Madam,
The Clostridium difficile restriction endonuclease analysis (REA) type BI [polymerase chain reaction (PCR) ribotype 027] strain, with cdtA/cdtB (genes for binary toxin CDT), an 18 bp tcdC deletion, elevated toxin A and B concentrations, and fluoroquinolone resistance, is possibly responsible for the increased Clostridium difficile infection (CDI) incidence, severity, and mortality observed recently.1–4 The historic BI strain, present at least since the early 1980s also contained the 18 bp deletion, cdtA/cdtB, and levofloxacin resistance, but has not previously been associated with outbreaks.5 The recent BI strain, however, has additionally acquired resistance to moxifloxacin and gatifloxacin.5
From 2002 to 2005, the number of cases of CDI at the University of Virginia Hospital (UVH) doubled and cases associated with ciprofloxacin use increased (LAP, personal communication). Previous work showed that high levels of fecal lactoferrin, a marker of leucocytes and intestinal inflammation, were associated with moderate to severe clinical CDI.6 Therefore, our aim was to determine whether the BI strain was present at UVH, and if there was a correlation between genotype, strain type, or moxifloxacin resistance patterns, and lactoferrin levels.
Fifty-two de-identified fecal specimens from inpatients at UVH with a clinical suspicion of CDI and testing positive for toxins A or B by enzyme-linked immunosorbent assay (ELISA) were collected. Thirty-four C. difficile strains were then successfully isolated from these faecal specimens. Moxifloxacin resistance was determined using E-test strips; only fully sensitive [minimum inhibitory concentration (MIC) <1 μg/mL] or fully resistant (MIC of >32 μg/mL) isolates were observed. DNA was extracted and amplified by PCR for the presence of gdh (glutamate dehydrogenase), tcdA (toxin A gene), tcdB (toxin B gene), tcdC, and cdtA/cdtB.
To determine if the BI strain was present, REA was performed on 33 of 34 isolates by the C. difficile Microbiology Reference Laboratory (MRL) at the Hines Veterans Administration Hospital. We further sequenced the tcdC gene of 12 representative isolates (10 resistant and two susceptible), to determine if our BI/027 strains contained a single nucleotide base pair deletion associated with toxin A and B hypersecretion.
Faecal specimens were evaluated for lactoferrin using the IBD Scan ELISA (TechLab, Inc.) and grouped according to lactoferrin concentration: >72.5 μg/g (10-fold greater than the set point for a positive test) or ≤72.5 μg/g.
Eighteen (53%) isolates tested positive for the tcdC deletion, 19 (56%) for cdtA/cdtB, and 26 (76%) were moxifloxacin resistant. Eighteen of 33 (55%) were REA type BI. All BI/027 isolates had the 18 bp tcdC deletion and cdtA/cdtB, and 16 (89%) were moxifloxacin resistant. All sequenced BI/027 strains contained a single base pair deletion at position 117.
Using χ2-analysis, we noted that faecal specimens with higher levels of lactoferrin were significantly associated with the presence of moxifloxacin-resistant isolates, and that susceptible isolates were associated with lower levels of lactoferrin (Figure 1a, P = 0.041). Elevated lactoferrin levels were not independently associated with the presence of the 18 bp tcdC deletion (Figure 1b) or cdtA/cdtB (data not shown).
Figure 1.
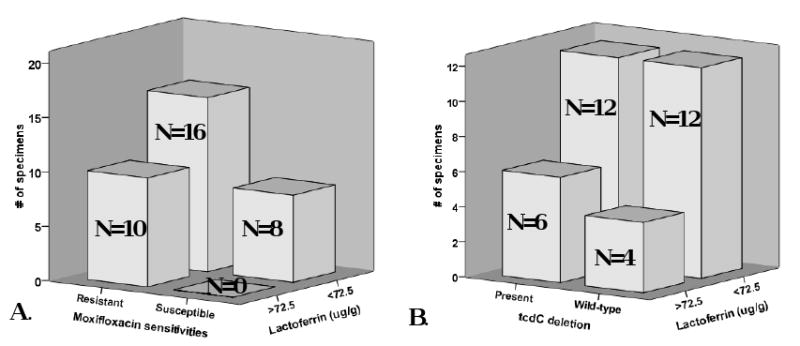
24 (71%) fecal specimens had lactoferrin levels ≤72.5 μg/g. A. Of the 10 specimens with lactoferrin levels >72.5, all were associated with the presence of moxifloxacin resistant isolates; in addition, all susceptible isolates were associated with lower lactoferrin levels (X2, p=0.041). B. There was no correlation between elevated lactoferrin levels and the presence of the 18-bp tcdC deletion (X2, p=0.44)
We report that higher levels of intestinal inflammation, as measured by quantitative fecal lactoferrin, are associated with moxifloxacin-resistant CDI. Notably, only moxifloxacin resistance, as opposed to the tcdC deletion or cdtA/cdtB, was significantly associated with lactoferrin levels >72.5 μg/g. We observed that the BI/027 strain is present at UVH and that moxifloxacin resistance, present in 76% of our strains, appears to be a factor leading to intestinal inflammation in patients with CDI.
This study does not aim to place a precise concentration indicating what constitutes elevated lactoferrin levels in CDI, but only to recognize that in our set of faecal specimens the highest lactoferrin levels, and thus the greatest levels of intestinal inflammation, were associated with moxifloxacin-resistant CDI, including but not limited to the BI/027 strain. A larger study looking at the correlation of faecal lactoferrin with patient characteristics, severity of disease, and presence of moxifloxacin-resistant CDI is needed to validate or refute this preliminary finding that fluoroquinolone resistance may be an important factor leading to more severe intestinal inflammation in CDI.
Acknowledgments
We thank J. Franasiak, Dr A. Samie, Dr G. Kolling, K. Nagaro, and A. Cheknis for their assistance in performing these experiments and Dr J.E. Sevilleja for his statistical analysis assistance.
Funding sources: This work was supported by NIH/NIAD T32 AI 55432-03, UO1 AI075526, UO1 AI070491 and the UVA Silvio O. Conte Digestive Health Research Center, Pilot Feasibility Award Program. REA typing was provided through grants from ViroPharma, Inc. and the Department of Veterans Affairs to D.N.G.
Footnotes
Conflict of interest statement: R.L.G. is licensed fecal lactoferrin to TechLab Inc. D.M.L., R.J.C., and C.W.G. are TechLab employees. D.N.G. consults for ViroPharma, Genzyme, Optimer, Salix, and Schering–Plough and holds research grants from ViroPharma, Optimer, Massachusetts Biological Laboratories, Cepheid, and GOJO Industries.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pepin J, Valiquette L, Alary ME, et al. Clostridium difficile-associated diarrhea in a region of Quebec from 1991 to 2003: a changing pattern of disease severity. Can Med Assoc J. 2004;171:466–472. doi: 10.1503/cmaj.1041104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Muto CA, Pokrywka M, Shutt K, et al. A large outbreak of Clostridium difficile-associated disease with an unexpected proportion of deaths and colectomies at a teaching hospital following increased fluoroquinolone use. Infect Control Hosp Epidemiol. 2005;26:273–280. doi: 10.1086/502539. [DOI] [PubMed] [Google Scholar]
- 3.McEllistrem MC, Carman RJ, Gerding DN, Genheimer CW, Zheng L. A hospital outbreak of Clostridium difficile disease associated with isolates carrying binary toxin genes. Clin Infect Dis. 2005;40:265–272. doi: 10.1086/427113. [DOI] [PubMed] [Google Scholar]
- 4.Warny M, Pepin J, Fang A, et al. Toxin production by an emerging strain of Clostridium difficile associated with outbreaks of severe disease in North America and Europe. Lancet. 2005;366:1079–1084. doi: 10.1016/S0140-6736(05)67420-X. [DOI] [PubMed] [Google Scholar]
- 5.McDonald LC, Killgore GE, Thompson A, et al. An epidemic, toxin gene-variant strain of Clostridium difficile. N Engl J Med. 2005;353:2433–2441. doi: 10.1056/NEJMoa051590. [DOI] [PubMed] [Google Scholar]
- 6.Steiner TS, Flores CA, Pizarro TT, Guerrant RL. Fecal lactoferrin, interleukin-1beta, and interleukin-8 are elevated in patients with severe Clostridium difficile colitis. Clin Diagn Lab Immunol. 1997;4:719–722. doi: 10.1128/cdli.4.6.719-722.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]


