Abstract
Human cytomegalovirus (HCMV), which infects the majority of the population worldwide, causes few, if any, symptoms in otherwise healthy people but is responsible for considerable morbidity and mortality in immuno-compromised patients and in congenitally infected newborns. The evolutionary success of HCMV depends in part on its ability to evade host defense systems. Here we review recent progress in elucidating the remarkable assortment of mechanisms employed by HCMV and the related β-herpesviruses, murine cytomegaloviruses (MCMV) and rhesus cytomegaloviruses (RhCMV), for counteracting the host interferon (IFN) response. Very early after infection, cellular membrane sensors such as the lymphotoxin β receptor initiate the production of antiviral cytokines including type I IFNs. However, virion factors, such as pp65 (ppUL83) and viral proteins made soon after infection including the immediate early gene 2 protein (pUL122), repress this response by interfering with steps in the activation of IFN regulatory factor 3 and NF-κB. CMVs then exert a multi-pronged attack on downstream IFN signaling. HCMV infection results in decreased accumulation and phosphorylation of the IFN signaling kinases Jak1 and Stat2, and the MCMV protein pM27 mediates Stat2 down-regulation, blocking both type I and type II IFN signaling. The HCMV immediate early gene 1 protein (pUL123) interacts with Stat2 and inhibits transcriptional activation of IFN-regulated genes. Infection also causes reduction in the abundance of p48/IRF9, a component of the ISGF3 transcription factor complex. Furthermore, CMVs have multiple genes involved in blocking the function of IFN-induced effectors. For example, viral double-stranded RNA-binding proteins are required to prevent the shutoff of protein synthesis by protein kinase R, further demonstrating the vital importance of evading the IFN response at multiple levels during infection.
Introduction
Human cytomegalovirus (HCMV), the prototype member of the β-herpesvirus subfamily, infects the majority of humans in most populations throughout the world (Mocarski and others 2007). In otherwise healthy adults, HCMV infection causes few if any symptoms, but it is often much more severe in patients with immune deficiency states and in congenitally infected newborns. Considerable recent interest has focused on a possible role of HCMV in a variety of other diseases, including atherosclerosis and glioblastomas (Streblow and others 2008; Miller 2009).
In addition to direct effects of the virus on cell growth and metabolism, indirect effects of HCMV on the host immune system may have a significant impact on overall disease outcome. For example, in hematopoietic stem cell transplant recipients, subclinical HCMV predisposes to an increased risk of bacterial and fungal infections (Nichols and others 2002). In HCMV seropositive patients, a remarkably high percentage of circulating T cells are specific for HCMV antigens and this number tends to increase with age, leading to the speculation that immunosenescence associated with aging might be connected to HCMV (Pawelec and others 2009). On the other hand, prior infection with murine cytomegalovirus (MCMV) can be protective against otherwise lethal infection with bacterial pathogens (Barton and others 2007). Much more work will be necessary to sort out the various viral and host factors that determine the outcome of CMV infections, but it is increasingly clear that the interactions between the host immune system, including the interferon (IFN) response, and viral genes that thwart these responses are key factors in the evolution of the virus and in disease pathogenesis.
HCMV is generally considered to be relatively resistant to IFN. In cell culture, however, IFN pretreatment of cells can inhibit viral production by at least 100- to 1,000-fold (Taylor and Bresnahan 2005). Mice lacking the IFN receptors suffer greatly increased mortality following MCMV infection (Presti and others 1998). On the other hand, IFN has not proven to be useful for treating HCMV infections. The reasons for its relatively poor clinical efficacy are complex, but include the ability of robust and interwoven viral mechanisms to counter the host IFN response.
CMV Replication
Cytomegaloviruses (CMVs) have large double-stranded DNA genomes with a coding capacity for ∼170 genes (Mocarski and others 2007). A core set of ∼40 genes have close homologs found in other herpesviruses and an additional ∼40 are shared among β-herpesviruses only (Brocchieri and others 2005; Murphy and Shenk 2008). The remaining genes are specific to individual or closely related members within the β subfamily. Studies over the past 2 decades have revealed that many CMV genes have roles in evading host defenses.
The HCMV virion is extremely complex, containing numerous viral and cellular proteins and RNAs in addition to the viral genome (Prichard and others 1998; Terhune and others 2004; Varnum and others 2004). As with other animal viruses, laboratory preparations of HCMV contain only a minority of plaque-forming units compared to the vast excess of particles that may bind to and enter cells and even express some genes yet are defective for productive replication. HCMV also produces distinctive particles known as dense bodies, which contain abundant viral tegument proteins within an envelope but without genomic DNA. Whether a similar abundance of noninfectious particles are produced during natural infections is not clear, but they certainly may complicate the interpretation of laboratory experiments, especially those involving the early events such as activation and repression of innate cellular defenses.
The mechanism of CMV entry into cells is a complex and still incompletely understood process (Isaacson and others 2008). As with several other viruses, an initial low-affinity interaction of virions with cell surface heparan sulfate proteoglycans is followed by a tighter association of viral envelope glycoproteins with their cellular receptors. Depending on the cell type, the virus then enters the cell by fusion at the plasma membrane or by an endosomal pathway. Identification of the specific cellular receptors for HCMV has been the subject of numerous studies and considerable controversy. The fact that HCMV can enter many cell types suggests that either the receptor is widespread or that there are multiple alternative receptors. Recent studies have suggested that integrins (α2β1, α6β1, and αVβ3), platelet-derived growth factor a receptor, and possibly the epidermal growth factor receptor can function as receptors for HCMV (Bentz and Yurochko 2008; Isaacson and others 2008; Soroceanu and others 2008), but which of these are important in which cell types in vivo is not yet certain. Nonetheless, it is quite clear that while the virus has evolved the ability to exploit cellular factors to enable its entry into the cell, the host cell has also evolved mechanisms for detecting the invading virus and initiating antiviral responses as described later.
Following infection, uncoating, and nuclear entry of the genome, the first wave of gene expression includes the major immediate early (IE) genes I and II (UL123 encoding IE72 and UL122 encoding IE86, respectively), which are complex regulatory proteins necessary for expression of later classes of genes (Mocarski and others 2007). More recent studies have revealed that these, as well as other immediate early genes IRS1 and TRS1, also contribute to the viral counterattack on the host cell antiviral defenses.
As for other herpesviruses, HCMV establishes a latent infection after the primary infection is brought under control. Hematopoietic mononuclear cells have received considerable attention as 1 site of latency but whether other sites exist and exactly how the latent state is maintained and how reactivation occurs is not known (Reeves and Sinclair 2008). The host immune system clearly contributes to the maintenance of latency since reactivation is most apparent in immuno-suppressed hosts.
Induction of IFN Following CMV Infection
Even as CMV is invading the host cell to start its replication cycle, cellular sensors are poised to detect the virus and initiate responses that help protect the cell and organism. These innate responses, which include the induction and secretion of IFNs as well as other cytokines, usually succeed in limiting the extent of viral replication and disease. However, the virus is also successful in that it usually establishes a lifelong latent or persistent infection with potential to reactivate and spread to new hosts at a later time. Studies of infections in cell culture and in genetically engineered mouse strains have revealed that the IFN response to CMVs is surprisingly complex, occurring in several temporal phases and involving multiple distinct mechanisms.
The cell detects HCMV infection very early and, by 4–8 h post-infection (hpi), responds by producing cytokines and an initial peak of IFN. Even in the absence of de novo viral gene expression, the virion-associated factors that are recognized by the cells as foreign pathogen-associated molecular patterns (PAMPs) activate IRF3, NF-κB, and an overall transcriptional profile similar to that observed following IFN treatment (Zhu and others 1997; Boyle and others 1999; Yurochko and Huang 1999; Browne and others 2001; Preston and others 2001; Abate and others 2004; Netterwald and others 2004). Viral gene expression blunts the induction of the IFN transcriptional program (Browne and others 2001), consistent with there being specific viral mechanisms for counteracting the IFN response. Toll-like receptor (TLR) 2 and CD14 play a role at this early stage in recognizing HCMV infection, likely through interactions with envelope proteins gB and gH, and lead to NF-κB activation and secretion of inflammatory cytokines but, interestingly, not IFN (Compton and others 2003; Boehme and others 2006; Juckem and others 2008).
Unlike other cytokines, IFN induction early after HCMV infection does not seem to rely on TLR signaling. HCMV infection induces similar levels of IFN-β even when signaling through TLR2, TLR3, TRL7, TLR8, or TLR9 is inhibited (Juckem and others 2008). MCMV infection of mice having deletions of TLR9 or MyD88, or TRIFLPS2/LPS2 mice (which cannot signal through TLR3) all activate IFN-β transcription early after infection to levels similar to those of wild-type mice (Schneider and others 2008). Conversely, inhibition of HMCV entry by disruption of lipid rafts or using entry inhibitors that act after binding but before fusion blocks IFN induction but does not affect production of several other cytokines. Thus, the induction of IFN is a distinct process from that of other cytokines.
Studies of both HCMV and MCMV have highlighted a major role for lymphotoxin β receptor (LTβR) signaling in mediating the early phase (4–8 hpi) induction of IFN. Addition of exogenous ligands that signal through LTβR induces IFN-β expression in HCMV-infected fibroblasts by a mechanism requiring the LTβR and activation of NF-κB (Benedict and others 2001). The resulting increase in IFN levels, primarily IFN-β, causes a marked but reversible repression of HCMV replication. In the MCMV system, deficiencies in LTβR signaling resulting from germ line mutations in LTβR, its ligands (lymphotoxin α or β), or NF-κB-inducing kinase (which signals downstream of the LTβR) or from expression of a soluble decoy LTβR, lead to inhibition of IFN induction, increased viral replication in the spleen, liver, and salivary glands, apoptotic death of T and B lymphocytes, and increased mortality (Benedict and others 2001; Banks and others 2005). Treatment of LTα-deficient mice with an LTβR agonist reverses the blunted IFN response, lymphocyte apoptosis, and increased mortality. Together, these studies demonstrate that the early induction of IFN mediated by signaling through the LTβR is necessary to limit viral replication and the severity of the disease.
Additional insights into the cell types involved in the protective effects of LTβR signaling have emerged from studies utilizing marrow transplantation and conditional knockout mice. The 4–8 hpi peak induction of IFN-α and IFN-β correlates with IFN transcription in the spleen and depends on LTβR expression in splenic stromal cells (Schneider and others 2008). Transplantation studies demonstrate that protection against the normal lymphocyte apoptotic response to MCMV infection requires that the ligand is present on hematopoietic cells (Banks and others 2005). Mice lacking B cells or having LTβ deleted specifically in B cells fail to mount the normal very early IFN response. Although the viral factors that mediate this early IFN response are not yet known, the observation that the magnitude of the IFN response is proportional to viral inoculum is consistent with the triggering ligand(s) being an as-yet-unidentified virion factor. Thus, these data suggest an intriguing model in which the early protective IFN response is mediated by naive B cells signaling through the LTβR on infected splenic stromal cells.
At least in mice, a second peak of IFN production begins at ∼36 hpi, and this phase depends on plasmacytoid dendritic cells (pDCs) and TLR signaling (Delale and others 2005; Schneider and others 2008). Infection of mice with mutations that eliminate MyD88 or TLR9 function results in markedly reduced levels of serum or pDC-produced type I IFN and of serum IFN-γ and IFN-γ-positive splenic NK and NK-T cells (Tabeta and others 2004; Zucchini and others 2008). Titers of MCMV in the spleens of these mice are 10,000-fold higher than in wild-type mice and the mortality rate is also significantly higher. Serum IFN-α levels are normal at 36 hpi in TLR9-null mice but these mice have decreased levels of IFN-γ (and other cytokines) and an increase in mortality that is similar to MyD88-null mice. TLR7-deficient mice also display a normal IFN-α response following MCMV infection, but mice lacking both TLR7 and TLR9 have a markedly reduced level of IFN-α production, similar to MyD88-null animals (Zucchini and others 2008). In contrast, TLR2, 3, and 4 null mice have normal serum IFN-α levels and survival (Edelmann and others 2004; Delale and others 2005). However, the role of TLR3 is controversial since 1 group reported that mice with deficiency of TLR3 or its downstream effector TRIF had lower serum concentrations of IFN-α/β and IFN-γ at 36 hpi, higher replication of MCMV in the spleen at 4–5 dpi, and increased mortality (Hoebe and others 2003; Tabeta and others 2004). Recent studies have shown that double-stranded RNA (dsRNA), the activator of TLR3 as well as other antiviral pathways such as RIG-I and MDA-5 that result in IFN production, is indeed produced during MCMV and HCMV infection, suggesting that TLR3 or cytoplasmic RNA helicase sensors might play a role in responding to CMV infection (Budt and others 2009; Marshall and others 2009). Together, these studies suggest that TLR signaling through either TLR7 or TLR9 is responsible for much of the IFN response at the 36 hpi with MCMV. Since pDC are not actively infected, triggering may occur by ingestion of infected cell debris including viral ssRNA and unmethylated CpG DNA, which are known triggers for TLR7 and TLR9, respectively.
Finally, there seems to be a distinct later phase of IFN production, beginning at ∼44 hpi in infected mice, that occurs even in MyD88-null mice and thus is not mediated by any of the MyD88-dependent TLRs, including TLR7 and TLR9 (Delale and others 2005). At this time, cells other than pDCs produce the IFN. Thus, the IFN response at 2–3 days post-MCMV infection depends first on TLR signaling in pDCs and later on IFN production in other cells by a TLR-independent mechanism.
Evasion of Viral Entry-Mediated IFN and IFN-Stimulated Gene (ISG) Induction
Experimental manipulations of the host that suppress or eliminate the IFN response enhance viral replication and disease severity. Similarly, evolution has provided the virus with natural mechanisms for blunting the IFN response at multiple steps (Figs. 1 and 2).
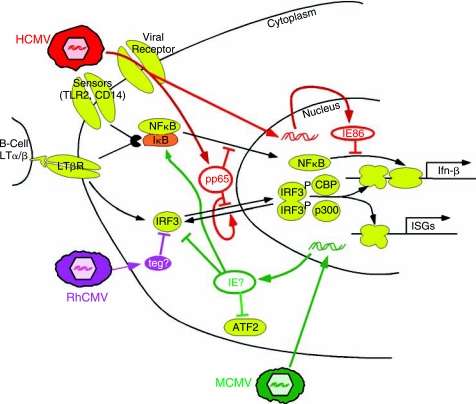
FIG. 1.
Early induction and evasion of interferons (IFNs) and IFN-stimulated genes (ISGs). As detailed in the text, initial infection activates several cellular signaling pathways leading to induction of IFN and ISGs (yellow factors, black arrows). However, human cytomegalovirus (HCMV) (red), rhesus cytomegaloviruses (RhCMV) (purple), and murine cytomegaloviruses (MCMV) (green) contain virion factors and encode immediate early or early proteins that interfere with these host defenses.
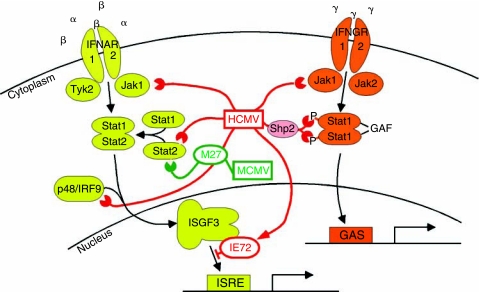
FIG. 2.
Cytomegalovirus (CMV) blockade of interferon (IFN) signaling. Binding of IFN type I (α, β) and type II (γ) receptors to their respective ligands triggers a cascade leading to activation of IFN-stimulated gene (ISG) transcription. Human cytomegalovirus (HCMV) (red) and murine cytomegalovirus (MCMV) (green) counteract these pathways at multiples steps as detailed in the text.
Among the viral mechanisms for limiting the impact of the IFN response is the blockade of IRF3 function. IRF3 is a common downstream target for signaling pathways that lead to transcriptional activation of several ISGs and IFN-β. IRF3 normally shuttles between the nucleus and cytoplasm but is primarily cytoplasmic when inactive. Signaling transduction through cellular sensors can result in IRF3 hyperphosphorylation by virally activated cellular kinases such as IKK-ɛ and TBK1 (Gravel and Servant 2005) and accumulation in the nucleus, where in combination with other transcription factors such as CREB-binding protein (CBP) and p300, it binds to and activates target promoters. IRF3 is a key mediator of ISG induction following HCMV infection since knockdown of IRF3 by siRNA or expression of a dominant negative IRF3 mutant counteracts the transcription induction of many ISGs (DeFilippis and others 2006).
CMVs, like other herpesviruses, appear to encode factors that block IRF3 function. Abate et al. reported that deletion of the gene encoding the tegument protein pp65 (ppUL83) resulted in a greater induction of an IFN-like transcriptional profile after HCMV infection (Abate and others 2004). IRF3 became hyperphosphorylated and translocated to the nucleus of HF or PBMC in the first few hours after infection with the pp65-mutant but not wild-type virus. Moreover, transduction of pp65 alone into HF blocked nuclear translocation of IRF3 following an activating stress. NF-κB relocalized to the nucleus after infection by either wild-type or pp65-deleted virus. Thus, these authors concluded that pp65 was a key for blocking IRF3 activation, either by preventing cytoplasmic phosphorylation and nuclear import or by promoting nuclear dephosphorylation and export.
Using the same pp65-deleted mutant virus, Browne and Shenk also found that transcription of IFN-β and several other ISGs was induced to a greater extent by the mutant compared to wild-type HCMV (Browne and Shenk 2003). In support of a role of pp65 in blocking the early ISG response, they found that adenoviral vector-mediated transduction of pp65 inhibited induction of MxA by IFN-α. However, in surprising contrast to the results of Abate et al., these authors, like several others (DeFilippis and Fruh 2005; Taylor and Bresnahan 2006b), found that IRF3 relocalized to the nucleus after infection with wild-type or pp65-mutant virus. Instead, the loss of pp65 resulted in induction of IRF1 expression and nuclear accumulation of both IRF1 and NF-κB.
Thus, 2 very similarly designed studies agree in concluding that pp65 delivered to the cell with the viral tegument functions to block activation of IFN pathway genes very early after entry (Browne and Shenk 2003; Abate and others 2004). However, whether the effect of pp65 is to block IRF3, IRF1, and/or NF-κB is not clear.
Further complicating matters, a subsequent study suggested that pp65 does not contribute to the evasion of the IFN response (Taylor and Bresnahan 2006b). A pp65 nonsense mutant virus, unlike the pp65 deletion virus, retained the ability to block the IFN-β induction, consistent with data showing that the impact of the pp65 deletion might be mediated by reduced expression of the neighboring pp71 gene, which in turn affects IE86 expression and IFN evasion (see below). Also, in contrast to the earlier studies, pp65 expression from an adenovirus vector did not block induction of IFN by UV-inactivated HCMV in this study.
The discrepancies among these studies could be due to any of several factors. One variable that is seldom considered in experimental work with HCMV is variation among viral stocks. In particular, the quantity of pp65 in virions and the abundance of pp65-rich dense bodies in viral preparations may vary widely (Jahn and others 1987). Thus, differences in the dose of pp65 or other virion factors delivered to the cells in different studies might underlie some of the conflicting results.
Rhesus CMV (RhCMV) also represses the very early ISG response but by a mechanism that differs from that used by HCMV. Infection of rhesus fibroblasts with live or UV-inactivated RhCMV did not stimulate ISG expression (DeFilippis and Fruh 2005). IRF3 remained an inactive cytoplasmic monomer after RhCMV infection with live or UV-inactivated RhCMV or after RhCMV infection in the presence of cycloheximide. In contrast, HCMV infection of these cells caused ISG induction and IRF3 dimerization and nuclear accumulation. Neither virus caused activation of NF-κB. Interestingly, prior RhCMV infection blocked nuclear accumulation of IRF3 induced by subsequent HCMV infection but not by polyI:C transfection. These results suggest that an as-yet-unidentified RhCMV virion factor blocks IRF3 activation at an early step in the IRF3 signaling pathway that is specific to a subset of activators, including an unidentified viral PAMP.
Role of de novo Viral Gene Expression in Blocking the Early IFN Response
Many groups have noted that the IFN-like response to HCMV infection is potentiated by infection with UV-inactivated virus or infection in the presence of cycloheximide (Browne and others 2001; Preston and others 2001; Simmen and others 2001; Browne and Shenk 2003; Abate and others 2004; Taylor and Bresnahan 2005; Taylor and Bresnahan 2006b). These observations suggest pp65 delivered by incoming HCMV virions by itself is insufficient to block the IFN response; rather de novo gene expression is required. The finding that IFN-β production is blocked very early after infection led Taylor and Bresnahan to focus on IE genes as possible mediators of the blockade (Taylor and Bresnahan 2005; Taylor and Bresnahan 2006b). Using both adenoviral vector-mediated expression as well as a mutant virus in which IE86 expression is diminished and delayed, they found that IE86 expression was necessary and sufficient to block IFN-β production induced by infection with UV-inactivated HCMV, by HCMV infection in the presence of cycloheximide, and even by infection with Sendai virus, another inducer of IFN-β.
Further studies suggested that IE86 acts by blocking the NF-κB pathway. IFN-β gene induction by HCMV requires activation of both IRF3 and NF-κB pathways (DeFilippis and others 2006; Taylor and Bresnahan 2006a). As in several other reports, Taylor and Bresnahan found that infection with live or UV-inactivated HCMV caused IRF3 phosphorylation, dimerization, nuclear relocalization, and transcriptional activation of a target gene, ISG15 (Taylor and Bresnahan 2006a). IE86 expression from an adenoviral vector did not block any of these indicators of IRF3 activation. However, like infection with live HCMV, IE86 expression did block the induction of NF-κB DNA binding and NF-κB-induced gene expression following infection with UV-inactivated virus or TNF-α treatment. IE86 could also block NF-κB binding to DNA stimulated by Sendai virus infection. Moreover, an IE86-deficient HCMV mutant was unable to block the induction of NF-κB DNA-binding activity. The block occurs quite late in the NF-κB activation pathway since IE86 did not prevent phosphorylation and degradation of IκB in response to TNF-α, nor did it prevent nuclear relocalization of NF-κB in response to infection with live or UV-inactivated HCMV. These experiments reveal that one of the multiple functions of IE86 is the blockade of NF-κB DNA binding in the nucleus by an unknown mechanism.
Like HCMV, MCMV relies on de novo viral gene expression to repress the IFN response within the first few hpi. Le et al. found that MCMV infection induced IFN-α and IFN-β expression by ∼2 hpi but the abundance of the transcripts declined starting by ∼6 hpi (Le and others 2008b). Prior infection with live but not UV-inactivated MCMV for 2–4 h was able to block the IFN-β induction that otherwise results from subsequent re-infection with MCMV or with Sendai virus, implicating an immediate early or early viral gene as an active repressor of the IFN induction pathway. IRF3 dimerization and nuclear localization occurred with the same kinetics as the transient peak of IFN expression, consistent with IRF3 contributing to the initial induction of IFN but then being inhibited by a viral antagonist. As well, IκB levels diminished at 2 hpi and were restored by 6 hpi, likely due to the prolongation of the IκBα half-life from 50′ to 6 h by ∼4 h after MCMV infection. ATF-2, another set of IFN-β gene transcriptional activating factors, followed a similar pattern of initial phosphorylation (activation) followed by dephosphorylation. At the later times, ATF-2 was unresponsive to another activating stimulus, suggesting that MCMV infection actively blocks the ATF-2 signaling pathway as well. Differences in the kinetics of the interference with IRF3, NF-κB, and ATF-2 activation pathways led the authors to conclude that more than 1 viral mechanism might be involved in blocking the induction of IFN-β. Deletion of M83 or M84, which are homologs of HCMV UL83 (pp65), or of M27 (see below), did not affect the pattern of IFN-β induction, indicating that these genes are not essential for the repression. The M45 gene, an inhibitor of apoptosis in MCMV-infected endothelial cells, blocks NF-κB activation as a result of its inhibitory effect on receptor-interacting protein kinase 1 (RIP1) signaling, but whether M45 impacts IFN or ISG production has not been reported (Mack and others 2008; Upton and others 2008). Thus, the MCMV factor(s) responsible for inhibiting the IFN pathway in MCMV is not yet known. Based on the studies of IE86 in the HCMV system, it would be worthwhile evaluating whether the MCMV immediate early genes can block the mouse IFN response to infection.
In summary, cells are poised to detect 1 or more CMV PAMPs and produce IFN and other cytokines and ISGs early after infection. However, a combination of virion-associated factors, such as pp65, and viral genes expressed very early after infection, such as IE86, blunt these responses. Seeming inconsistencies among reports that suggest the viral mechanisms involve blocking IRF3, NF-κB, or both might be due to any of several experimental variables, including the specific cells, viruses, and reagents used by different groups. In addition to variation in the viral stocks noted earlier, genetic polymorphisms among the primary cells used for most work with HCMV could affect the innate immune responses detected in different labs. The serum used for cell cultivation undoubtedly varies in levels of cytokines and other factors that could influence these kinds of experiments. For example, interleukin-1 can potentiate the production of IFN-β from infected cells (Randolph-Habecker and others 2002). The co-evolution of CMVs with their host species undoubtedly has lead to considerable variation in the specific strategies that have evolved in HCMV vs. RhCMV vs. MCMV. Regardless, these studies reveal that CMVs have evolved at least several mechanisms to blunt the early IFN-like response to infection. One or more of these mechanisms may predominate, depending on the particular setting, in order to enable the virus to establish a productive or latent infection.
Evasion of IFN Signaling
As illustrated in Figure 2, IFN-α/β signal through a heterodimeric receptor (IFNAR) consisting of 2 subunits, IFNAR1 and IFNAR2. Upon ligand binding, receptor-associated Tyk2 and Jak1 are activated and phosphorylate Stat2. Phosphorylated Stat2 recruits Stat1, and the phosphorylated Stat1/2 complex combines with p48 (also known as IRF9) to form the heterotrimeric IFN-stimulated gene factor 3 (ISGF3) complex, which translocates to the nucleus. The ISGF3 complex binds to IFN-stimulated response element (ISRE) sequences, activating transcription of IFN-α/β-stimulated genes. IFN-α/β signaling can also result in Stat1 homodimers that activate transcription of ISGF3-independent genes, including IRF1 (Li and others 1996).
IFN-γ also signals through a heterodimeric receptor, consisting of IFNGR1 and IFNGR2 associated with Jak1 and Jak2, respectively. Receptor triggering results in the phosphorylation of Stat1 homodimers, which translocate to the nucleus as the IFN-γ activation factor (GAF) and activate transcription from genes containing γ-activated sequence (GAS) elements. Although the IFN-α/β and IFN-γ systems share some components, including Jak1 and Stat1, they perform different functions in the immune response and induce different sets of IFN-response genes, and thus are not redundant.
CMVs interfere with these IFN-signaling pathways at several points, starting with the degradation of Jak1 (Miller and others 1998; Miller and others 1999). HCMV-infected cells have decreased levels of Jak1 compared to uninfected cells, and this decrease in Jak1 correlates with inhibition of IFN-α-stimulated signal transduction. Levels of phosphorylated IFNAR1, Stat2, Stat1α, and Tyk2 are all impaired in HCMV-Towne-infected fibroblasts, although Jak1 is the only protein whose overall levels are decreased in this system (Miller and others 1999). Jak1 mRNA levels remain constant during HCMV infection, and experiments using the inhibitor Z-L3VS demonstrated that Jak1 degradation is mediated by the proteosome (Miller and others 1998). Although it remains unknown which HCMV gene or genes is responsible for the degradation of Jak1, DNA polymerase inhibitors do not interfere, implying that immediate early or early, but not late, gene expression is required (Miller and others 1998).
The loss of Jak1-mediated signal transduction leads to inhibition of both ISGF3-dependent and -independent gene expression. IRF1 transcription does not rely on ISGF3 but does require Jak1-mediated Stat phosphorylation, and thus the degradation of Jak1 results in decreased IRF1 mRNA levels during HCMV infection (Miller and others 1999). Similarly, levels of 2′,5,-oligoadenylate synthetase (OAS) and MxA, both IFN-α/ISGF3-stimulated genes are, in some reports, undetectable at the RNA level during infection of both fibroblasts and endothelial cells (Miller and others 1999). However, these results conflict with other data, including microarray studies, showing up-regulation of these genes following infection in fibroblasts (Zhu and others 1997; Zhu and others 1998; Boyle and others 1999; Simmen and others 2001; Browne and Shenk 2003; Abate and others 2004). In addition to Jak1 degradation, levels of p48 are also decreased in HCMV infection, further disrupting the formation of the ISGF3 complex (Miller and others 1999). These results imply that HCMV-mediated degradation of Jak1 and p48 weakens the ability of infected cells to up-regulate cellular defense genes in response to IFN-α stimulation.
Not surprisingly, Jak1 degradation is also linked to inhibition of IFN-γ-stimulated genes. Similar to results with IFN-α stimulation, IFN-γ-mediated phosphorylation of Stat1α, IFNγR1, Jak2, and Jak1 are abrogated during HCMV infection, although only Jak1 protein levels are lowered appreciably (Miller and others 1998). GAF induction, which is dependent on upstream signaling and phosphorylation events, is impaired in HCMV-infected cells, resulting in decreased expression of the class II transactivator (CIITA), which is required for up-regulation of MHC class II (Miller 98). The interference with Jak1 expression may thus contribute to the down-regulation of MHC class II expression seen during HCMV infection of endothelial cells and fibroblasts (Scholz and others 1992; Sedmak and others 1994; Knight and others 1997).
In addition to degrading Jak1, CMV infections also interfere with Stat protein function at several levels. Stat2 is degraded during HCMV infection of fibroblasts, and this degradation is strain-specific, with many clinical isolates and the lab strain AD169 causing degradation, but another lab strain (Towne) not affecting Stat2 levels (Le and others 2008a). Although the HCMV gene(s) responsible for Stat2 degradation remain unknown, experiments by Le et al. suggest that an early gene is responsible. Stat2 mRNA is actually up-regulated at the transcriptional level during HCMV infection, but HCMV then controls Stat2 protein levels by proteasomal degradation. Interestingly, experiments comparing the IFN-α, β, or γ sensitivity of HCMV AD169, which degrades Stat2, and Towne, which does not, demonstrated only a modest benefit of Stat2 degradation for HCMV replication (Le and others 2008a).
In an apparently redundant effort to control IFN responses, HCMV interferes directly with Stat2 signaling as well as its expression. The viral immediate early protein IE72 interacts directly with Stat2 and blocks association of the activated ISGF3 complex with ISRE elements in the nucleus, preventing the up-regulation of IFN-α/β-stimulated genes including ISG54 and MxA (Paulus and others 2006). Interestingly, IE72 does not interfere with Stat2 protein levels, phosphorylation, or formation of the ISGF3 complex, but acts after nuclear translocation of ISGF3. Recent studies demonstrated that an acidic domain near the C-terminus of IE72 is responsible for the interaction with Stat2, and this domain is required for efficient viral growth, particularly in IFN-treated cells (Huh and others 2008). Experiments comparing wild-type virus to a mutant virus expressing IE72 lacking this acidic domain established that Stat2 translocation is independent of IE72 binding, but chromatin immunoprecipitation analysis demonstrated that IE72 binding resulted in reduced loading of Stat2 onto ISRE sites. Sumoylation of IE72 in the acidic domain prevents IE72:Stat2 binding and reverses the repressive effect of IE72 on IFN-regulated gene expression (Huh and others 2008).
During MCMV infection, pM27 mediates Stat2 down-regulation, and loss of pM27 renders the virus more susceptible to both type I and type II IFNs (Zimmermann and others 2005). Stat1, but not Stat2, is thought to be required for IFN-γ signaling, but pM27 appears to be important for resistance to both types of IFN. Although there is synergy between type I and type II IFN, ΔM27-MCMV was more sensitive than wild-type virus to IFN-γ even in the absence of type I IFN, suggesting that Stat2 might also directly activate IFN-γ responses. In fact, Zimmermann et al. demonstrated that in INFAR1−/− cells, but not INFGR1−/− cells, IFN-γ induced phosphorylation of Stat2, and this could be blocked by MCMV infection (Zimmermann and others 2005). The phosphorylated Stat2 formed active ISGF3 complexes capable of binding ISRE sites. pM27 is required for MCMV replication in vivo (Abenes and others 2001; Zimmermann and others 2005), likely due to its role in blocking both IFN-α/β and IFN-γ responses by interfering with Stat2 and preventing synergy between the 2 arms of the IFN response. Recently, Le et al. demonstrated that pUL27, the HCMV homolog of pM27 (Rawlinson and others 1996), does not induce Stat2 down-regulation and is not required for viral growth in cell culture (Le and others 2008a), suggesting that the HCMV mechanisms for Stat2 interference are UL27-independent.
HCMV takes a different tack for interference with IFN-γ signaling by impairing Stat1 phosphorylation. Upon IFN-γ stimulation, levels of phosphorylated Stat1 are decreased in HCMV-infected cells, and activation of GAS-mediated transcription is similarly diminished (Baron and Davignon 2008). These authors demonstrated that the degradation of Jak1 is not responsible for this inhibition as Jak1 levels do not decline until later in infection. Instead, HCMV activates phosphorylation of the tyrosine phosphatase Shp2, which then binds to and dephosphorylates Stat1. Further experiments should determine if this Shp2-mediated Stat1 dephosphorylation also plays a role in preventing ISRE-driven expression, as would seem likely due to the involvement of Stat1 in the ISGF3 complex.
CMVs have developed effective strategies for evasion of many steps of the IFN pathway. The overall effect of this interference with IFN signaling is a dampening of the host immune response to viral infection. The degradation of Jak1, which regulates both type I and type II IFN signaling, might seem to be sufficient for blocking the IFN response as it prevents efficient Stat phosphorylation and dimerization. However, because Jak1 degradation may not be complete in all cell types and under all conditions, and because it does not occur early in infection, CMVs also tamper with the downstream Stat protein function by activating cellular Stat phosphatases, degrading the Stat proteins, and preventing their binding to target sequences. Thus not only do CMVs interfere with IFN production, they also block IFN signal transduction at multiple steps.
Evasion of ISG Effector Function
Type I IFN signaling results in transcriptional up-regulation of hundreds of IFN-stimulated genes (ISGs) (Der and others 1998), many of which encode proteins important for the amplification of the IFN response, including Stat and IRF family members. Although the role of many ISGs has not yet been established, several are important effectors of the antiviral response. In addition to blocking IFN and ISG production, the CMVs have evolved mechanisms to limit ISG function, demonstrating that even if the blockades described earlier are incomplete or ineffective under some conditions, the viruses still have other tools to ensure success in evading the IFN system. MHC class I and II are expressed constitutively on some cells but also are induced to higher levels and expressed in expanded cell types as part of the IFN response. Interference with IFN-signaling pathways and Stat-mediated transcriptional activation can result in decreased expression of MHC class I and II, and thus is likely responsible, at least in part, for the down-regulation of antigen presentation observed during CMV infection. In addition to preventing transcription of MHC genes, the CMVs have multiple strategies for blocking formation of, and peptide presentation by, MHC complexes. Several viral proteins play a role in this MHC down-regulation, as reviewed recently elsewhere (Lin and others 2007; Wiertz and others 2007; Powers and others 2008). Since the role of ISGs involved in the direct establishment of an antiviral state in CMV-infected cells has been less extensively studied, we will discuss recent progress in understanding viral antagonism of the host cell response to dsRNA.
dsRNA is detectable in cells by 24 hpi during HCMV infection and by 16 hpi during MCMV infection (Budt and others 2009; Marshall and others 2009). The origin of dsRNA during CMV infection remains unknown but it may result from breakdown in polyadenylation efficiency late in infection that allows synthesis of long transcripts from both strands of the genome that can anneal in the cytoplasm. Infection with UV-inactivated MCMV does not yield dsRNA, indicating that viral gene expression is required for dsRNA production (Budt and others 2009). Accumulation of dsRNA during HCMV infection occurs in the presence of ganciclovir (Marshall and others 2009), suggesting that late gene expression is not required. Since CMV genomes are quite GC-rich, another source of “dsRNA” might be mRNAs with sufficient secondary structure to activate dsRNA-mediated antiviral responses.
Protein kinase R (PKR) and OAS are 2 ISGs that sense dsRNA and activate cellular pathways, which result in the shutdown of protein synthesis. PKR dimerizes as a result of a conformational change induced by binding to dsRNA, then autophosphorylates and phosphorylates translation initiation factor eIF2α on serine 51. Phosphorylated eIF2α inhibits guanine nucleotide exchange factor eIF2B, thereby limiting restoration of the eIF2α–tRNAmet–GTP ternary complex and preventing translation initiation (Dever and others 2007). The OAS family of proteins catalyze the formation of novel 2′,5′-oligoadenylates, which activate latent ribonuclease RNase L. RNase L degrades rRNA and both cellular and viral mRNA (Silverman 2007). Viruses depend on the protein synthetic capabilities of the host, so together the PKR and OAS pathways create an antiviral environment.
The presence of basal levels of PKR in cells even in the absence of IFN induction coupled with the accumulation of dsRNA during infection suggests that CMV replication might depend on mechanisms to counteract the PKR pathway directly. Indeed, the HCMV proteins, pIRS1 and pTRS1, and MCMV proteins, pm142 and pm143, serve this function. pIRS1 and pTRS1, which are encoded partly in the inverted repeats flanking the unique short region of the HCMV genome, are present in the virion and expressed from immediate early times throughout infection (Romanowski and Shenk 1997). Either one of these genes is sufficient to rescue replication of a vaccinia virus mutant lacking its own dsRNA-binding protein antagonist of PKR and OAS, E3L (Child and others 2004). The combination of m142 and m143 can also rescue VVΔE3L replication (Child and others 2006). Infection with HCMV lacking both IRS1 and TRS1, or MCMV lacking either m142 or m143, results in phosphorylation of eIF2α that triggers a profound shutoff of protein synthesis, and entirely eliminates viral replication (Valchanova and others 2006; Budt and others 2009; Marshall and others 2009). Thus HCMV has 2 genes that serve a redundant essential function associated with blocking PKR. The findings that MCMV has 2 genes, both of which are necessary to block PKR and allow viral replication, along with results of coimmunoprecipitation and colocalization experiments, suggest that the pm142 and pm143 proteins function as a complex (Hanson and others 2005; Budt and others 2009; Child and Geballe 2009). The phenotype of HCMV lacking IRS1 and TRS1 can be reversed by introduction of E3L (Marshall and others 2009). Similarly, the replication and protein synthesis defects of the MCMV m142 and m143 mutant viruses can be reversed at least in part by TRS1, E3L, or HSV-1 γ34.5 (Valchanova and others 2006; Budt and others 2009). Finally, IRS1 and TRS1 can complement herpes simplex type I mutant that lacks the γ34.5 gene (Cassady 2005), further supporting the conclusion that these proteins all serve a similar critical function in blocking PKR (Cassady 2005; Valchanova and others 2006).
Like E3L and several other viral antagonists of PKR, pIRS1 and pTRS1 bind to dsRNA (Hakki and Geballe 2005). The dsRNA-binding domain maps to a region in the identical N-terminus of these 2 proteins that is not homologous to any other known dsRNA-binding proteins. Deletion of this region eliminates the ability of pTRS1 to rescue replication of VVΔE3L, suggesting the dsRNA binding is critical to pIRS1 and pTRS1 function. The combination of pm142 and pm143 proteins also bind to dsRNA, although whether dsRNA binding is required for inhibition of PKR has not been established (Child and others 2006).
pIRS1 and pTRS1 each bind directly to PKR as well, as do several other virally encoded dsRNA-binding protein antagonists of PKR (Mohr and others 2007). The binding requires a region in their divergent C-terminal region, which is also necessary for rescuing replication of VVΔE3L (Hakki and others 2006). The MCMV pm142 and pm143 proteins also bind to PKR based on coimmunoprecipitation data (Budt and others 2009; Child and Geballe 2009). However, cell fractionation studies showed that pm142 and pm143 form a stable heterotetrameric complex consisting of 2 molecules each of pm142 and pm143, but containing little if any PKR. It is possible that a pm142:pm143:PKR complex in the cells is unstable and dissociates under the experimental conditions used. Alternatively, transient interactions between pm142:pm143 and PKR may be sufficient to inactivate PKR or sequester it into compartments where it cannot shut off protein synthesis. Indeed, both HCMV and MCMV cause an unusual relocalization of PKR into the nucleus and into insoluble cytoplasmic aggregates during infection (Hakki and others 2006; Child and Geballe 2009), which may explain the inability of PKR to block translation during HCMV and MCMV infection. However, the role of this relocalization is unclear since E3L, which in the context of VV infection, does not cause PKR relocalization, enables replication of HCMV lacking both IRS1 and TRS1 (Marshall and others 2009). Regardless of the exact mechanism, these studies demonstrate that dsRNA-binding PKR-binding proteins are essential for CMV replication, likely at least partly because of their ability to inactivate PKR-mediated translational repression.
Although expression of the antiviral ISG OAS is induced by CMV binding and entry and its activating ligand, dsRNA, accumulates in infected cells (Zhu and others 1998; Boyle and others 1999; Budt and others 2009), the OAS/RNase L pathway is not activated during infection, even with the HCMV mutant lacking both IRS1 and TRS1 or with MCMV lacking m142 and m143 (Budt and others 2009; Marshall and others 2009). These observations suggest that OAS/RNase L pathway may not play a role during CMV infection. The dsRNA produced by the CMVs may not be sufficient in quantity or composition to activate OAS. Alternatively, the failure to detect RNase L activation might be due to a CMV-mediated blockade downstream of OAS activation but upstream of RNase L-mediated RNA degradation.
Studies of dsRNA-binding protein mutant CMVs stress the importance of evading ISG-mediated antiviral pathways. Although these viruses disturb many steps of the IFN production and signaling pathways, those evasive maneuvers are by themselves insufficient to allow for viral replication without additional interference with IFN effectors like PKR, further illustrating the complex arsenal that CMVs have evolved to block the innate immune response.
Conclusion
CMVs have been coevolving with their hosts for millions of years. Remarkably, host defenses have been at least partially successful in thwarting CMV replication as well as that of other viruses, most of which have the potential for much more rapid evolution than their hosts. The success of the IFN response appears to have driven the evolution of myriad interwoven CMV countermeasures designed to interfere with the initial induction of IFN, the amplification of the response by effects on IFN signaling, and the actions of antiviral effectors. Although much progress has been made in defining some components at the interface of the CMVs and the IFN system, the continually emerging surprises and complexities of this research engender a sense that we are still only scratching the surface and that many more discoveries are awaiting.
Acknowledgments
We thank Ann Campbell (Eastern Virginia Medical School) for critical review of our manuscript. This work was supported by NIH grant AI26672.
References
- Abate DA. Watanabe S. Mocarski ES. Major human cytomegalovirus structural protein pp65 (ppUL83) prevents interferon response factor 3 activation in the interferon response. J Virol. 2004;78(20):10995–11006. doi: 10.1128/JVI.78.20.10995-11006.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abenes G. Lee M. Haghjoo E. Tong T. Zhan X. Liu F. Murine cytomegalovirus open reading frame M27 plays an important role in growth and virulence in mice. J Virol. 2001;75(4):1697–1707. doi: 10.1128/JVI.75.4.1697-1707.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banks TA. Rickert S. Benedict CA. Ma L. Ko M. Meier J. Ha W. Schneider K. Granger SW. Turovskaya O. Elewaut D. Otero D. French AR. Henry SC. Hamilton JD. Scheu S. Pfeffer K. Ware CF. A lymphotoxin-IFN-beta axis essential for lymphocyte survival revealed during cytomegalovirus infection. J Immunol. 2005;174(11):7217–7225. doi: 10.4049/jimmunol.174.11.7217. [DOI] [PubMed] [Google Scholar]
- Baron M. Davignon JL. Inhibition of IFN-gamma-induced STAT1 tyrosine phosphorylation by human CMV is mediated by SHP2. J Immunol. 2008;181(8):5530–5536. doi: 10.4049/jimmunol.181.8.5530. [DOI] [PubMed] [Google Scholar]
- Barton ES. White DW. Cathelyn JS. Brett-McClellan KA. Engle M. Diamond MS. Miller VL. Virgin HWT. Herpesvirus latency confers symbiotic protection from bacterial infection. Nature. 2007;447(7142):326–329. doi: 10.1038/nature05762. [DOI] [PubMed] [Google Scholar]
- Benedict CA. Banks TA. Senderowicz L. Ko M. Britt WJ. Angulo A. Ghazal P. Ware CF. Lymphotoxins and cytomegalovirus cooperatively induce interferon-beta, establishing host-virus detente. Immunity. 2001;15(4):617–626. doi: 10.1016/s1074-7613(01)00222-9. [DOI] [PubMed] [Google Scholar]
- Bentz GL. Yurochko AD. Human CMV infection of endothelial cells induces an angiogenic response through viral binding to EGF receptor and beta1 and beta3 integrins. Proc Natl Acad Sci USA. 2008;105(14):5531–5536. doi: 10.1073/pnas.0800037105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehme KW. Guerrero M. Compton T. Human cytomegalovirus envelope glycoproteins B and H are necessary for TLR2 activation in permissive cells. J Immunol. 2006;177(10):7094–7102. doi: 10.4049/jimmunol.177.10.7094. [DOI] [PubMed] [Google Scholar]
- Boyle KA. Pietropaolo RL. Compton T. Engagement of the cellular receptor for glycoprotein B of human cytomegalovirus activates the interferon-responsive pathway. Mol Cell Biol. 1999;19(5):3607–3613. doi: 10.1128/mcb.19.5.3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brocchieri L. Kledal TN. Karlin S. Mocarski ES. Predicting coding potential from genome sequence: application to betaherpesviruses infecting rats and mice. J Virol. 2005;79(12):7570–7596. doi: 10.1128/JVI.79.12.7570-7596.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne EP. Shenk T. Human cytomegalovirus UL83-coded pp65 virion protein inhibits antiviral gene expression in infected cells. Proc Natl Acad Sci USA. 2003;100(20):11439–11444. doi: 10.1073/pnas.1534570100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne EP. Wing B. Coleman D. Shenk T. Altered cellular mRNA levels in human cytomegalovirus-infected fibroblasts: viral block to the accumulation of antiviral mRNAs. J Virol. 2001;75(24):12319–12330. doi: 10.1128/JVI.75.24.12319-12330.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budt M. Niederstadt L. Valchanova RS. Jonjic S. Brune W. Specific inhibition of the PKR-mediated antiviral response by the murine cytomegalovirus proteins m142 and m143. J Virol. 2009;83(3):1260–1270. doi: 10.1128/JVI.01558-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassady KA. Human cytomegalovirus TRS1 and IRS1 gene products block the double-stranded-RNA-activated host protein shutoff response induced by herpes simplex virus type 1 infection. J Virol. 2005;79(14):8707–8715. doi: 10.1128/JVI.79.14.8707-8715.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Child SJ. Geballe AP. Binding and relocalization of protein kinase R by murine cytomegalovirus. J Virol. 2009;83(4):1790–1799. doi: 10.1128/JVI.01484-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Child SJ. Hakki M. De Niro KL. Geballe AP. Evasion of cellular antiviral responses by human cytomegalovirus TRS1 and IRS1. J Virol. 2004;78(1):197–205. doi: 10.1128/JVI.78.1.197-205.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Child SJ. Hanson LK. Brown CE. Janzen DM. Geballe AP. Double-stranded RNA binding by a heterodimeric complex of murine cytomegalovirus m142 and m143 proteins. J Virol. 2006;80(20):10173–10180. doi: 10.1128/JVI.00905-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compton T. Kurt-Jones EA. Boehme KW. Belko J. Latz E. Golenbock DT. Finberg RW. Human cytomegalovirus activates inflammatory cytokine responses via CD14 and Toll-like receptor 2. J Virol. 2003;77(8):4588–4596. doi: 10.1128/JVI.77.8.4588-4596.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFilippis V. Fruh K. Rhesus cytomegalovirus particles prevent activation of interferon regulatory factor 3. J Virol. 2005;79(10):6419–6431. doi: 10.1128/JVI.79.10.6419-6431.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFilippis VR. Robinson B. Keck TM. Hansen SG. Nelson JA. Fruh KJ. Interferon regulatory factor 3 is necessary for induction of antiviral genes during human cytomegalovirus infection. J Virol. 2006;80(2):1032–1037. doi: 10.1128/JVI.80.2.1032-1037.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delale T. Paquin A. Asselin-Paturel C. Dalod M. Brizard G. Bates EE. Kastner P. Chan S. Akira S. Vicari A. Biron CA. Trinchieri G. Briere F. MyD88-dependent and -independent murine cytomegalovirus sensing for IFN-alpha release and initiation of immune responses in vivo. J Immunol. 2005;175(10):6723–6732. doi: 10.4049/jimmunol.175.10.6723. [DOI] [PubMed] [Google Scholar]
- Der SD. Zhou A. Williams BR. Silverman RH. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc Natl Acad Sci USA. 1998;95(26):15623–15628. doi: 10.1073/pnas.95.26.15623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dever TE. Dar AC. Sicheri F. The eIF2alpha kinases. In: Matthews MB, editor; Sonenberg N, editor; Hershey JWB, editor. Translational control in biology and medicine. Cold Spring Harbor, NY: Cold Spring Harbor Press; 2007. pp. 319–344. [Google Scholar]
- Edelmann KH. Richardson-Burns S. Alexopoulou L. Tyler KL. Flavell RA. Oldstone MB. Does Toll-like receptor 3 play a biological role in virus infections? Virology. 2004;322(2):231–238. doi: 10.1016/j.virol.2004.01.033. [DOI] [PubMed] [Google Scholar]
- Gravel SP. Servant MJ. Roles of an IkappaB kinase-related pathway in human cytomegalovirus-infected vascular smooth muscle cells: a molecular link in pathogen-induced proatherosclerotic conditions. J Biol Chem. 2005;280(9):7477–7486. doi: 10.1074/jbc.M410392200. [DOI] [PubMed] [Google Scholar]
- Hakki M. Geballe AP. Double-stranded RNA binding by human cytomegalovirus pTRS1. J Virol. 2005;79(12):7311–7318. doi: 10.1128/JVI.79.12.7311-7318.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakki M. Marshall EE. De Niro KL. Geballe AP. Binding and nuclear relocalization of protein kinase R by human cytomegalovirus TRS1. J Virol. 2006;80(23):11817–11826. doi: 10.1128/JVI.00957-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson LK. Dalton BL. Cageao LF. Brock RE. Slater JS. Kerry JA. Campbell AE. Characterization and regulation of essential murine cytomegalovirus genes m142 and m143. Virology. 2005;334(2):166–177. doi: 10.1016/j.virol.2005.01.046. [DOI] [PubMed] [Google Scholar]
- Hoebe K. Du X. Georgel P. Janssen E. Tabeta K. Kim SO. Goode J. Lin P. Mann N. Mudd S. Crozat K. Sovath S. Han J. Beutler B. Identification of Lps2 as a key transducer of MyD88-independent TIR signalling. Nature. 2003;424(6950):743–748. doi: 10.1038/nature01889. [DOI] [PubMed] [Google Scholar]
- Huh YH. Kim YE. Kim ET. Park JJ. Song MJ. Zhu H. Hayward GS. Ahn JH. Binding STAT2 by the acidic domain of human cytomegalovirus IE1 promotes viral growth and is negatively regulated by SUMO. J Virol. 2008;82(21):10444–10454. doi: 10.1128/JVI.00833-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaacson MK. Juckem LK. Compton T. Virus entry and innate immune activation. Curr Top Microbiol Immunol. 2008;325:85–100. doi: 10.1007/978-3-540-77349-8_5. [DOI] [PubMed] [Google Scholar]
- Jahn G. Scholl BC. Traupe B. Fleckenstein B. The two major structural phosphoproteins (pp65 and pp150) of human cytomegalovirus and their antigenic properties. J Gen Virol. 1987;68(Pt 5):1327–1337. doi: 10.1099/0022-1317-68-5-1327. [DOI] [PubMed] [Google Scholar]
- Juckem LK. Boehme KW. Feire AL. Compton T. Differential initiation of innate immune responses induced by human cytomegalovirus entry into fibroblast cells. J Immunol. 2008;180(7):4965–4977. doi: 10.4049/jimmunol.180.7.4965. [DOI] [PubMed] [Google Scholar]
- Knight DA. Waldman WJ. Sedmak DD. Human cytomegalovirus does not induce human leukocyte antigen class II expression on arterial endothelial cells. Transplantation. 1997;63(9):1366–1369. doi: 10.1097/00007890-199705150-00030. [DOI] [PubMed] [Google Scholar]
- Le VT. Trilling M. Wilborn M. Hengel H. Zimmermann A. Human cytomegalovirus interferes with signal transducer and activator of transcription (STAT) 2 protein stability and tyrosine phosphorylation. J Gen Virol. 2008a;89(Pt 10):2416–2426. doi: 10.1099/vir.0.2008/001669-0. [DOI] [PubMed] [Google Scholar]
- Le VT. Trilling M. Zimmermann A. Hengel H. Mouse cytomegalovirus inhibits beta interferon (IFN-beta) gene expression and controls activation pathways of the IFN-beta enhanceosome. J Gen Virol. 2008b;89(Pt 5):1131–1141. doi: 10.1099/vir.0.83538-0. [DOI] [PubMed] [Google Scholar]
- Li X. Leung S. Qureshi S. Darnell JE., Jr Stark GR. Formation of STAT1-STAT2 heterodimers and their role in the activation of IRF-1 gene transcription by interferon-alpha. J Biol Chem. 1996;271(10):5790–5794. doi: 10.1074/jbc.271.10.5790. [DOI] [PubMed] [Google Scholar]
- Lin A. Xu H. Yan W. Modulation of HLA expression in human cytomegalovirus immune evasion. Cell Mol Immunol. 2007;4(2):91–98. [PubMed] [Google Scholar]
- Mack C. Sickmann A. Lembo D. Brune W. Inhibition of proinflammatory and innate immune signaling pathways by a cytomegalovirus RIP1-interacting protein. Proc Natl Acad Sci USA. 2008;105(8):3094–3099. doi: 10.1073/pnas.0800168105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall EE. Bierle CJ. Brune W. Geballe AP. Essential role for either TRS1 or IRS1 in human cytomegalovirus replication. J Virol. 2009;83(9):4112–4120. doi: 10.1128/JVI.02489-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DM. Rahill BM. Boss JM. Lairmore MD. Durbin JE. Waldman JW. Sedmak DD. Human cytomegalovirus inhibits major histocompatibility complex class II expression by disruption of the Jak/Stat pathway. J Exp Med. 1998;187(5):675–683. doi: 10.1084/jem.187.5.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DM. Zhang Y. Rahill BM. Waldman WJ. Sedmak DD. Human cytomegalovirus inhibits IFN-alpha-stimulated antiviral and immunoregulatory responses by blocking multiple levels of IFN-alpha signal transduction. J Immunol. 1999;162(10):6107–6113. [PubMed] [Google Scholar]
- Miller G. Brain cancer. A viral link to glioblastoma? Science. 2009;323(5910):30–31. doi: 10.1126/science.323.5910.30. [DOI] [PubMed] [Google Scholar]
- Mocarski ES. Shenk T. Pass RF. Cytomegaloviruses. In: Knipe DM, editor; Howley PM, editor. Fields virology. Philadelphia: Lippincott Williams & Wilkins; 2007. pp. 2702–2772. [Google Scholar]
- Mohr IJ. Pe'ery T. Mathews MB. Protein synthesis and translational control during viral infection. In: Matthews MB, editor; Sonenberg N, editor; Hershey JWB, editor. Translational control in biology and medicine. Cold Spring Harbor, NY: Cold Spring Harbor Press; 2007. pp. 545–599. [Google Scholar]
- Murphy E. Shenk T. Human cytomegalovirus genome. Curr Top Microbiol Immunol. 2008;325:1–19. doi: 10.1007/978-3-540-77349-8_1. [DOI] [PubMed] [Google Scholar]
- Netterwald JR. Jones TR. Britt WJ. Yang SJ. McCrone IP. Zhu H. Postattachment events associated with viral entry are necessary for induction of interferon-stimulated genes by human cytomegalovirus. J Virol. 2004;78(12):6688–6691. doi: 10.1128/JVI.78.12.6688-6691.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols WG. Corey L. Gooley T. Davis C. Boeckh M. High risk of death due to bacterial and fungal infection among cytomegalovirus (CMV)-seronegative recipients of stem cell transplants from seropositive donors: evidence for indirect effects of primary CMV infection. J Infect Dis. 2002;185(3):273–282. doi: 10.1086/338624. [DOI] [PubMed] [Google Scholar]
- Paulus C. Krauss S. Nevels M. A human cytomegalovirus antagonist of type I IFN-dependent signal transducer and activator of transcription signaling. Proc Natl Acad Sci USA. 2006;103(10):3840–3845. doi: 10.1073/pnas.0600007103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawelec G. Derhovanessian E. Larbi A. Strindhall J. Wikby A. Cytomegalovirus and human immunosenescence. Rev Med Virol. 2009;19(1):47–56. doi: 10.1002/rmv.598. [DOI] [PubMed] [Google Scholar]
- Powers C. DeFilippis V. Malouli D. Fruh K. Cytomegalovirus immune evasion. Curr Top Microbiol Immunol. 2008;325:333–359. doi: 10.1007/978-3-540-77349-8_19. [DOI] [PubMed] [Google Scholar]
- Presti RM. Pollock JL. Dal Canto AJ. O'Guin AK. Virgin HWT. Interferon gamma regulates acute and latent murine cytomegalovirus infection and chronic disease of the great vessels. J Exp Med. 1998;188(3):577–588. doi: 10.1084/jem.188.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston CM. Harman AN. Nicholl MJ. Activation of interferon response factor-3 in human cells infected with herpes simplex virus type 1 or human cytomegalovirus. J Virol. 2001;75(19):8909–8916. doi: 10.1128/JVI.75.19.8909-8916.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prichard MN. Jairath S. Penfold ME. St Jeor S. Bohlman MC. Pari GS. Identification of persistent RNA-DNA hybrid structures within the origin of replication of human cytomegalovirus. J Virol. 1998;72(9):6997–7004. doi: 10.1128/jvi.72.9.6997-7004.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randolph-Habecker J. Iwata M. Geballe AP. Jarrahian S. Torok-Storb B. Interleukin-1-mediated inhibition of cytomegalovirus replication is due to increased IFN-beta production. J Interferon Cytokine Res. 2002;22(7):765–772. doi: 10.1089/107999002320271350. [DOI] [PubMed] [Google Scholar]
- Rawlinson WD. Farrell HE. Barrell BG. Analysis of the complete DNA sequence of murine cytomegalovirus. J Virol. 1996;70(12):8833–8849. doi: 10.1128/jvi.70.12.8833-8849.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves M. Sinclair J. Aspects of human cytomegalovirus latency and reactivation. Curr Top Microbiol Immunol. 2008;325:297–313. doi: 10.1007/978-3-540-77349-8_17. [DOI] [PubMed] [Google Scholar]
- Romanowski MJ. Shenk T. Characterization of the human cytomegalovirus irs1 and trs1 genes: a second immediate-early transcription unit within irs1 whose product antagonizes transcriptional activation. J Virol. 1997;71(2):1485–1496. doi: 10.1128/jvi.71.2.1485-1496.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider K. Loewendorf A. De Trez C. Fulton J. Rhode A. Shumway H. Ha S. Patterson G. Pfeffer K. Nedospasov SA. Ware CF. Benedict CA. Lymphotoxin-mediated crosstalk between B cells and splenic stroma promotes the initial type I interferon response to cytomegalovirus. Cell Host Microbe. 2008;3(2):67–76. doi: 10.1016/j.chom.2007.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholz M. Hamann A. Blaheta RA. Auth MK. Encke A. Markus BH. Cytomegalovirus- and interferon-related effects on human endothelial cells. Cytomegalovirus infection reduces upregulation of HLA class II antigen expression after treatment with interferon-gamma. Hum Immunol. 1992;35(4):230–238. doi: 10.1016/0198-8859(92)90004-7. [DOI] [PubMed] [Google Scholar]
- Sedmak DD. Guglielmo AM. Knight DA. Birmingham DJ. Huang EH. Waldman WJ. Cytomegalovirus inhibits major histocompatibility class II expression on infected endothelial cells. Am J Pathol. 1994;144(4):683–692. [PMC free article] [PubMed] [Google Scholar]
- Silverman RH. Viral encounters with 2′,5′-oligoadenylate synthetase and RNase L during the interferon antiviral response. J Virol. 2007;81(23):12720–12729. doi: 10.1128/JVI.01471-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmen KA. Singh J. Luukkonen BG. Lopper M. Bittner A. Miller NE. Jackson MR. Compton T. Fruh K. Global modulation of cellular transcription by human cytomegalovirus is initiated by viral glycoprotein B. Proc Natl Acad Sci USA. 2001;98(13):7140–7145. doi: 10.1073/pnas.121177598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soroceanu L. Akhavan A. Cobbs CS. Platelet-derived growth factor-alpha receptor activation is required for human cytomegalovirus infection. Nature. 2008;455(7211):391–395. doi: 10.1038/nature07209. [DOI] [PubMed] [Google Scholar]
- Streblow DN. Dumortier J. Moses AV. Orloff SL. Nelson JA. Mechanisms of cytomegalovirus-accelerated vascular disease: induction of paracrine factors that promote angiogenesis and wound healing. Curr Top Microbiol Immunol. 2008;325:397–415. doi: 10.1007/978-3-540-77349-8_22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabeta K. Georgel P. Janssen E. Du X. Hoebe K. Crozat K. Mudd S. Shamel L. Sovath S. Goode J. Alexopoulou L. Flavell RA. Beutler B. Toll-like receptors 9 and 3 as essential components of innate immune defense against mouse cytomegalovirus infection. Proc Natl Acad Sci USA. 2004;101(10):3516–3521. doi: 10.1073/pnas.0400525101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor RT. Bresnahan WA. Human cytomegalovirus immediate-early 2 gene expression blocks virus-induced beta interferon production. J Virol. 2005;79(6):3873–3877. doi: 10.1128/JVI.79.6.3873-3877.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor RT. Bresnahan WA. Human cytomegalovirus IE86 attenuates virus- and tumor necrosis factor alpha-induced NFkappaB-dependent gene expression. J Virol. 2006a;80(21):10763–10771. doi: 10.1128/JVI.01195-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor RT. Bresnahan WA. Human cytomegalovirus immediate-early 2 protein IE86 blocks virus-induced chemokine expression. J Virol. 2006b;80(2):920–928. doi: 10.1128/JVI.80.2.920-928.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terhune SS. Schroer J. Shenk T. RNAs are packaged into human cytomegalovirus virions in proportion to their intracellular concentration. J Virol. 2004;78(19):10390–10398. doi: 10.1128/JVI.78.19.10390-10398.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upton JW. Kaiser WJ. Mocarski ES. Cytomegalovirus M45 cell death suppression requires receptor-interacting protein (RIP) homotypic interaction motif (RHIM)-dependent interaction with RIP1. J Biol Chem. 2008;283(25):16966–16970. doi: 10.1074/jbc.C800051200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valchanova RS. Picard-Maureau M. Budt M. Brune W. Murine cytomegalovirus m142 and m143 are both required to block protein kinase R-mediated shutdown of protein synthesis. J Virol. 2006;80(20):10181–10190. doi: 10.1128/JVI.00908-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varnum SM. Streblow DN. Monroe ME. Smith P. Auberry KJ. Pasa-Tolic L. Wang D. Camp DG., II Rodland K. Wiley S. Britt W. Shenk T. Smith RD. Nelson JA. Identification of proteins in human cytomegalovirus (HCMV) particles: the HCMV proteome. J Virol. 2004;78(20):10960–10966. doi: 10.1128/JVI.78.20.10960-10966.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiertz EJ. Devlin R. Collins HL. Ressing ME. Herpesvirus interference with major histocompatibility complex class II-restricted T-cell activation. J Virol. 2007;81(9):4389–4396. doi: 10.1128/JVI.01525-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurochko AD. Huang ES. Human cytomegalovirus binding to human monocytes induces immunoregulatory gene expression. J Immunol. 1999;162(8):4806–4816. [PubMed] [Google Scholar]
- Zhu H. Cong JP. Mamtora G. Gingeras T. Shenk T. Cellular gene expression altered by human cytomegalovirus: global monitoring with oligonucleotide arrays. Proc Natl Acad Sci USA. 1998;95(24):14470–14475. doi: 10.1073/pnas.95.24.14470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H. Cong JP. Shenk T. Use of differential display analysis to assess the effect of human cytomegalovirus infection on the accumulation of cellular RNAs: induction of interferon-responsive RNAs. Proc Natl Acad Sci USA. 1997;94(25):13985–13990. doi: 10.1073/pnas.94.25.13985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann A. Trilling M. Wagner M. Wilborn M. Bubic I. Jonjic S. Koszinowski U. Hengel H. A cytomegaloviral protein reveals a dual role for STAT2 in IFN-{gamma} signaling and antiviral responses. J Exp Med. 2005;201(10):1543–1553. doi: 10.1084/jem.20041401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zucchini N. Bessou G. Traub S. Robbins SH. Uematsu S. Akira S. Alexopoulou L. Dalod M. Cutting edge: Overlapping functions of TLR7 and TLR9 for innate defense against a herpesvirus infection. J Immunol. 2008;180(9):5799–5803. doi: 10.4049/jimmunol.180.9.5799. [DOI] [PubMed] [Google Scholar]