Abstract
Substance P (SP), a neurotransmitter in stress pathways, exerts its effects mainly through the neurokinin-1 receptor (NK1R). Genetic and pharmacologic studies show that binding of ligands to NK1R decreases anxiety-related behaviors and therefore self-administration of alcohol in mice and craving for alcohol in humans. As genetic variants may result in differential expression of the receptor through various molecular mechanisms, we examined whether allelic variations in the NK1R gene are associated with alcohol dependence (AD) by genotyping 11 single nucleotide polymorphisms (SNPs) across NK1R in alcoholic (n=271) and healthy control (n=337) subjects of Caucasian descent. The AD was diagnosed using the Structured Clinical Interview for Diagnostic and Statistical Manual of Mental Disorders, 4th edition. Associations of the SNPs with AD were assessed at both the individual SNP and haplotype levels. We found that genotype and allele frequencies of rs6715729, a synonymous SNP in exon 1, differed significantly in alcoholics and controls (p=0.0006; OR=6.13; 95% CI = 4.06, 9.23). Haplotype analyses indicated two risk haplotypes for AD in the 5’ end of the gene, formed by the three-SNP combination rs6715729-rs735668-rs6741029. Taken together, we conclude that polymorphisms of NK1R are significantly associated with the development of AD in Caucasian individuals. Additional studies are needed to replicate these results in other samples and to elucidate the mechanism(s) by which these polymorphisms affect NK1R function in the brain.
Keywords: Association, SNPs, Alcohol Dependence, Addiction, NK1R
Introduction
Alcohol dependence (AD) is a psychiatric disorder that is the 3rd most common preventable cause of death and morbidity in the US (Johnson, 2008). The World Health Organization (WHO) estimated the number of alcohol users to be 2 billion worldwide in the year 2002. In an estimate of factors responsible for global disease burden, alcohol contributed 3.2% of all deaths in the year 2000 (http://www.who.int/substance_abuse/facts/global_burden/en/).
Despite advances in AD research, treatment completion and relapse rates remain substantial (Garbutt et al, 2005). The etiology of AD is complex: once use is initiated, the vulnerability to become dependent on alcohol is modulated by genetic and environmental factors as well as by their interactions. The contribution of genetic factors to AD is substantial, with a heritability of 50–64% (Yang et al, 2007). Thus, it is important to identify genes involved in the neurobiological pathways mediating alcohol’s reinforcing effects and associated with its abuse liability.
Dampening of the stress response is one of the main mechanisms by which alcohol exerts its reinforcing effects (Sinha et al, 1998). Animal and human studies suggest a central role for substance P (SP) in mediating the stress response (Holmes et al, 2003). The biological actions of SP are exerted primarily through neurokinin receptor subtype 1 (NK1R; also called tachykinin receptor 1; TACR1), which is densely expressed in the brain structures implicated in stress pathways (Giegling et al, 2007), as well as in non-neuronal peripheral tissues including gut, joints, muscle, and cutaneous tissues (Santarelli et al, 2001; Almeida et al, 2004). Antagonists of NK1R are effective in treating depression, anxiety, and post-operative and chemotherapy-induced emesis (Randolph et al, 2004). Recently, a reduction in the craving for alcohol in anxious alcoholics was reported in response to the NK1R blocker LY686017 (George et al, 2008).
The NK1R protein is encoded by NK1R, located on chromosome 2p13.1 spanning about 151 kb with five exons. According to the NCBI database (http://www.ncbi.nlm.nih.gov), NK1R is transcribed into at least four mRNA variants, resulting from alternative promoter usage, differential splicing, or both. The mRNA variants are putatively translated into four functional NK1R isoforms that may possess different affinities for SP. Additionally, the expression of these isoforms appears to be tissue specific. For example, so far, only the two longest isoforms of mRNA have been found in the human brain (Caberlotto et al, 2003; Lai et al, 2008). Therefore, naturally occurring polymorphisms at or near promoter or splice regions may determine NK1R density, altering SP-mediated signaling.
The objective of the current study was to determine whether genetic variants of NK1R are associated with AD by performing a case-control association study in a sample of Caucasian AD and control subjects.
Materials and Methods
Participants
A total of 608 unrelated Caucasian subjects were recruited from central-southern USA. Participants provided written informed consent on a form approved by all involved institutional review boards. Of these subjects, 271 were treatment-seeking persons with AD, and 337 were controls. Numbers of subjects recruited at each recruitment site are provided in Table 1. All AD subjects were recruited as part of pharmacotherapy trials for the treatment of alcohol dependence. Alcohol dependence was diagnosed using the Structured Clinical Interview (First et al, 1994) for Diagnostic and Statistical Manual of Mental Disorders, 4th edition (American Psychiatric Association, 1994) Axis I Disorders. In addition, all the AD subjects had a score of ≥ 8 on the Alcohol Use Disorders Identification Test (AUDIT) (Babor et al., 1992), which assesses the personal and social harm after alcohol consumption, reported drinking of ≥ 21 standard drinks/week for women and ≥ 30 standard drinks/week for men during the 90 days prior to enrollment, and negative urine toxicologic screen for narcotics, amphetamines, or sedative-hypnotics at enrollment. Abstinence was not an entry criterion. Subjects who had current Axis I psychiatric diagnoses other than alcohol or nicotine dependence; significant alcohol withdrawal symptoms [revised clinical institute withdrawal assessment for alcohol scale (Sullivan et al, 1989) score >15]; clinically significant physical abnormalities identified at physical examination, electrocardiogram recording, hematologic assessment, biochemistry tests including serum bilirubin concentration and urinalysis; pregnant or lactating state; treatment for alcohol dependence ≤30 days prior to enrollment; and mandated incarceration or employment loss for not receiving alcohol treatment were excluded. All control subjects were selected from a large genetic study on tobacco smoking to match ethnicity and age of the AD subjects and were recruited primarily from the Mid-South States during 1999–2005 (Beuten et al, 2005; Li et al, 2006). All control individuals were screened for a history of substance use disorders and other Axis I psychiatric disorders, and subjects who were positive were excluded. Additionally, in the control group, none of the subjects reported having consumed > 4 standard drinks in total, during the 90 days prior to enrollment. Self-reported ethnicity data was collected for both control and AD individuals at study entry. The demographic characteristics of both AD and control subjects are presented in Table 1.
Table 1.
Demographics of AD and control samples used in this study
| AD subjects | Control subjects | |
|---|---|---|
| Total number | 271 | 337 |
| San Antonio, Texas | 189 | 4 |
| Charlottesville, Virginia | 82 | 0 |
| Jackson, Mississippi | 0 | 333 |
| Male/female | 196/75 | 127/210 |
| Mean age (yr ± SD) | 46.16 ± 13.09 | 46.43 ± 14.58 |
| Mean age of onset of AD (yr ± SD) | 30.13 ± 11.49 | NA |
DNA extraction, SNP selection and genotyping
Ten milliliters of blood was drawn from each subject to obtain white blood cells. The DNA was extracted using a Gentra Puregene® kit (QIAGEN Inc., Valencia, CA). We selected 11 SNPs from the National Center for Biotechnology Information (NCBI) dbSNP database (http://www.ncbi.nlm.nih.gov/SNP/) on the basis of high heterozygosity (minor allele frequency ≥ 0.05) and chromosomal position for maximum uniform coverage of the gene. Detailed information on SNP locations, chromosomal positions, allelic variants, minor allele frequency, and primer/probe sequences is summarized in Table 2. Additionally, 24 Ancestry Informative Markers (AIMs) were genotyped in all AD and control subject DNA samples to test for population stratification. The primary reasons for us to use these AIMs are that they have been demonstrated to have high frequency differences for South American/European ancestry and European/West African ancestry based on a survey study for a genome-wide ancestry marker panel and are suggested to be essential markers for population stratification investigation (Mao et al., 2007). Information on these marker SNPs are given in Supplementary Table 1.
Table 2.
Information on NK1R SNPs genotyped in the current study
| SNP number | dbSNP ID | Location within the gene | Physical Location | Alleles | ABI SNP assay ID |
|---|---|---|---|---|---|
| 1 | rs2111375 | Intergenic at 5′ end | 54,243,815 | G/A | C_2679170_10 |
| 2 | rs6715729 | Silent mutation on Exon 1 | 54,241,665 | G/A | C_25473413_10 |
| 3 | rs3771860 | Intron 1 | 54,231,673 | T/G | C_27514344_10 |
| 4 | rs3821319 | Intron 1 | 54,205,876 | C/G | C_26138472_10 |
| 5 | rs735668 | Intron 1 | 54,178,981 | C/A | C_2679114_10 |
| 6 | rs6741029 | Intron 2 | 54,161,132 | G/T | C_2974531_20 |
| 7 | rs4853105 | Intron 2 | 54,140,523 | A/T | C_29680163_10 |
| 8 | rs4439987 | Intron 2 | 54,103,043 | A/G | C_1227902_10 |
| 9 | rs727156 | Intron 3 | 54,096,144 | T/C | C_580584_10 |
| 10 | rs1106855 | Intron 4 | 54,093,924 | A/G | C_1227897_10 |
| 11 | rs881 | 3′-UTR | 54,092,366 | C/G | C_11466393_10 |
All SNP assays were carried out with the TaqMan® SNP genotyping assay in the 384-well microplate format (Applied Biosystems, Foster City, CA). Polymerase chain reaction (PCR) conditions were 50°C for 2 min, 95°C for 10 min, 30 cycles of 95°C for 25 s, and 60°C for 1 min. Each reaction was carried out with 10ng of DNA from each subject, and SNP-specific control samples were added to each 384-well plate to ensure the quality of the assays. Allelic discrimination analyses were performed with an ABI PRISM® 7900HT instrument (Applied Biosystems) using sequence detection software.
Statistical Analysis
We assessed linkage disequilibrium (LD) and haplotype blocks with all 11 SNPs in cases and controls separately, using Haploview (v. 4.1) (Gabriel et al., 2000). The associations between individual SNPs and AD were analyzed with Unphased (v. 3.0.13) software (Dudbridge, 2008) which calculates a maximum-likelihood ratio statistic allowing for covariate adjustments. An association of a SNP with AD was considered significant only if its p value was less than the corrected p value based on the SNP spectral decomposition (SNPSpD) approach for multiple testing (Nyholt, 2004). As calculated by SNPSpD, the corrected statistically significant p value for 11 SNP analyses was set at p=0.006. The associations between haplotypes and AD were analyzed with Haplo Stats (v. 1.2.1) software (http://mayoresearch.mayo.edu/mayo/research/biostat/schaid.cfm). We defined a “major haplotype” as one with a frequency of more than 5% in either the AD or the control samples. Bonferroni correction was used to calculate the corrected p value for the haplotype analyses. In both individual SNP and haplotype association analyses with AD, using Unphased and Haplo Stats programs respectively, age and sex were used as modifiers and site IDs where each AD and control subject was recruited initially, were used as confounders in all our association analyses. Furthermore, to test for possible moderating effects of each covariate on the observed associations between AD and individual SNPs, we employed a general linear model (GLM) in SAS (v. 9.1) that included age, sex and center IDs as main effects as well as their interactions with individual SNPs.
The program Structure 2.2 (http://www.stats.ox.ac.uk/~pritch/home.html) was used to assess the population stratification in the subjects enrolled at the three sites. Simulation parameters were set to 10,000 burn-ins, 10,000 iterations and K was set to 3 (for three recruitment sites).
Results
Individual SNP association analyses with AD
Genotypic distributions of all 11 SNPs conformed to the Hardy-Weinberg equilibrium with the smallest p value being 0.102. The genotypic and allelic frequencies for each SNP in both case and control samples are shown in Table 3. Individual SNP-based association analyses performed with Unphased (v. 3.0.13) software, using a statistical model incorporating age, sex and site IDs as covariates, revealed that the genotypic and allelic frequencies were significantly different in the case and control samples for SNPs rs2111375 (p=0.0454) and rs6715729 (p=0.0012). After correction for multiple testing, only SNP rs6715729 remained significant. When the frequency of the G allele, which is the minor allele for rs6715729 in the AD sample, was compared with the frequency of the G allele in the control group, the difference was statistically significant (p=0.0006) with an odds ratio (OR) of 6.13 (95% CI= 4.06, 9.23). As revealed by SAS GLM model, the interaction between rs6715729 and site IDs was significant at p < 0.0001; notably, the individual SNP rs6715729 effect on AD also remained significant (p= 0.0002) in this GLM model, confirming the genetic effect of rs6715729 on AD.
Table 3.
Genotype and allele frequencies of NK1R SNPs for alcohol-dependent and control subjects
| db SNP ID | Frequencies from HapMap-CEU data set |
Genotype count (frequency) from the present study |
Allele count (frequency) |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genotype | Allele | AD | Controls | P* value | AD | Controls | P† value | ||||||||
| 1/1 | 1 /2 | 2/2 | 1/1 | 1/2 | 2 /2 | 1 | 2 | 1 | 2 | ||||||
| rs211137 | |||||||||||||||
| 5 | GG=0.518 | ||||||||||||||
| 1 = G | GA=0.393 | G=0.714 | 147 | 102 | 17 | 183 | 126 | 21 | 0.0454 | 396 | 136 | 492 | 168 | 0.0155 | |
| 2 = A | AA=0.089 | A=0.286 | (0.553) | (0.383) | (0.064) | (0.555) | (0.382) | (0.064) | (0.744) | (0.256) | (0.745) | (0.255) | |||
| rs671572 | |||||||||||||||
| 9 | ‡GG=0.130 | ||||||||||||||
| 1 = G | GA=0.391 | ‡G=0.326 | 63 | 129 | 73 | 92 | 164 | 76 | 0.0012 | 255 | 275 | 348 | 316 | 0.0006 | |
| 2 = A | AA=0.478 | A=0.674 | (0.238) | (0.487) | (0.275) | (0.277) | (0.494) | (0.229) | (0.481) | (0.519) | (0.524) | (0.476) | |||
| rs377186 | |||||||||||||||
| 0 | TT=0.491 | ||||||||||||||
| 1 = T | TG=0.404 | T=0.693 | 153 | 91 | 18 | 185 | 132 | 16 | 0.4853 | 397 | 127 | 502 | 164 | 0.2544 | |
| 2 = G | GG=0.105 | G=0.307 | (0.584) | (0.347) | (0.069) | (0.556) | (0.396) | (0.048) | (0.758) | (0.242) | (0.754) | (0.246) | |||
| rs382131 | |||||||||||||||
| 9 | CC=0.186 | ||||||||||||||
| 1 = C | CG=0.475 | C=0.424 | 30 | 98 | 137 | 50 | 148 | 136 | 0.6243 | 158 | 372 | 248 | 420 | 0.4708 | |
| 2 = G | GG=0.339 | G=0.576 | (0.113) | (0.370) | (0.517) | (0.150) | (0.443) | (0.407) | (0.298) | (0.702) | (0.371) | (0.629) | |||
| rs735668 | |||||||||||||||
| 1 = C | CC=0.281 | ||||||||||||||
| 2 = A | CA=0.474 | C=0.518 | 73 | 123 | 68 | 77 | 165 | 93 | 0.2129 | 269 | 259 | 319 | 351 | 0.1393 | |
| AA=0.246 | A=0.482 | (0.277) | (0.466) | (0.258) | (0.230) | (0.493) | (0.278) | (0.509) | (0.491) | (0.476) | (0.524) | ||||
| rs674102 | |||||||||||||||
| 9 | GG=0.458 | ||||||||||||||
| 1 = G | GT=0.373 | G=0.644 | 100 | 112 | 43 | 138 | 154 | 41 | 0.3023 | 312 | 198 | 430 | 236 | 0.2172 | |
| 2 = T | TT=0.169 | T=0.356 | (0.392) | (0.439) | (0.169) | (0.414) | (0.462) | (0.123) | (0.611) | (0.388) | (0.646) | (0.354) | |||
| rs485310 | |||||||||||||||
| 5 | TT=0.167 | ||||||||||||||
| 1 = T | TA=0.333 | T=0.333 | 32 | 102 | 128 | 34 | 149 | 148 | 166 | 358 | 217 | 445 | 0.0665 | ||
| 2 = A | AA=0.500 | A=0.667 | (0.122) | (0.389) | (0.489) | (0.103) | (0.450) | (0.447) | 0.1212 | (0.317) | (0.683) | (0.328) | (0.672) | ||
| rs443998 | |||||||||||||||
| 7 | GG=0.220 | ||||||||||||||
| 1 = G | GA=0.542 | G=0.492 | 58 | 133 | 62 | 69 | 142 | 121 | 0.4375 | 249 | 257 | 280 | 384 | 0.3739 | |
| 2 = A | AA=0.237 | A=0.508 | (0.229) | (0.526) | (0.245) | (0.208) | (0.428) | (0.364) | (0.492) | (0.508) | (0.422) | (0.578) | |||
| rs727156 | TT=0.768 | ||||||||||||||
| 1 = T | TC=0.196 | T=0.866 | 187 | 67 | 9 | 219 | 96 | 16 | 0.7694 | 441 | 85 | 534 | 128 | 0.7225 | |
| 2 = C | CC=0.036 | C=0.134 | (0.711) | (0.255) | (0.034) | (0.662) | (0.290) | (0.048) | (0.838) | (0.162) | (0.807) | (0.193) | |||
| rs110685 | |||||||||||||||
| 5 | GG=0.593 | ||||||||||||||
| 1 = G | AG=0.339 | G=0.763 | 149 | 90 | 21 | 196 | 117 | 22 | 0.8343 | 388 | 132 | 509 | 161 | 0.8255 | |
| 2 = A | AA=0.068 | A=0.237 | (0.573) | (0.346) | (0.081) | (0.585) | (0.349) | (0.066) | (0.746) | (0.254) | (0.760) | (0.240) | |||
| rs881 | |||||||||||||||
| 1 = G | GG=0.719 | ||||||||||||||
| 2 = C | GC=0.263 | G=0.851 | 184 | 59 | 10 | 221 | 93 | 19 | 0.9394 | 427 | 79 | 535 | 131 | 0.7564 | |
| CC=0.018 | C=0.149 | (0.727) | (0.233) | (0.040) | (0.664) | (0.279) | (0.057) | (0.844) | (0.156) | (0.803) | (0.197) | ||||
HapMap-CEU - Utah residents with ancestry from northern and western Europe, studied in the International HapMap project.
European American descent, submitted to NCBI dbSNP data base from Perlegen Sciences Inc.
- Chi-square P value for the statistical model that compared genotypic frequencies of alcoholic and control groups with covariate adjustments.
- Chi-square P value for the statistical model that compared minor allele frequency in alcoholic group and frequency of the same allele in control group with covariate adjustments.
Statistically significant P values (before correcting for multiple testing) are shown in bold letters, and P values significant after correction for multiple testing are shown in bold italics.
Haplotype analyses
We employed Haploview to calculate pair-wise correlation coefficients (r2, the squared total linkage disequilibrium divided by the product of the allelic frequencies at both loci) to detect haplotype blocks among all 11 SNPs according to the criteria defined by Gabriel et al. (2002). As shown in Figure 1, we detected three haplotype blocks in the control sample. These blocks, each comprising two-SNP combinations, were located at the 5’ end of the gene, around exon 2, and at the 3’ end of the gene: rs2111375–rs6715729, 1 kb; rs735668-rs6741029, 17kb; rs1106855-rs881, 2kb. In contrast, we did not detect any haplotype blocks in the AD sample.
Figure 1.
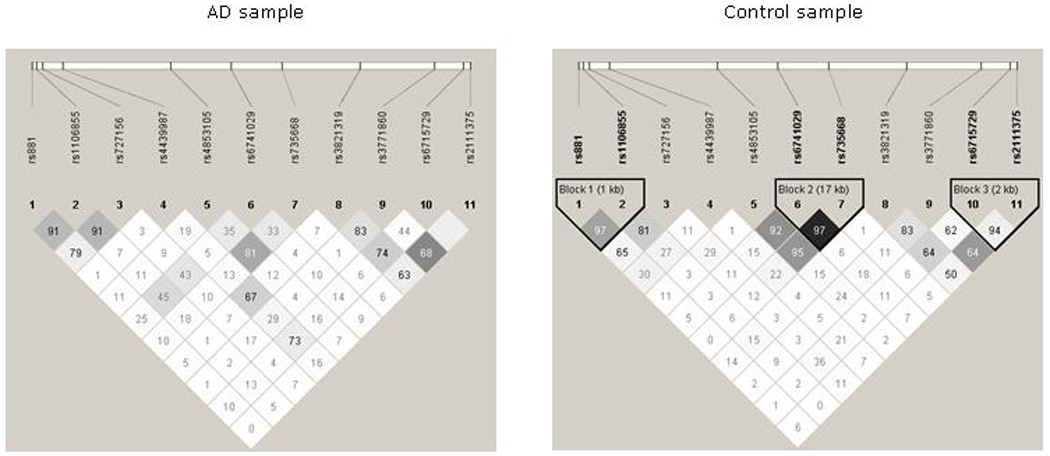
Linkage disequilibrium analysis of NK1R SNPs in Caucasian alcohol dependent and control samples. Values in squares are r2 numbers.
Next, using Haplo Stats, we performed haplotype-based association analyses in several ways: First, within each Haploview defined LD block comprising of 2 SNPs as shown in Figure 1; and secondly, with all possible three-SNP combinations within the three Haploview defined LD blocks. Also, we performed a haplotype analysis with all 27 possible three-SNP haplotype combinations formed by the 11 SNPs that were genotyped in this study. The haplotype analyses within the three 2-SNP LD blocks, detected only one Haploview-defined LD block (rs735668 and rs6741029) to be significantly associated with AD (global association test p=1.439×10−9). Our second approach, the haplotype analyses with all 6 SNPs within the 3 Haploview defined LD blocks, yielded four major haplotypes formed by a three-SNP combination of rs6715729-rs7356680-rs6741029 (Table 4). These three SNPs were located in adjacent LD blocks within a distance of 80,533bp in the NK1R gene and comprised of SNP rs6715729 that was significantly associated with AD at individual level. Therefore we considered 3-SNP combination formed by rs6715729-rs7356680-rs6741029 to be more informative than the 2-SNP combination formed by rs7356680 and rs6741029 alone. Of the four major haplotypes formed by rs6715729-rs7356680-rs6741029, two were negatively associated with AD (G-C-G, G-A-G), whereas the other two were positively associated with AD (A-A-G, G-C-T). After correcting for multiple testing, only the two positively associated haplotypes remained significant. (The corrected statistically significant p value for a three-SNP haplotype analysis was set at p=0.006.) The A-A-G and G-C-T haplotypes together accounted for 17.5% of the pooled sample (29.4% of the AD sample and 8.8% of the control sample). The observed frequency differences for haplotypes A-A-G (18.5% in AD and 8.1% in controls) and G-C-T (10.9% in AD and 0.8% in controls) in the AD and control samples were significantly different (χ2 =2.8953, p=0.0038 and χ2 =6.0062, p=1.8992×10−9, respectively) with corresponding ORs of 1.89 (95% CI=1.16, 3.11) for A-A-G and 11.31 (95% CI= 3.62, 35.35) for G-C-T. Therefore, the odds of becoming dependent on alcohol were more than 1 in subjects carrying either of these two haplotypes. Hence, they were named “risk haplotypes.” The SNPs that formed the two risk haplotypes were located in exon 1 and intronic regions on either side of exon 2 spanning about 82 kb.
Table 4.
Association results of major haplotypes formed by SNPs rs6715729, rs735668, and rs6741029 with AD
| Haplotype | Hapo-score | P value | Haplotype frequency |
Odds ratio |
||||
|---|---|---|---|---|---|---|---|---|
| Pooled | Controls | AD | Lower OR | OR | Upper OR | |||
| G-C-G | −2.5405 | 0.0111 | 0.2051 | 0.2346 | 0.1580 | 0.4824 | 0.7641 | 1.2103 |
| G-A-G | −2.3559 | 0.0184 | 0.0737 | 0.0995 | 0.0478 | 0.3415 | 0.6192 | 1.1227 |
| A-A-T | −1.5829 | 0.1134 | 0.1301 | 0.1598 | 0.0921 | 0.4229 | 0.7184 | 1.2203 |
| G-A-T | −1.3526 | 0.1761 | 0.1778 | 0.1826 | 0.1657 | 0.5411 | 0.8205 | 1.2441 |
| A-C-G | 0.2575 | 0.7968 | 0.2265 | 0.2303 | 0.2222 | NA | 1.0000 | NA |
| A-A-G | 2.8953 | 0.0038 | 0.1267 | 0.0805 | 0.1846 | 1.1552 | 1.8938 | 3.1045 |
| G-C-T | 6.0062 | 1.8992×10−9 | 0.0486 | 0.0075 | 0.1092 | 3.6194 | 11.3113 | 35.3498 |
global-stat = 59.2790; df = 7; p-val = 2.1024 ×10−10
Statistically significant P values (after correcting for multiple testing) for individual haplotype frequency differences in AD and control sample are shown in bold italics.
Population stratification for subjects enrolled at the three study sites was assessed with 24 AIM SNP markers. Results from program Structure showed no evidence for the presence of population admixture in both case and control subjects included in this study. Therefore, all 608 subjects were included in association analyses with AD, both at individual and haplotype levels.
Discussion
In this study, we provide the first evidence for an association of NK1R polymorphisms with AD in Caucasian individuals. After genotyping for 11 SNPs spanning the gene, we detected SNP rs6715729 and two haplotypes, A-A-G and G-C-T formed by rs6715729-rs7356680-rs6741029, that were significantly associated with AD. The allele frequencies of rs6715729, a synonymous SNP in exon 1, differed significantly in the AD and control samples. In the AD sample, allele A of rs6715729 was significantly more frequent than that in the control sample. The frequency of the A allele in our control sample was in agreement with the findings of an independent study by Randolph et al. (2004) that showed the A allele of rs6715729 to be less frequent than the G allele in a sample of 100 healthy individuals of Caucasian descent.
Moreover, we detected two haplotypes, G-C-T and A-A-G, formed by rs6715729-rs735668-rs6741029, that were significantly associated with AD; and both showed a significant risk effect on the development of AD. The first risk haplotype, A-A-G, was two-fold more common (OR=1.89; 95% CI=1.16, 3.11) in the AD than in the control sample. The second risk haplotype, G-C-T, was present predominantly in the AD sample (10.92% compared with 0.75% of the control sample) with a likelihood of about 11-fold higher in AD subjects than in controls (OR=11.31; 95% CI: 3.62, 35.35). By comparing allele composition in the two haplotypes, we found that unlike the A-A-G haplotype, the G-C-T risk haplotype G allele at rs6715729 showed a negative association with AD individually. On the other hand, the other two SNPs in the two haplotypes, rs735668 and rs6741029, did not show a significant association with AD individually. This suggests that the risk effects of these two haplotypes are conferred by a combination of alleles among the three SNPs. Indeed, haplotype analyses identify a combination(s) of SNPs that modulate each others’ individual effects to shape the overall modest effects caused by single genes in the etiology of complex traits such as AD (Zhang et al, 2008). In this context, another possibility is the presence of an unknown functional cis-acting polymorphism(s) in the region extending from SNPs rs735668 to rs6741029 that is associated with AD.
To our knowledge, this is the first study to investigate associations between AD and NK1R SNPs. The strength of our findings is enhanced by the differences in LD structures in the cases and controls. In the control sample, the SNPs that constitute the two risk haplotypes were in adjacent haplotype blocks; interestingly, these blocks were not seen in the AD sample, suggesting that this region may have undergone selection. Furthermore, the region of NK1R extending from rs6715729 to rs6741029 corresponds to the 5’ end of NK1R primary mRNA, which is translated into the N-terminal region through the end of the 2nd extracellular loop of NK1 receptors (Almeida et al, 2004). The NK1 receptors are 7-transmembrane domain G-protein coupled with three extracellular loops, three intracellular loops, an intracellular C terminus, and an extracellular N terminus. Among the many properties of the 5’ end portion of NK1 receptors important in SP signal transduction, the presence of a binding site for SP has been reported involving the 2nd extracellular loop (Lequin et al, 2002). Although none of the SNPs included in the two risk haplotypes affect the amino acid sequence of a protein, they could affect SP signal transduction by altering receptor density through alternative splicing of NK1R mRNA. Given the large size of the gene and the relatively low density of the SNPs we have genotyped, future genotyping studies should be aimed at identifying causative loci within this region.
There are several limitations to this study. First, the controls and cases were recruited at three geographic locations. To reduce the potential geographic effects on our association results, we included center ID as a covariate in all the association analyses. Additionally, we performed an analysis for genetic structure with 24 unlinked AIMs to exclude ethnic admixture within the Caucasian sample studied. Although the number of AIMs genotyped in our study was not as large as we wish, we do think they are sufficient for our purpose here. This is because: 1) 24 AIMs have been reported to be as effective in estimating continental population ancestry as identified with more than 90 AIMs (Kosoy et al., 2008) and 2) self-reported ethnicity usually is consistent with classification based on molecular markers (Yang et al, 2005). The second main limitation in our study is that the control sample had a relatively higher female population than the AD sample. This issue was addressed by including sex as a covariate in all the analyses. Third, our cases were a population of AD individuals who were motivated to decrease or cease their alcohol consumption. Therefore, this cohort may be more motivated and perhaps less dense in drinking pathophysiology than AD individuals derived from community samples (Wrase et al, 2006).
Numerous studies have affirmed that the product of NK1R, the receptor, together with its substrate SP, modulates mesocorticolimbic dopamine (DA) release, which is essential for the reward involved in AD (Nikolaev et al, 2003; Goodman, 2008). The SP-receptor system also is implicated in the etiology of other psychiatric disorders such as anxiety, depression, or dysfunctional behavioral patterns observable prior to the onset of AD (Goodman, 2008). Although NK1Rs play a vital role in the etiology of these psychiatric disorders that are comorbid with AD, to date, association studies on NK1R polymorphisms with psychiatric disorders in general are sparse. In a cohort of German subjects, Giegling et al (2007) showed that four SNPs located across NK1R were not associated with suicidal behavior but may modulate aggression features. Nominal associations were found with panic disorder (Hodges et al, 2009) and bipolar disorder (Perlis et al, 2008); lack of an association of NK1R SNPs was reported with autism (Marui et al, 2007) and dyslexia (Peyrard-Janvid et al, 2004). To our knowledge, associations of NK1R polymorphisms with any substance use disorders have not been examined.
In summary, we detected SNP rs6715729 and two haplotypes in NK1R that were significantly associated with AD in Caucasians. These results strongly indicate that NK1R is involved in the etiology of AD. In consideration of the relatively small sample size, more replication is greatly needed in a larger sample of both Caucasian and other ethnic samples. Furthermore, more molecular studies are needed to identify the mechanism(s) by which these genetic variants affect the development of AD.
Supplementary Material
ACKNOWLEDGEMENTS
This study was funded by grants from the National Institute on Alcohol Abuse and Alcoholism to B.A.J. (Grants 7 U10 AA011776-10, 1 N01 AA001016-000, 7 R01 AA010522-12, and 5 R01 AA012964-06) and N.A.-D. (Grant 5 K23 AA000329-06) and from the National Institute on Drug Abuse to M.D.L. (Grants R01 DA012844 and 5 R01 DA013783).
We appreciate the skilled technical assistance of the staff at the South Texas Addiction Research and Technology Center, Department of Psychiatry, The University of Texas Health Science Center at San Antonio, and the Department of Psychiatry and Neurobehavioral Sciences, University of Virginia. We also are grateful to Dr. Xiang-Yang Lou for his expert advice and Eva Jenkins-Mendoza, BS, for her service as project coordinator.
Footnotes
Disclosure/Conflict of Interest
Related to this report, authors Seneviratne, Ait-Daoud, Ma, Chen and Li have nothing to disclosure. Prof. Johnson currently serves as a consultant to Johnson & Johnson (Ortho-McNeil Janssen Scientific Affairs, LLC), Transcept Pharmaceuticals, Inc., Organon, ADial Pharmaceuticals, and Eli Lilly and Company.
REFERENCE
- Almeida TA, Rojo J, Nieto PM, Pinto FM, Hernandez M, Martin JD, Candenas ML. Tachykinins and tachykinin receptors: structure and activity relationships. Curr Med Chem. 2004;11:2045–2081. doi: 10.2174/0929867043364748. [DOI] [PubMed] [Google Scholar]
- American Psychiatric Association. Diagnostic and statistical manual of mental disorders. 4th edition. Washington, D.C.: American Psychiatric Association; 1994. [Google Scholar]
- Babor TF, de la Fuente JR, Saunders J, Grant M. AUDIT: The alcohol use disorders identification test. Geneva, Switzerland: World health organization; 1992. [Google Scholar]
- Beuten J, Ma JZ, Payne TJ, Dupont RT, Crews KM, Somes G, Williams NJ, Elston RC, Li MD. Single- and Multilocus Allelic Variants within the GABAB Receptor Subunit 2 (GABAB2) Gene Are Significantly Associated with Nicotine Dependence. Am. J. Hum. Genet. 2005;76:859–864. doi: 10.1086/429839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caberlotto L, Hurd YL, Murdock P, Wahlin JP, Melotto S, Corsi M, Carletti R. Neurokinin 1 receptor and relative abundance of the short and long isoforms in the human brain. Eur J Neurosci. 2003;17:1736–1746. doi: 10.1046/j.1460-9568.2003.02600.x. [DOI] [PubMed] [Google Scholar]
- Dudbridge F. Likelihood-based association analysis for nuclear families and unrelated subjects with missing genotype data. Hum Hered. 2008;66:87–98. doi: 10.1159/000119108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- First MB, Spitzer RL, Gibbon M, Williams JBW. Structured Clinical Interview for DSM-IV Axis I Disorders - Patient Edition (SCID-I/P, Version 2.0) New York: New York State Psychiatric Institute, Biometrics Research Department; 1994. [Google Scholar]
- Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, Liu-Cordero SN, Rotimi C, Adeyemo A, Cooper R, Ward R, Lander ES, Daly MJ, Altshuler D. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- Garbutt JC, Kranzler HR, O'Malley SS, Gastfriend DR, Pettinati HM, Silverman BL, Loewy JW, Ehrich EW. Efficacy and tolerability of long-acting injectable naltrexone for alcohol dependence: a randomized controlled trial. JAMA. 2005;293:1617–1625. doi: 10.1001/jama.293.13.1617. [DOI] [PubMed] [Google Scholar]
- George DT, Gilman J, Hersh J, Thorsell A, Herion D, Geyer C, Peng X, Kielbasa W, Rawlings R, Brandt JE, Gehlert DR, Tauscher JT, Hunt SP, Hommer D, Heilig M. Neurokinin 1 receptor antagonism as a possible therapy for alcoholism. Science. 2008;319:1536–1539. doi: 10.1126/science.1153813. [DOI] [PubMed] [Google Scholar]
- Giegling I, Rujescu D, Mandelli L, Schneider B, Hartmann AM, Schnabel A, Maurer K, De Ronchi D, Moller HJ, Serretti A. Tachykinin receptor 1 variants associated with aggression in suicidal behavior. Am J Med Genet B Neuropsychiatr Genet. 2007;144B:757–761. doi: 10.1002/ajmg.b.30506. [DOI] [PubMed] [Google Scholar]
- Goodman A. Neurobiology of addiction. An integrative review. Biochem Pharmacol. 2008;75:266–322. doi: 10.1016/j.bcp.2007.07.030. [DOI] [PubMed] [Google Scholar]
- Holmes A, Heilig M, Rupniak NM, Steckler T, Griebel G. Neuropeptide systems as novel therapeutic targets for depression and anxiety disorders. Trends Pharmacol Sci. 2003;24:580–588. doi: 10.1016/j.tips.2003.09.011. [DOI] [PubMed] [Google Scholar]
- Johnson BA. Update on neuropharmacological treatments for alcoholism: scientific basis and clinical findings. Biochem Pharmacol. 2008;75:34–56. doi: 10.1016/j.bcp.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosoy R, Nassir R, Tian C, White PA, Butler LM, Silva G, Kittles R, Alarcon-Riquelme ME, Gregersen PK, Belmont JW, De La Vega FM, Seldin MF. Ancestry Informative Marker Sets for Determining Continental Origin and Admixture Proportions in Common Populations in America. Human Mutation. 2008;30:69–78. doi: 10.1002/humu.20822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai JP, Cnaan A, Zhao H, Douglas SD. Detection of full-length and truncated neurokinin-1 receptor mRNA expression in human brain regions. J Neurosci Methods. 2008;168:127–133. doi: 10.1016/j.jneumeth.2007.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges LM, Weissman MM, Haghighi F, Costa R, Bravo O, Evgrafov O, Knowles JA, Fyer AJ, Hamilton SP. Association and linkage analysis of candidate genes GRP, GRPR, CRHR1, and TACR1 in panic disorder. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:65–73. doi: 10.1002/ajmg.b.30773. [DOI] [PubMed] [Google Scholar]
- Lequin O, Bolbach G, Frank F, Convert O, Girault-Lagrange S, Chassaing G, Lavielle S, Sagan S. Involvement of the second extracellular loop (E2) of the neurokinin-1 receptor in the binding of substance P. Photoaffinity labeling and modeling studies. J Biol Chem. 2002;277:22386–22394. doi: 10.1074/jbc.M110614200. [DOI] [PubMed] [Google Scholar]
- Li MD, Payne TJ, Ma JZ, Lou XY, Zhang D, Dupont RT, Crews KM, Somes G, Williams NJ, Elston RC. A genomewide search finds major susceptibility Loci for nicotine dependence on chromosome 10 in african americans. Am J Hum Genet. 2006;79:745–751. doi: 10.1086/508208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao X, Bigham AW, Mei R, Gutierrez G, Weiss KM, Brutsaert TD, Leon-Velarde F, Moore LG, Vargas E, McKeigue PM, Shriver MD, Parra EJ. A Genomewide Admixture Mapping Panel for Hispanic/Latino Populations. Am J Hum Genet. 2007;80:1171–1178. doi: 10.1086/518564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marui T, Funatogawa I, Koishi S, Yamamoto K, Matsumoto H, Hashimoto O, Nanba E, Nishida H, Sugiyama T, Kasai K, Watanabe K, Kano Y, Kato N, Sasaki T. Tachykinin 1 (TAC1) gene SNPs and haplotypes with autism: a case-control study. Brain Dev. 2007;29:510–513. doi: 10.1016/j.braindev.2007.01.010. [DOI] [PubMed] [Google Scholar]
- Nikolaev SV, Bychkov ER, Lebedev AA, Dambinova SA. Mechanisms of the influences of the central administration of substance P on ethanol consumption in chronically alcoholic rats. Neurosci Behav Physiol. 2003;33:905–909. doi: 10.1023/a:1025905023554. [DOI] [PubMed] [Google Scholar]
- Nyholt DR. A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am J Hum Genet. 2004;74:765–769. doi: 10.1086/383251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlis RH, Purcell S, Fagerness J, Kirby A, Petryshen TL, Fan J, Sklar P. Family-based association study of lithium-related and other candidate genes in bipolar disorder. Arch Gen Psychiatry. 2008;65:53–61. doi: 10.1001/archgenpsychiatry.2007.15. [DOI] [PubMed] [Google Scholar]
- Peyrard-Janvid M, Anthoni H, Onkamo P, Lahermo P, Zucchelli M, Kaminen N, Hannula-Jouppi K, Nopola-Hemmi J, Voutilainen A, Lyytinen H, Kere J. Fine mapping of the 2p11 dyslexia locus and exclusion of TACR1 as a candidate gene. Hum Genet. 2004;114:510–516. doi: 10.1007/s00439-004-1103-0. [DOI] [PubMed] [Google Scholar]; Randolph GP, Simon JS, Arreaza MG, Qiu P, Lachowicz JE, Duffy RA. Identification of single-nucleotide polymorphisms of the human neurokinin 1 receptor gene and pharmacological characterization of a Y192H variant. Pharmacogenomics J. 2004;4:394–402. doi: 10.1038/sj.tpj.6500276. [DOI] [PubMed] [Google Scholar]
- Santarelli L, Gobbi G, Debs PC, Sibille ET, Blier P, Hen R, Heath MJ. Genetic and pharmacological disruption of neurokinin 1 receptor function decreases anxiety-related behaviors and increases serotonergic function. Proc Natl Acad Sci U S A. 2001;98:1912–1917. doi: 10.1073/pnas.041596398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha R, Robinson J, O'Malley S. Stress response dampening: effects of gender and family history of alcoholism and anxiety disorders. Psychopharmacology (Berl) 1998;137:311–320. doi: 10.1007/s002130050624. [DOI] [PubMed] [Google Scholar]
- Sullivan JT, Sykora K, Schneiderman J, Naranjo CA, Sellers EM. Assessment of alcohol withdrawal: the revised clinical institute withdrawal assessment for alcohol scale (CIWA-Ar) Br J Addict. 1989;84:1353–1357. doi: 10.1111/j.1360-0443.1989.tb00737.x. [DOI] [PubMed] [Google Scholar]
- SAS Institute Inc. SAS version 9.1. Cary, NC: SAS Institute Inc.; 2003. [Google Scholar]
- Wrase J, Reimold M, Puls I, Kienast T, Heinz A. Serotonergic dysfunction: brain imaging and behavioral correlates. Cogn Affect Behav Neurosci. 2006;6:53–61. doi: 10.3758/cabn.6.1.53. [DOI] [PubMed] [Google Scholar]
- Yang BZ, Kranzler HR, Zhao H, Gruen JR, Luo X, Gelernter J. Association of haplotypic variants in DRD2, ANKK1, TTC12 and NCAM1 to alcohol dependence in independent case control and family samples. Hum Mol Genet. 2007;16:2844–2853. doi: 10.1093/hmg/ddm240. [DOI] [PubMed] [Google Scholar]
- Yang BZ, Zhao H, Kranzler HR, Gelernter J. Practical population group assignment with selected informative markers: characteristics and properties of Bayesian clustering via STRUCTURE. Genet Epidemiol. 2005;28:302–312. doi: 10.1002/gepi.20070. [DOI] [PubMed] [Google Scholar]
- Zhang H, Kranzler HR, Yang BZ, Luo X, Gelernter J. The OPRD1 and OPRK1 loci in alcohol or drug dependence: OPRD1 variation modulates substance dependence risk. Mol Psychiatry. 2008;13:531–543. doi: 10.1038/sj.mp.4002035. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


