functions in NADPH metabolism of Escherichia coli. J Biol Chem. 2004;279:6613–6619. doi: 10.1074/jbc.M311657200. [DOI] [PubMed] [Google Scholar]
- Canonaco F, Hess TA, Heri S, Wang TT, Szyperski T, Sauer U. Metabolic flux response to phosphoglucose isomerase knock-out in Escherichia coli and impact of overexpression of the soluble transhydrogenase UdhA. FEMS Microbiol Lett. 2001;204:247–252. doi: 10.1111/j.1574-6968.2001.tb10892.x. [DOI] [PubMed] [Google Scholar]
- Bizouarn T, Althage M, Pedersen A, Tigerstrom A, Karlsson J, Johansson C, Rydstrom J. The organization of the membrane domain and its interaction with the NADP(H)-binding site in proton-translocating transhydrogenase from E. coli. Biochimica Et Biophysica Acta-Bioenergetics. 2002;1555:122–127. doi: 10.1016/S0005-2728(02)00266-9. [DOI] [PubMed] [Google Scholar]
- Arner ESJ, Holmgren A. Physiological functions of thioredoxin and thioredoxin reductase. Eur J Biochem. 2000;267:6102–6109. doi: 10.1046/j.1432-1327.2000.01701.x. [DOI] [PubMed] [Google Scholar]
- Durnin G, Clomburg J, Yeates Z, Alvarez PJJ, Zygourakis K, Campbell P, Gonzalez R. Understanding and harnessing the microaerobic metabolism of glycerol in Escherichia coli. Biotechnol Bioeng. 2009;103:148–161. doi: 10.1002/bit.22246. [DOI] [PubMed] [Google Scholar]
- Boonstra B, French CE, Wainwright I, Bruce NC. The udhA gene of Escherichia coli encodes a soluble pyridine nucleotide transhydrogenase. J Bacteriol. 1999;181:1030–1034. doi: 10.1128/jb.181.3.1030-1034.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen LC, Atkinson DE. Regulation of pyruvate dehydrogenase from Escherichia coli. Interactions of adenylate energy charge and other regulatory parameters. J Biol Chem. 1970;245:5974–5978. [PubMed] [Google Scholar]
- Kim Y, Ingram LO, Shanmugam KT. Dihydrolipoamide dehydrogenase mutation alters the NADH sensitivity of pyruvate dehydrogenase complex of Escherichia coli K-12. J Bacteriol. 2008;190:3851–3858. doi: 10.1128/JB.00104-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen NT, Maurus R, Stokell DJ, Ayed A, Duckworth HW, Brayer GD. Comparative analysis of folding and substrate binding sites between regulated hexameric type II citrate synthases and unregulated dimeric type I enzymes. Biochemistry. 2001;40:13177–13187. doi: 10.1021/bi010408o. [DOI] [PubMed] [Google Scholar]
- Maurus R, Nguyen NT, Stokell DJ, Ayed A, Hultin PG, Duckworth HW, Brayer GD. Insights into the evolution of allosteric properties. The NADH binding site of hexameric type II citrate synthases. Biochemistry. 2003;42:5555–5565. doi: 10.1021/bi020622s. [DOI] [PubMed] [Google Scholar]
- Smith MW, Neidhardt FC. 2-oxoacid dehydrogenase complexes of Escherichia coli: cellular amounts and patterns of synthesis. J Bacteriol. 1983;156:81–88. doi: 10.1128/jb.156.1.81-88.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Affymetrix http://www.affymetrix.com
- Smyth GK. Limma: Linear models for microarray data. In: Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W, editor. Bioinformatics and computational biology solutions using R and Bioconductor. New York: Springer; 2005. pp. 397–420. [Google Scholar]
- Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article 3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- R http://www.r-project.org
- Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: a practical and powerful approach to multiple testing. J Royal Stat Soc Ser B. 1995;57:289–300. [Google Scholar]
- Dennis G, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biology. 2003;4:Article R60. [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:D277–D280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
-
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the
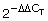 method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar] - Bernofsky C, Swan M. An improved cycling assay for nicotinamide adenine dinucleotide. Anal Biochem. 1973;53:452–458. doi: 10.1016/0003-2697(73)90094-8. [DOI] [PubMed] [Google Scholar]
- Gibon Y, Larher F. Cycling assay for nicotinamide adenine dinucleotides: Nacl precipitation and ethanol solubilization of the reduced tetrazolium. Anal Biochem. 1997;251:153–157. doi: 10.1006/abio.1997.2283. [DOI] [PubMed] [Google Scholar]
- Walton AZ, Stewart JD. Understanding and improving NADPH-dependent reactions by nongrowing Escherichia coli cells. Biotechnol Prog. 2004;20:403–411. doi: 10.1021/bp030044m. [DOI] [PubMed] [Google Scholar]
- Chin JW, Khankal R, Monroe CA, Maranas CD, Cirino PC. Analysis of NADPH supply during xylitol production by engineered Escherichia coli. Biotechnol Bioeng. 2009;102:209–220. doi: 10.1002/bit.22060. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Signal values for the complete probe set data for the E. coli K-12 genome.
Table S2. Expression levels of the genes that are differentially expressed between WT G and WT conditions (presented in Figure 1). Genes highlighted in green have a CRP binding site close to their start codon.
Table S3. Expression levels of the genes that are differentially expressed between CRP* G and CRP* conditions (presented in Figure 2). Genes highlighted in green have a CRP binding site close to their start codon.
Table S4. Expression levels of the genes that respond differently to the presence of glucose in W3110 compared to PC05, (CRP* G/CRP*)/(WT G/WT) (presented in Figure 3).
Table S5. Expression levels of the genes that are differentially expressed between CRP* G and WT G conditions (presented in Figure 4).
Table S6. Expression levels of the genes that are differentially expressed between CRP* and WT conditions.
Table S7. Expression levels of the genes that are differentially expressed between CRP* G and WT conditions.
Table S8. a) Primer and probe sequences used for RT-PCR. b) Comparison between microarray and real-time reverse transcription PCR results. Data are presented as fold changes (signal ratio) for all conditions tested.
The file entitled "sup_code.doc" contains the written code in R, used for data analysis.