Abstract
Brr2 is a DExD/H-box helicase responsible for U4/U6 unwinding during spliceosomal activation. Brr2 contains two helicase-like domains, each of which is followed by a Sec63 domain with unknown function. We determined the crystal structure of the second Sec63 domain, which unexpectedly resembles domains 4 and 5 of DNA helicase Hel308. This, together with sequence similarities between Brr2’s helicase-like domains and domains 1–3 of Hel308, led us to hypothesize that Brr2 contains two consecutive Hel308-like modules (Hel308-I and II). Our structural model and mutagenesis data suggest that Brr2 shares a similar helicase mechanism with Hel308. We demonstrate that Hel308-II interacts with Prp8 and Snu114 in vitro and in vivo. We further find that the C-terminal region of Prp8 (Prp8-CTR) facilitates the binding of the Brr2/Prp8-CTR complex to U4/U6. Our results have important implications for the mechanism and regulation of Brr2’s activity.
Keywords: Pre-mRNA splicing, Brr2, Sec63, Hel308, Prp8, Snu114
Pre-mRNA splicing is carried out by the spliceosome, which contains five small nuclear RNAs (snRNAs U1, U2, U4, U5, and U6) and over 100 different proteins. The spliceosome typically assembles on pre-mRNA in a step-wise manner 1. In the first step of spliceosomal assembly, the 5’ splice site (ss), Branch Point Sequence (BPS), and 3’ss of pre-mRNA are recognized by the U1 snRNP, SF1/BBP, and U2AF65/35, respectively. Subsequently, U2 snRNP joins the spliceosome, followed by the joining of the U4/U6.U5 tri-snRNP. Extensive structural rearrangements occur at this stage 2. For example, the base-pairing between the 5’ss and U1 snRNA is disrupted and the 5’ss interacts with U6 instead. The base-pairing between U4 and U6 is also disrupted and new interactions between U2 and U6 are formed. These rearrangements help convert the spliceosome to the catalytically active complex, which subsequently splices out the intron and ligate the two exons.
At least eight DExD/H-box proteins are involved at various stage of the splicing cycle 3. DExD/H-box proteins contain many RNA helicases and belong to helicase superfamily 2 (SF2). All superfamily 1 (SF1) and 2 helicases contain the minimal helicase domain (two RecA domains with ~400 amino acids encompassing at least eight conserved helicase motifs) and sometimes additional domains 4,5. Motifs I and II are highly conserved among all SF1 and SF2 helicases but other motifs are less so and are often used to classify SF1 or SF2 helicases into smaller families. DExD/H-box proteins can be further divided into DEAD, DEAH, Ski2-like DExH, and a few other families 4. Multiple DExD/H-box proteins in the spliceosome have been demonstrated to have weak helicase activity in vitro 6–10. While their precise molecular targets remain largely unknown, the DExD/H-box proteins are thought to facilitate the many conformational rearrangements required for the successful assembly and activation of the spliceosome 3.
Brr2 is a large DExD/H-box protein (2,163 amino acids in yeast) and a stable component of the U5 snRNP 7,11,12. Early experiments identified a role for Brr2 in the unwinding of U4/U6, a critical step in spliceosomal activation 7,11,12. Recent work supports a role for Brr2 in unwinding of U2/U6 during spliceosomal disassembly 13. As an integral component of the U5 snRNP, tri-snRNP, and spliceosome 14, regulation of Brr2’s helicase activity is particularly important to ensure the correct timing of spliceosomal activation or disassembly. Prp8 and Snu114 (both components of the U5 snRNP, tri-snRNP, and spliceosome) have been implicated in regulating the activity of Brr2 2,13,15–17. For example, Brr2’s in vitro ATPase and helicase activities are modulated by the C-terminal region of Prp8, although the site of modulation on Brr2 and the mechanism of modulation are not yet known 18.
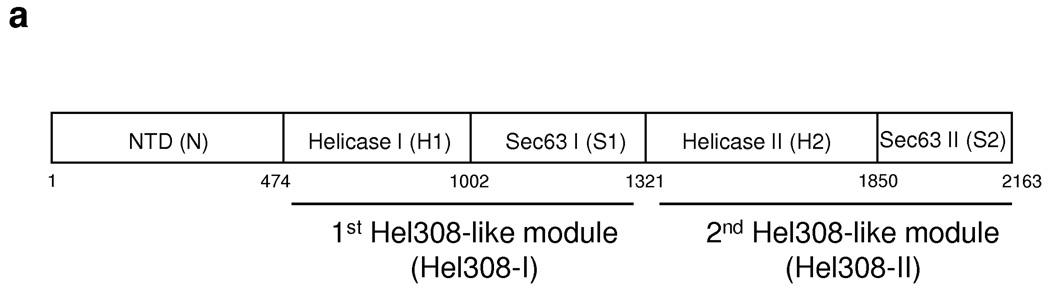
To our knowledge, Brr2 is a the only DExD/H-box protein that contains two putative helicase domains, with the second helicase-like domain deviating more from the prototypical helicase motifs 14,19,20. Whereas the motifs in the first helicase-like domain of S. cerevisiae Brr2 (yBrr2) have been shown to be critical for ATPase activity, U4/U6 unwinding, and cell viability, the motifs in the second helicase domain can be disrupted with little consequence 12. Brr2 also contains multiple other domains, including an N-terminal domain and two Sec63 domains. The five domains are arranged as shown in Figure 1a and are abbreviated as N, H1, S1, H2, and S2. The Sec63 domain is defined by sequence similarity with Sec63 protein, a central component of the protein translocation apparatus on the ER membrane, although the structure and function of the Sec63 domain are unknown 21. With the exception of the H1 domain which is likely involved in RNA unwinding, the function of the other Brr2 domains is unclear.
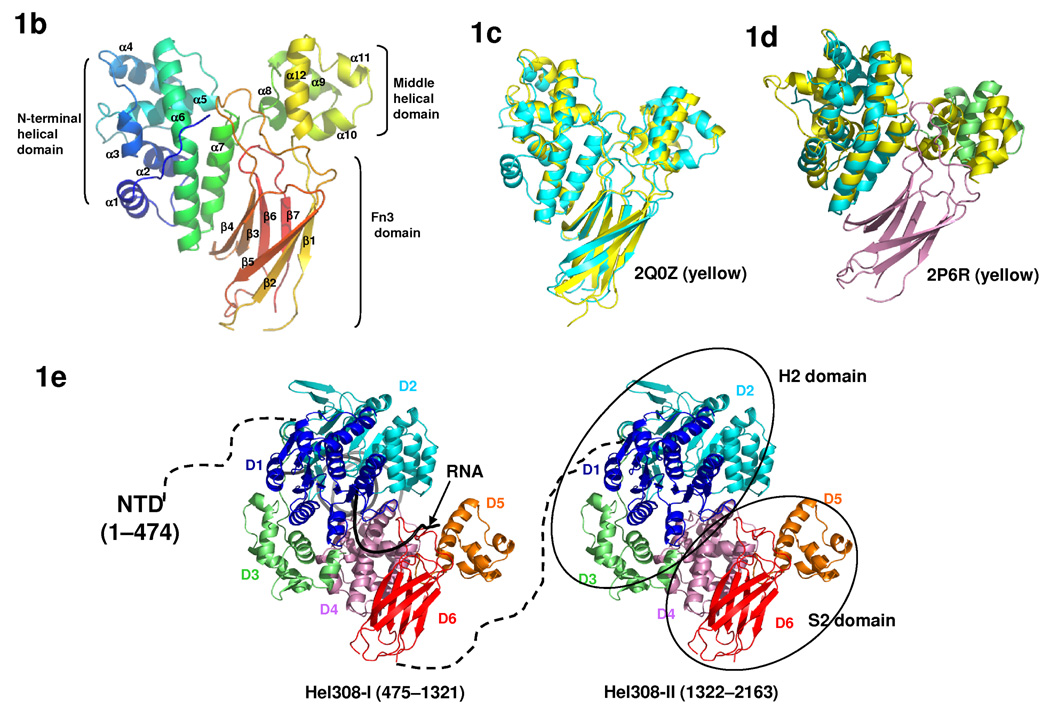
Fig. 1.
The structure of yBrr2. (a) A schematic representation of the domain organization (abbreviations shown in parenthesis) of yBrr2. (b) Overall structure of the S2 domain, colored in rainbow spectrum from the N-terminus (blue) to the C-terminus (red). (c) Superimposition of the yeast (cyan) and human (yellow, PDB ID 2Q0Z) S2 domain structures. (d) Superimposition of the S2 domain and domains 4 and 5 of Hel308 (yellow, PDB ID 2P6R 23). The N-terminal helical domain, the middle helical domain, and the C-terminal Fn3 domain in S2 are colored blue, green, and purple, respectively. (e) A backbone model (by combining the Hel308 and S2 structure) depicting the overall structure of Brr2 consisting the NTD, Hel308-I, and Hel308-II. In each Hel308-like module, the D1-6 domains are colored in dark blue, light blue, green, purple, orange, and red, respectively. A partially unwound DNA duplex (black) as seen in the Hel308 structure was modeled in Hel308-I to represent the RNA substrate. The traditional H2 and S2 domain were indicated in ovals on the structural model.
In this paper, we determined the crystal structure of S2 of yBrr2, and find unexpected structural similarity between S2 and domains 4 & 5 of Hel308 (a Ski2-type SF2 DNA helicase implicated in DNA repair and recombination). This structural similarity, in combination with sequence analyses, led us to hypothesize that the entire Brr2 protein is composed of an N-terminal domain and two consecutive Hel308-like modules. This model offers our first glimpse of the overall structure of this large and unique spliceosomal ATPase and helicase. The structural similarity with Hel308 suggests mechanistic similarities between Brr2 and Hel308, which are consistent with mutagenesis data. We further demonstrated that the second Hel308-like module interacts with Prp8 and Snu114 in vitro and in vivo. We also show that the C-terminal region of Prp8 (Prp8-CTR, residues 1822–2395) facilitates the binding of the Brr2/Prp8-CTR complex to U4/U6, and discuss the implications of this result for the activity and regulation of Brr2.
Results
The crystal structure of S2 suggests two consecutive Hel308 modules in Brr2
We determined the crystal structure of S2 to 2.0 Å resolution, which shows that S2 is composed of three domains, an N-terminal helical domain, a middle helical domain, and a C-terminal beta domain with the fibronectin type 3 (Fn3) fold (Fig. 1b, Supplementary Fig. 1). The overall structure of yeast Brr2 S2 is highly similar to the human Brr2 S2 domain (PDB ID 2Q0Z, Northeast Structural Genomics Consortium) (Fig. 1c). The root mean square deviation of 269 superimposed main chain atoms between the two structures is 1.6Å, consistent with the sequence similarity between the two S2 domains (Supplementary Fig. 2). All of our subsequent discussion will use the yBrr2 S2 structure we determined.
In spite of the lack of obvious sequence similarity, a structural homology search using the Dali server 22 revealed that the two helical domains of S2 have exactly the same topology as domains 4 and 5 of Hel308, a Ski2-type SF2 DNA helicase 23,24 (PDB ID 2P6R) (Fig. 1d). Hel308 also contains two RecA domains (domain 1 and 2) and another helical domain 3 (86 residues). The H2 regions of Brr2 share sequence similarity with the two RecA domains common to all SF2 helicases including Hel308 14,20. In addition, we noticed that there are ~120 residues immediately downstream of the second RecA domain in H2 that is all helical, based on secondary structure predictions 25. Therefore, the H2 region of Brr2 could resemble domains 1–3 of Hel308 in three-dimensional structures and the H2+S2 region of Brr2 resembles the entire Hel308 with an additional Fn3 domain (Fig. 1e).
The S1 domain of Brr2 likely forms a similar structure to the S2 domain, considering their sequence similarities. Secondary structure predictions of S1 and S2 match well with the actual secondary structures observed in the S2 structure (Supplementary Fig. 1), supporting the hypothesis that S1 forms a similar structure to S2. It is also known that the H1 domain has substantial sequence similarity to the Ski2-type helicase domains (higher than that between H2 and these helicases) 14. It is, therefore, highly likely that the H1+S1 region also forms a structure similar to Hel308. Taken together, we propose that the overall structural of Brr2 consists of an N-terminal domain and two consecutive Hel308-like modules (Hel308-I and II) (Fig. 1e).
The structural model of Hel308-I suggests possible helicase mechanisms
Hel308-I likely serves the unwinding function based on previous mutagenesis studies 12. Hel308-I has the highest sequence conservation among different species, with 35% identity among yeast, human, C. elegans, Drosophila, and Arabidopsis Brr2, while 9% of the residues in the N-terminal domain and 18% of the residues in Hel308-II are absolutely conserved (MULTALIN 26, data not shown). The high degree of sequence conservation in Hel308-I is likely a reflection of the critical importance of the helicase activity of this module.
A model of Brr2 Hel308-I can be built based on the crystal structure of Hel308 in complex with a partially unwound DNA duplex (15bp dsDNA with a 10nt 3’ overhang) 23 (Fig. 1e). The proposed unwinding mechanism of Hel308 is substantially different from many well-studied DEAD-box RNA helicases. A prominent β-hairpin between motifs V and VI in domain 2 of Hel308 disrupts two base pairs of the dsDNA (Supplementary Fig. 3a) and is thought to be important for strand separation 23. DEAD-box RNA helicases such as eIF4A 27, UAP56 28,29, and Vasa 30 do not have a similar β-hairpin and use local RNA bending as a different unwinding mechanism 30. Sequence alignment shows that motifs V and VI of Brr2 are similar to Hel308, but are drastically different from eIF4A (Supplementary Fig. 3b). Brr2 Hel308-I contains a similar number of residues between motifs V and VI as in Hel308 (Supplementary Fig. 3b), making it feasible for Hel308-I to also form a similar β-hairpin in this region, although it is often difficult to accurately predict a short β-hairpin based on secondary structure predictions. The sequence between motifs V and VI in Brr2 and Hel308 are not well conserved, but it does not necessarily conflict with the potential functional importance of this region, since many different amino acid compositions may form a β-hairpin.
As a first step toward exploring the functional importance of the putative β-hairpin region, we generated a brr2-3GS mutant by replacing residues 860–865 (WEQLSP, downstream of an existing Gly-Ser pair) with three additional sets of Gly-Ser (Supplementary Fig. 3b). Gly-Ser residues have often been used to create flexible linkers in protein engineering 31. We reason that this stretch of four Gly-Ser residues will likely disrupt any potential β-hairpin structure in this region, although we realize that the 3GS mutant could potentially be too drastic and disrupt the overall structure of Brr2. We generated brr2-3GS on the pGPD-BRR2-TAP vector (containing a C-terminal Protein A tag) and shuffled it into the yTB105 strain (endogenous brr2 deleted and WT brr2 on an URA3-marked plasmid) 18. brr2-3GS grows much slower than WT at 30 °C and 18 °C and does not grow at 37 °C (Supplementary Fig. 3c). We showed that the Brr2 protein levels in the WT and brr2-3GS strains are similar by pulling down Brr2 proteins from cell extracts using IgG resin followed by Western blot analyses using an anti-Brr2 antibody 32 (Supplementary Fig. 3c). We also demonstrated that Brr2-3GS pulls down similar amounts of Prp8 and Snu114 as WT Brr2 (Supplementary Fig. 3c), suggesting that the 3GS mutant has not caused major disruption of the overall structure of Brr2. We then purified WT Brr2 and Brr2-3GS proteins using IgG resin and cleaved off the Protein A tag with TEV protease. As we purified away Brr2-associated proteins, Brr2-3GS became more prone to degradation and resulted in less full-length protein than WT Brr2 (~20% of WT level) (Supplementary Fig. 3d). The full-length Brr2-3GS no longer has in vitro ATPase and helicase activity 18 (data not shown). Although the loss of helicase activity could be a combination of the effect of the putative β-hairpin itself and other structural changes caused by the 3GS mutation (noting that Brr2-3GS is more prone to degradation in purification and also lost ATPase activity), these results do suggest that the putative β-hairpin region is important for the structure and/or function of Brr2. Further structural, mutagenesis (for example, scanning single-site mutants in the putative β-hairpin region), and biochemical analyses will unambiguously reveal whether Brr2 contains a β-hairpin and utilizes an unwinding mechanism similar to Hel308.
Hel308 is also a processive DNA helicase, in contrast with typical DExD/H-box RNA helicases. In the Hel308+DNA structure, the ssDNA goes through the enclosure formed by domains 1, 3 & 4 and also interacts with domains 2 and 5 (ref. 23). It was suggested that the presence of domains 3 & 4, as well as a central ratchet helix in domain 4, contribute to the processivity of Hel308. Our structural model suggests that similar domains and the ratchet helix also exist in Hel308-I of Brr2, suggesting Brr2 may be more processive than other typical DExD/H-box proteins involved in splicing.
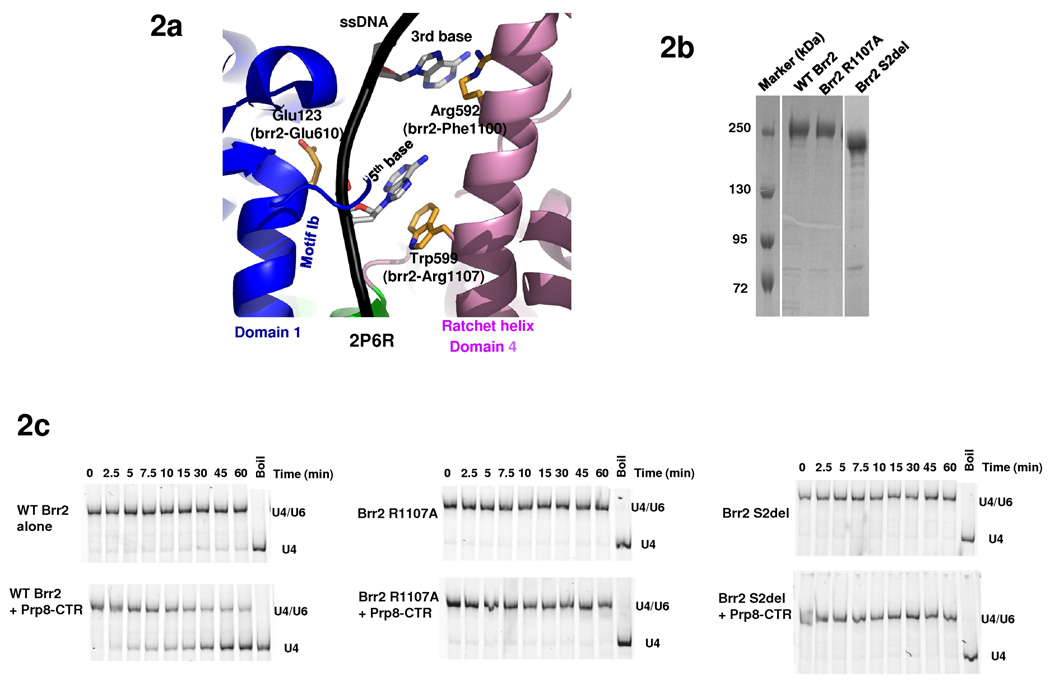
Two previously identified brr2 mutants are located in the Hel308-I module. Brr2-1 (E610G) and Brr2-R1107A are cs mutants that are defective in U4/U6 unwinding 11 and/or spliceosome disassembly 13. Residue E610 is located in motif Ib (TPEK in Brr2) of domain 1, which is typically involved in RNA or DNA substrate binding 23,30. The equivalent E123 residue of Hel308 is indeed on a helix right next to the ssDNA in the Hel308+DNA structure 23 (Fig. 2a). Interestingly, Hel308-W599 (the equivalent of Brr2-R1107, Supplementary Fig. 1) is on the midpoint of the ratchet helix of domain 4, across from motif 1b (Brr2-E610) on the opposite side of the same ssDNA (Fig. 2a). Hel308-W599 forms a stacking interaction with the 5th base of the ssDNA. Two helical-turns away, Hel308-R592 on the same helix interacts with the 3rd base of the ssDNA. In addition, the N-terminus of the ratchet helix interacts with motif IVa of domain 2.
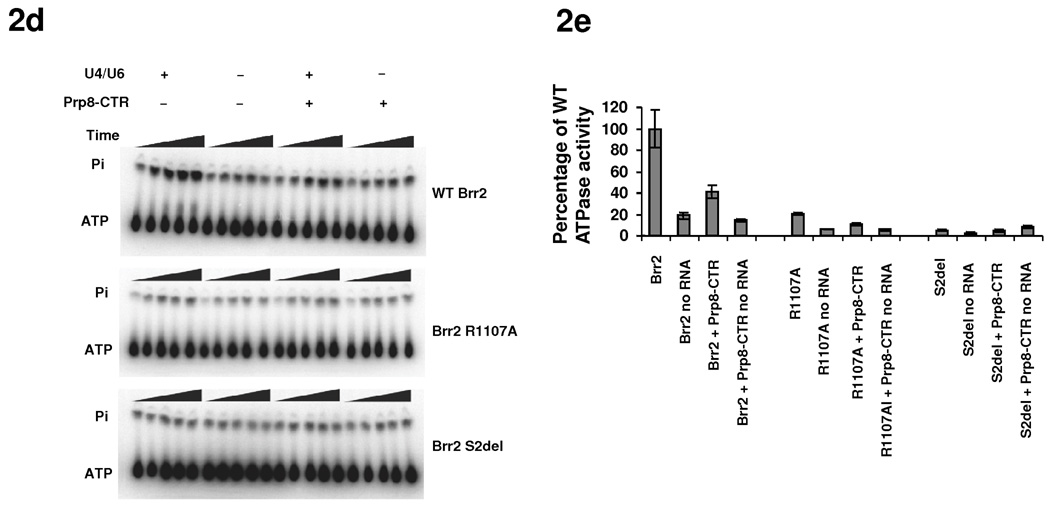
Fig. 2.
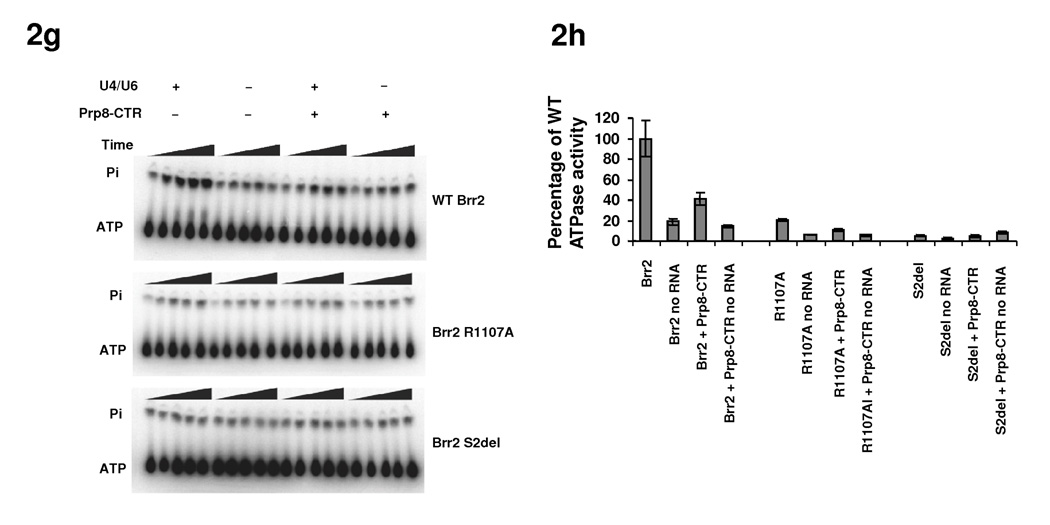
ATPase and helicase activities of Brr2 mutants. (a) Locations of residues corresponding to brr2-1 (E610G) and brr2-R1107A on the Hel308 structure (PDB ID 2P6R 23) and their relationship with the ssDNA (black). Color scheme is the same as 1e. (b) SDS PAGE of purified Brr2 WT, R1107A, and Brr2 S2del mutants. (c) Helicase assay of WT Brr2, Brr2 R1107A, and Brr2 S2del, in the absence and presence of Prp8-CTR. (d) ATPase activity of WT Brr2, Brr2 R1107A, and Brr2 S2del in the absence and presence of Prp8-CTR and U4/U6. (e) Quantification of the data in panel g. Error bars indicates s.e.m.
Therefore, ATP-dependent movement of domain 2 was thought to modulate the position of the ratchet helix and facilitate strand translocation 23. Brr2-F1100 (the equivalent of Hel308-R592) in combination with Brr2-R1107 (the equivalent of Hel308-W599) can potentially perform a similar function to Hel308-R592 and Hel308-W599 in strand translocation and processivity. Mutations of Brr2-E610 (i.e., brr2-1) and Brr2-R1107 (i.e., brr2-R1107A) may conceivably affect RNA binding and/or strand translocation, and consequently affect U4/U6 unwinding and spliceosomal disassembly. Indeed, Brr2-1 does not have detectable unwinding activity in an in vitro helicase assay 18. To test the role for R1107 in Brr2 function, we constructed the R1107A mutant on the pGPD-BRR2-TAP vector and shuffled the mutant plasmid into the yTB105 strain. We were able to purify a similar quantity of Brr2 R1107A and Brr2 WT (Fig. 2b) for ATPase and helicase assays. As previously observed 18, WT Brr2 demonstrates weak helicase activity on its own, but the activity is greatly stimulated by Prp8-CTR (the previously used Prp8 region consists of residues 1806–2413 and is highly similar to our Prp8-CTR consisting of residues 1822–2395) (Fig. 2c). WT Brr2 also demonstrates RNA stimulated ATPase activity which is inhibited by Prp8-CTR (Fig. 2d, e). We found that the Brr2 R1107A mutant has greatly reduced ATPase activity and no detectable helicase activity under the current assay conditions (Fig. 2c–e). Both Brr2-1 and R1107A likely retain weak helicase activity in vivo since yeast strains containing these mutants grow similarly to WT at 30 °C. In general, these results support the mechanistic similarity between Brr2 and Hel308.
Domain deletions in Hel308-II are lethal or detrimental to growth
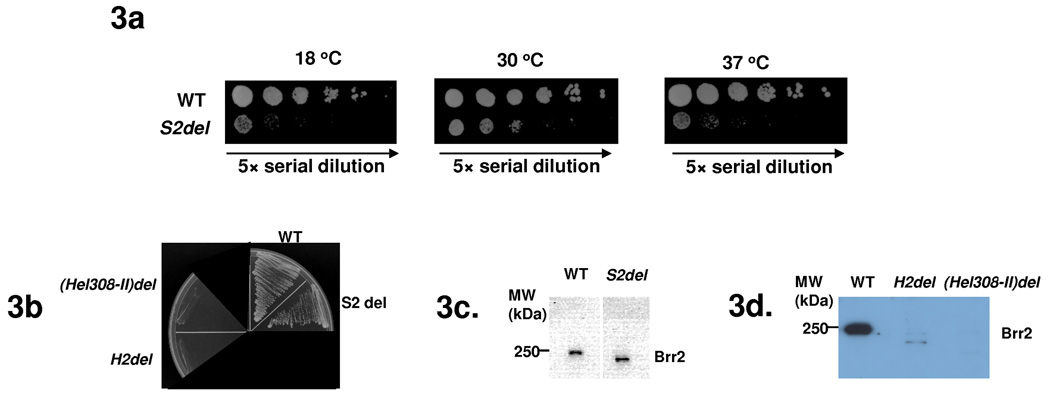
In contrast to the likely unwinding function of Hel308-I, the Hel308-II module of Brr2 does not have ATPase activity and is unlikely to have helicase activity 12. We focused our subsequent studies on understanding the function of Hel308-II. We generated yeast strains carrying Brr2 with the H2, S2, or Hel308-II (H2+S2) domains deleted. These deletions were constructed on pPR150 (carrying a C-terminal polyoma tag) 11 and were shuffled into yeast strain yJPS996 (endogenous brr2 deleted and WT brr2 on an URA3-marked plasmid) 13. The S2-deletion strain grows much slower than WT at all three temperatures (Fig. 3a). All other deletion strains do not grow on 5-FOA plates (Fig. 3b), indicating that these deletions are lethal. We then performed pull down experiments using yeast extract and an anti-polyoma antibody, followed by Western blotting with an anti-Brr2 antibody 32. There are similar quantities of Brr2 protein in both the WT and S2-deletion strain (Fig. 3c), indicating that the slow growth phenotype of the S2-deletion is not caused by the lack of Brr2 protein. We also performed similar pull down experiments using yeast strains prior to 5-FOA shuffling. The H2 and Hel308-II deletion strains have much less Brr2 protein than the WT strain, indicating that these deletions destabilize the Brr2 protein (Fig. 3d).
Fig. 3.
Deletions of domains in Hel308-II lead to growth and splicing defects. (a) S2 deletion grows slower than the WT brr2 strain in all three temperatures. Five-fold Serial dilutions were plated on YEPD plates and grown at 30 °C and 37 °C for 2 days and at 18 °C for 5 days. (b) A 5-FOA plate demonstrating that the S2-deletion is viable but H2 and Hel308-II deletions are lethal. (c) Pull down experiments using yeast extract and an anti-polyoma antibody, followed by Western analyses using an anti-Brr2 antibody, showed that the cellular Brr2 protein levels are similar in the WT and S2-deletion strains. (d) Similar pull down experiments using strains prior to 5-FOA shuffling showed that H2 or Hel308-II deletions led to dramatically reduced Brr2 protein levels. (e) Microarray and quantitative RT-PCR analyses of the genome-wide splicing phenotype and snRNA levels of the S2-deletion strain. Each horizontal line describes the behavior of a single intron-containing gene. T, P, and M represent total mRNA, pre-mRNA, and mature mRNA, respectively. Yellow indicates increased and blue indicates decreased RNA levels compared to the WT. (f) Total snRNA levels assessed using quantitative RT-PCR indicates that the S2 deletion leads to an accumulation of the U1, U2 and U5 snRNAs. Error bars indicate s.d.
The S2 deletion affects the splicing of many genes
To examine the molecular phenotype of the S2 deletion, we performed splicing-specific microarray experiments. These microarrays contain three probes for each of the ~300 intron-containing genes in yeast: one targeting a region of the intron to measure pre-mRNA levels, one targeting the junction between exons 1 and 2 to measure the mature mRNA, and one targeting a region of either exon 1 or 2 to measure changes in total mRNA 33. As a control, we simultaneously examined the behavior of the brr2-1 strain 11. The microarray data show that the S2 deletion affects the splicing of the vast majority of intron-containing genes, as demonstrated by the accumulation of pre-mRNA and reduction of spliced mRNA for these genes (Fig. 3e).
The defects seen in the S2 deletion, both in terms of the number of transcripts that are affected, and the magnitude to which their splicing is affected, are similar in scale to the defects seen in the brr2-1 strain (Fig. 3e), indicating a strong defect in pre-mRNA splicing. Interestingly, the S2 deletion, but not the brr2-1 mutation, also showed accumulation of U1, U2, and U5 snRNAs (1.9, 3.5, and 2.6 fold, respectively) through quantitative RT-PCR analyses (Fig. 3f). The mechanism and significance of this accumulation await further studies.
Hel308-II does not bind several RNAs tested in vitro
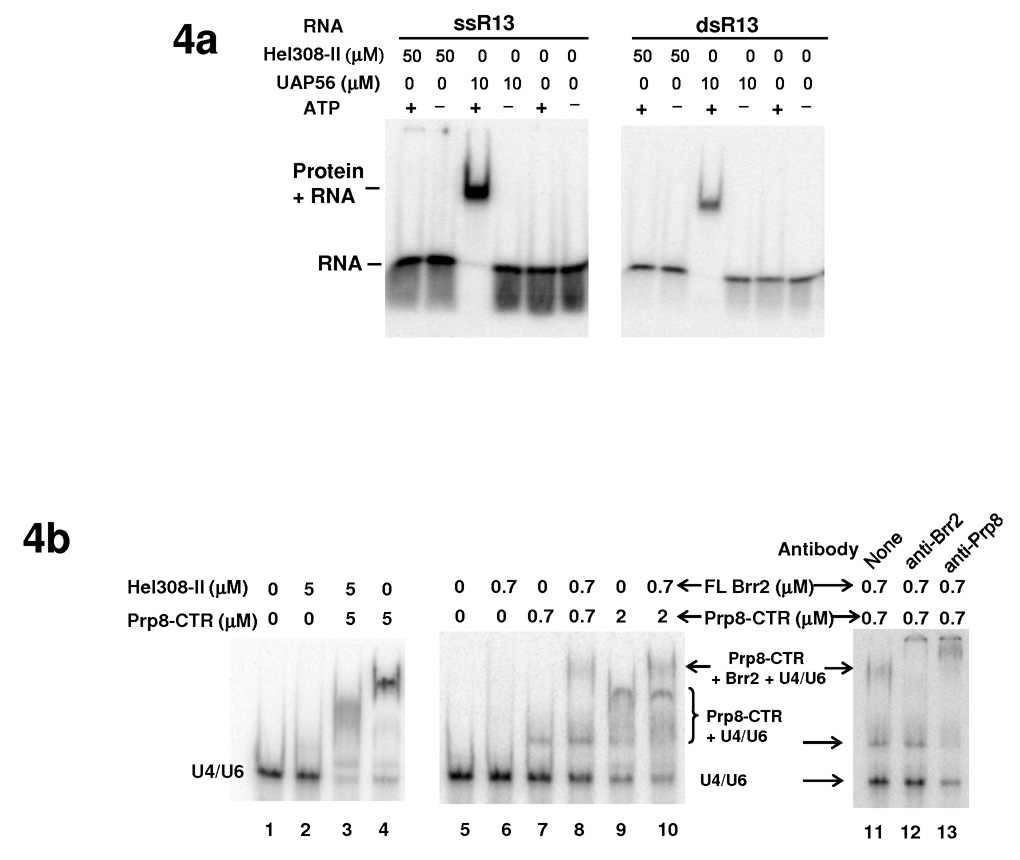
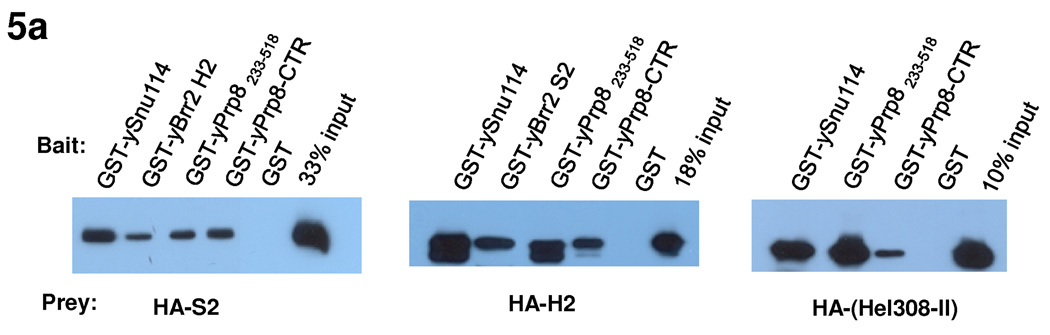
We next examined whether Hel308-II retains the ability to bind to RNA, even though it lacks ATPase and helicase activities. We demonstrated using Electrophoresis Mobility Shift Assay (EMSA) that purified Hel308-II does not bind a 13nt ss or ds RNA (GAGAUUUAUUUCG, arbitrary sequence, designated as ssR13 and dsR13) even at a concentration of 50 µM, in the absence or presence of ATP (Fig. 4a). Under similar conditions, the positive control UAP56 (a splicing factor and DEAD -box protein) clearly binds RNA at 10 µM concentration in the presence of ATP (ATP is known to increase the RNA binding affinity of many DEAD-box proteins including eIF4A 30,34) (Fig. 4a). Nor does Hel308-II have detectable binding to U4/U6 at 5 µM protein concentration in the presence or absence of ATP (Fig. 4b, lanes 1 and 2).
Fig. 4.
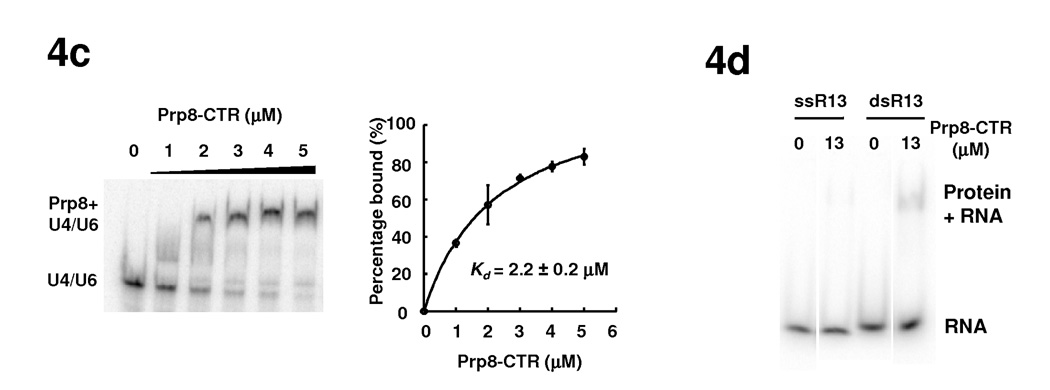
The interaction of Brr2 with RNA and the effect of Prp8-CTR on these interactions. (a) EMSA demonstrates that Hel308-II does not bind a 13nt ss or dsRNA with arbitrary sequence (GAGAUUUAUUUCG) in the absence or presence of ATP. UAP56, another DExD/H-box protein involved in splicing, is used as a positive control and binds both ss and dsRNA in the presence of ATP. (b) Hel308 (lanes 1–4) and full-length Brr2 (lanes 5–10) do not bind U4/U6 at indicated concentrations but the presence of Prp8-CTR generate a complex that binds U4/U6 better. Supershift experiments using anti-Brr2 and anti-Prp8 antibodies confirm the identity of the shifted bands (lanes 11–13). (c) Titration experiments indicate that Prp8-CTR binds U4/U6 with a Kd of 2.2±0.2 µM. Error bars and errors in Kd are s.d. d) Prp8-CTR binds ss or dsR13 very weakly.
Hel308-II interacts with Prp8 and Snu114 in vitro
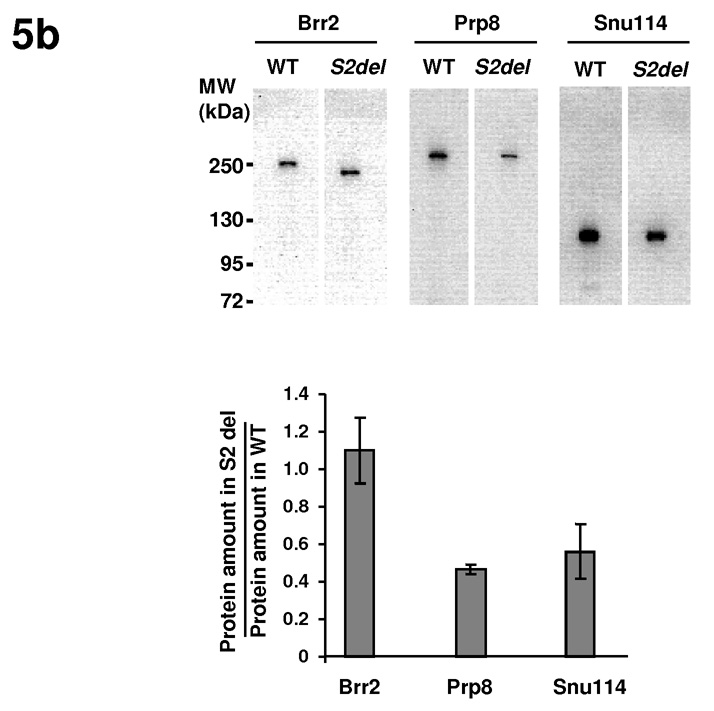
We then evaluated whether the Hel308-II module can potentially function as a protein interaction domain and mediate interactions with other spliceosomal proteins. We performed in vitro GST pull down experiments using GST-fused versions of several Brr2 domains, Prp8 domains, and Snu114 as baits, and HA-tagged Brr2 H2, S2, or Hel308-II domains as prey (Fig. 5a). These experiments demonstrate that the H2 and S2 domain of Brr2 interact with each other, consistent with the idea that the H2 and S2 domains come together to form a Hel308 module. Furthermore, the H2, S2, and Hel308-II domains all interact with both the N-terminal (residues 233–518) and C-terminal (Prp8-CTR, residues 1822–2395) regions of Prp8, as well as with Snu114. These interactions are specific as no interaction is detected with GST alone. Our results are consistent with previous observations using yeast two-hybrid analyses that the C-terminal region of yBrr2 (a construct containing the combination of residues 112–356 and 1184–2163) is responsible for the vast majority of interactions between Brr2 and many spliceosomal proteins including Prp8 (ref. 35). The interactions between the H2 domain of hBrr2 and hPrp8 as well as hSnu114 have also been observed previously using yeast two-hybrid analyses 36,37.
Fig. 5.
Hel308-II interacts with Prp8 and Snu114 in vitro and in vivo. (a) GST pull down experiments demonstrate that the H2, S2, and Hel308-II domains interact with Prp8 and Snu114. (b) Co-immunoprecipitation experiments demonstrate that the S2-deletion strain has similar level of Brr2 but lower level of Prp8 and Snu114 than WT strains. Brr2 in yeast extract was pulled down using anti-polyoma antibody and probed in Western blot using anti-Brr2, anti-Prp8 and anti-Snu114 antibodies. Error bars indicate s.d.
The S2 deletion decreases Brr2’s association with Prp8 and Snu114 in vivo
We took advantage of the fact that the S2-deletion strain is viable to examine whether the S2 domain also interacts with Prp8 and Snu114 in vivo. We immunoprecipitated Brr2 from extracts of the WT and S2-deletion strains using an anti-polyoma antibody. We then performed Western blot analyses on the immunoprecipitated sample using anti-Brr2 32, anti-Prp8 38, and anti-Snu114 39 antibodies. The level of Brr2 protein in the immunoprecipitated sample from the S2-deletion strain is similar to WT, but the level of Prp8 and Snu114 are much less in the S2-deletion strain (about 50% of the WT level) (Fig. 5b). These experiments demonstrate that S2 deletion is defective in Prp8 and Snu114 binding, suggesting that the S2 domain indeed participates in Prp8 and Snu114 interactions in vivo.
The S2 deletion reduces the ATPase and helicase activity of Brr2 in vitro
To evaluate the effects of the S2 deletion on the ATPase and helicase activity of Brr2, we replaced the WT brr2 in the pGPD-Brr2-TAP vector 18 with the S2-deleted brr2. We expressed the full-length and S2-deleted Brr2 from yeast strain yTB105 16 and purified proteins using IgG resin and cleaved off the Protein A tag with TEV protease. We obtained similar quantities of purified proteins (Fig. 2b) for ATPase and helicase assays. The S2 deletion reduces Brr2’s ATPase and helicase activities to levels that are undetectable in these assays (Fig. 2c–e). The S2-deleted Brr2 is likely to still have weak ATPase and helicase activities in vivo, since the S2-deletion strain is viable even though it grows much slower than the WT strain. These results suggest that the S2 domain and Hel308-I interact/communicate with each other, and the S2 domain potentially stabilizes the structure and conformation of Hel308-I. The S2-deletion does not seem to lead to a complete mis-folding of Brr2, since the S2-deletion strain is still viable and we can purify similar quantities of soluble S2-deleted Brr2 protein as WT Brr2 protein (Fig. 2b). However, the S2-deletion probably leads to certain conformational changes in Hel308-I, rendering it less active in ATPase and helicase assays.
Prp8-CTR facilitates the binding of the Brr2/Prp8-CTR complex to U4/U6
We then examined the effect of Prp8-CTR on the RNA binding property of Brr2, using both Hel308-II and full-length Brr2. We showed using EMSA that Brr2 Hel308-II does not bind U4/U6 appreciably at 5 µM concentration (Fig. 4b, lane 2). Prp8-CTR, on the other hand, essentially binds and shifts all of the U4/U6 at 5 µM concentration (Fig. 4b, lane 4). We performed a titration experiment and determined that the Kd of Prp8-CTR to U4/U6 is 2.2±0.2 µM (Fig. 4c). This affinity is substantially higher than the affinity of Prp8-CTR to ssR13 and dsR13, where 13 µM Prp8-CTR shows essentially no binding to ssR13 and very little binding to dsR13 (Fig. 4d). When Brr2 Hel308-II and Prp8-CTR are incubated together with U4/U6, nearly all of the U4/U6 shifts to a position that is different from the band formed by Prp8-CTR alone + U4/U6 (Fig. 4b, compare lane 3 with lane 4). We interpreted this new gel-shift band as the Prp8-CTR + Hel308-II + U4/U6 complex (Fig. 4b, lanes 3).
The full-length Brr2 does not show appreciable binding to U4/U6 at 0.7 µM concentration (Fig. 4b, lanes 5 and 6). The addition of 0.7 µM Prp8-CTR in lane 8 forms a band at the same position as the Prp8-CTR alone + U4/U6 band in lane 7, and importantly, a new slower migrating band that likely corresponds to the Prp8-CTR + Brr2 + U4/U6 complex. Supershift experiments using anti-Prp8 38 and anti-Brr2 32 antibodies (Fig. 4b, lanes 11–13) confirmed our assignment of the faster migrating band as Prp8-CTR alone + U4/U6 and the slower migrating band as Prp8-CTR + Brr2 + U4/U6. The addition of more Prp8-CTR (lane 10) continues to form a band corresponding to the Prp8-CTR alone + U4/U6 band in lane 9 and a new band that likely represents the Prp8-CTR + Brr2 + U4/U6 complex. Note the positions of the Prp8-CTR alone + U4/U6 bands are somewhat different at 0.7 µM and 2 µM Prp8-CTR concentrations (compare lane 7 with 9) for reasons we do not yet fully understand, and this phenomenon is also observed in the Kd determination experiment in Fig. 4c. All binding reactions are highly reproducible and were performed both in the absence and presence of ATP, yielding identical results (data not shown). These results indicate that Prp8-CTR can facilitate the binding of the Brr2/Prp8-CTR complex to U4/U6.
Discussion
Our structural studies in combination with sequence analyses suggest that the H2+S2 domain of Brr2 resembles the entire Hel308 and that the full length Brr2 protein is composed of an N-terminal domain and two consecutive Hel308-like modules (Hel308-I and II). A recent structure of another Ski2-type DNA helicase Hjm also revealed a similar domain structure and organization as Hel308 40. This raises the possibility that many Ski2-type helicases may share structural (and potentially mechanistic) similarities in regions beyond the helicase domains, in spite of the lack of recognizable sequence similarities in these regions.
The structural resemblance between Brr2 and Hel308 suggests possible helicase mechanisms for Brr2. Our brr2-3GS mutant suggests that the putative β-hairpin region in Brr2 Hel308-I is structurally and/or functionally important. Further structural and mutational analyses will reveal whether Brr2 indeed contains a β-hairpin and utilizes an unwinding mechanism similar to Hel308 30. Furthermore, our structural mapping and biochemical analyses of the brr2-1 and brr2-R1107A mutants support the structural and mechanistic similarity between Brr2 and Hel308. These results suggest the possibility that Brr2 is more processive than other DExD/H-box proteins involved in pre-mRNA splicing. This potential processivity is attractive considering that yBrr2 needs to unwind U4/U6, which contains long stem regions and is highly stable in yeast 41.
Brr2 is the only known helicase that contains two helicase-like modules. The function of the second helicase-like module has long been elusive and intriguing. There are several substantial deviations between the putative helicase motifs in Hel308-II and the canonical helicase motifs. For example, the typical DExD/H residues in motif II are replaced with DDAH in Hel308-II (Supplementary Fig. 4). The Glu of motif II (DExD/H) has been postulated to be the key catalytic residue which activates a water molecule to hydrolyze ATP in DExD/H-box proteins and other helicases 5,30,42. Likewise, the Ser-Ala-Thr (SAT) residues in motif III are replaced with SNC in Hel308-II (Supplementary Fig. 4). The SAT residues in motif III do not interact with ATP or RNA but participate in inter-domain interactions between the N-terminal and C-terminal domains upon ATP and RNA binding and are thought to be important for the unwinding activity of DExD/H-box proteins 30. Hel308-II also lacks obvious motifs IV – VI (Supplementary Fig. 4). Substantial deviations from the canonical helicase motifs in Hel308-II probably have led to the lack of ATPase and helicase activities of this module.
We demonstrated that Hel308-II interacts with Prp8 and Snu114 in vitro and in vivo (Fig. 5a, 7). We do not rule out the possibility that Prp8 and Snu114 also interact with the Hel308-I module, although we cannot yet test this hypothesis since Hel308-I alone is unstable. It is worth noting that protein-tag labeling and antibody recognition approaches have mapped the C-terminus of Brr2 to be somewhat distant from the C-terminus of Prp8 in the EM projection structure of the yeast tri-snRNP, while the C-termini of Prp8 and Snu114 are in close proximity to each other 43. However, these labeling methods are designed to map the extreme C-terminus of a protein. The longest dimension of Hel308 is about 80Å and the Prp8-CTR can reach similar dimensions depending on the relative orientation of its C-terminal and β-finger domains 37,44–47. This dimension can easily span the distance between the C-termini of Brr2 and Prp8 observed in the EM structure 43. Therefore, the main body of Brr2 Hel308-II and Prp8-CTR can overlap and interact with each other even if the extreme C-termini of Brr2 and Prp8 are far from each other. Our results, in general, represent the first example of a helicase-like structural fold serving as a major protein-interaction platform.
The Hel308-II module can potentially play a role in mediating the regulation of Brr2 activity. Recently, it was shown that the C-terminal fragment of Prp8 (residues 1806–2413) greatly stimulates Brr2’s helicase activity but inhibits its ATPase in vitro 18. Here we showed that the deletion of S2 results in a dramatic reduction of Brr2’s ATPase and helicase activity (Fig. 2c–e). We also found that the deletion of Hel308-II drastically destabilizes the protein (Fig. 3d). Both results suggest that the S2 domain as well as the entire Hel308-II module interact and communicate with Hel308-I. It is foreseeable that the interaction between Prp8 and Hel308-II can potentially affect the structure and/or stability of Hel308-I and consequently the ATPase and helicase activity of Brr2. We do not rule out the possibility that Prp8 may directly interact with Hel308-I to modulate Brr2’s activity. Either directly (through Hel308-I) or indirectly (through Hel308-II), Prp8 can potentially affect Brr2’s ATPase and helicase activities through increasing RNA binding affinity, stabilizing a favorable conformational change, increasing processivity, or a combination of the above. In analogous situations, detailed kinetic analyses suggest that eIf4B, eIF4H, and eIF4G stimulate eIF4A’s ATPase and helicase activity through one or more of the above mechanisms 48. Likewise, Ntr1 was recently shown to stimulate the activity of Prp43 (another DExD/H-box protein involved in spliceosome disassembly) and was thought to affect Prp43’s processivity 49.
Our observation of the effect of Prp8-CTR on Brr2’s RNA binding property raises another interesting possibility for the regulation of Brr2’s activity. We showed that Prp8-CTR binds U4/U6 with a much higher affinity than arbitrary 13nt ss or ds RNAs (Fig. 4c, d). When Prp8-CTR and Brr2 (Hel308-II or full-length protein) are combined, the complex binds U4/U6 much better than Brr2 alone (Fig. 4b). We cannot differentiate at this point whether Prp8-CTR increases Brr2’s intrinsic affinity to U4/U6 or the increased binding is solely attributed to Prp8-CTR in the complex. However, in either case, the interaction between Prp8-CTR and Brr2 clearly helps the complex bind U4/U6 better. This increased affinity provides a possible mechanism for how Prp8-CTR helps Brr2 to confer specificity toward U4/U6. Further studies will reveal the extent of this specificity, such as how stringent the Prp8-CTR is toward the sequence and/or structure of U4/U6. It is worth noting that Ritchie et al. found that the N-terminal RNase H domain of the human Prp8-CTR has a much higher binding affinity with a U2/U6 mimic than other RNAs 46, consistent with the possibility that Prp8 may also help Brr2 confer specificity toward U2/U6 to facilitate Brr2’s role in U2/U6 unwinding. The higher local concentration of U4/U6 brought to Brr2 by Prp8-CTR could also serve as an additional mechanism for the stimulation effect by Prp8-CTR we observed in vitro. There are other enzymes (such as collagen prolyl 4-hydroxylase, HIV integrase, and endonuclease NaeI) that have separate substrate binding and catalytic domains, especially when the substrate is a polypeptide or oligonucleotide 50–52. Prp8 can potentially serve as an auxiliary substrate-binding and specificity domain for Brr2.
Our results also lead to reflections on the function of the Sec63 domain in general. Our structural result shows that the so-called Sec63 domain is in fact made of three domains, two helical and one all-beta Fn3 domains. The Sec63 domain has at least two functions. It can serve as a major RNA binding and processivity domain as in the case of Hel308-I. It can also serve as a major protein interaction domain as in the case of Hel308-II. The Fn3 fold in the Sec63 domain belongs to the immunoglobulin-like superfamily whose members are almost always involved in binding functions 53. Deletion of the Sec63 domain in the Sec63 protein leads to impaired higher-order complex formation 54. This Sec63 domain may also play a role in protein interaction, indicating the generality of the Sec63 domain serving as a protein interaction domain.
Methods
Yeast strain and plasmids
The brr2-3GS, bbr2-1, and R1107A mutations were generated on the pGPD-Brr2-TAP vector 18 using the QuikChange Site-Directed Mutagenesis Kit (Stratagene) and all mutations were confirmed by DNA sequencing of the entire Brr2 coding region. The mutated plasmid was shuffled into pTB105 and its growth phenotype was compared to WT brr2 on the same plasmid in pTB105. Brr2 deletion constructs on pPR150 11 were generated by amplifying the pPR150 plasmid without the deletion region, using PCR primers containing a common restriction site for subsequent digestion and ligation. A similar strategy was used to generate the S2-deleted brr2 on the pGPD-Brr2-TAP vector. The WT and truncated pPR150 plasmids were shuffled into yeast strain yJPS996 13 (gift of J. Staley, University of Chicago) for growth phenotype or pull down analyses.
Protein expression and purification
The Brr2 S2 domain was sub-cloned into the pGEX-6p1 vector (GE Healthcare) and expressed in E. coli strain XA90 as a GST-fusion protein. The fusion protein was first purified using glutathione Sepharose resin and cleaved using PreScission protease. The resultant S2 domain was further purified on a Superdex-200 (S200) gel-filtration column (GE Healthcare) and concentrated to 10 mg ml−1 for crystallization trials. Se-Met substituted S2 domain was expressed in minimal medium containing Se-Met. The protein was then purified using glutathione resin, a Resource Q column (GE Healthcare), followed by the S200 column. The purified protein was concentrated to 12 mg ml−1 for crystallization trials.
The Brr2 H2 and H2+S2 domain used for RNA binding experiments and the HA-tagged H2, S2, and H2+S2 domains used for GST-pull down experiments were also sub-cloned into the pGEX-6p1 vector, expressed and purified similarly as the S2 domain. The GST-fused Brr2, Prp8 and Snu114 proteins used in GST-pull down experiments were obtained similarly without the PreScission cleavage and gel-filtration purification.
WT and mutated Brr2 were expressed in yeast strains yTB105 containing the pGPD-BRR2-TAP or pGPD-BRR2-mutant-TAP plasmids and were purified as previously described 18 with the following exceptions. The frozen cell paste was homogenized with a SPEX SamplePrep 6870 Freezer Mill. The proteins were cleaved off the IgG resin using TEV and dialyzed into the storage buffer (20 mM Hepes pH7.9, 100 mM NaCl, 5 mM BME, 0.2 mM EDTA, 0.01% (v/v) NP-40, 20% (v/v) glycerol) for subsequent enzymatic and EMSA analyses.
Crystallization, data collection, and structure determination
The Brr2 S2 domain was crystallized by the hanging drop vapor diffusion method using a well solution containing 0.1 M Sodium Citrate (pH 6.0), 17% PEG8000, and 0.2 M NaCl. All crystallographic data were collected at 100K using the Molecular Biology Consortium beamline 4.2.2 of the Advanced Light Source at Lawrence Berkeley National Laboratory. Data were processed using the d*trek package 55 and data statistics are shown in Table 1.
Table 1.
Data collection, phasing and refinement statistics.
| Brr2 S2 1 | |||
|---|---|---|---|
| Data collection | |||
| Space group | P212121 | ||
| Cell dimensions | |||
| a, b, c (Å) | 55.15, 75.68, 83.96 | ||
| α,β,γ (°) | 90, 90, 90 | ||
| Peak | Inflection | Remote | |
| Wavelength (Å) | 0.979 | 0.9789 | 0.964 |
| Resolution (Å) | 40-2.0 (2.07-2.00) 2 | 40-2.0 (2.07-2.00) | 40-2.0 (2.07-2.00) |
| Rsym or Rmerge | 0.084 (0.414) | 0.076 (0.422) | 0.082 (0.494) |
| I / σI | 10.7 (3.9) | 12.0 (3.9) | 10.3 (2.9) |
| Completeness (%) | 100.0 (100.0) | 100.0 (100.0) | 100.0 (100.0) |
| Redundancy | 7.2 (7.3) | 7.2 (7.1) | 7.2 (7.1) |
| Refinement | |||
| Resolution (Å) | 30-2.0 | ||
| No. reflections | 45,120 | ||
| Rwork / Rfree | 0.246/0.257 | ||
| No. atoms | |||
| Protein | 2,482 | ||
| Ligand/ion | 0 | ||
| Water | 192 | ||
| B-factors | |||
| Protein | 36.28 | ||
| Ligand/ion | |||
| Water | 36.27 | ||
| R.m.s deviations | |||
| Bond lengths (Å) | 0.008 | ||
| Bond angles (°) | 1.674 | ||
One crystal was used for data collection at all three wavelengths.
Values in parentheses are for highest-resolution shell.
The structure of S2 was determined using the Se-Met MAD method and programs Solve and Resolve 56. Model building was carried out using O 57. Refinement was performed using CNS 58 and the peak data set. Refinement statistics are shown in Table 1. 90.2% and 9.8% of the residues fall into the most favored and additionally allowed regions and no residues are in the generously allowed and disallowed regions of the Ramachandran plot.
ATPase and helicase assays
ATPase and helicase assays were performed as previously described 18 except that 250 nM Brr2 proteins and 1 µM U4/U6 were used in ATPase assays. The helicase assay contains 25 nM Brr2, 250 nM Prp8-CTR, and 200 nM U4/U6.
Splicing microarray
To monitor pre-mRNA splicing defects on a global scale, whole genome splicing microarrays analyses were performed as described 59. Samples for both the brr2-ΔS2 strain and its matched wild type strain were collected at 30°C for splicing microarray analyses. Samples for the brr2-1 strain and its matched wild type were collected after shifting to 16°C for 10 minutes.
Quantitative RT-PCR
Quantitative RT-PCR was performed as described 59. The reaction was performed using a Roche LightCycler 480 and the following primers. Sample loading was normalized according to the composite behavior of the YMR286w and YAL010c genes.
| GENE | FORWARD Primer | REVERSE primer |
|---|---|---|
| U1 | TGACTACTTTTCTCTAGCGTGCC | CATAACGGGAACGAGCAAAGTTG |
| U2 | AACTGAAATGACCTCAATGAGGCTC | AGACCTGACATTAGCGGAAAACAAC |
| U4 | ATCCTTATGCACGGGAAATACG | AAAGGTATTCCAAAAATTCCCTAC |
| U5 | CAAGCAGCTTTACAGATCAATGG | AGTTCCAAAAAATATGGCAAGCC |
| U6 | GTTCGCGAAGTAACCCTTCG | AAAACGAAATAAATCTCTTTGTAAAAC |
| YMR286w | GTTGAGTAGGTCGCTTATCGGTGT | CTTTTACTTTAGCTAGGGAGCCAGC |
| YAL010c | ATAGCTACGAGGATATAACGGCCA | AATTGCTGTGCATCGGAGTATAAAT |
Protein and RNA interaction using Electrophoresis Mobility Shift Assay (EMSA)
Short 13nt RNA oligos (ssR13 and its complementary sequence) were ordered from Integrated DNA Technologies. ssR13 was 5’-end-labeled with 32P-γ-ATP using Optikinase (USB Corporation). dsR13 was generated by combining labeled ssR13 and its complementary sequence, boiling for three minutes, and cooling to room temperature in about three hours. U4 was in vitro transcribed from pT7U4 linearized with Sty1 using T7 RNA polymerase and in the presence of α-32P-UTP 60. U6 was transcribed from pT7U6(ΔU) (first two Us removed from U6 to increase transcription efficiency) linearized with Dra1 8. Both RNAs were purified by extraction from a 6% (w/v) Urea-TBE denaturing polyacrylamide gel, phenol:chloroform extraction, and ethanol precipitation. The U4/U6 duplex were generated by combining U4 and U6 RNAs in 40 mM Tris-HCl pH7.4 and 100 mM Sodium Acetate, boiling for three minutes, and cooling to room temperature in about three hours.
RNA was incubated on ice with protein (UAP56, Prp8-CTR, and various Brr2 proteins) in 20 µl of 10 mM Hepes, pH7.6, 1 mM MgCl2, 100 mM NaCl, 5% glycerol, and in the absence or presence of 1 mM ATP for 60 minutes. In supershift experiments, anti-Brr2 or anti-Prp8 antibody was added in the binding reaction after 30 minutes and was incubated for another 30 minutes on ice. The reaction mixture was separated on a 4% native polyacrylamide gel in TBE buffer (for short 13nt RNAs) or Hepes buffer (20 mM Hepes, pH7.9 for U4/U6), visualized and quantified using a phosphorimager.
GST-pull down experiments
GST-fused Snu114, Brr2 and Prp8 fragments were incubated with glutathione resin at 4 °C for 2 hours in the binding buffer (50 mM Tris-HCl, pH7.5, 120 mM NaCl, 0.2% NP40, 10% glycerol, 2 mM EDTA, 1 mM DTT, 1 mg ml−1 BSA) and washed with the wash buffer (50 mM Tris-HCl, pH8, 100 mM NaCl, 0.5% NP40, and 1 mM EDTA) for three times. Resin carrying equal quantity of fusion protein were incubated with HA-tagged Brr2 domains (H2, S2, or H2+S2) at 4 °C for 2 hours. The resin were washed four times with the low salt buffer (50 mM Tris-HCl, pH7.5, 120 mM NaCl, 0.2% NP40, 10% glycerol, 2 mM EDTA, 1 mM DTT) and three more times with the high salt buffer (same as the low salt buffer with the exception of containing 500 mM NaCl and 0.5% NP40). The resin was then analyzed using Western blot with an anti-HA antibody.
Co-Immunoprecipitation
Yeast strains were grown to OD600 of 1.0. 20 OD cells were harvested by centrifugation, washed with 1 ml of buffer A (10 mM HEPES, pH 7.9, 100 mM NaCl, 5 mM Benzamidine, 2 mM PMSF, 5 mM NEM, 2 mM EDTA), and resuspended in 200 µl of buffer B (0.1% Deoxycholic acid, 1 mM EDTA, 50 mM HEPES-KOH, pH7.5, 140 mM NaCl, 1% (w/v) Triton X-100, 1 mM PMSF, 1×protease inhibitors cocktail (Roche). Cells were lysed using the bead beater and separated using centrifugation. The supernatant containing 1.6 mg total proteins was incubated with 5ul anti-polyoma antibody (Covance) overnight at 4 °C. 30 µl protein G agarose (GE Healthcare) was added to the mixture and incubated at 4 °C for 2 hours. The resin was washed with buffer B four times. The resin was then run on a 6% SDS polyacrylamide gel and analyzed using Western blot with specific antibodies.
Supplementary Material
Acknowledgments
We are grateful to J. Staley (University of Chicago) and S. Stevens (University of Texas Austin) for providing yeast strains and plasmids; J. Beggs (University of Edinburgh) for anti-Brr2 antibody; and P. Fabrizio (Max Planck Institute for Biophysical Chemistry) for anti-Snu114 antibody. We thank Brandi Chong for help generating preliminary crystals. We thank the X-ray facility (supported in part by the University of Colorado Cancer Center) at the University of Colorado Denver. R. Z. is a Kimmel Scholar. This work was supported by a grant from the National Institutes of Health (NIH) to R.Z. (GM080334), an American Cancer Society Research Scholar grant to R.Z. (RSG-06-165-01-GMC), a NIH grant to C.G. (GM21119), an American Heart Association postdoctoral fellowship to L.Z. (0820036Z), and a National Institutes of General Medical Sciences postdoctoral fellowship to C.M. (F32GM077844).
Footnotes
Accession codes
The structure of yeast Brr2 S2 is deposited in the Protein Data Bank with accession code 3HIB. The splicing microarray data is deposited at NCBI’s Gene Expression Omnibus with accession code GSE16135.
References
- 1.Burge CB, Tuschl TH, Sharp PA. Splicing of precursors to mRNAs by the spliceosome. In: Gesteland RF, Cech T, Atkins JF, editors. The RNA World. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1999. pp. 525–560. [Google Scholar]
- 2.Brow DA. Allosteric cascade of spliceosome activation. Annu Rev Genet. 2002;36:333–360. doi: 10.1146/annurev.genet.36.043002.091635. [DOI] [PubMed] [Google Scholar]
- 3.Staley JP, Guthrie C. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell. 1998;92:315–326. doi: 10.1016/s0092-8674(00)80925-3. [DOI] [PubMed] [Google Scholar]
- 4.Jankowsky E, Fairman ME. RNA helicases--one fold for many functions. Curr Opin Struct Biol. 2007;17:316–324. doi: 10.1016/j.sbi.2007.05.007. [DOI] [PubMed] [Google Scholar]
- 5.Cordin O, Banroques J, Tanner NK, Linder P. The DEAD-box protein family of RNA helicases. Gene. 2006;367:17–37. doi: 10.1016/j.gene.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 6.Wagner JD, Jankowsky E, Company M, Pyle AM, Abelson JN. The DEAH-box protein PRP22 is an ATPase that mediates ATP-dependent mRNA release from the spliceosome and unwinds RNA duplexes. Embo J. 1998;17:2926–2937. doi: 10.1093/emboj/17.10.2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Laggerbauer B, Achsel T, Luhrmann R. The human U5-200kD DEXH-box protein unwinds U4/U6 RNA duplices in vitro. Proc Natl Acad Sci U S A. 1998;95:4188–4192. doi: 10.1073/pnas.95.8.4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Y, Wagner JD, Guthrie C. The DEAH-box splicing factor Prp16 unwinds RNA duplexes in vitro. Curr Biol. 1998;8:441–451. doi: 10.1016/s0960-9822(98)70178-2. [DOI] [PubMed] [Google Scholar]
- 9.Tanaka N, Schwer B. Mutations in PRP43 that uncouple RNA-dependent NTPase activity and pre-mRNA splicing function. Biochemistry. 2006;45:6510–6521. doi: 10.1021/bi052656g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shen J, Zhang L, Zhao R. Biochemical characterization of the ATPase and helicase activity of UAP56, an essential pre-mRNA splicing and mRNA export factor. J Biol Chem. 2007 doi: 10.1074/jbc.M702304200. [DOI] [PubMed] [Google Scholar]
- 11.Raghunathan PL, Guthrie C. RNA unwinding in U4/U6 snRNPs requires ATP hydrolysis and the DEIH-box splicing factor Brr2. Curr Biol. 1998;8:847–855. doi: 10.1016/s0960-9822(07)00345-4. [DOI] [PubMed] [Google Scholar]
- 12.Kim DH, Rossi JJ. The first ATPase domain of the yeast 246-kDa protein is required for in vivo unwinding of the U4/U6 duplex. Rna. 1999;5:959–971. doi: 10.1017/s135583829999012x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Small EC, Leggett SR, Winans AA, Staley JP. The EF-G-like GTPase Snu114p regulates spliceosome dynamics mediated by Brr2p, a DExD/H box ATPase. Mol Cell. 2006;23:389–399. doi: 10.1016/j.molcel.2006.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lauber J, et al. The HeLa 200 kDa U5 snRNP-specific protein and its homologue in Saccharomyces cerevisiae are members of the DEXH-box protein family of putative RNA helicases. Embo J. 1996;15:4001–4015. [PMC free article] [PubMed] [Google Scholar]
- 15.Grainger RJ, Beggs JD. Prp8 protein: at the heart of the spliceosome. Rna. 2005;11:533–557. doi: 10.1261/rna.2220705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brenner TJ, Guthrie C. Genetic analysis reveals a role for the C terminus of the Saccharomyces cerevisiae GTPase Snu114 during spliceosome activation. Genetics. 2005;170:1063–1080. doi: 10.1534/genetics.105.042044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bartels C, Klatt C, Luhrmann R, Fabrizio P. The ribosomal translocase homologue Snu114p is involved in unwinding U4/U6 RNA during activation of the spliceosome. EMBO Rep. 2002;3:875–880. doi: 10.1093/embo-reports/kvf172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maeder C, Kutach AK, Guthrie C. ATP-dependent unwinding of U4/U6 snRNAs by the Brr2 helicase requires the C terminus of Prp8. Nat Struct Mol Biol. 2009;16:42–48. doi: 10.1038/nsmb.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu D, Nouraini S, Field D, Tang SJ, Friesen JD. An RNA-dependent ATPase associated with U2/U6 snRNAs in pre-mRNA splicing. Nature. 1996;381:709–713. doi: 10.1038/381709a0. [DOI] [PubMed] [Google Scholar]
- 20.Noble SM, Guthrie C. Identification of novel genes required for yeast pre-mRNA splicing by means of cold-sensitive mutations. Genetics. 1996;143:67–80. doi: 10.1093/genetics/143.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ponting CP. Proteins of the endoplasmic-reticulum-associated degradation pathway: domain detection and function prediction. Biochem J. 2000;351(Pt 2):527–535. [PMC free article] [PubMed] [Google Scholar]
- 22.Holm L, Sander C. Dali: a network tool for protein structure comparison. Trends Biochem Sci. 1995;20:478–480. doi: 10.1016/s0968-0004(00)89105-7. [DOI] [PubMed] [Google Scholar]
- 23.Buttner K, Nehring S, Hopfner KP. Structural basis for DNA duplex separation by a superfamily-2 helicase. Nat Struct Mol Biol. 2007;14:647–652. doi: 10.1038/nsmb1246. [DOI] [PubMed] [Google Scholar]
- 24.Richards JD, et al. Structure of the DNA repair helicase hel308 reveals DNA binding and autoinhibitory domains. J Biol Chem. 2008;283:5118–5126. doi: 10.1074/jbc.M707548200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rost B. PHD: predicting one-dimensional protein structure by profile-based neural networks. Methods Enzymol. 1996;266:525–539. doi: 10.1016/s0076-6879(96)66033-9. [DOI] [PubMed] [Google Scholar]
- 26.Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Caruthers JM, Johnson ER, McKay DB. Crystal structure of yeast initiation factor 4A, a DEAD-box RNA helicase. Proc Natl Acad Sci U S A. 2000;97:13080–13085. doi: 10.1073/pnas.97.24.13080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao R, Shen J, Green MR, MacMorris M, Blumenthal T. Crystal structure of UAP56, a DExD/H-box protein involved in pre-mRNA splicing and mRNA export. Structure (Camb) 2004;12:1373–1381. doi: 10.1016/j.str.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 29.Shi H, Cordin O, Minder CM, Linder P, Xu RM. Crystal structure of the human ATP-dependent splicing and export factor UAP56. Proc Natl Acad Sci U S A. 2004;101:17628–17633. doi: 10.1073/pnas.0408172101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sengoku T, Nureki O, Nakamura A, Kobayashi S, Yokoyama S. Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa. Cell. 2006;125:287–300. doi: 10.1016/j.cell.2006.01.054. [DOI] [PubMed] [Google Scholar]
- 31.Trinh R, Gurbaxani B, Morrison SL, Seyfzadeh M. Optimization of codon pair use within the (GGGGS)3 linker sequence results in enhanced protein expression. Mol Immunol. 2004;40:717–722. doi: 10.1016/j.molimm.2003.08.006. [DOI] [PubMed] [Google Scholar]
- 32.Boon KL, et al. prp8 mutations that cause human retinitis pigmentosa lead to a U5 snRNP maturation defect in yeast. Nat Struct Mol Biol. 2007;14:1077–1083. doi: 10.1038/nsmb1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pleiss JA, Whitworth GB, Bergkessel M, Guthrie C. Transcript specificity in yeast pre-mRNA splicing revealed by mutations in core spliceosomal components. PLoS Biol. 2007;5:e90. doi: 10.1371/journal.pbio.0050090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rogers GW, Jr, Komar AA, Merrick WC. eIF4A: the godfather of the DEAD box helicases. Prog Nucleic Acid Res Mol Biol. 2002;72:307–331. doi: 10.1016/s0079-6603(02)72073-4. [DOI] [PubMed] [Google Scholar]
- 35.van Nues RW, Beggs JD. Functional contacts with a range of splicing proteins suggest a central role for Brr2p in the dynamic control of the order of events in spliceosomes of Saccharomyces cerevisiae. Genetics. 2001;157:1451–1467. doi: 10.1093/genetics/157.4.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu S, Rauhut R, Vornlocher HP, Luhrmann R. The network of protein-protein interactions within the human U4/U6.U5 tri-snRNP. Rna. 2006;12:1418–1430. doi: 10.1261/rna.55406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pena V, Liu S, Bujnicki JM, Luhrmann R, Wahl MC. Structure of a multipartite protein-protein interaction domain in splicing factor prp8 and its link to retinitis pigmentosa. Mol Cell. 2007;25:615–624. doi: 10.1016/j.molcel.2007.01.023. [DOI] [PubMed] [Google Scholar]
- 38.Brenner TJ, Guthrie C. Assembly of Snu114 into U5 snRNP requires Prp8 and a functional GTPase domain. Rna. 2006;12:862–871. doi: 10.1261/rna.2319806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bartels C, Urlaub H, Luhrmann R, Fabrizio P. Mutagenesis suggests several roles of Snu114p in pre-mRNA splicing. J Biol Chem. 2003;278:28324–28334. doi: 10.1074/jbc.M303043200. [DOI] [PubMed] [Google Scholar]
- 40.Oyama T, et al. Atomic structures and functional implications of the archaeal RecQ-like helicase Hjm. BMC Struct Biol. 2009;9:2. doi: 10.1186/1472-6807-9-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brow DA, Guthrie C. Spliceosomal RNA U6 is remarkably conserved from yeast to mammals. Nature. 1988;334:213–218. doi: 10.1038/334213a0. [DOI] [PubMed] [Google Scholar]
- 42.Caruthers JM, McKay DB. Helicase structure and mechanism. Curr Opin Struct Biol. 2002;12:123–133. doi: 10.1016/s0959-440x(02)00298-1. [DOI] [PubMed] [Google Scholar]
- 43.Hacker I, et al. Localization of Prp8, Brr2, Snu114 and U4/U6 proteins in the yeast tri-snRNP by electron microscopy. Nat Struct Mol Biol. 2008;15:1206–1212. doi: 10.1038/nsmb.1506. [DOI] [PubMed] [Google Scholar]
- 44.Zhang L, et al. Crystal structure of the C-terminal domain of splicing factor Prp8 carrying retinitis pigmentosa mutants. Protein Sci. 2007;16:1024–1031. doi: 10.1110/ps.072872007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang K, Zhang L, Xu T, Heroux A, Zhao R. Crystal structure of the beta-finger domain of Prp8 reveals analogy to ribosomal proteins. Proc Natl Acad Sci U S A. 2008;105:13817–13822. doi: 10.1073/pnas.0805960105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ritchie DB, et al. Structural elucidation of a PRP8 core domain from the heart of the spliceosome. Nat Struct Mol Biol. 2008;15:1199–1205. doi: 10.1038/nsmb.1505. [DOI] [PubMed] [Google Scholar]
- 47.Pena V, Rozov A, Fabrizio P, Luhrmann R, Wahl MC. Structure and function of an RNase H domain at the heart of the spliceosome. Embo J. 2008;27:2929–2940. doi: 10.1038/emboj.2008.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rogers GW, Jr, Richter NJ, Lima WF, Merrick WC. Modulation of the helicase activity of eIF4A by eIF4B, eIF4H, and eIF4F. J Biol Chem. 2001;276:30914–30922. doi: 10.1074/jbc.M100157200. [DOI] [PubMed] [Google Scholar]
- 49.Tanaka N, Aronova A, Schwer B. Ntr1 activates the Prp43 helicase to trigger release of lariat-intron from the spliceosome. Genes Dev. 2007;21:2312–2325. doi: 10.1101/gad.1580507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pekkala M, et al. The peptide-substrate-binding domain of collagen prolyl 4-hydroxylases is a tetratricopeptide repeat domain with functional aromatic residues. J Biol Chem. 2004;279:52255–52261. doi: 10.1074/jbc.M410007200. [DOI] [PubMed] [Google Scholar]
- 51.Chiu TK, Davies DR. Structure and function of HIV-1 integrase. Curr Top Med Chem. 2004;4:965–977. doi: 10.2174/1568026043388547. [DOI] [PubMed] [Google Scholar]
- 52.Colandene JD, Topal MD. The domain organization of NaeI endonuclease: separation of binding and catalysis. Proc Natl Acad Sci U S A. 1998;95:3531–3536. doi: 10.1073/pnas.95.7.3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stutz F, et al. REF, an evolutionary conserved family of hnRNP-like proteins, interacts with TAP/Mex67p and participates in mRNA nuclear export. Rna. 2000;6:638–650. doi: 10.1017/s1355838200000078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jermy AJ, Willer M, Davis E, Wilkinson BM, Stirling CJ. The Brl domain in Sec63p is required for assembly of functional endoplasmic reticulum translocons. J Biol Chem. 2006;281:7899–7906. doi: 10.1074/jbc.M511402200. [DOI] [PubMed] [Google Scholar]
- 55.Pflugrath JW. The finer things in X-ray diffraction data collection. Acta Crystallogr D Biol Crystallogr. 1999;55:1718–1725. doi: 10.1107/s090744499900935x. [DOI] [PubMed] [Google Scholar]
- 56.Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr D Biol Crystallogr. 1999;55(Pt 4):849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jones TA, Zou JY, Cowan SW, Kjeldgaard Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 58.Brunger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54(Pt 5):905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 59.Inada M, Pleiss JA. Genome-wide approaches to monitor pre-mRNA splicing. Methods in Enzymology. doi: 10.1016/S0076-6879(10)70003-3. (In press) [DOI] [PubMed] [Google Scholar]
- 60.Salghetti SE, Kim SY, Tansey WP. Destruction of Myc by ubiquitin-mediated proteolysis: cancer-associated and transforming mutations stabilize Myc. Embo J. 1999;18:717–726. doi: 10.1093/emboj/18.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.