Abstract
Centrioles perform the dual functions of organizing both centrosomes and cilia. The biogenesis of nascent centrioles is an essential cellular event that is tightly coupled to the cell cycle so that each cell contains only two or four centrioles at any given point in the cell cycle. The assembly of centrioles and their analogs, basal bodies, is well characterized at the ultrastructural level whereby structural modules are built into a functional organelle. Genetic studies in model organisms combined with proteomic, bioinformatic, and identifying ciliary disease gene orthologs have revealed a wealth of molecules requiring further analysis to determine their roles in centriole duplication, assembly, and function. Nonetheless, at this stage our understanding of how molecular components interact to build new centrioles and basal bodies is limited. The ciliates, Tetrahymena and Paramecium, historically have been the subject of cytological and genetic study of basal bodies. Recent advances in the ciliate genetic and molecular toolkit have placed these model organisms in a favorable position to study the molecular mechanisms of centriole and basal body assembly.
Keywords: ciliate, Tetrahymena, Paramecium, basal body, centriole, microtubule, cilia, centrosome
Introduction
Since their discovery in the late 1800s, centrosome research has largely focused on chromosome segregation and the implications of defective segregation in tumorigenesis (1, 2). Centrosomes are microtubule organizing centers (MTOCs) that are responsible for organizing the spindle poles that direct bipolar spindle assembly for mitotic chromosome segregation. In addition, centrosomes serve to direct vesicle and organelle transport, regulate cytokinesis, and function as a signaling platform for proper cell cycle progression. At the centrosome core, a centriole pair organizes the surrounding matrix (pericentriolar material (PCM)) for microtubule nucleation and organization. Centrioles are nearly identical to basal bodies which are required to assemble the axonemes of cilia and flagella (3). Contemporary studies propose that their function as basal bodies is at least as important as their centrosomal function ((4), rev (5)). Consistent with this, basal body and ciliary dysfunction is found in human diseases that exhibit maladies including kidney cysts, obesity, hypertension, altered left-right asymmetry of the body, and renal and mucus clearance anomalies, collectively known as ciliopathies (5–9).
Basal bodies that organize cilia are derived through multiple mechanisms of assembly (10). In the case of primary cilia, centrioles migrate to the cell cortex and are converted to basal bodies to nucleate and assemble cilia (11, 12). Alternatively, multi-ciliated epithelial cells undergo a massive amplification of centrioles which then migrate to the cell cortex to become basal bodies that organize many cilia (10). The differences between centrioles and basal bodies lie in their distinct accessory structures that facilitate pericentriolar material (PCM) recruitment and nucleation of microtubules for centrioles, or cortical localization and cilia organization for basal bodies. Thus, while the organization (assembly process and core structure) of centrioles and basal bodies is likely to be identical, critical modifications enable their divergent functions within the cell (10).
The molecular events leading to the assembly of new centrioles and basal bodies are incompletely understood (13, 14). Early structural studies in a number of model systems defined many of the conserved morphological stages leading to new organelles (reviewed in (13)). In addition, genetic studies predominantly in Chlamydomonas reinhardtii and Caenorhabditis elegans, bioinformatic and proteomic studies, and human disease genes found to affect basal bodies and cilia have identified many molecules that now require detailed cell biology studies to understand their function. In this review, we focus on the potential the ciliates Tetrahymena and Paramecium have as model systems for increasing our understanding of basal body and centriole assembly.
Ciliates
Tetrahymena and Paramecium are single celled, ciliated protists belonging to the Alveolates group that also contains Dinoflagellates and Apicomplexans (15). Although both ciliates are related, they are distinct enough to be classified in separate ciliate subclasses (16). While comparisons between the two are helpful, it is not uncommon to observe experimental differences (17), including some relevant to basal bodies. Moreover, Paramecium, but not Tetrahymena, has experienced recent genome duplications in which duplicated genes have been maintained, adding to the complexity in comparing these two organisms (18, 19). Because these cells have highly organized, microtubule intensive cytoskeletons with many basal bodies, they provide opportunities for cell biologists to study the mechanisms of basal body assembly and ciliogenesis.
Tetrahymena has proven to be a valuable model system, the study of which has contributed to the discovery of dynein, telomere sequences, telomerase, ribozymes, and the detailed analysis of post-translational modifications of both histones and tubulin. Tetrahymena has several characteristics that make it a successful model system (Table 1). The combination of molecular techniques with advantageous cytology is beneficial for the study of basal body duplication and assembly. Paramecium also provides excellent cytology combined with the use of RNAi to selectively inhibit translation.
Table 1.
Tetrahymena biology
| CHARACTERISTICS OF TETRAHYMENA BIOLOGY AND STUDY | |
|---|---|
| • | Approximately 3 hour cell doubling time in axenic culture at 30° C. |
| • | Growth to cell density of approximately 106 cells / mL 750 basal bodies per cell |
| • | Nuclear dimorphism (Deleterious mutations can be maintained, without phenotype, in the inactive germline micronucleus before mating to produce mutant phenotype in the transcriptionally active macronucleus during development) |
| • | RNAi (Controlled expression) |
| • | Antisense gene silencing and anti-sense libraries for forward genetic approaches Homologous recombination for genetic knock-ins and knock-outs |
| • | Transformation of both nuclei (by injection, electroporation, and biolistics) |
| • | Tagged genes (endogenous and exogenous) Regulated gene control |
| • | Long term storage (liquid nitrogen) |
| • | Strain resources (Tetrahymena Stock Center; http://tetrahymena.vet.cornell.edu/) |
| • | Clonal genetic strains |
| • | Mendelian genetic inheritance |
| • | Synchronized mass mating |
| • | Genome sequence |
Adapted from (102).
Basal body structure and organization
Ciliate basal bodies primarily function to organize motile cilia. Unlike vertebrate centrioles, they do not organize centrosomes for chromosome segregation but are dedicated to the task of supporting cellular motility and cortical organization. Tetrahymena cells are 50 µm long and contain roughly 750 basal bodies at the outset of the cell cycle, while the larger Paramecium cells are 120 µm long and contain approximately 4,000 basal bodies (Figure 1; (17, 20, 21)). Basal bodies in both ciliates are found in two cellular locations. Cortical rows of basal bodies (ciliary rows or kineties) run the length of the cells and nucleate cilia for motility. A second population of basal bodies is tightly organized within the oral apparatus or cellular feeding structure (22). In Tetrahymena, there are 18–21 longitudinal rows of basal bodies and 150 basal bodies in the oral apparatus (Figure 1). In Paramecium, there are approximately 70 longitudinal rows and and approximately 1000 basal bodies in the oral apparatus (21, 23).
Figure 1. New basal body formation during the late stages of Tetrahymena cell division.
Tetrahymena thermophila cells with basal bodies labeled by staining with anti-centrin (upper panels) and combined basal bodies (anti-centrin (green)) and DNA (Hoerscht 33342 (red)) (lower panels). (A) Interphase cell with relatively uniform basal body spacing along the ciliary rows. Arrow denotes mature oral apparatus at the cell anterior. DNA staining shows both the larger macronucleus (red arrow) and the smaller, dense micronucleus (white arrowhead). (B) Basal bodies in the ciliary rows increase in density as a result of new basal body assembly. The development of the oral primordium (stomatogenesis) is initiated (arrowhead) at the cell median in preparation for cell division. (C) New basal body assembly occurs at ciliary rows (most of the new basal body assembly is positioned immediately posterior to the division plane) and the oral primordium. The micronucleus begins to elongate into a prolate spheroid or rugby ball shape. (D) The oral primordium organizes into the three membranelles and the crescent shaped undulating membrane. The micronucleus has divided. (E) The division plan is evident with both the micronucleus and macronucleus separated between the mother and daughter cells. Scale bar, 10 µm.
Ciliate basal bodies are comprised of the same conserved structure found for centrioles and basal bodies in other eukaryotes. Nine triplet microtubule blades are arranged in a barrel configuration (Figure 2), which extend from the proximal end, or base, containing the nine-fold symmetric hub and spoke structures (cartwheels) to the transition zone where triplet microtubules are converted to the doublet microtubules of the ciliary axoneme. A mature basal body is approximately 200 nm in diameter and 550 nm in length (24) and the lumen is filled with an electron dense column of material that is approximately 75 nm in diameter and 400 nm in length (Figure 2). Cartwheels in many organisms are lost upon centriole and basal body maturation (13, 25), however ciliates maintain robust basal body cartwheels through their lifecycle (24, 26, 27).
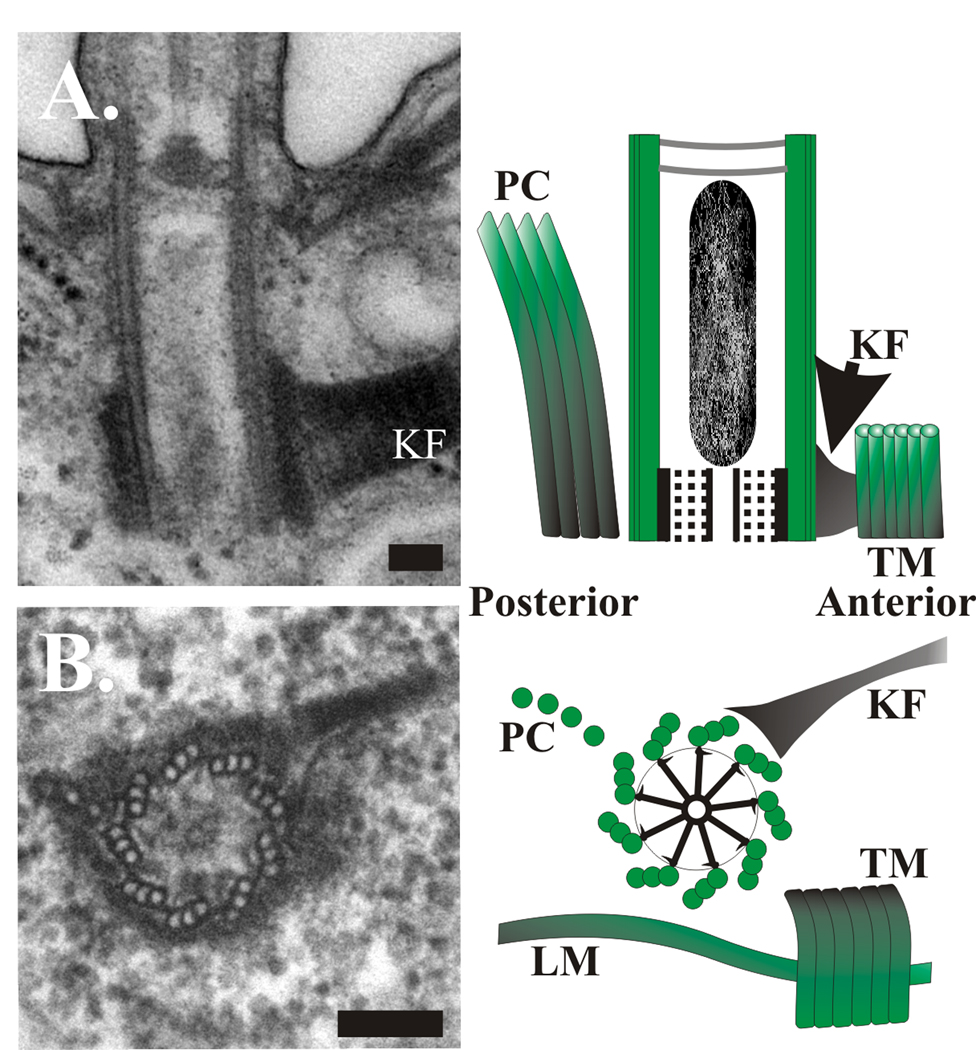
Figure 2. Tetrahymena basal body structure.
(A) Longitudinal thin section EM through a single basal body showing basal body cylinder structure and kinetodesmal fiber (KF). The cartoon identifies a longitudinal basal body section with accessory structures. The cartwheel structure is at the base of the organelle. Several of the accessory structures surrounding the basal bodies are indicated. (B) Cross thin section EM through the basal body structure, kinetodesmal fiber, and post-ciliary microtubles. The cartoon reveals a cross section through the cartwheel structure and several of the accessory structures surrounding basal bodies. Labeled structures are as follows: LM, longitudinal microtubule. PC, post-ciliary microtubule bands. KF, kinetodesmal fibers. TM, transverse microtubules. The anterior and posterior positioning relative to the cellular geometry is indicated. Scale bars, 100 nm.
While ciliate basal bodies contain the same core structure found in most centrioles and basal bodies, there are differences in the accessory structures that facilitate their function in supporting ciliary motility and organization of the cell cortex. These accessory structures are composed of both microtubule and non-microtubule components that are arranged in a polarized array for cortical organization (17, 27, 28). Three major accessory structures include the kinetodesmal fiber or striated rootlet and two microtubule bundles that are uniformly positioned at the posterior end (postciliary microtubules) and at the side (transverse microtubules) of basal bodies (Figure 1B; (17)). These structures are found for cortical basal bodies of the ciliary rows, whereas the densely packed basal bodies in the oral apparatus contain a modified version of these accessory structures (17, 28). Cortical basal bodies are also involved in the nucleation of the centrin-based infraciliary lattice of Paramecium (29), and the more limited but probably homologous apical filamentous band of Tetrahymena (30).
Basal body assembly
In mammalian systems, the formation of new centrioles / basal bodies can be generalized into two modes: centriolar and acentriolar. The distinction between each of these mechanisms is generally classified by the fate of the basal bodies, depending on whether they organize a single primary cilium or many cilia in multi-ciliated epithelia. In the centriolar pathway, centriole duplication is tightly coupled with the cell cycle so that during DNA synthesis each centriole pair separates, and a new daughter centriole forms immediately adjacent to each mother centriole. The centriolar pathway maintains a constant number of centrosomes in each phase of the cell cycle, one in G1 and two in G2 and M, each containing a centriole pair. Here the duplicated centriole pair functions to organize centrosomes and then migrates to the cell cortex where the mature, mother centriole acts as a basal body for primary cilia formation (11). Alternatively, multi-ciliated epithelial cells require a massive amplification of basal bodies that occurs by a combination of centriolar and acentriolar pathways. In contrast to the centriolar pathway described above, where only one new centriole is formed adjacent to the mother, several centrioles may form at a given time in close proximity to the mother centriole (10). The second acentriolar pathway is the de novo assembly of multiple centrioles around a matrix of electron dense fibrous granules called deuterosomes (10). This second pathway enables massive centriole/basal body amplification. For both mechanisms, duplication and assembly occurs when centrioles are resident within the cell interior before migrating to the cell cortex where they become basal bodies. In addition, the two pathways share structural assembly intermediates and molecular requirements suggesting that they are conserved (13, 31, 32).
Consistent with centriolar assembly in mammalian cells, Tetrahymena and Paramecium basal bodies are assembled adjacent to an existing parent organelle. However, ciliate basal bodies duplicate and remain at the cell cortex. A significant advantage to studying new basal body assembly in Tetrahymena and Paramecium is the spatially constrained pattern by which new basal bodies are repeatedly assembled in ciliary rows (33, 34). In these ciliates, 750 and 4,000 basal bodies, respectively, must be assembled prior to each cell division to maintain the constant number of these organelles. Both circumferential and anterior-posterior gradients of proliferation exist at the cell cortex, so that new basal bodies are generally assembled in the medial region of Tetrahymena and Paramecium cells (21, 33, 34). Furthermore, new basal bodies are always positioned anterior to and in close proximity to the existing organelle, creating pairs of old and new basal bodies (Figure 1; (21, 24, 26)). As basal bodies mature, they separate and move in the anterior direction away from the mother basal body along the ciliary row. Most of these basal bodies eventually become ciliated (21, 30), with a variable lag in ciliogenesis that is dependent upon where they are located in the cell (33, 35, 36). Furthermore, newly assembled basal bodies have the capacity to nucleate new basal body assembly soon after, within a single cell cycle, suggesting that the maturation into an organelle that is competent to form daughters can occur fairly rapidly in ciliates (21, 27). Thus, the competence to assemble individual basal bodies is not tightly coupled to the cell cycle, but instead the time and place of new basal body formation is subject to regional controls of basal body populations (21).
The centriolar duplication of mammalian centrioles occurs exactly once during the cell cycle, such that a licensing mechanism that is coupled to the cell cycle must exist for tight regulation of new duplication. Models suggest that the linkage between the orthogonally paired new and old centrioles is important for tight control of assembly (37). Furthermore, defects in centrioles and centrosomes activate a checkpoint response, suggesting that centriole duplication and activity is tightly coupled to the cell cycle (38). The timing of basal body assembly in ciliates is consistent with the cell cycle. The majority of Tetrahymena basal bodies are formed as cells prepare for cell division (33, 39) and, similarly, in Paramecium new duplication occurs in the final 30 minutes of the cell cycle (40). However, the regulation of duplication does not appear to be constrained by the cell cycle as found for the cell cycle observed for mammalian cells in which newly assembled centrioles do not reduplicate until the following cell cycle (41). The lack of either a checkpoint response to basal body impairment, or cell cycle based regulation at the level of individual basal bodies is an advantage for dissecting the mechanisms of basal body assembly in ciliates (42).
In addition to basal body assembly along the ciliary rows, new basal bodies are rapidly amplified at the oral primordium, which will become the oral apparatus of the daughter cell (23, 43). Here new basal bodies are assembled in “clusters” either close to (Paramecium) or distant from (Tetrahymena) the mature parental oral apparatus. In Tetrahymena, these basal bodies duplicate from basal bodies contained in the ciliary rows before becoming grouped into four discrete clusters that make up the oral apparatus (Figure 1). While this amplification requires prolific assembly of new basal bodies, no acentriolar or de novo assembly has been observed in either Tetrahymena or Paramecium (24, 44). However, de novo basal body assembly does occur during excystment and likely during during oral development in the Spirotrich ciliate, Oxytricha (45, 46). Most of the basal bodies of the new oral apparatus nucleate cilia immediately during oral apparatus development (23, 43).
Structural order of assembly
Upon initiation of duplication, the assembly intermediates leading to these structurally complex organelles has been thoroughly reviewed (13, 14, 32, 47) and the current belief is that the cartwheel defines the nine-fold symmetry that is the hallmark of centrioles, basal bodies, and cilia (13) (Figure 3). In Tetrahymena, new basal bodies are assembled by the formation of a new organelle perpendicular and at the base or proximal end of an existing, mature basal body (mother) (Figure 3A). The initial observed structure contains short singlet microtubules and the cartwheel (24). At the base of the cartwheel is an amorphous electron dense disk, known as the generative disk. As the new basal body matures into a pro-basal body, doublet and triplet microtubules are assembled. It is important to note that the early stages leading to cartwheel and singlet microtubules were not captured by this work. In Paramecium, Dippell described a detailed series of intermediate stages suggesting that the first structure responsible for the nine-fold symmetry is the generative disk and not the cartwheel. Singlet or “A” microtubules develop from the generative disk in a sequential and clockwise fashion (26). Then, in a non-sequential manner, doublet or “B” tubules form coincidentally with cartwheel assembly. Finally triplet microtubules are formed. Thus, cartwheels may not be the morphological prerequisite for the nine-fold symmetry. Subsequent work, however, suggests that cartwheels assemble prior to the A-tubules in Paramecium (27) (Figure 3B). The effort to resolve this and understand the stages of assembly in greater detail has been hampered by a deficiency of knowledge about molecules that function in this pathway. Work in Chlamydomonas and C. elegans has advanced the field through forward genetic analysis and RNAi screens which identified new proteins important for centriole and basal body assembly (47, 48). Mutations in some of these genes affect cartwheels and also disrupt the nine-fold symmetry in Chlamydomonas and Drosophila centrioles (49–51). More recently, the development of powerful molecular biology techniques and sequenced genomes in ciliates has opened up this unique genetic and cytological system for detailed basal body studies. Advances in electron microscopy, namely tomography, may enable a detailed morphological analysis of normal stages of assembly and establish structural precursors that lead to the observed nine-fold symmetry. This technology has significantly advanced C. elegans centriole research (52). Finally, the ability to assemble basal body cartwheels from Tetrahymena basal body lysates, in vitro, suggests that ciliates could become a formidable model system in defining the key structures and molecules for cartwheel assembly (53).
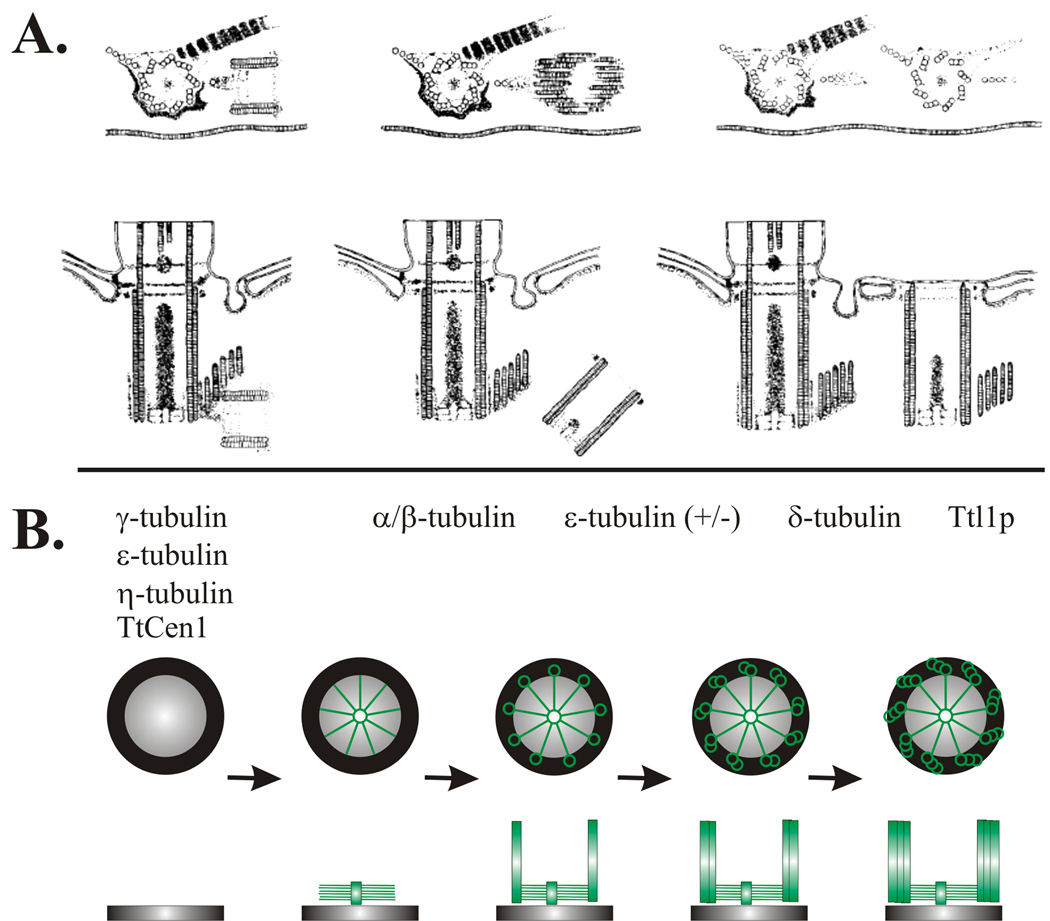
Figure 3. Stages of early basal body assembly in Tetrahymena.
Drawing depicts longitudinal sections of basal body formation. Basal bodies form on the anterior side (respective to the cellular geometry) of the parent organelle. Nascent assembly is initiated at the proximal end and perpendicular to the existing parent basal body to form a short pro-basal body. The pro-basal body then separates and tilts parallel to the parent organelle and becomes inserted into the plasma membrane (figure courtesy of The Journal of Cell Biology).
Stages of early basal body assembly. (A) Drawing depicts longitudinal sections of basal body formation in Tetrahymena. Basal bodies form on the anterior side (respective to the cellular geometry) of the parent organelle. Nascent assembly is initiated at the proximal end and perpendicular to the existing parent basal body to form a short pro-basal body. The pro-basal body then separates and tilts parallel to the parent organelle and becomes inserted into the plasma membrane (permission from The Journal of Cell Biology). (B) Cartoon depicts the steps to new basal body assembly based on combined studies from Tetrahymena and Paramecium. Assembly begins with the formation of the generative disk, followed by cartwheel assembly, the first structural evidence for nine-fold symmetry. Singlet microtubules are assembled at the ends of each cartwheel spoke followed by the B- and C-tubules to generate doublet and triplet microtubules, respectively. Genes involved in each transition stage are indicated, based upon the molecular components described in this review. (+/−) indicates partial ɛ-tubulin depletion. Note that this is not an exhaustive list of the known molecular components. Top panels, cross section. Bottom panels, longitudinal section.
From structure to molecules
EM studies of centrioles and basal bodies, including those of ciliates, show the conserved fine structure and intermediate stages of assembly of these organelles. More recent investigations in a number of model systems have identified conserved and non-conserved molecules that comprise centrioles and basal bodies. In addition, the identification of proteins associated with human ciliary disease brings an extensive number of new and important targets for functional analysis, with respect to basal body assembly and function.
Basal body proteome
Proteomic, genomic, and bioinformatic studies have defined many of the molecular components that comprise centrosomes, centrioles, basal bodies, and cilia (54–62). The use of multiple strategies and organisms allows for the assembly of a comprehensive list of both overlapping and unique components. The large number of putative basal body, centriolar, and ciliary proteins indicates the complexity of MTOCs and cilia.
To identify the molecules that comprise centrioles and basal bodies, two analogous proteomic studies were performed in Chlamydomonas and Tetrahymena (54, 63). The extensive list of candidate Tetrahymena basal body proteins was reduced to a manageable list by using a comparative strategy. Proteins were prioritized based on whether they contained protein motifs associated with microtubules, are conserved to vertebrate proteins, are present in other similar proteomes, and / or are known to have human disease relevance (63). High priority proteins were GFP tagged to determine whether they localize to Tetrahymena basal bodies. 24 novel Tetrahymena basal body proteins localized to basal bodies in this study. Finally, proteins were localized using immuno-electron microscopy (IEM) to define their position within the ultrastructure of the basal bodies. The localization of basal body components to discrete domains provides significant information in determining how these proteins may function at basal bodies. For example, proteins that localize to the distal portions are likely responsible for ciliary formation and function while proteins that reside at the generative disk or cartwheel structures may be early assembling components critical for new basal body assembly. Indeed, the Tetrahymena Sas6a protein, which is essential for new centriole and basal body assembly in all organisms studied thus far, localized to the central hub of the cartwheel (63). Three novel cartwheel proteins were also identified in this study (Poc1, Bbc29, and Bbc82).
Molecular studies of ciliate basal body assembly
Tubulins
Tubulin modifications
The major structural unit of centrioles and basal bodies is the triplet microtubule structure comprised of αβ-tubulin dimers. The Tetrahymena genome contains one α-tubulin gene and two redundant β-tubulin genes while Paramecium contains five α-tubulin genes and three β-tubulin genes, with minimal divergence between corresponding genes within each of these ciliates (64, 65). Ciliates build a large number of complex tubulin containing structures with varying degrees of microtubule stability and a wide range of functions using this limited number of tubulin genes (66). Tubulin post-translational modifications may be responsible for establishing different classes of microtubules within these structures by affecting microtubule structure and/or the binding of microtubule associated proteins (67–70). Specifically, centriole and basal body microtubules must be regulated to form a stable structure that is conservatively maintained once it is assembled ((71, 72), C.G.P. and M.W., unpublished results).
Mammalian centrioles are disrupted by antibodies that target glutamylated tubulin suggesting that this modification is required to stabilize the triplet microtubules (73). Furthermore, tubulin glutamylation is found at both Tetrahymena and Paramecium basal bodies (74, 75). To test the possibility that glutamylation is required for new basal body assembly and stabilization, Wloga and colleagues generated mutants in the Tetrahymena α-tubulin glutamylase that is specific to basal bodies, Ttll1p (76). While the mutants had no effect on new basal body assembly, glutamylation was required for proper basal body maturation, stabilization, and insertion into the cortex (76). In addition to glutamylation, tubulin acetylation, phosphorylation, tyrosination / detyrosination, and glycylation of α- and/or β- tubulin have been identified in ciliates (reviewed in (67, 68)). However, the functional significance of these modifications for basal body assembly remains unclear. As histone modifications have proven to be important for nucleosome function, tubulin modifications may also be important for proper basal body assembly, maintenance, and function (69).
Tubulin isoforms
Beyond the canonical α- and β-tubulins and their modifications, a new frontier in MTOC study was initiated by the discovery of additional tubulin family members. γ-tubulin was found as an extragenic suppressor of a heat sensitive β-tubulin mutation that causes stable microtubules in the mold Aspergillus (77). In ciliates, Tetrahymena contain one γ-tubulin while Paramecium has two nearly identical γ-tubulins that are essential for viability and basal body assembly. Utilizing the advantage of well defined spatial patterns of basal body duplication that is not tightly coupled to the cell cycle, Ruiz and colleagues found that new basal body assembly is lost in Paramecium when the two γ-tubulin genes are disrupted (78). In ciliates, these defects can be identified by the lack of basal body pairs assembled near the cell equator prior to cell division (Figure 4). Tetrahymena also requires γ-tubulin for basal body duplication and for maintenance of existing organelles (79). Reintroduction of γ-tubulin expression following its depletion results in the rapid assembly of new basal bodies even though existing organelles appear not to be present. This surprising result suggests that de novo basal body formation might be able to occur in Tetrahymena cells without basal bodies. Two important experiments may further define how γ-tubulin functions in basal body assembly and maintenance and whether de novo assembly can occur in Tetrahymena. First, the localization of γ-tubulin within the ultrastructure of newly assembling and existing basal bodies would define where this protein acts in Tetrahymena basal body assembly. Second, detailed electron microscopy of cells depleted of γ-tubulin and thus basal bodies may determine whether residual elements of the basal bodies remain that may function to nucleate new basal body assembly. Alternatively, these basal bodies may form and organize in the absence of existing structures as previously found for de novo assembly in other organisms, including certain ciliates (45). Finally, a novel mechanism for γ-tubulin regulation of basal body assembly was identified using site directed mutagenesis in Tetrahymena (80). γ-tubulin mutations commonly resulted in conditional phenotypes with defective basal body assembly. However, mutations targeted to the nucleotide binding domain (NBD) were typically lethal, except for two that caused excessive basal body proliferation outside of the normal cortical organization as well as in the cytoplasm (80). These results suggest that γ-tubulin is an important initiator of basal body duplication in a manner regulated by the nucleotide binding domain.
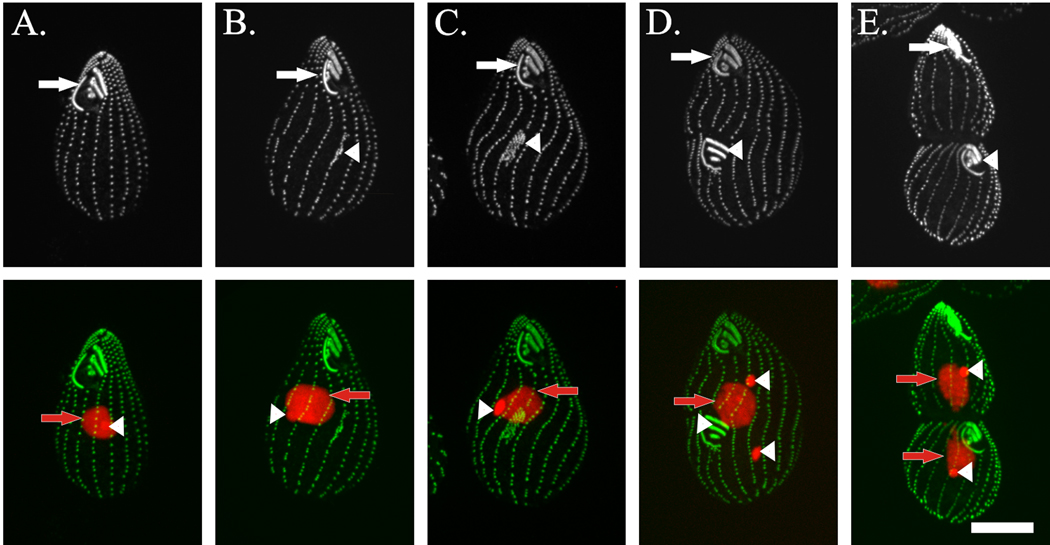
Figure 4. Loss of basal body duplication in Paramecium cells.
(A) The cartoon represents a dividing Paramecium cell at the stages visualized and box indicates the anterior region of the dorsal side taken from both wild-type (B) and ɛ-tubulin depleted (C) cells. Basal bodies (and some cilia) are stained with anti-α-tubulin antibodies (ID5). (B) In wild-type cells, basal body doublets are easily visualized in close proximity to each other. Each doublet contains a mature basal body (arrow, posterior with associated cilia detectable at some basal bodies) and an immature basal body (arrowhead, anterior). (C) Cells depleted of ɛ-tubulin by RNAi for 72 hours do not have pairs of duplicated basal bodies (arrow). Images were kindly provided by Dr. Pascale Dupuis-Williams (Université Evry Val d’Essonne, France). Scale bar, 2 µm.
Additional tubulins that are responsible for proper centriole and basal body assembly have been identified using both forward genetic and genome database searches (reviewed in (81)). Intensive work in Chlamydomonas, as well as in Paramecium has contributed to our functional understanding of these alternative tubulin proteins (81). First localized at mammalian centrosomes (82), ɛ-tubulin in Paramecium localizes to cartwheels and along the length of the microtubule triplets of the basal body (42). Loss of ɛ-tubulin is lethal and inhibits centriole and basal body duplication (42, 83). Partial depletion of ɛ-tubulin causes a loss of B- and C- tubules in both Chlamydomonas and Paramecium (42, 84), suggesting that B- and C- tubule formation are required to form and stabilize new centrioles and basal bodies.
Paramecium δ-tubulin is not required for basal body duplication, but is responsible for either the formation or maintenance of the C-tubule of the microtubule triplet (85, 86). Accessory structures required to maintain cortical organization are lost in the absence of δ-tubulin, suggesting that C-tubules are responsible for linking these structures to the basal body cylinder. Basal bodies without C-tubules maintain the capacity to nucleate cilia or flagella.
Finally, forward genetic strategies identified a Paramecium specific tubulin superfamily member, η-tubulin that was originally found as a conditional mutation (sm19) that, at restrictive temperature, inhibited basal body duplication (87). η-tubulin is a divergent tubulin that genetically interacts with β-tubulin in a manner suggesting that the proteins directly interact (88). Furthermore, γ-tubulin, is mislocalized in η-tubulin mutants (87). Thus, η-tubulin may be important for tethering γ-tubulin to the basal body for proper basal body assembly.
Centrin
First identified in green algae, centrin is responsible for contraction of calcium sensitive striated flagellar rootlets and for centriole, basal body, and spindle pole body duplication (89). The related budding yeast centrin, Cdc31p, is required for the initiation of yeast spindle pole body duplication (90, 91). There are an extensive number of centrin-like proteins, especially in ciliates.
Divergent results are reported for whether or not centrins are required for centriole and basal body assembly. Two human centrins, Centrin2 and Centrin3, are reported to be required for centrosome duplication (92, 93). Similarly, loss of centrin function is found to abrogate centriole and basal body duplication in Chlamydomonas, Marsilea vestita, Schizosaccharomyces pombe, Leishmania, and Tetrahymena (93–98). These studies indicate centrin is responsible for centriole and basal body assembly, whether using the centriolar or the acentriolar assembly pathways. However, recent RNAi experiments in human osteosarcoma (U2OS) cells found no role for Centrin2 (CrVfl2) or Centrin3 (ScCdc31) in normal or Plk4 induced pro-centriole assembly (99, 100). Ciliate studies also contribute to this discrepancy. Paramecium Centrin2 (HsCen2/CrVfl2) or Centrin3 (HsCen3/ScCdc31) depletion by RNAi feeding suggests that, at early timepoints, within the first cell cycle of RNAi treatment, these proteins are primarily responsible for basal body positioning and not duplication (101). New basal body assembly occurs normally, however basal bodies are not deposited into the cell cortex causing basal bodies to become internalized. This results in a dilution of the total number of basal bodies before the terminal phenotype of small, rounded cells with internalized basal bodies (101). In contrast, a complete genome knockout of the Tetrahymena centrin, TtCen1 (HsCen2/CrVfl2), inhibits both basal body duplication and maintenance so that basal bodies cannot be detected by either immuno-fluorescence or electron microscopy (98). The answers to two lingering questions could explain this discrepancy. First, it is not known whether or not the total cellular centrin protein levels are significantly reduced in the Paramecium studies. A low level of Tetrahymena centrin remains detectable even 96 hours after induction of the knockout allele suggesting that centrin is stable for multiple cell cycles (98). Following centrin RNAi in Paramecium, old basal bodies retain centrin and it is possible that this is enough to facilitate new basal body assembly. Second, ciliates have a number of centrin-like proteins raising the possibility that there are redundant functions for centrins to facilitate basal body assembly (98, 101).
Conclusions
The utility of ciliates for the detailed structural and functional analyses of nascent basal body assembly is well documented. The importance of extending this work beyond tubulins and centrins is more apparent as the tools required for detailed molecular study in ciliates becomes available. In particular, analysis of cartwheel proteins identified by proteomics studies may determine how early stages of basal body assembly occurs. Now that many basal body proteins have been identified (many of which are conserved (Table 2)), and techniques are readily available to either reduce their abundance, or introduce null, mutant, or tagged alleles, ciliates are well positioned as a model organism to lead to insights into the functions of these proteins in basal body biology.
Table 2.
Proteins for centriole and / or basal body duplication and assembly
| PROTEIN | PHENOTYPE* | CILIATE CONSERVATION** |
|---|---|---|
| SAK / PLK4 | No duplication or reduplication | NO |
| ZYG1 | No duplication or reduplication | NO |
| MPS1 | No SPB or centrosome duplication*** | NO |
| PLK1 | No overduplication in S-phase arrest | YES (01443850) |
| PLK2 | No overduplication in S-phase arrest | YES (00191790) |
| SPD-2 / CEP192 | No duplication*** | NO |
| SAS-6 | No duplication or reduplication | YES (00388200 / 00137600) |
| SAS-5 | No duplication | NO |
| SAS-4 / CPAP | No duplication | YES (00382220) |
| γ-tubulin | No duplication | YES (00079520) |
| δ-tubulin | C-tubule formation disrupted (doublets) | YES (00335970) |
| ɛ-tubulin | No duplication; (singlets) | YES (00118700) |
| BLD10 / CEP135 | No duplication | YES (01164140) |
| SFI1 | No duplication | YES (00999030) |
| Centrin2 / VFL2 | No duplication*** | YES (00384910) |
| Centrin3 / CDC31 | No duplication *** | YES (00523060) |
| SCF complex (SKP1) | Increased centrosome number | YES (00426320) |
| p53 | Centrosome amplification | NO |
| CP110 | No duplication | NO |
| Centrobin | No duplication | NO |
| Nucleophosmin | No duplication | NO |
| Separase | No centriole separation | YES (00297160) |
| Ana1 | Reduced centrosome number | NO |
| Ana2 | Reduced centrosome number | NO |
| Ana3 | Reduced centrosome number | NO |
Conservation is predicted based on reciprocal BLAST scores and sequence alignments. Accession numbers for Tetrahymena thermophila (TTHERM_#) are provided. Links to the Tetrahymena genome site (ciliate.org), can be used to identify the Paramecium orthologs.
Proteins where a defect in centriole or centrosome duplication has been observed but contradictory results are also observed in the literature.
One possibility for the dissonance between the structural studies of basal body assembly in ciliates, is that the early stages of basal body assembly may not proceed through a linear pathway, but are rather accomplished through modular assembly of distinct components that ultimately leads to a functional organelle. We still do not know how the structures or components assemble together to generate the nine-fold symmetric structure that is important for ciliary motility. To begin to understand this process, initial studies have focused on the gross morphology to determine those proteins that are essential to build the pro-centriole/basal body structure. As we understand the assembly in greater detail, such research will benefit by determining subtle defects and changes in the overall protein assembly dynamics of the basal body and centriole. Ciliates, with their increasingly sophisticated techniques for molecular analysis are in an opportune position for detailed study of basal body assembly and function.
ACKNOWLEDGEMENTS
We are indebted to Joseph Frankel for providing a wonderful and detailed critique of this review. We thank Pascale Dupuis-Williams, Maria Dziadosz, and Alex Stemm-Wolf for critical reading and discussions. Pascale Dupuis-Williams, Tom Giddings and R.D. Allen generously provided images for figures. C.G. Pearson is supported by the Damon Runyon Cancer Research Foundation (1879-05) and our Tetrahymena work is supported by the NIH (GM074746) and the March of Dimes Birth Defects Foundation (1-FY07-520) to M. Winey.
REFERENCES
- 1.Boveri T. Germany: Jena; 1914. Zur frage der entstehung maligner tumoren. [Google Scholar]
- 2.Wilson E. 3rd ed. New York: Macmillan; 1925. The Cell in Development and Heredity. [Google Scholar]
- 3.Bornens M, Azimzadeh J. Origin and evolution of the centrosome. Advances in experimental medicine and biology. 2007;607:119–129. doi: 10.1007/978-0-387-74021-8_10. [DOI] [PubMed] [Google Scholar]
- 4.Basto R, Lau J, Vinogradova T, Gardiol A, Woods CG, Khodjakov A, Raff JW. Flies without centrioles. Cell. 2006;125(7):1375–1386. doi: 10.1016/j.cell.2006.05.025. [DOI] [PubMed] [Google Scholar]
- 5.Marshall WF, Nonaka S. Cilia: tuning in to the cell's antenna. Curr Biol. 2006;16(15):R604–R614. doi: 10.1016/j.cub.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 6.Beales PL. Lifting the lid on Pandora's box: the Bardet-Biedl syndrome. Curr Opin Genet Dev. 2005;15(3):315–323. doi: 10.1016/j.gde.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 7.Hagiwara H, Ohwada N, Takata K. Cell biology of normal and abnormal ciliogenesis in the ciliated epithelium. Int Rev Cytol. 2004;234:101–141. doi: 10.1016/S0074-7696(04)34003-9. [DOI] [PubMed] [Google Scholar]
- 8.Badano JL, Mitsuma N, Beales PL, Katsanis N. The Ciliopathies: An Emerging Class of Human Genetic Disorders. Annu Rev Genomics Hum Genet. 2006;7:125–148. doi: 10.1146/annurev.genom.7.080505.115610. [DOI] [PubMed] [Google Scholar]
- 9.Pan J, Wang Q, Snell WJ. Cilium-generated signaling and cilia-related disorders. Laboratory investigation; a journal of technical methods and pathology. 2005;85(4):452–463. doi: 10.1038/labinvest.3700253. [DOI] [PubMed] [Google Scholar]
- 10.Dawe HR, Farr H, Gull K. Centriole/basal body morphogenesis and migration during ciliogenesis in animal cells. J Cell Sci. 2007;120(sPt 1):7–15. doi: 10.1242/jcs.03305. [DOI] [PubMed] [Google Scholar]
- 11.Sorokin S. Centrioles and the formation of rudimentary cilia by fibroblasts and smooth muscle cells. J Cell Biol. 1962;15:363–377. doi: 10.1083/jcb.15.2.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sorokin SP. Reconstructions of centriole formation and ciliogenesis in mammalian lungs. J Cell Sci. 1968;3(2):207–230. doi: 10.1242/jcs.3.2.207. [DOI] [PubMed] [Google Scholar]
- 13.Strnad P, Gonczy P. Mechanisms of procentriole formation. Trends Cell Biol. 2008;18(8):389–396. doi: 10.1016/j.tcb.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 14.Bettencourt-Dias M, Glover DM. Centrosome biogenesis and function: centrosomics brings new understanding. Nat Rev Mol Cell Biol. 2007;8(6):451–463. doi: 10.1038/nrm2180. [DOI] [PubMed] [Google Scholar]
- 15.Cavalier-Smith T. Kingdom protozoa and its 18 phyla. Microbiological reviews. 1993;57(4):953–994. doi: 10.1128/mr.57.4.953-994.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lynn DH, Small EB. Phylum Ciliophora. In: Lee PCB JJ, Leedale GF, editors. An Illustrated Guide to the Protozoa. Lawrence, KS: Society of Protozoologists; 2002. pp. 371–676. [Google Scholar]
- 17.Frankel J. Cell biology of Tetrahymena thermophila. Methods Cell Biol. 2000;62:27–125. doi: 10.1016/s0091-679x(08)61528-9. [DOI] [PubMed] [Google Scholar]
- 18.Aury JM, Jaillon O, Duret L, Noel B, Jubin C, Porcel BM, Segurens B, Daubin V, Anthouard V, Aiach N, Arnaiz O, Billaut A, Beisson J, Blanc I, Bouhouche K, et al. Global trends of whole-genome duplications revealed by the ciliate Paramecium tetraurelia. Nature. 2006;444(7116):171–178. doi: 10.1038/nature05230. [DOI] [PubMed] [Google Scholar]
- 19.Eisen JA, Coyne RS, Wu M, Wu D, Thiagarajan M, Wortman JR, Badger JH, Ren Q, Amedeo P, Jones KM, Tallon LJ, Delcher AL, Salzberg SL, Silva JC, Haas BJ, et al. Macronuclear genome sequence of the ciliate Tetrahymena thermophila, a model eukaryote. PLoS biology. 2006;4(9):286. doi: 10.1371/journal.pbio.0040286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beisson J, Jerka-Dziadosz M. Polarities of the centriolar structure: morphogenetic consequences. Biology of the cell / under the auspices of the European Cell Biology Organization. 1999;91(4–5):367–378. [PubMed] [Google Scholar]
- 21.Iftode F, Cohen J, Ruiz F, Rueda AT, Chen-Shan L, Adoutte A, Beisson J. Development of surface pattern during divisionin Paramecium I. Mapping of duplication and reorganization of cortical cytoskeletal structures in the wild type. 1989:191–211. [Google Scholar]
- 22.Nilsson JR, Williams NE. An electron microscope study of the oral apparatus of Tetrahymena pyriformis. Comptes-rendus des travaux du Laboratoire Carlsberg. 1966;35(7):119–141. [PubMed] [Google Scholar]
- 23.Iftode F Fleury, Adoutte A. Development of the surface pattern during division in Paramecium III. Study of stomatogenesis in the wild type using antitubulin antibodies and confocal microscopy. Europ J Protistol. 1997;33:145–167. [Google Scholar]
- 24.Allen RD. The morphogenesis of basal bodies and accessory structures of the cortex of the ciliated protozoan Tetrahymena pyriformis. J Cell Biol. 1969;40(3):716–733. doi: 10.1083/jcb.40.3.716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Azimzadeh J, Bornens M. Structure and duplication of the centrosome. J Cell Sci. 2007;120(Pt 13):2139–2142. doi: 10.1242/jcs.005231. [DOI] [PubMed] [Google Scholar]
- 26.Dippell RV. The development of basal bodies in paramecium. Proc Natl Acad Sci U S A. 1968;61(2):461–468. doi: 10.1073/pnas.61.2.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iftode F, Fleury-Aubusson A. Structural inheritance in Paramecium: ultrastructural evidence for basal body and associated rootlets polarity transmission through binary fission. Biol Cell. 2003;95(1):39–51. doi: 10.1016/s0248-4900(03)00005-4. [DOI] [PubMed] [Google Scholar]
- 28.Beisson J, Jerka-Dziadosz M. Polarities of the centriolar structure: morphogenetic consequences. Biol Cell. 1999;91(4–5):367–378. [PubMed] [Google Scholar]
- 29.Beisson J, Clerot JC, Fleury-Aubusson A, Garreau de Loubresse N, Ruiz F, Klotz C. Basal body-associated nucleation center for the centrin-based cortical cytoskeletal network in Paramecium. Protist. 2001;152(4):339–354. doi: 10.1078/1434-4610-00072. [DOI] [PubMed] [Google Scholar]
- 30.Jerka-Dziadosz M. Cytoskeleton-related structures in tetrahymena thermophila: microfilaments at the apical and division-furrow rings. J Cell Sci. 1981;51:241–253. doi: 10.1242/jcs.51.1.241. [DOI] [PubMed] [Google Scholar]
- 31.Vladar EK, Stearns T. Molecular characterization of centriole assembly in ciliated epithelial cells. J Cell Biol. 2007;178(1):31–42. doi: 10.1083/jcb.200703064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rodrigues-Martins A, Riparbelli M, Callaini G, Glover DM, Bettencourt-Dias M. From centriole biogenesis to cellular function: centrioles are essential for cell division at critical developmental stages. Cell Cycle. 2008;7(1):11–16. doi: 10.4161/cc.7.1.5226. [DOI] [PubMed] [Google Scholar]
- 33.Nanney DL. Patterns of basal body addition in ciliary rows in Tetrahymena. J Cell Biol. 1975;65(3):503–512. doi: 10.1083/jcb.65.3.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaczanowski A. Gradients of proliferation of ciliary basal bodies and the determination of the position of the oral primordium in Tetrahymena. J Exp Zool. 1978;204(3):417–430. doi: 10.1002/jez.1402040313. [DOI] [PubMed] [Google Scholar]
- 35.Frankel J, Nelsen EM, Martel E. Development of the ciliature of Tetrahymena thermophila. II. Spatial subdivision prior to cytokinesis. Dev Biol. 1981;88(1):39–54. doi: 10.1016/0012-1606(81)90217-7. [DOI] [PubMed] [Google Scholar]
- 36.Thazhath R, Jerka-Dziadosz M, Duan J, Wloga D, Gorovsky MA, Frankel J, Gaertig J. Cell context-specific effects of the beta-tubulin glycylation domain on assembly and size of microtubular organelles. Mol Biol Cell. 2004;15(9):4136–4147. doi: 10.1091/mbc.E04-03-0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsou MF, Stearns T. Controlling centrosome number: licenses and blocks. Curr Opin Cell Biol. 2006;18(1):74–78. doi: 10.1016/j.ceb.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 38.Mikule K, Delaval B, Kaldis P, Jurcyzk A, Hergert P, Doxsey S. Loss of centrosome integrity induces p38-p53-p21-dependent G1-S arrest. Nature cell biology. 2007;9(2):160–170. doi: 10.1038/ncb1529. [DOI] [PubMed] [Google Scholar]
- 39.Nelsen EM, Frankel J, Martel E. Development of the ciliature of TetrahymenathermophilaI Temporal coordination with oral development. Dev Biol. 1981;88(1):27–38. doi: 10.1016/0012-1606(81)90216-5. [DOI] [PubMed] [Google Scholar]
- 40.Ruiz F, Garreau de Loubresse N, Beisson J. A mutation affecting basal body duplication and cell shape in Paramecium. J Cell Biol. 1987;104(3):417–430. doi: 10.1083/jcb.104.3.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tsou MF, Stearns T. Mechanism limiting centrosome duplication to once per cell cycle. Nature. 2006;442(7105):947–951. doi: 10.1038/nature04985. [DOI] [PubMed] [Google Scholar]
- 42.Dupuis-Williams P, Fleury-Aubusson A, de Loubresse NG, Geoffroy H, Vayssie L, Galvani A, Espigat A, Rossier J. Functional role of epsilon-tubulin in the assembly of the centriolar microtubule scaffold. J Cell Biol. 2002;158(7):1183–1193. doi: 10.1083/jcb.200205028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bakowska J, Frankel J, Nelsen EM. Regulation of the pattern of basal bodies within the oral apparatus of Tetrahymena thermophila. Journal of embryology and experimental morphology. 1982;69:83–105. [PubMed] [Google Scholar]
- 44.Sonneborn TM. Gene action in development. Proceedings of the Royal Society of London Series B, Containing papers of a Biological character. 1970;176(44):347–366. doi: 10.1098/rspb.1970.0054. [DOI] [PubMed] [Google Scholar]
- 45.Grimes GW. Origin and development of kinetosomes in Oxytricha fallax. J Cell Sci. 1973;13(1):43–53. doi: 10.1242/jcs.13.1.43. [DOI] [PubMed] [Google Scholar]
- 46.Grimes GW. Morphological discontinuity of kinetosomes during the life cycle of Oxytricha fallax. J Cell Biol. 1973;57(1):229–232. doi: 10.1083/jcb.57.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dutcher SK. Finding treasures in frozen cells: new centriole intermediates. Bioessays. 2007;29(7):630–634. doi: 10.1002/bies.20594. [DOI] [PubMed] [Google Scholar]
- 48.Dutcher SK. Elucidation of basal body and centriole functions in Chlamydomonas reinhardtii. Traffic. 2003;4(7):443–451. doi: 10.1034/j.1600-0854.2003.00104.x. [DOI] [PubMed] [Google Scholar]
- 49.Nakazawa Y, Hiraki M, Kamiya R, Hirono M. SAS-6 is a Cartwheel Protein that Establishes the 9-Fold Symmetry of the Centriole. Curr Biol. 2007;17(24):2169–2174. doi: 10.1016/j.cub.2007.11.046. [DOI] [PubMed] [Google Scholar]
- 50.Hiraki M, Nakazawa Y, Kamiya R, Hirono M. Bld10p constitutes the cartwheel-spoke tip and stabilizes the 9-fold symmetry of the centriole. Curr Biol. 2007;17(20):1778–1783. doi: 10.1016/j.cub.2007.09.021. [DOI] [PubMed] [Google Scholar]
- 51.Rodrigues-Martins A, Bettencourt-Dias M, Riparbelli M, Ferreira C, Ferreira I, Callaini G, Glover DM. DSAS-6 Organizes a Tube-like Centriole Precursor, and Its Absence Suggests Modularity in Centriole Assembly. Curr Biol. 2007;17(17):1465–1472. doi: 10.1016/j.cub.2007.07.034. [DOI] [PubMed] [Google Scholar]
- 52.Pelletier L, O'Toole E, Schwager A, Hyman AA, Muller-Reichert T. Centriole assembly in Caenorhabditis elegans. Nature. 2006;444(7119):619–623. doi: 10.1038/nature05318. [DOI] [PubMed] [Google Scholar]
- 53.Gavin RH. In vitro reassembly of basal body components. J Cell Sci. 1984;66:147–154. doi: 10.1242/jcs.66.1.147. [DOI] [PubMed] [Google Scholar]
- 54.Keller LC, Romijn EP, Zamora I, Yates JR, 3rd, Marshall WF. Proteomic analysis of isolated chlamydomonas centrioles reveals orthologs of ciliary-disease genes. Curr Biol. 2005;15(12):1090–1098. doi: 10.1016/j.cub.2005.05.024. [DOI] [PubMed] [Google Scholar]
- 55.Andersen JS, Wilkinson CJ, Mayor T, Mortensen P, Nigg EA, Mann M. Proteomic characterization of the human centrosome by protein correlation profiling. Nature. 2003;426(6966):570–574. doi: 10.1038/nature02166. [DOI] [PubMed] [Google Scholar]
- 56.Ostrowski LE, Blackburn K, Radde KM, Moyer MB, Schlatzer DM, Moseley A, Boucher RC. A proteomic analysis of human cilia: identification of novel components. Mol Cell Proteomics. 2002;1(6):451–465. doi: 10.1074/mcp.m200037-mcp200. [DOI] [PubMed] [Google Scholar]
- 57.Avidor-Reiss T, Maer AM, Koundakjian E, Polyanovsky A, Keil T, Subramaniam S, Zuker CS. Decoding cilia function: defining specialized genes required for compartmentalized cilia biogenesis. Cell. 2004;117(4):527–539. doi: 10.1016/s0092-8674(04)00412-x. [DOI] [PubMed] [Google Scholar]
- 58.Li JB, Gerdes JM, Haycraft CJ, Fan Y, Teslovich TM, May-Simera H, Li H, Blacque OE, Li L, Leitch CC, Lewis RA, Green JS, Parfrey PS, Leroux MR, Davidson WS, et al. Comparative genomics identifies a flagellar and basal body proteome that includes the BBS5 human disease gene. Cell. 2004;117(4):541–552. doi: 10.1016/s0092-8674(04)00450-7. [DOI] [PubMed] [Google Scholar]
- 59.Pazour GJ, Agrin N, Leszyk J, Witman GB. Proteomic analysis of a eukaryotic cilium. J Cell Biol. 2005;170(1):103–113. doi: 10.1083/jcb.200504008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Smith JC, Northey JG, Garg J, Pearlman RE, Siu KW. Robust method for proteome analysis by MS/MS using an entire translated genome: demonstration on the ciliome of Tetrahymena thermophila. Journal of proteome research. 2005;4(3):909–919. doi: 10.1021/pr050013h. [DOI] [PubMed] [Google Scholar]
- 61.Stolc V, Samanta MP, Tongprasit W, Marshall WF. Genome-wide transcriptional analysis of flagellar regeneration in Chlamydomonas reinhardtii identifies orthologs of ciliary disease genes. Proc Natl Acad Sci U S A. 2005;102(10):3703–3707. doi: 10.1073/pnas.0408358102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Inglis PN, Boroevich KA, Leroux MR. Piecing together a ciliome. Trends Genet. 2006;22(9):491–500. doi: 10.1016/j.tig.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 63.Kilburn CL, Pearson CG, Romijn EP, Meehl JB, Giddings TH, Jr, Culver BP, Yates JR, 3rd, Winey M. New Tetrahymena basal body protein components identify basal body domain structure. J Cell Biol. 2007;178(6):905–912. doi: 10.1083/jcb.200703109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Aury JM, Jaillon O, Duret L, Noel B, Jubin C, Porcel BM, Segurens B, Daubin V, Anthouard V, Aiach N, Arnaiz O, Billaut A, Beisson J, Blanc I, Bouhouche K, et al. Global trends of whole-genome duplications revealed by the ciliate Paramecium tetraurelia. Nature. 2006;444(7116):171–178. doi: 10.1038/nature05230. [DOI] [PubMed] [Google Scholar]
- 65.Eisen JA, Coyne RS, Wu M, Wu D, Thiagarajan M, Wortman JR, Badger JH, Ren Q, Amedeo P, Jones KM, Tallon LJ, Delcher AL, Salzberg SL, Silva JC, Haas BJ, et al. Macronuclear genome sequence of the ciliate Tetrahymena thermophila, a model eukaryote. PLoS biology. 2006;4(9):286. doi: 10.1371/journal.pbio.0040286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gaertig J, Thatcher TH, McGrath KE, Callahan RC, Gorovsky MA. Perspectives on tubulin isotype function and evolution based on the observation that Tetrahymena thermophila microtubules contain a single alpha- and beta-tubulin. Cell Motil Cytoskeleton. 1993;25(3):243–253. doi: 10.1002/cm.970250305. [DOI] [PubMed] [Google Scholar]
- 67.Gaertig J. Molecular mechanisms of microtubular organelle assembly in Tetrahymena. J Eukaryot Microbiol. 2000;47(3):185–190. doi: 10.1111/j.1550-7408.2000.tb00037.x. [DOI] [PubMed] [Google Scholar]
- 68.Libusova L, Draber P. Multiple tubulin forms in ciliated protozoan Tetrahymena and Paramecium species. Protoplasma. 2006;227(2–4):65–76. doi: 10.1007/s00709-005-0152-0. [DOI] [PubMed] [Google Scholar]
- 69.Verhey KJ, Gaertig J. The tubulin code. Cell Cycle. 2007;6(17):2152–2160. doi: 10.4161/cc.6.17.4633. [DOI] [PubMed] [Google Scholar]
- 70.McKean PG, Vaughan S, Gull K. The extended tubulin superfamily. J Cell Sci. 2001;114(Pt 15):2723–2733. doi: 10.1242/jcs.114.15.2723. [DOI] [PubMed] [Google Scholar]
- 71.Kochanski RS, Borisy GG. Mode of centriole duplication and distribution. J Cell Biol. 1990;110(5):1599–1605. doi: 10.1083/jcb.110.5.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Grimes GW, Gavin RH. Ciliary protein conservation during development in the ciliated protozoan, Oxytricha. J Cell Biol. 1987;105(6 Pt 1):2855–2859. doi: 10.1083/jcb.105.6.2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bobinnec Y, Khodjakov A, Mir LM, Rieder CL, Edde B, Bornens M. Centriole disassembly in vivo and its effect on centrosome structure and function in vertebrate cells. J Cell Biol. 1998;143(6):1575–1589. doi: 10.1083/jcb.143.6.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bre MH, de Nechaud B, Wolff A, Fleury A. Glutamylated tubulin probed in ciliates with the monoclonal antibody GT335. Cell motility and the cytoskeleton. 1994;27(4):337–349. doi: 10.1002/cm.970270406. [DOI] [PubMed] [Google Scholar]
- 75.Kann ML, Soues S, Levilliers N, Fouquet JP. Glutamylated tubulin: diversity of expression and distribution of isoforms. Cell Motil Cytoskeleton. 2003;55(1):14–25. doi: 10.1002/cm.10107. [DOI] [PubMed] [Google Scholar]
- 76.Wloga D, Rogowski K, Sharma N, Van Dijk J, Janke C, Edde B, Bre MH, Levilliers N, Redeker V, Duan J, Gorovsky MA, Jerka-Dziadosz M, Gaertig J. Glutamylation on alpha-tubulin is not essential but affects the assembly and functions of a subset of microtubules in Tetrahymena thermophila. Eukaryot Cell. 2008;7(8):1362–1372. doi: 10.1128/EC.00084-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Oakley CE, Oakley BR. Identification of gamma-tubulin, a new member of the tubulin superfamily encoded by mipA gene of Aspergillus nidulans. Nature. 1989;338(6217):662–664. doi: 10.1038/338662a0. [DOI] [PubMed] [Google Scholar]
- 78.Ruiz F, Beisson J, Rossier J, Dupuis-Williams P. Basal body duplication in Paramecium requires gamma-tubulin. Curr Biol. 1999;9(1):43–46. doi: 10.1016/s0960-9822(99)80045-1. [DOI] [PubMed] [Google Scholar]
- 79.Shang Y, Li B, Gorovsky MA. Tetrahymena thermophila contains a conventional gamma-tubulin that is differentially required for the maintenance of different microtubule-organizing centers. J Cell Biol. 2002;158(7):1195–1206. doi: 10.1083/jcb.200205101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shang Y, Tsao CC, Gorovsky MA. Mutational analyses reveal a novel function of the nucleotide-binding domain of gamma-tubulin in the regulation of basal body biogenesis. J Cell Biol. 2005;171(6):1035–1044. doi: 10.1083/jcb.200508184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dutcher SK. Long-lost relatives reappear: identification of new members of the tubulin superfamily. Curr Opin Microbiol. 2003;6(6):634–640. doi: 10.1016/j.mib.2003.10.016. [DOI] [PubMed] [Google Scholar]
- 82.Chang P, Stearns T. Delta-tubulin and epsilon-tubulin: two new human centrosomal tubulins reveal new aspects of centrosome structure and function. Nat Cell Biol. 2000;2(1):30–35. doi: 10.1038/71350. [DOI] [PubMed] [Google Scholar]
- 83.Chang P, Giddings TH, Jr, Winey M, Stearns T. Epsilon-tubulin is required for centriole duplication and microtubule organization. Nat Cell Biol. 2003;5(1):71–76. doi: 10.1038/ncb900. [DOI] [PubMed] [Google Scholar]
- 84.Dutcher SK, Morrissette NS, Preble AM, Rackley C, Stanga J. Epsilon-tubulin is an essential component of the centriole. Mol Biol Cell. 2002;13(11):3859–3869. doi: 10.1091/mbc.E02-04-0205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Garreau de Loubresse N, Ruiz F, Beisson J, Klotz C. Role of delta-tubulin and the C-tubule in assembly of Paramecium basal bodies. BMC cell biology. 2001;2:4. doi: 10.1186/1471-2121-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dutcher SK, Trabuco EC. The UNI3 gene is required for assembly of basal bodies of Chlamydomonas and encodes delta-tubulin, a new member of the tubulin superfamily. Mol Biol Cell. 1998;9(6):1293–1308. doi: 10.1091/mbc.9.6.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ruiz F, Krzywicka A, Klotz C, Keller A, Cohen J, Koll F, Balavoine G, Beisson J. The SM19 gene, required for duplication of basal bodies in Paramecium, encodes a novel tubulin, eta-tubulin. Curr Biol. 2000;10(22):1451–1454. doi: 10.1016/s0960-9822(00)00804-6. [DOI] [PubMed] [Google Scholar]
- 88.Ruiz F, Dupuis-Williams P, Klotz C, Forquignon F, Bergdoll M, Beisson J, Koll F. Genetic evidence for interaction between eta- and beta-tubulins. Eukaryot Cell. 2004;3(1):212–220. doi: 10.1128/EC.3.1.212-220.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Salisbury JL, Baron A, Surek B, Melkonian M. Striated flagellar roots: isolation and partial characterization of a calcium-modulated contractile organelle. J Cell Biol. 1984;99(3):962–970. doi: 10.1083/jcb.99.3.962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Byers B. Cytology of the yeast life cycle. In: Strathern JN, Jones EW, Broach JR, editors. The Molecular Biology of the yeast Saccharomyces: Life Cycle and inheritance. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory; 1981. pp. 59–96. [Google Scholar]
- 91.Baum P, Furlong C, Byers B. Yeast gene required for spindle pole body duplication: homology of its product with Ca2+-binding proteins. Proc Natl Acad Sci U S A. 1986;83(15):5512–5516. doi: 10.1073/pnas.83.15.5512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Middendorp S, Kuntziger T, Abraham Y, Holmes S, Bordes N, Paintrand M, Paoletti A, Bornens M. A role for centrin 3 in centrosome reproduction. J Cell Biol. 2000;148(3):405–416. doi: 10.1083/jcb.148.3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Salisbury JL, Suino KM, Busby R, Springett M. Centrin-2 is required for centriole duplication in mammalian cells. Curr Biol. 2002;12(15):1287–1292. doi: 10.1016/s0960-9822(02)01019-9. [DOI] [PubMed] [Google Scholar]
- 94.Koblenz B, Schoppmeier J, Grunow A, Lechtreck KF. Centrin deficiency in Chlamydomonas causes defects in basal body replication, segregation and maturation. J Cell Sci. 2003;116(Pt 13):2635–2646. doi: 10.1242/jcs.00497. [DOI] [PubMed] [Google Scholar]
- 95.Klink VP, Wolniak SM. Centrin is necessary for the formation of the motile apparatus in spermatids of Marsilea. Mol Biol Cell. 2001;12(3):761–776. doi: 10.1091/mbc.12.3.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Selvapandiyan A, Debrabant A, Duncan R, Muller J, Salotra P, Sreenivas G, Salisbury JL, Nakhasi HL. Centrin gene disruption impairs stage-specific basal body duplication and cell cycle progression in Leishmania. The Journal of biological chemistry. 2004;279(24):25703–25710. doi: 10.1074/jbc.M402794200. [DOI] [PubMed] [Google Scholar]
- 97.Paoletti A, Bordes N, Haddad R, Schwartz CL, Chang F, Bornens M. Fission yeast cdc31p is a component of the half-bridge and controls SPB duplication. Molecular biology of the cell. 2003;14(7):2793–2808. doi: 10.1091/mbc.E02-10-0661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Stemm-Wolf AJ, Morgan G, Giddings TH, Jr, White EA, Marchione R, McDonald HB, Winey M. Basal body duplication and maintenance require one member of the Tetrahymena thermophila centrin gene family. Mol Biol Cell. 2005;16(8):3606–3619. doi: 10.1091/mbc.E04-10-0919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kleylein-Sohn J, Westendorf J, Le Clech M, Habedanck R, Stierhof Y-D, Nigg EA. Plk4-Induced Centriole Biogenesis in Human Cells. Developmental Cell. 2007;13(2):190–202. doi: 10.1016/j.devcel.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 100.Strnad P, Leidel S, Vinogradova T, Euteneuer U, Khodjakov A, Gonczy P. Regulated HsSAS-6 Levels Ensure Formation of a Single Procentriole per Centriole during the Centrosome Duplication Cycle. Developmental Cell. 2007;13(2):203–213. doi: 10.1016/j.devcel.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ruiz F, Garreau de Loubresse N, Klotz C, Beisson J, Koll F. Centrin deficiency in Paramecium affects the geometry of basal-body duplication. Curr Biol. 2005;15(23):2097–2106. doi: 10.1016/j.cub.2005.11.038. [DOI] [PubMed] [Google Scholar]
- 102.Turkewitz AP. Out with a bang! Tetrahymena as a model system to study secretory granule biogenesis. Traffic. 2004;5(2):63–68. doi: 10.1046/j.1600-0854.2003.00155.x. [DOI] [PubMed] [Google Scholar]