To the editor
Reversible protein phosphorylation is a universal process that is involved in the control of most biological processes. The comprehensive and quantitative analysis of the protein phosphorylation patterns of cells at different states is therefore of considerable and general interest. Over the past years, mass spectrometry has become the method of choice for the analysis of protein phosphorylation and impressive gains have been realized in the isolation of phosphorylated peptides from complex samples as well as their mass spectrometric and computational analysis. In such studies hundreds to thousands of phosphopeptides and phosphorylation sites are now routinely identified.
Currently, several databases exist which store and disseminate protein phosphorylation data obtained from large scale studies, however, several factors limit their utility. First, and most importantly, the current phosphopeptide databases are human and/or rodent centric. Examples include the human proteome reference database (HPRD)1, PhosphoElm2, Phosida3 and PhosphoSitePlus (www.phosphosite.org). Extensive phosphoproteome data sets for model organisms organized in databases are still missing. Second, the lack of phosphorylation data from diverse species precludes comparative studies, e.g. those that assess whether specific phosphorylation sites or perturbation induced phosphopeptide patterns are conserved between species. For example, the analysis of the evolutionary conservation of the human phosphorylation sites of the Phosida3 database relies on amino acid sequence conservation, but not on observed phosphorylation sites in other species. Third, none of these databases provides sufficient information to validate, identify and quantify the presented phosphorylation sites by mass spectrometry in independent experiments.
To address these issues and to complement existing protein databases for life science research we describe the PhosphoPep v2.0 database (www.phosphopep.org)4 which is a significant extension of its first version, PhosphoPep v1.0. In its initial implementation the database contained 12,756 assigned phosphorylation sites identified in D. melanogaster Kc167 cells, the tandem mass spectra that led to their assignment4, 5 and a suite of associated software tools supporting the interactive use of the data contained in the database for further experiments and meta-analysis.
PhosphoPep v2.0 significantly extends the contents and utilities of the database compared to v1.0. First, PhosphoPep now includes phosphoproteome data from the four species yeast (S. cerevisiae), worm (C. elegans), fly (D. melanogaster) and human (H. sapiens) (see Table 1). These data also represent the first large scale phosphorylation data set for C. elegans. Second, we implemented a novel function to analyze the conservation of the identified phosphorylation sites across species (Figure 1). Third, for every phosphorylation site we provide, in downloadable form, a mass spectrometric assay based on multiple reaction monitoring to support further experimentation, including accurate quantification of the respective site in complex samples. Fourth, we implemented a dedicated help page which explains all displayed parameters and a downloadable tutorial for those scientists who intend to use the resource but are not trained in the analysis of mass spectrometry data. Specifically, the tutorial describes how the quality of a phosphopeptide and phosphorylation site identification based on fragment ion spectra can be assessed (see Supplementary Material and Methods), and fifth, the pre-existing software tools were adapted for use with the data from all four species. Collectively, these advances significantly expand the available data, support a wider range of queries and make the resource accessible to a wider range of scientists.
Table 1.
| Organism | Phosphopeptides with P>0.8a | Total phosphorylation sites | Phosphopeptides with assigned phosphorylation site(s)b |
|---|---|---|---|
| D. melanogaster | 16,875 | 16,608 | 12,756 |
| S. cerevisiae | 9,554 | 8,901 | 5,890 |
| C. elegans | 5,444 | 4,986 | 3,545 |
| Human | 3,784 | 3,980 | 2,810 |
PeptideProphet Score as computed by PeptideProphet12
A phosphopeptide was considered to have an unassigned/assigned site if a dCn threshold was not reached/exceeded (See Supplementary Material and Methods)
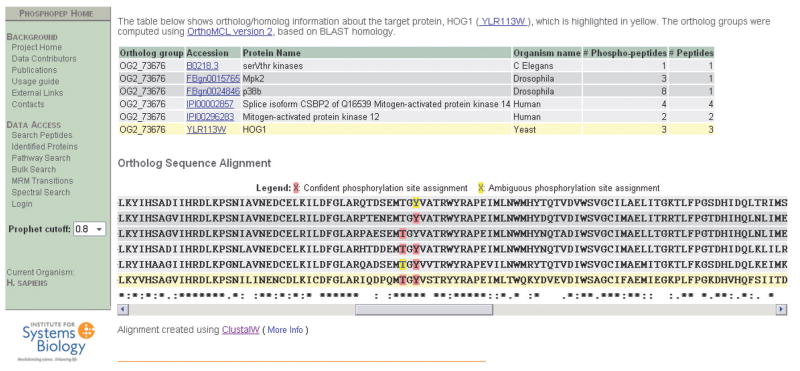
Figure 1.
Figure 1A. Analysis of phosphorylation site conservation. As an example the yeast protein Hog1 is used. In the upper half, the orthologous proteins of Hog1 in worm, fly and human are displayed10. In the lower half, the amino acid sequences of the proteins are shown and the identified phosphorylation sites are highlighted11. It can be seen that the phosphorylation of the TXY motif, which is known to activate MAP kinases, is conserved between all species.
The new data added were obtained from focused phosphoproteome mapping experiments carried out in our lab and to some extent by contributions from laboratories making their phosphoproteomic data generously available6–9. For the data collected in our group we followed the data collection strategy described for D. melanogaster (See Supplementary Material and Methods). For C. elegans this generated 5,444 unique high confidence phosphopeptides that could be assigned to 2,959 gene products, comprising 3,545 assigned unique phosphorylation sites. For S. cerevisiae, using the same strategy and combining the in house data with a published data set9 we identified at high confidence 9,554 phosphopeptides that could be assigned to 2,071 gene products, comprising 5,890 assigned unique phosphorylation sites. The assigned proteins cover nearly one third of the predicted yeast proteome with no bias in the range of protein abundance (Supplementary Figure 1A) but with a bias towards proteins involved in signal transduction (Supplementary Figure 1B). For human, we used previously published data from cancer and HELA cells6–8 that were made accessible to identify at high confidence 3,784 unique phosphopeptides that could be assigned to 5,160 gene product, comprising 2,810 assigned phosphorylation sites. Finally, the contents of the D. melanogaster data set include 16,875 phosphopeptides that could be assigned to 5,347 gene products, comprising 12,756 assigned phosphorylation sites.
The ability to support cross species comparisons arose from the inclusion of phosphopeptide data from four species and is a significant new and unique feature of PhosphoPep v2.0. In a first step the user can view the orthologous phosphoproteins (if known) between the species starting from any protein information page10 (Figure 1). In a second step, the amino acid sequences of the orthologous proteins are aligned and the phosphorylation sites which are stored in PhosphoPep are highlighted on the alignment. In addition, the level of conservation is displayed for each site11 (Figure 1). This new function will help to assess the conservation of signaling networks and the assignment of phosphorylation sites across species.
In summary, the novel model organism datasets and the unique set of software tools implemented in PhosphoPep v.2.0 support the analysis of single phosphoproteins, the detection of quantitative changes in the state of phosphorylation of whole signaling pathways at different cellular states and the investigations into the evolution of signaling networks from yeast, worm, fly to human. The system has been designed to enable the rapid iterative cycles of experimentation and analysis that are the basis of systems biology research and should therefore find wide application in basic and applied research.
Supplementary Material
Acknowledgments
We want to acknowledge Steve Gygi, Sean Beausoleil and Cell Signaling Technology for providing us with phosphorylation data as published6–9. We also want to thank the Functional Genomics Center Zurich for the generous support with mass spectrometry resources. This project has been funded in part by ETH Zurich, the Swiss National Science Foundation under grant No. 31000-10767, with Federal (US) funds from the National Heart, Lung, and Blood Institute, National Institutes of Health, under contract No. N01-HV-28179, and by the Center for Model Organism Proteomics of SystemsX.ch, the Swiss initiative for systems biology. Work at the FGCZ has been supported by the University Research Priority Program Systems Biology and Functional Genomics of the University of Zurich. RA was supported in part by a grant from F Hoffmann-La Roche Ltd (Basel, Switzerland) provided to the Competence Center for Systems Physiology and Metabolic Disease. BB is the recipient of a fellowship by the Boehringer Ingelheim Fonds.
Footnotes
Data availability
All data presented in this study are available from PhosphoPep (www.phosphopep.org).
References
- 1.Peri S, et al. Development of human protein reference database as an initial platform for approaching systems biology in humans. Genome Res. 2003;13:2363–2371. doi: 10.1101/gr.1680803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Diella F, et al. Phospho. ELM: a database of experimentally verified phosphorylation sites in eukaryotic proteins. BMC Bioinf. 2004;5:79. doi: 10.1186/1471-2105-5-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gnad F, et al. PHOSIDA (phosphorylation site database): management, structural and evolutionary investigation, and prediction of phosphosites. Genome Biol. 2007;8:R250. doi: 10.1186/gb-2007-8-11-r250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bodenmiller B, et al. PhosphoPep--a phosphoproteome resource for systems biology research in Drosophila Kc167 cells. Mol Syst Biol. 2007;3:139. doi: 10.1038/msb4100182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bodenmiller B, Mueller LN, Mueller M, Domon B, Aebersold R. Reproducible isolation of distinct, overlapping segments of the phosphoproteome. Nat Methods. 2007;4:231–237. doi: 10.1038/nmeth1005. [DOI] [PubMed] [Google Scholar]
- 6.Beausoleil SA, Villen J, Gerber SA, Rush J, Gygi SP. A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat Biotechnol. 2006;24:1285–1292. doi: 10.1038/nbt1240. [DOI] [PubMed] [Google Scholar]
- 7.Rikova K, et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131:1190–1203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 8.Beausoleil SA, et al. Large-scale characterization of HeLa cell nuclear phosphoproteins. P Natl Acad Sci USA. 2004;101:12130–12135. doi: 10.1073/pnas.0404720101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li X, et al. Large-scale phosphorylation analysis of alpha-factor-arrested Saccharomyces cerevisiae. J Proteome Res. 2007;6:1190–1197. doi: 10.1021/pr060559j. [DOI] [PubMed] [Google Scholar]
- 10.Chen F, Mackey AJ, Vermunt JK, Roos DS. Assessing performance of orthology detection strategies applied to eukaryotic genomes. PLoS ONE. 2007;2:e383. doi: 10.1371/journal.pone.0000383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 12.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.