Abstract
Background
KCNE1 represents the regulatory beta-subunit of the slowly activating delayed rectifier potassium channel (IKs). Variants of KCNE1 have repeatedly been linked to the long-QT syndrome (LQTS), a disorder which predisposes to deafness, ventricular tachyarrhythmia, syncope, and sudden cardiac death.
Results
We here analyze the evolution of the common Gly38Ser variant (rs1805127), using genomic DNAs, complementary DNAs, and HEK293-expressed variants of altogether 19 mammalian species. The between species comparison reveals that the human-specific Gly38Ser polymorphism evolved under strong positive Darwinian selection, probably in adaptation to specific challenges in the fine-tuning of IKs channels. The involved amino acid exchanges (Asp > Gly, Gly > Ser) are moderately radical and do not induce apparent changes in posttranslational modification. According to population genetic analyses (HapMap phase II) a heterozygote advantage accounts for the maintenance of the Gly38Ser polymorphism in humans. On the other hand, the expression of the 38Ser allele seems to be disadvantageous under certain conditions, as suggested by the sporadic deficiency of 38Ser-coding mRNAs in heterozygote Central Europeans and the depletion of homozygotes 38Ser in the Yoruban sample.
Conclusion
We speculate that individual differences in genomic imprinting or genomic recoding might have contributed to conflicting results of recent association studies between Gly38Ser polymorphism and QT phenotype. The findings thus highlight the relevance of mRNA data in future association studies of genotypes and clinical disorders. To the best of our knowledge, they moreover provide first time evidence for a unique pattern; i.e. coincidence of positive Darwinian selection and polymorphism with a sporadically suppressed expression of one allele.
Background
The potassium voltage-gated channel, Isk-related family, member 1 (KCNE1)-gene is located on chromosome 21q22.12 and stretches over three exons, with the coding 381 bp being confined to the third and last exon [1]. The encoded protein (KCNE1, also termed minK) is a single-span membrane protein of about 14.4 kDa that coassembles with KCNQ1 (potassium voltage-gated channel, KQT-like subfamily, member 1), thus forming the slowly activating delayed rectifier potassium (IKs) channel of heart muscle, inner ear, and a variety of other tissues [2-4]. Upon docking of KCNE1 to KCNQ1, activation and deactivation kinetics of IKs channels are slowed down by nearly two orders of magnitude [5]. This renders KCNE1 an interesting candidate for studying the causes of cardiovascular diseases and congenital as well as aquired hearing loss. Particularly, the inherited long-QT syndrome (LQTS) has repeatedly been linked to polymorphisms of KCNE1 [e.g. [6-8]]. LQTS occurs with or without accompanying deafness and is characterized by delayed ventricular repolarization, visible as a prolonged electrocardiographic QT interval. The dysfunction predisposes patients to ventricular tachyarrhythmia, syncope, and sudden cardiac death. As sudden death may occur with the first syncope and LQTS diagnosis based solely on electrocardiography is neither sensitive nor specific enough, it is of outmost importance to screen subjects at risk for a genetic predisposition to LQTS.
Several population-wide screens have recently been carried out to check for the relevance of the supposed association between genomic KCNE1 variants and QT length. Two of these studies identified QT-modifying single nucleotide polymorphisms (SNPs) in non-coding regions of the KCNE1 locus, in particular in intron 2 [9] and in a putative promoter region [10]. Family based association analysis showed a significant association between QT length and an A/G-SNP at nucleotide position 112 (A112G; rs1805127) that codes for a glycine/serine polymorphism at codon 38 (Gly38Ser) [11]. The same SNP is undisputably associated with mortality and T-wave alternans in electrocardiograms [12] as well as with high noise susceptibility and noise-induced hearing loss [13]. On the other hand, an association between Gly38Ser and QT length has been questioned by other association studies [14-16]. Different genetic backgrounds of the studied populations have been discussed to account for the conflicting results. For example, KCNE1 may be in linkage disequilibrium to adjacent causal variants only in some ethnic groups, but not in others [15]. However, since all available association studies exclusively used genomic information, some additional factors might have had a confounding effect on the results of association studies. Firstly, one of both alleles might have been silenced in some individuals, due to genomic imprinting or null mutations in the regulatory region. Secondly, genomic recoding at the mRNA level might have contributed to the observed patterns. According to this, an adenosine deaminase acting on RNA (ADAR) might sporadically have recoded the adenosine at KCNE1 codon 38 (AGU) to inosine (IGU), which is equivalent to guanosine (GGU) for the translational machinery [17]. To pick up this thought we compared allele frequencies of genomic (gDNAs) and complementary DNAs (cDNAs) from a total of 43 human individuals. Furthermore to get an idea about the selective forces behind KCNE1 evolution we analyzed the available human population genetic data (HapMap phase II) as well as orthologs (gDNAs and cDNAs) from 18 non-human mammals (Eutheria).
Results
Genotyping of human samples and allele frequencies in the gDNA/cDNA comparison
Direct sequencing of KCNE1-coding gDNAs revealed that 19 out of 29 lymphoblastoid cell lines were heterozygous for Gly38Ser, while six and four cell lines were homozygote for 38Ser and 38Gly, respectively. KCNE1 sequencing of gDNAs of 14 brain samples (frontal cortices) added another five heterozygotes Gly38Ser, four homozygotes 38Ser, and five homozygotes 38Gly. All genotypes were confirmed once (brain samples) and twice (cell lines) using a reverse pyrosequencing assay (Tables 1 and 2). We take the wide agreement of the results from direct sequencing and pyrosequencing and particularly the exclusive detection of one allele in homozygotes as evidence for the reliability of the present reverse pyrosequencing assay. Twofold parallel pyrosequencing of cDNAs coding for KCNE1 widely reflected the patterns described for gDNAs (Tables 1 and 2): heterozygotes displayed an about 50:50 distribution of 38Gly- and 38Ser-coding alleles. Moreover, when homozygotes had been analysed, the pyrosequencing assay revealed an extensive (mostly absolute) excess of either the 38Gly- or the 38Ser-coding allele. In contrast to the general agreement of the allele frequencies inferred from gDNAs and cDNAs, we found exclusively 38Gly-coding alleles in the cDNAs of three individuals (cell lines 24 and 26; brain sample 11) that had been genotyped as heterozygotes Gly38Ser. Brain sample 11 was conspicuous on the gDNA level, too: though clearly heterozygous, pyrosequencing persistently detected less 38Ser- (12–22.2%) than 38Gly-coding alleles (88–77.8%) in a total of eight parallel measurements.
Table 1.
Frequencies of 38Ser- and 38Gly-coding KCNE1 alleles (%112A/%112G) in gDNAs and cDNAs of lymphoblastoid cell lines, as inferred by pyrosequencing
| Lymphoblastoid cell line |
gDNA (%112A/%112G) | cDNA (%112A/%112G) | ||
| 1st Run | 2nd Run | 1st Run | 2nd Run | |
| 1 | 56.4/43.6 | 54.7/45.3 | 54.7/45.3 | 59.8/50.2 |
| 2 | 54.7/45.3 | 52.7/47.3 | 40.6/59.4 | 59.2/40.8 |
| 3 | 100.0/0.0 | 100.0/0.0 | 100.0/0.0 | 100.0/0.0 |
| 4 | 48.7/51.3 | 46.8/53.2 | 39.5/60.5 | 45.1/54.9 |
| 5 | 56.6/43.4 | 57.2/42.8 | 61.6/38.4 | 54.2/45.8 |
| 6 | 100.0/0.0 | 100.0/0.0 | 100.0/0.0 | 100.0/0.0 |
| 7 | 57.5/42.5 | 53.0/47.0 | 60.8/39.2 | 56.2/43.8 |
| 8 | 56.5/43.5 | 55.3/44.7 | 58.3/41.7 | 59.4/40.6 |
| 9 | 0.0/100.0 | 0.0/100.0 | 0.0/100.0 | 0.0/100.0 |
| 10 | 0.0/100.0 | 0.0/100.0 | 0.0/100.0 | 0.0/100.0 |
| 11 | 0.0/100.0 | 0.0/100.0 | 0.0/100.0 | 5.9/94.1 |
| 12 | 0.0/100.0 | 0.0/100.0 | 0.0/100.0 | 0.0/100.0 |
| 13 | 53.1/46.9 | 55.2/44.8 | 60.0/40.0 | 55.1/44.9 |
| 14 | 56.3/43.7 | 54.9/45.1 | 45.9/54.1 | 62.5/37.5 |
| 15 | 56.7/43.3 | 55.7/44.3 | 59.3/40.7 | 69.2/30.8 |
| 16 | 56.0/44.0 | 54.9/45.1 | 59.3/40.7 | 53.1/46.9 |
| 17 | 57.6/42.4 | 51.7/48.3 | 63.6/36.4 | 56.5/43.5 |
| 18 | 54.7/45.3 | 50.1/49.9 | 60.0/40.0 | 54.6/45.4 |
| 19 | 52.9/47.1 | 56.5/43.5 | 51.2/48.8 | 51.5/48.5 |
| 20 | 54.4/45.6 | 52.8/47.2 | 52.7/47.3 | 51.4/48.6 |
| 21 | 100.0/0.0 | 100.0/0.0 | 96.6/3.4 | 100.0/0.0 |
| 22 | 100.0/0.0 | 100.0/0.0 | 100.0/0.0 | 100.0/0.0 |
| 23 | 56.2/43.8 | 56.0/44.0 | 62.6/37.4 | 52.2/47.8 |
| 24 | 55.0/45.0 | 53.2/46.8 | 0.0/100.0 | 0.0/100.0 |
| 25 | 100.0/0.0 | 100.0/0.0 | 100.0/0.0 | 96.8/3.2 |
| 26 | 54.1/45.9 | 54.7/45.3 | 0.0/100.0 | 0.0/100.0 |
| 27 | 100.0/0.0 | 100.0/0.0 | 100.0/0.0 | 97.1/2.9 |
| 28 | 49.6/50.4 | 52.0/48.0 | 59.7/40.3 | 59.3/40.7 |
| 29 | 52.3/47.7 | 50.5/49.5 | 52.1/47.9 | 48.4/51.6 |
Lymphoblastoid cell lines with an absolute depletion of 38Ser-coding alleles (112A) at the cDNA level are highlighted with bold.
Table 2.
Frequencies of 38Ser- and 38Gly-coding KCNE1 alleles (%112A/%112G) in gDNAs and cDNAs of brain samples (frontal cortex), as inferred by pyrosequencing
| Brain sample |
gDNA (%112A/%112G) | cDNA (%112A/%112G) | |
| 1st Run | 2nd Run | ||
| 1 | 0.0/100.0 | - | - |
| 2 | 0.0/100.0 | - | - |
| 3 | 0.0/100.0 | - | - |
| 4 | 100.0/0.0 | 100.0/0.0 | 100.0/0.0 |
| 5 | 51.8/48.2 | 60.0/40.0 | 50.1/49.9 |
| 6 | 0.0/100.0 | - | - |
| 7 | 0.0/100.0 | - | - |
| 8 | 52.5/47.5 | 52.6/47.4 | 53.6/46.4 |
| 9 | 100.0/0.0 | 100.0/0.0 | 100.0/0.0 |
| 10 | 100.0/0.0 | 100.0/0.0 | 100.0/0.0 |
| 11 | 22.2/77.8* | 0.0/100.0 | 0.0/100.0 |
| 12 | 61.8/38.2 | 48.1/51.9 | 56.4/43.6 |
| 13 | 100.0/0.0 | 100.0/0.0 | 100.0/0.0 |
| 14 | 54.5/45.5 | 59.6/40.4 | 60.0/40.0 |
*The genotype of brain sample 11 has been verified by a total of eight parallel measurements (not shown). It is marked with bold as it exerts an absolute depletion of 38Ser-coding alleles (112A) at the cDNA level.
We additionally pyrosequenced the highly heterozygous A/G-SNP rs11254413 of the housekeeping gene tRNA aspartic acid methyltransferase 1 (TRDMT1). Based on gDNAs and cDNAs we found no evidence for a complete depletion of one allele in the cDNAs of 18 heterozygous lymphoblastoid cell lines. Instead, all heterozygotes were confirmed on the cDNA level. This suggests that neither RNA extraction nor specific conditions of some of the samples affected the allele quantification of the KCNE1-SNP rs1805127.
Population genetic analyses of HapMap phase II data
As can be seen from Table 3, the Gly38Ser polymorphism is present not only in our own sample, but also in the genepools of those populations that had been genotyped in the course of phase II of the HapMap project (Central Europeans, Han-Chinese from Beijing, Japanese from Tokyo, Yoruba from Ibadan/Nigeria). Of particular interest is the finding of both alleles in sub-Saharan Yoruba, as this population is commonly regarded as the deepest coalescing among the sampled ones (see e.g. [18]). Thereafter, it is very likely that the A/G-SNP at KCNE1 codon 38 was already present before the divergence of modern humans. Table 3 moreover illustrates a general trend for selection in favour of heterozygotes: compared to the expectation, there are persistently less observed homozygotes (38Ser, 38Gly) and more observed heterozygotes (Gly38Ser) in Central Europeans, Han Chinese, and Yoruba. Consequently, Fstat calculates a negative fixation index, Fis, for these samples. In case of the Yoruban sample, homozygotes 38Ser are even absent, though 11 homozygotes 38Ser are to be expected given the allele frequencies. The Yoruban sample is also specific in that probability test statistic (Fstat) indicates a significant (p-value = 0.003) heterozygote excess exclusively in this sample, even when the p-value has been adjusted for multiple testing (strict Bonferroni correction). Finally, it is worth to note that Japanese from Tokyo represent the only exception from the general trend for selection in favour of heterozygotes: expected and observed genotype counts are the same with respect to homozygotes and nearly identical regarding the heterozygotes. We expect that a future extension of the HapMap dataset will change some details of the picture outlined above. For instance, it appears probable to us that additional data on the Yoruban population will contribute homozygotes 38Ser. It may moreover happen that the reported extent of heterozygote excess in Central Europeans, Han Chinese, and Yoruba will change with additional data. However, we are confident that the general trends inferred from the present dataset (heterozygote excess in the one or other population) will be confirmed in the future.
Table 3.
Population genetic analyses of the A112G-SNP (rs1805127) encoding the Gly38Ser polymorphism of KCNE1
| HapMap-sample | Genotype counts | Allele frequency | Test statistic | ||||
| AA | GA | GG | |||||
| (number of individuals) | Homozygotes 38Ser | Heterozygotes Gly38Ser | Homozygotes 38Gly | A | G | Fis | p-value |
| Central Euro-peans (120) | O = 14 E = 18 |
O = 64 E = 57 |
O = 42 E = 46 |
0.383 | 0.617 | -0.124 | 0.110 |
| Han Chinese, Beijing (90) |
O = 12 E = 14 |
O = 48 E = 43 |
O = 30 E = 32 |
0.400 | 0.600 | -0.106 | 0.223 |
| Japanese, Tokyo (88) | O = 8 E = 8 |
O = 36 E = 37 |
O = 44 E = 44 |
0.295 | 0.705 | +0.023 | 0.660 |
| Yoruba, Nigeria (120) | O = 0 E = 11 |
O = 72 E = 51 |
O = 48 E = 59 |
0.300 | 0.700 | -0.425 | 0.003 |
Comparison of the observed (O) and expected (E) genotype counts reveals a general trend for less observed homozygotes and more observed heterozygotes than expected. In case of the Yoruban sample, the heterozygote excess is even significant (see fixation index, Fis, and probability for deviation from Hardy-Weinberg equilibrium, p-value).
Evolution of the KCNE1 gene in eutherian mammals
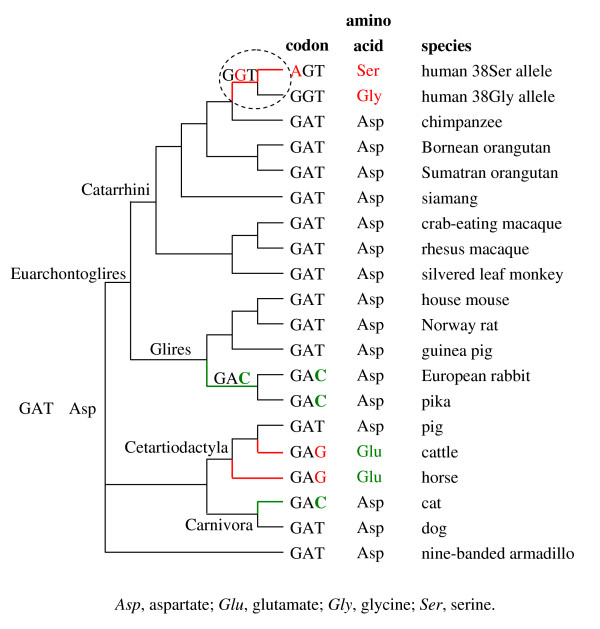
The gDNAs and cDNAs of the 18 non-human species either encode aspartate (Asp) or glutamate (Glu) at the codon position homolog to codon 38 in human KCNE1. There was no evidence for a polymorphism of the codon under scrutiny in any of the non-human species for which SNP data were available at the time of the study (chimpanzee, Bornean orangutan, house mouse, Norway rat, and dog). Neither did databank mining reveal any hint for genomic recoding of this particular codon in the non-human sample, as indicated by identical triplets in the gDNA/cDNA comparison in Bornean orangutan, house mouse, Norway rat, guinea pig, European rabbit, cattle, and cat (see Table 4 for scientific species names and accession numbers).
Table 4.
Species and KCNE1 accession numbers in the order of their appearence in Figure 1
| Scientific name (common name) | Accession numbers (GenBank and ENSEMBL) of gDNAs (g) and cDNAs (c) |
| Homo sapiens, 38Ser variant (human) | EF514881 (g, ne, pa) |
| H. sapiens, 38Ser variant (human) | EF514882 (c, ne) |
| H. sapiens, 38Gly variant (human) | EF514883 (c, ne) |
| H. sapiens, 38Gly variant (human) | EF514884 (g, ne, pa) |
| Pan troglodytes (chimpanzee) | ENSPTRT00000025885 (g, pa) |
| Pongo pygmaeus (Bornean orangutan) | ENSPPYT00000013220 (g), EF514885 (c, ne, pa) |
| Pongo abelii (Sumatran orangutan) | NM_001132604 (c, pa), CR854432 (c) |
| Symphalangus syndactylus (siamang) | EF514886 (c, ne, pa) |
| Macaca fascicularis (crab-eating macaque) | EF514887 (c, ne, pa) |
| Macaca mulatta (rhesus macaque) | ENSMMUT00000042620 (g, pa) |
| Presbytis cristata (silvered leaf monkey) | EF514888 (c, ne, pa) |
| Mus musculus (house mouse) | ENSMUST00000051705 (g), X60457 (c, pa), NM_008424 (c), BX518225 (c), BY708822 (c), AI956381 (c), BB613272 (c), AJ507048 (c), BF540248 (c), AA667912 (c), AA667785 (c), AK028907 (c), BC094276 (c) |
| Rattus norvegicus (Norway rat) | ENSRNOT00000002717 (g), NM_012973 (c, pa), M36461 (c), BQ194830 (c), CX569446 (c) |
| Cavia porcellus (guinea pig) | ENSCPOT00000011328 (g), L20462 (c, pa) |
| Oryctolagus cuniculus (European rabbit) | L41659 (g, pa), EU008571 (c), EU008572 (c), NM_001109822 (c) |
| Ochotona princeps (pika) | ENSOPRT00000007254 (g, pa) |
| Sus scrofa (pig) | NM_214165 (c, pa) |
| Bos taurus (cattle) | ENSBTAT00000001530 (g), NM_001077977 (c, pa), BE486735 (c), DV843524 (c), CK849850 (c) |
| Equus caballus (horse) | ENSECAT00000004027 (g, pa) |
| Felis catus (cat) | ENSFCAT00000006761 (g), NM_001009206 (c, pa) |
| Canis familiaris (dog) | ENSCAFT00000015169 (g, pa) |
| Dasypus novemcinctus (nine-banded armadillo) | ENSDNOT00000001508 (g, pa) |
Accession numbers of new entries are highlighted by "ne". Sequences that have been used for PAML analyses are labeled with "pa".
In line with the overall prevalence of GAT (Asp) at the site corresponding to codon 38 in human KCNE1, baseml suggests GAT as the ancestral character state at the base of the tree (see Figure 1). Mapping the extant as well as the ancestral character states onto the phylogeny in Figure 1 suggests that only few evolutionary steps occurred since the last common ancestor of the studied species. Thus, two independent synonymous substitutions from GAT to GAC can be assumed for the terminal branch to cat and for the internal branch uniting pika and European rabbit. Moreover, the cattle and horse branches are characterized by a parallel non-synonymous substitution changing the coding from GAT to GAG. However, the associated exchange from Asp to Glu is conservative since both amino acids are negatively charged (for amino acid classification see, e.g. [19]). The functional consequences of the Asp/Glu shift can thus be regarded as neglegible.
Figure 1.
Remarkable variability at codon 38 of human KCNE1. Extant and baseml-reconstructed tripletts (and the encoded amino acids) were mapped onto the widely accepted phylogeny among catarrhine primates [49] and Eutheria [47,48]. The discontinuous line encircles branches defined as foreground in branch-site analysis (codeml model A). Red indicates non-synonymous nucleotide substitutions and radical amino acid replacements; green highlights synonymous nucleotide substitutions and conservative amino acid exchanges. Only newly arisen tripletts are labelled along the internal branches.
The human stem lineage represents the only exception from the wide phylogenetic conservation of codon 38 and its counterparts (Figure 1). According to the presence of the Gly38Ser polymorphism in all human populations studied (e.g. [9,11,12,14-16] and present study), two non-synonymous transitions can be assumed for the branch leading to modern humans: 1) a change from GAT to GGT, and, thus, from Asp to glycine (Gly), and 2) a shift from GGT to AGT, which corresponds to a Gly to Ser replacement at the amino acid level (Figure 1). Both, the Asp/Gly shift (negatively charged/hydrophobic) and the Gly/Ser shift (hydrophobic/polar) are moderately radical. The moderately radical character of the amino acid exchanges argues for their effectiveness without disrupting protein function (for amino acid classification see, e.g. [19]). In accordance with the human-specificity of the Ser38Gly polymorphism, branch-site analysis (codeml) provides highly significant support (p-value = 0.01) for the branch-site test of positive selection across the human foreground. Not surprisingly, codon 38 of human KCNE1 belongs to the specific site class (p-value = 0.970) that is under positive selection across the human foreground (ω2 = 187), but is under negative selection (ω2a = 0.073) or neutral evolution (ω2b = 1) across the remaining phylogeny (Table 5; Figure 1). The other codon sites belonging to this specific site class are codons 105 and 115. The high ω-value assigned to the specific site class (= 187) most likely results from the short branch leading to modern humans. Irrespectively, there is no doubt about the nature of the selective force acting particularly on codon 38 of human KCNE1, i.e. strong positive Darwinian selection (Figure 1).
Table 5.
Parameter estimates from modified branch-site model A [50]
| Site class | Proportion | Remaining phylogeny ω | Human foreground ω |
| 0 | 0.709 | 0.073 | 0.073 |
| 1 | 0.267 | 1.000 | 1.000 |
| 2a | 0.017 | 0.073 | 187 |
| 2b | 0.006 | 1.000 | 187 |
Codon sites pinpointed as positively selected across the human foreground: 38 (p-value = 0.970), 105 (p-value = 0.686), and 115 (p-value = 0.962).
Motif search and Western blotting
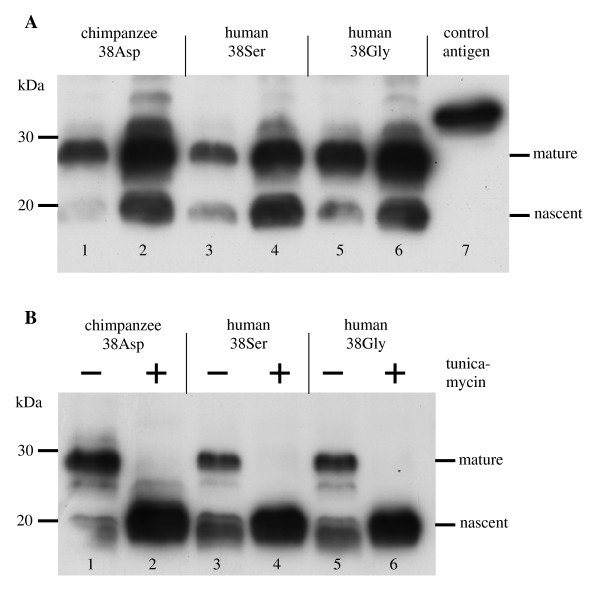
According to results from PROSITE motif search the human 38Gly variant is characterized by a glycosaminoglycan attachment motif (SerGlyXGly) spanning from amino acid positions 37–40 that is absent in the 38Ser variant and the non-human KCNE1 orthologs. Motiv search moreover found motivs for N-glycosylation, protein kinase C phosphorylation, and N-myristoylation offside the glycosaminoglycan attachment motif (not shown). To adress the question whether the evolution of the Gly38Ser polymorphism in humans entailed differences in glycosylation, we expressed different KCNE1 variants in human HEK293 cells and analysed the KCNE1 proteins by Western blotting (Figure 2A and 2B). Beside the 15 kDa KCNE1 proteins, most probably representing the nascent forms (Figure 2A, n) we could also detect slower migrating KCNE1 proteins, representing the mature, posttranslationally modified proteins (Figure 2A, m). The mobility shift resulted from N-glycosylation as addition of the N-glycosylation inhibitor tunicamycin resulted in complete loss of slower migrating KCNE1 variants (Figure 2B, lanes 2, 4 and 6). However, both human variants and the chimpanzee ortholog of KCNE1 (representing the ancestral 38Asp situation) revealed no obvious differences in mobility. This shows that all KCNE1 variants are highly N-glycosylated and suggests that the 38Gly variant does not confer an extra glucosaminoglycan modification. Therefore, we conclude that the presumed functional differences linked to the specific evolution of KCNE1 codon 38 in humans are not due to differences in posttranslational modification. Instead, they seem to be caused exclusively by the moderately radical character of the involved amino acid exchanges (Asp > Gly, Gly > Ser).
Figure 2.
KCNE1 proteins from chimpanzee and human are highly N-glycosylated. A HEK293 cells were transfected with 1 μg (lanes 1, 3, and 5) or 3 μg (lanes 2, 4, and 6) expression plasmids for the indicated KCNE1 variants. Nascent (n) proteins and mature (m) proteins were detected by Western blotting. B HEK293 cells transfected with 1 μg expression plasmids were treated without (lanes 1, 3, and 5) and with tunicamycin (lanes 2, 4, and 6). Addition of the N-glycosylation inhibitor tunicamycin results in loss of the glycosylated KCNE1 variants. A, B 15 μg total protein were loaded per lane.
Discussion
Deviating allelic copy numbers in one heterozygote Gly38Ser brain sample
As outlined above, we consider the presented pyrosequencing results as very reliable, because all gDNA based runs confirmed the genotypes inferred from direct sequencing. This applies particularly to brain sample 11 whose deviating allelic copy numbers had even been validated by a total of eight measurements. It can thus be excluded that technical problems account for the about 20:80 distribution of 38Ser- and 38Gly-coding alleles in the gDNA of brain sample 11. Accumulating experimental evidence suggests that neural progenitor cells often display loss and/or gain of chromosomes. Consequently, a considerable percentage of aneuploid neurons populate the mature brain [20]. Chromosome painting studies, for instance, revealed approximately 4% chromosome 21 aneuploid cells (both postmitotic neurons and non-neural cells) in normal adult human brain [21]. These aneuploid neurons are projected into the appropriate anatomical areas and appear to be functionally active [22]. In this light, genetic mosaicism must be considered a frequently observable feature of the human brain. The most likely explanation for the uneven distribution of chromosome 21 alleles in brain sample 11 is gain and/or loss of a chromosome 21 copy in a relatively high proportion of cells in the dissected cortex area. Another possibility could be a uniparental disomy for chromosome 21 (UPD21), since the sampled man was phenotypically normal, what is a general feature of UPD21 [23,24]. However, as the question for the underlying mechanism is second ranked in the present context we did not further investigate the alternatives herein.
Sporadic deficiency of 38Ser-coding mRNAs in heterozygote humans
Four different mechanisms could principally explain the observed absence of 38Ser-coding alleles in cDNAs from three individuals with a heterozygous genotype (two lymphoblastoid cell lines and brain sample 11): i) monoallelic expression of the 38Gly-coding allele as a consequence of null mutations in the regulatory region of the 38Ser-coding allele, ii) allelic dropout of the 38Ser-coding allele, iii) monoallelic expression of the 38Gly-coding allele due to silencing of the 38Ser-coding allele by genomic imprinting, and iv) genomic recoding of 38Ser-coding transcripts to 38Gly-coding alleles by A to I editing. We consider it very unlikely that the observed pattern results from deleterious mutations in the regulatory region since null mutations should affect not only the 38Ser-coding allele. Allelic dropout can further be ruled out as the 38Ser allele did not drop out when analysing the other 21 heterozygote samples. One possible explanation for the deviating allele frequencies in the three genomically heterozygous individuals is genomic imprinting, even though KCNE1 does not belong to the known imprinted genes, neither in humans nor in mouse [25,26]. Alternatively, genomic recoding at the mRNA level might account for the observed pattern. According to this, an ADAR sporadically recodes the adenosine at KCNE1 codon 38 (AGU) to inosine (IGU), which is equivalent to guanosine (GGU) for the translational machinery [17]. This assumption is additionally corroborated by a hint for A to I editing at KCNE1 codon 38 in GenBank entry BC046224 ("G in cDNA is A in the human genome" [27]). If one further considers evidence for genomic recoding of the human potassium voltage-gated channel, shaker-related subfamily, member 1 (KCNA1) [28] as well as of ion channels in diverse non-primate taxa such as rodents [29,30], insects [31,32], and molluscs [33,34], genomic recoding might indeed be the reason for the observed specific allele frequencies in three humane heterozygotes. Future investigations will finally clarify whether genomic imprinting or genomic recoding are causative for the sporadic occurence of deviating allele frequencies in the gDNA/cDNA comparison in humans. Irrespectively, the present data provide first time evidence for the coincidence of positive selection at a polymorph codon site and the sporadic absence of one allele in the mRNA of heterozygotes.
Functional considerations in the light of KCNE1 variance and divergence
The finding that the Gly38Ser polymorphism is a human-specific feature that emerged under positive selection poses the question for its adaptive value (see Figure 1). Given the relevance of IKs channels for inner ear homeostasis and the function of muscles and neurons it is tempting to speculate that the evolution of the Gly38Ser polymorphism is connected to the physiological challenges of specific human features such as speech perception, upright and enduring walking, and advanced cognitive capabilities. Regardless of these speculations, the evolution of the human-specific Gly38Ser polymorphism probably conferred some functional advantage at the cellular level. This conclusion agrees well with the results of a recent study showing an involvement of the Gly38Ser polymorphism in the fine-tuning of IKs channels [35]. As we found no evidence for changes in glycosylation between the chimpanzee protein and the two human KCNE1 variants (see Figure 2), the functional relevance of the Gly38Ser polymorphism most probably results from the moderately radical character of the amino acid exchanges involved (Asp > Gly, Gly > Ser). While amino acid position 38 belongs to the extracellular N-terminus of human KCNE1, the two other amino acid sites pinpointed herein as positively selected (105 and 115) belong to the intracellular C-terminus [1]. As such they might be involved in the interaction with KCNQ1, namely in the regulation of channel assembly, open-state destabilization, and kinetics of channel deactivation [36].
According to the population genetic data a subtle heterozygote advantage accounts for the maintenance of the Gly38Ser polymorphism in humans. However, the extent of such an heterozygote advantage might depend on the environment, since we found a significant heterozygote excess solely in Yoruba from Nigeria, but nearly no differences between the observed and expected genotype counts in Japanese from Tokyo (see Table 3). The depletion of 38Ser homozygotes in the Yoruban sample and particularly the sporadic absence of 38Ser-coding mRNAs in Central Europeans further suggest that the expression of the 38Ser allele is disadvantageous under certain conditions. 38Ser to 38Gly recoding or monoallelic expression of the 38Gly allele might then represent mechanisms to be functionally homozygote 38Gly when required by specific physiological conditions. Beyond this, there might be individual differences in the capability of genomic recoding or genomic imprinting. Such a scenario could explain why we found a depletion of 38Ser-coding alleles only in two out of 19 heterozygous cell lines and one out of five heterozygous brain samples.
Conclusion
It is well established that aberrant editing patterns can lead to diseases such as epilepsy [30], depression [37], amyotrophic lateral sclerosis [38] and malignant gliomas [39]. Likewise, genomic imprinting is assumed to play a role in the susceptibility to diverse diseases [40,41]. In extension of this, individual differences in the capability of suppressing the expression of the 38Ser variant might predispose 38Ser carriers more or less to LQTS. For instance, sex-specific differences in the depletion of 38Ser-coding mRNAs could explain why the Gly38Ser polymorphism correlates significantly with QT length in men from North Israel, but not in women [11]. Furthermore, sporadic 38Ser deficiency might have obscured the recognition of an associations between Gly38Ser and LQTS in diverse Central European cohorts (see [9,14-16]). However, these are speculations since it remains an open question whether the present conclusions drawn from lymphoblastoid cell lines and brain samples can be transferred to heart tissue. Provided that future investigations will confirm a sporadic depletion of 38Ser- coding mRNAs in heart of Gly38Ser heterozygotes and 38Ser homozygotes, this would underline the relevance of mRNA data in future association studies between Gly38Ser variant and LQTS. For apparent reasons heart tissue can only be collected post mortem. Another important question would thus be if peripheral blood lymphocytes reflect the recoding or imprinting pattern in heart tissue. Only under such circumstances peripheral blood lymphocytes could be taken as a predictor for the situation in heart. Irrespective of these considerations, the present evolutionary approach reveals an unusual, if not unique, pattern at the codon level, i.e. coincidence of positive Darwinian selection and polymorphism with a sporadically suppressed expression of one allele. This further corroborates the previously demonstrated functional relevance of the Gly38Ser polymorphism of KCNE1 [35].
Methods
Human sample
The present sample comprised lymphoblastoid cell lines of 29 individuals and brain samples (frontal cortex) of another 14 individuals. All individuals were Central Europeans. Anonymized brain samples were collected during pathomorphological diagnostics and immediately shock-frozen in liquid nitrogene. Lymphoblastoid cell lines were generated by Epstein-Barr virus transformation of peripheral blood leucocytes propagated according to standard procedures [42]. We extracted gDNAs using standard procedures. Total RNAs were extracted using TRIzol Reagent (Invitrogen). cDNAs were synthesized from total RNAs with random primers and SuperScript III (Invitrogen) following the supplier's protocol. Use of the brain samples was in accordance with the Helsinki Declaration and approved by the local ethics committee [Aerztekammer Rheinland-Pfalz, Decisions 837.103.04 (4261) and 837.073.07 (5608)].
Direct sequencing of human samples
For direct sequencing of gDNAs, PCR was carried out using FastStart Taq DNA Polymerase (Roche) and normal hot start PCR. The primers used bound to exon 3, in particular -18 and +26 bp upstream and downstream of start and stop codon, respectively (forward primer: 5'-TGCCTGGGAAGTTTGAGCTG-3'; reverse primer: 5'-CAGGTTGCCAGGCAGGATG-3'). PCR conditions were as follows: 2 min, 94°C; 35× (40 s at 94°C, 40 s at 65°C, 40 s at 72°C); 5 min, 72°C. Following exonuclease I and shrimp alkaline phosphatase digestion, dye terminator cycle sequencing of the PCR products was performed using the PCR primers and the CEQ DTCS Quick Start Kit (Beckman Coulter). Sequencing products were separated on a Beckman Coulter CEQ 8000 Genetic Analysis System. SCF-files were analyzed by visual inspection using BioEdit [43].
Quantitative Allele-Specific Expression Analysis of human samples
To more accurately measure the A versus G allele ratios of the A112G (Gly38Ser) variant in gDNAs and cDNAs, Quantification of Allele-Specific Expression by Pyrosequencing (QUASEP) was used [44]. QUASEP allows to accurately quantify the relative amount of one allele to the other, if their sequences differ in at least one base pair. The primer sequences for QUASEP were designed using the Pyrosequencing Assay Design Software (Biotage AB).
PCR products for QUASEP-analyses of the KCNE1-A/G-SNP rs1805127 were generated from gDNAs and cDNAs of lymphoblastoid cell lines and brain samples using FastStart Taq DNA Polymerase (Roche) and normal hot start PCR with the forward primer 5'-AGAGGGCCTCCAGCTTGC-3' and 5' biotinylated reverse primer 5'-GCAGGGTGGCAACATGTC-3' according to standard protocols. Pyrosequencing was performed on a PSQ™96MA Pyrosequencing System (Biotage) with the PyroGold SQA reagent kit (Biotage) using the forward sequencing primer 5'-CCAGCTTGCCGTCAC-3'. The PSQ™96MA 2.1.1 software (Biotage) was used for data analysis.
In order to assess whether RNA extraction or specific conditions of some of the samples affect the results of QUASEP-analyses of the KCNE1-SNP rs1805127 we performed analogue QUASEP-analyses of the highly heterozygous A/G-SNP rs11254413 of the housekeeping gene TRDMT1. Hot start PCR was carried out using the forward primer 5'-TGGCTATCCTCTACAAAATGACAA-3' and 5' biotinylated reverse primer 5'-CGGCAGGGTGATATGACTGAT-3'. For pyrosequencing we took the forward sequencing primer 5'-CCTTGGGAGAATATCTAGAA-3'. QUASEP of TRDMT1-SNP rs11254413 was confined to gDNAs and cDNAs of lymphoblastoid cell lines.
Interspecies comparison
To assess the evolution of KCNE1 across Eutheria, we complemented the human KCNE1 data with gDNA and cDNA data from non-human species (Mammalia, Eutheria), using NCBI and ENSEMBL databases. New sequences were generated from four non-human primates, i.e Bornean orangutan, siamang, crab-eating macaque, and silvered leaf monkey. Total RNA was extracted from testis (crab-eating macaque) and lymphoblastoid cell lines (all others) using TRIzol Reagent (Invitrogen). cDNAs were synthesized from total RNA with SuperScript II reverse transcriptase (Invitrogen), using the reverse gene-specific primer 5'-TCAGGTTGCCAGGCAGGAT-3'. The target cDNA encompassing the entire KCNE1-coding sequence was amplified with the forward gene-specific primer 5'-AGCCAAGGATATTCAGAGGT-3' and the reverse gene-specific primer mentioned above by standard PCR under the following conditions: 3 min at 94°C, 30 × (30 s at 94°C, 30 s at 58°C, 40 s at 72°C), 5 min at 72°C. Direct sequencing was carried out as described for the human samples. In a final step, we completed the dataset with GenBank entries from another 14 eutherian representatives, thus generating a dataset comprising orthologs of human and 18 non-human species (see bold accession numbers in Table 4). The sequences were translated, ClustalW aligned, and re-translated using BioEdit [43]. The non-human sequences were moreover scrutinized for polymorphisms of the codon site ortholog to human KCNE1 38, using ENSEMBL data. We finally checked the respective codon site of non-human KCNE1 sequences for hints of genomic recoding at the mRNA level. Therefore, we compared gDNAs retrieved from ENSEMBL and NCBI with the actual set of cDNAs deposited in the Nucleotide collection and EST databases at NCBI (see Table 4).
Statistical analyses
Population genetic analyses were carried out using allele frequencies and genotype counts of the geographically distinct human samples covered by HapMap phase II (120 Central Europeans, 90 Han Chinese from Beijing, 88 Japanese from Tokyo, 120 Yoruba from Ibadan/Nigeria). Expected genotype counts were calculated using Fstat 2.9.3 [45]. We engaged the same software to infer the fixation index, Fis. Fis is a measure of the heterozygote deficit within subpopulations. Consequently, positive Fis-values point to a heterozygote deficit, while negative Fis-values indicate a heterozygote excess. Fstat has moreover been used to test for deviations from Hardy-Weinberg equilibrium (400 randomisations). We corrected for multiple testing by strict Bonferroni adjustment, thus lowering the 5% level of significance to 0.0125.
The evolutionary history of the codon position corresponding to codon 38 in human KCNE1 was traced back using ancestral sequences reconstructed by baseml (PAML 3.15 [46]). The dataset comprised the non-human KCNE1 orthologs plus the two human alleles. The intree used is shown in Figure 1 and represents the commonly accepted phylogeny among mammals [47,48] and primates [49].
We furthermore assessed the evolutionary regime acting on KCNE1 as a whole using the ratio of non-synonymous (amino acid altering) to synonymous (silent) nucleotide substitution rates (= dn/ds = ω). Rate ratios > 1 are commonly accepted as a conservative measure of positive selection (= adaptive evolution) while rate ratios = 1 and < 1 are indicative of neutral evolution and negative selection, respectively. To study the evolution of KCNE1 across the human branches (foreground, see Figure 1) in comparison to the remaining phylogeny, we employed the modified version of branch-site model A [50] of codeml (PAML 4 [46]). Model A assumes four site classes: site class 0 integrates codon sites that are conserved throughout the entire phylogeny (foreground plus remaining phylogeny) with 0 < ω0 < 1. Site class 1 comprises codon positions that evolve neutrally throughout the entire phylogeny with ω1 = 1. Site classes 2a and 2b include codon sites that undergo positive selection on the foreground, but are under negative selection or neutral evolution across the remaining phylogeny. The model involves four free parameters: the proportion (p0) and the ω estimate (ω0) of site class 0; the proportion estimate of site class 1 (p1) and the ω estimate of the codon sites that evolve under positive selection across the foreground (ω2). The codon frequency was estimated from a 3 × 4 contingency table. Ambiguity data were not removed from the data (cleandata = 0). The Bayes empirical Bayes approach was used to identify codon sites under positive selection along the (human) foreground branches [51]. To test for the significance of the results, the branch-site test of positive selection (test 2 in [50]) was carried out. Therefore, we compared twice the log likelihood difference between model A with ω2 > 1, estimated from the data, and model A with ω2 = 1 fixed with the critical values from chi-square distribution with 1 degree of freedom.
Motif search
Motif search was carried out using the PROSITE database as implemented in the PredictProtein server [52].
Expression of KCNE1 in HEK293 cells and Western blotting
HEK293 cells (ATCC CRL 1573) were grown at 37°C under 5% CO2 in Dulbecco's Modified Eagle Medium (Gibco) supplemented with 10% fetal calf serum. 2.5 * 105 cells were transfected in six well plates (Greiner) with 1 μg or 3 μg of expression plasmids pcDNA3.1-chimpanzee_KCNE1_GAT, pcDNA3.1-human_KCNE1_AGT or pcDNA3.1-human_KCNE1_GGT by calcium phosphate coprecipitation. To assess the extent of N-glycosylation, tunicamycin (Sigma) was added to the medium to a concentration of 5 μg/ml 10 hours after transfection. For Western blotting, whole cell lysates were prepared 24 hours after transfection. Cells were washed three times in phosphate-buffered saline and pelleted by centrifugation at 300 g. The pellet was resuspended in 5 volumes of ice-cold NP-40 (Fluka) lysis buffer consisting of 137 mM KCl, 20 mM HEPES pH 7.9, 1% Nonidet P-40, 5 mM NaF, 10 μg/ml aprotinin, 10 μg/ml leupeptin, 1 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride, and incubated on ice for 40 min. The lysate was cleared by centrifugation at 80,000 g for 30 min. Protein concentrations were determined using the Bradford method (Bio-Rad). Cell lysates were separated by SDS-PAGE and proteins were transferred to polyvinylidene difluoride membranes (Millipore) for 20 min at 150 mA, using a Tris-glycine buffer system. After blocking, membranes were incubated with the primary antibody directed against KCNE1 (Calbiochem). As secondary antibody, a 1:5,000 dilution of peroxidase-conjugated donkey, anti-rabbit antibody (GE healthcare) was used. The control antigen corresponds to residues 67 to 129 of human KCNE1 fused to glutathione S-transferase (Calbiochem).
Sequence data
New sequences reported in this manuscript have been submitted to GenBank under accession numbers EF514881 to EF514888.
Abbreviations
cDNA: complementary DNA; dn/ds = ω: ratio of non-synonymous (amino acid altering) to synonymous (silent) nucleotide substitution rates; gDNA: genomic DNA; IKs: slowly activating delayed rectifier potassium channel; KCNA1: potassium voltage-gated channel, shaker-related subfamily, member 1; KCNE1: potassium voltage-gated channel, Isk-related family, member 1; KCNQ1: potassium voltage-gated channel, KQT-like subfamily, member 1; LQTS: long-QT syndrome; SNP: single nucleotide polymorphism; TRDMT1: tRNA aspartic acid methyltransferase 1; UPD21: uniparental disomy for chromosome 21.
Authors' contributions
HH and HZ initiated the study on the basis of preliminary data. HH contributed to the laboratory work, carried out databank mining, performed evolutionary and population genetic analyses, and wrote and edited the manuscript. UZ and TH provided human and non-human samples, and carried out direct and pyrosequencing. FO conducted HEK293-expression of KCNE1 and subsequent protein work. AP contributed to the population genetic analyses. UZ, FO, AP, HZ, and TH participated in the data interpretation and contributed to the writing of the manuscript. The experimental design was jointly conceived by all of the authors. All authors read and approved the final manuscript.
Acknowledgments
Acknowledgements
Special thanks go to Katja Mengel from the Institute of Anthropology, University of Mainz, and Daniela Weise from the Institute of Human Genetics, University of Mainz, for technical assistance. Furthermore, we gratefully acknowledge technical support from Dr. Ying Yue from the Massachusetts Institute of Technology. We are grateful to Dr. Andreas Hapke from the Institute of Anthropology, University of Mainz, for suggestions on the population genetic analyses. This work was supported by research grants from the German Research Foundation (DFG) to FO (SFB 518, A18) and TH (1374/7-1).
Contributor Information
Holger Herlyn, Email: herlyn@uni-mainz.de.
Ulrich Zechner, Email: zechner@humgen.klinik.uni-mainz.de.
Franz Oswald, Email: franz.oswald@uni-ulm.de.
Arne Pfeufer, Email: arne.pfeufer@web.de.
Hans Zischler, Email: zischler@uni-mainz.de.
Thomas Haaf, Email: haaf@humgen.klinik.uni-mainz.de.
References
- Splawski I, Shen J, Timothy KW, Vincent GM, Lehmann MH, Keating MT. Genomic structure of three long QT syndrome genes KVLQT1, HERG, and KCNE1. Genomics. 1998;51:86–97. doi: 10.1006/geno.1998.5361. [DOI] [PubMed] [Google Scholar]
- Barhanin J, Lesage F, Guillemare E, Fink M, Lazdunski M, Romey G. KvLQT1 and IsK (minK) proteins associate to form the IKs cardiac potassium channel. Nature. 1996;384:78–80. doi: 10.1038/384078a0. [DOI] [PubMed] [Google Scholar]
- Sanguinetti MC, Curran ME, Zou A, Shen J, Spector PS, Atkinson DL, Keating MT. Coassembly of KvLQT1 and minK (IsK) proteins to form cardiac IKs potassium channel. Nature. 1996;384:80–83. doi: 10.1038/384080a0. [DOI] [PubMed] [Google Scholar]
- Kang C, Tian C, Sönnichsen FD, Smith JA, Meiler J, George AL, Vanoye CG, Kim HJ, Sanders CR. Structure of KCNE1 and implications for how it modulates the KCNQ1 potassium channel. Biochemistry. 2008;47:7999–8006. doi: 10.1021/bi800875q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melman YF, Um SY, Krumerman A, Kagan A, McDonald TV. KCNE1 binds to the KCNQ1 pore to regulate potassium channel activity. Neuron. 2004;42:927–937. doi: 10.1016/j.neuron.2004.06.001. [DOI] [PubMed] [Google Scholar]
- Ma L, Lin C, Teng S, Chai Y, Bähring R, Vardanyan V, Li L, Pongs O, Hui R. Characterization of a novel Long QT syndrome mutation G52R KCNE1 in a Chinese family. Cardiovasc Res. 2003;59:612–619. doi: 10.1016/S0008-6363(03)00510-8. [DOI] [PubMed] [Google Scholar]
- Tian C, Vanoye CG, Kang C, Welch RC, Kim HJ, George AL, Sanders CR. Preparation, functional characterization, and NMR studies of human KCNE1, a voltage-gated potassium channel accessory subunit associated with deafness and long QT syndrome. Biochemistry. 2007;46:11459–11472. doi: 10.1021/bi700705j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S, Zankov DP, Yoshida H, Tsuji K, Makiyama T, Itoh H, Akao M, Hancox JC, Kita T, Horie M. N- and C-terminal KCNE1 mutations cause distinct phenotypes of long QT syndrome. Heart Rhythm. 2007;4:332–340. doi: 10.1016/j.hrthm.2006.11.004. [DOI] [PubMed] [Google Scholar]
- Aydin A, Bahring S, Dahm S, Guenther UP, Uhlmann R, Busjahn A, Luft FC. Single nucleotide polymorphism map of five long-QT genes. J Mol Med. 2005;83:159–165. doi: 10.1007/s00109-004-0595-3. [DOI] [PubMed] [Google Scholar]
- Pfeufer A, Jalilzadeh S, Perz S, Mueller JC, Hinterseer M, Illig T, Akyol M, Huth C, Schöpfer-Wendels A, Kuch B, Steinbeck G, Holle R, Näbauer M, Wichmann HE, Meitinger T, Kääb S. Common variants in myocardial ion channel genes modify the QT interval in the general population: results from the KORA study. Circ Res. 2005;96:693–701. doi: 10.1161/01.RES.0000161077.53751.e6. [DOI] [PubMed] [Google Scholar]
- Friedlander Y, Vatta M, Sotoodehnia N, Sinnreich R, Li H, Manor O, Towbin JA, Siscovick DS, Kark JD. Possible association of the human KCNE1 (minK) gene and QT interval in healthy subjects: evidence from association and linkage analyses in Israeli families. Ann Hum Genet. 2005;69:645–656. doi: 10.1046/j.1529-8817.2005.00182.x. [DOI] [PubMed] [Google Scholar]
- Koskela J, Kähönen M, Fan M, Nieminen T, Lehtinen R, Viik J, Nikus K, Niemelä K, Kööbi T, Turjanmaa V, Pörsti I, Lehtimäki T. Effect of common KCNE1 and SCN5A ion channel gene variants on T-wave alternans, a marker of cardiac repolarization, during clinical exercise stress test: the Finnish Cardiovascular Study. Transl Res. 2008;152:49–58. doi: 10.1016/j.trsl.2008.06.003. [DOI] [PubMed] [Google Scholar]
- Van Laer L, Carlsson PI, Ottschytsch N, Bondeson ML, Konings A, Vandevelde A, Dieltjens N, Fransen E, Snyders D, Borg E, Raes A, Van Camp G. The contribution of genes involved in potassium-recycling in the inner ear to noise-induced hearing loss. Hum Mutat. 2006;27:786–795. doi: 10.1002/humu.20360. [DOI] [PubMed] [Google Scholar]
- Gouas L, Nicaud V, Berthet M, Forhan A, Tiret L, Balkau B, Guicheney P, the D.E.S.I.R. Study Group Association of KCNQ1, KCNE1, KCNH2 and SCN5A polymorphisms with QTc interval length in a healthy population. KCNE1. 2005;13:1213–1222. doi: 10.1038/sj.ejhg.5201489. [DOI] [PubMed] [Google Scholar]
- Akyol M, Jalilzadeh S, Sinner MF, Perz S, Beckmann B, Gieger C, Illig T, Wichmann H-E, Meitinger T, Kääb S, Pfeufer A. The common non-synonymous variant G38S of the KCNE1-(minK)-gene is not associated to QT interval in Central European Caucasians: results from the KORA study. Eur Heart J. 2007;28:305–309. doi: 10.1093/eurheartj/ehl460. [DOI] [PubMed] [Google Scholar]
- Chevalier P, Bellocq C, Millat G, Piqueras E, Potet F, Schott JJ, Baró I, Lemarec H, Barhanin J, Rousson R, Rodriguez-Lafrasse C. Torsades de pointes complicating atrioventricular block: evidence for a genetic predisposition. Heart Rhythm. 2007;4:170–174. doi: 10.1016/j.hrthm.2006.10.004. [DOI] [PubMed] [Google Scholar]
- Keegan LP, Leroy A, Sproul D, O'Connell MA. Adenosine deaminases acting on RNA (ADARs): RNA-editing enzymes. Genome Biol. 2004;5:209. doi: 10.1186/gb-2004-5-2-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingman M, Kaessmann H, Pääbo S, Gyllensten U. Mitochondrial genome variation and the origin of modern humans. Nature. 2000;408:708–713. doi: 10.1038/35047064. [DOI] [PubMed] [Google Scholar]
- Livingstone CD, Barton GJ. Protein sequence alignments: a strategy for the hierarchical analysis of residue conservation. CABIOS. 1993;9:745–756. doi: 10.1093/bioinformatics/9.6.745. [DOI] [PubMed] [Google Scholar]
- Kaushal D, Contos JJ, Treuner K, Yang AH, Kingsbury MA, Rehen SK, McConnell MJ, Okabe M, Barlow C, Chun J. Alteration of gene expression by chromosome loss in the postnatal mouse brain. J Neurosci. 2003;23:5599–5606. doi: 10.1523/JNEUROSCI.23-13-05599.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehen SK, Yung YC, McCreight MP, Kaushal D, Yang AH, Almeida BS, Kingsbury MA, Cabral KM, McConnell MJ, Anliker B, Fontanoz M, Chun J. Constitutional aneuploidy in the normal human brain. J Neurosci. 2005;25:2176–2180. doi: 10.1523/JNEUROSCI.4560-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsbury MA, Friedman B, McConnell MJ, Rehen SK, Yang AH, Kaushal D, Chun J. Aneuploid neurons are functionally active and integrated into brain circuitry. Proc Natl Acad Sci USA. 2005;102:6143–6147. doi: 10.1073/pnas.0408171102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogan PK, Sabol DW, Punnett HH. Maternal uniparental disomy of chromosome 21 in a normal child. Am J Med Genet. 1999;83:69–71. doi: 10.1002/(SICI)1096-8628(19990305)83:1<69::AID-AJMG14>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Bán Z, Nagy B, Papp C, Beke A, Tóth-Pál E, Papp Z. Recurrent trisomy 21 and uniparental disomy 21 in a family. Fetal Diagn Ther. 2003;18:454–458. doi: 10.1159/000073142. [DOI] [PubMed] [Google Scholar]
- Luedi PP, Hartemink AJ, Jirtle RL. Genome-wide prediction of imprinted murine genes. Genome Res. 2005;15:875–884. doi: 10.1101/gr.3303505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luedi PP, Dietrich FS, Weidman JR, Bosko JM, Jirtle RL, Hartemink AJ. Computational and experimental identification of novel human imprinted genes. Genome Res. 2007;17:1723–1730. doi: 10.1101/gr.6584707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strausberg RL, Feingold EA, Grouse LH, Derge JG, Klausner RD, Collins FS, Wagner L, Shenmen CM, Schuler GD, Altschul SF, Zeeberg B, Buetow KH, Schaefer CF, Bhat NK, Hopkins RF, Jordan H, Moore T, Max SI, Wang J, Hsieh F, Diatchenko L, Marusina K, Farmer AA, Rubin GM, Hong L, Stapleton M, Soares MB, Bonaldo MF, Casavant TL, Scheetz TE, et al. Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc Natl Acad Sci USA. 2002;99:16899–16903. doi: 10.1073/pnas.242603899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalla T, Rosenthal JJ, Holmgren M, Reenan R. Control of human potassium channel inactivation by editing of a small mRNA hairpin. Nat Struct Mol Biol. 2004;11:950–956. doi: 10.1038/nsmb825. [DOI] [PubMed] [Google Scholar]
- Lomeli H, Mosbacher J, Melcher T, Hoger T, Geiger JR, Kuner T, Monyer H, Higuchi M, Bach A, Seeburg PH. Control of kinetic properties of AMPA receptor channels by nuclear RNA editing. Science. 1994;266:1709–1713. doi: 10.1126/science.7992055. [DOI] [PubMed] [Google Scholar]
- Brusa R, Zimmermann F, Koh DS, Feldmeyer D, Gass P, Seeburg PH, Sprengel R. Early-onset epilepsy and postnatal lethality associated with an editing-deficient GluR-B allele in mice. Science. 1995;270:1677–1680. doi: 10.1126/science.270.5242.1677. [DOI] [PubMed] [Google Scholar]
- Hanrahan CJ, Palladino MJ, Ganetzky B, Reenan RA. RNA editing of the Drosophila para Na+ channel transcript: evolutionary conservation and developmental regulation. Genetics. 2000;155:1149–1160. doi: 10.1093/genetics/155.3.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoopengardner B, Bhalla T, Staber C, Reenan R. Nervous system targets of RNA editing identified by comparative genomics. Science. 2003;301:832–836. doi: 10.1126/science.1086763. [DOI] [PubMed] [Google Scholar]
- Patton DE, Silva T, Bezanilla F. RNA editing generates a diverse array of transcripts encoding squid Kv2K1 channels with altered functional properties. Neuron. 1997;19:711–722. doi: 10.1016/S0896-6273(00)80383-9. [DOI] [PubMed] [Google Scholar]
- Rosenthal JJ, Bezanilla F. Extensive editing of mRNAs for the squid delayed rectifier K+ channel regulates subunit tetramerization. Neuron. 2002;34:743–757. doi: 10.1016/S0896-6273(02)00701-8. [DOI] [PubMed] [Google Scholar]
- Ehrlich JR, Zicha S, Coutu P, Hebert TE, Nattel S. Atrial fibrillation-associated minK38G/S polymorphism modulates delayed rectifier current and membrane localization. Cardiovasc Res. 2005;67:520–528. doi: 10.1016/j.cardiores.2005.03.007. [DOI] [PubMed] [Google Scholar]
- Chen J, Zheng R, Melman YF, McDonald TV. Functional interactions between KCNE1 C-terminus and the KCNQ1 channel. PloS ONE. 2009;4:e5143. doi: 10.1371/journal.pone.0005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevich I, Tamir H, Arango V, Dwork AJ, Mann JJ, Schmauss C. Altered editing of serotonin 2C receptor pre-mRNA in the prefrontal cortex of depressed suicide victims. Neuron. 2002;34:349–356. doi: 10.1016/S0896-6273(02)00660-8. [DOI] [PubMed] [Google Scholar]
- Kawahara Y, Ito K, Sun H, Aizawa H, Kanazawa I, Kwak S. Glutamate receptors: RNA editing and death of motor neurons. Nature. 2004;427:801. doi: 10.1038/427801a. [DOI] [PubMed] [Google Scholar]
- Maas S, Patt S, Schrey M, Rich A. Underediting of glutamate receptor GluR-B mRNA in malignant gliomas. Proc Natl Acad Sci USA. 2001;98:14687–14692. doi: 10.1073/pnas.251531398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millington GWM. Genomic imprinting and dermatological disease. Clin Exp Dermatol. 2006;31:681–688. doi: 10.1111/j.1365-2230.2006.02233.x. [DOI] [PubMed] [Google Scholar]
- Tang WY, Ho SM. Epigenetic reprogramming and imprinting in origins of disease. Rev Endocr Metab Disord. 2007;8:173–182. doi: 10.1007/s11154-007-9042-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neitzel H. A routine method for the establishment of permanent growing lymphoblastoid cell lines. Hum Genet. 1986;73:320–326. doi: 10.1007/BF00279094. [DOI] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 94/95/NT. Nucl Acids Symp. 1999;41:95–98. [Google Scholar]
- Ruf N, Bähring S, Galetzka D, Pliushch G, Luft FC, Nürnberg P, Haaf T, Kelsey G, Zechner U. Sequence-based bioinformatic prediction and QUASEP identify genomic imprinting of the KCNK9 potassium channel gene in mouse and human. Hum Mol Genet. 2007;16:2591–2599. doi: 10.1093/hmg/ddm216. [DOI] [PubMed] [Google Scholar]
- Goudet J. FSTAT (Version 1.2): a computer program to calculate F-statistics. J Hered. 1995;86:485–486. [Google Scholar]
- Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- Murphy WJ, Eizirik E, O'Brian SJ, Madsen O, Scally M, Douady CJ, Teeling E, Ryder OA, Stanhope MJ, de Jong WW, Springer MS. Resolution of the early placental mammal radiation using Bayesian phylogenetics. Science. 2001;294:2348–2351. doi: 10.1126/science.1067179. [DOI] [PubMed] [Google Scholar]
- Kriegs JO, Churakov G, Kiefmann M, Jordan U, Brosius J, Schmitz J. Retroposed elements as archives for the evolutionary history of placental mammals. PLoS Biology. 2006;4:e91. doi: 10.1371/journal.pbio.0040091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RJ, Cheverud JM. Scaling of sexual dimorphism in body mass: a phylogenetic analysis of Rensch's Rule in primates. Int J Primatol. 2002;23:1095–1135. doi: 10.1023/A:1019654100876. [DOI] [Google Scholar]
- Zhang J, Nielsen R, Yang Z. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol Biol Evol. 2005;22:2472–2479. doi: 10.1093/molbev/msi237. [DOI] [PubMed] [Google Scholar]
- Yang Z, Wong WSW, Nielsen R. Bayes empirical Bayes inference of amino acid sites under positive election. Mol Biol Evol. 2005;22:1107–1118. doi: 10.1093/molbev/msi097. [DOI] [PubMed] [Google Scholar]
- Rost B, Liu J. The PredictProtein server. Nucleic Acids Res. 2003;31:3300–3304. doi: 10.1093/nar/gkg508. [DOI] [PMC free article] [PubMed] [Google Scholar]