Abstract
Twenty-five (27 %) of 92 clinically normal macaques were found to have β-haemolytic Escherichia coli isolated from their faeces. Five of six isolates chosen for further characterization had multiple antibiotic resistance and were PCR-positive for cytotoxic necrotizing factor 1 (cnf1) with a demonstrated cytopathic effect in vitro. By repetitive element sequence-based PCR genotyping, genetic similarity was established for selected isolates. We believe this to be the first report of E. coli strains producing CNF1 in non-human primates.
INTRODUCTION
Escherichia coli strains that produce cytotoxic necrotizing factors (CNFs) belong to the pathotype necrotoxigenic E. coli (NTEC) and are associated with intestinal and extra-intestinal infections in both humans and animals (Kaper et al., 2004). The majority of toxin-producing isolates produce either the chromosomally encoded CNF1 (Falbo et al., 1993) or the plasmid-encoded CNF2 (Oswald et al., 1989); more recently, a third type has been isolated from sheep and goats and named CNF3 (Orden et al., 2007). CNF1 is a 115 kDa protein with lethal and necrotizing activity in vivo. In vitro, affected cells undergo multinucleation and membrane ruffling and form focal adhesions and actin stress fibres. The mechanism involves activation of Rho GTPases, a family of molecular switches with multiple cellular functions, resulting in reorganization of the actin cytoskeleton. CNF1 shares a pathogenicity island with α-haemolysin and P fimbriae (De Rycke et al., 1999). CNF1-producing strains most notably cause urinary tract infections in humans. These strains have also been isolated from healthy and diseased animal species. The list includes weaned pigs (Tóth et al., 2000) and dogs (Starcic et al., 2002) with diarrhoea; cats (Féria et al., 2001) and dogs (Johnson et al., 2003; Siqueira et al., 2009; Yuri et al., 1998) with urinary tract infections; ferrets with diarrhoea and extra-intestinal infections (Marini et al., 2004); and birds and mink with suspected colibacillosis (Rodriguez-Siek et al., 2005) and colisepticaemia (Tibbetts et al., 2003). NTEC-2 strains, although commonly reported in ruminants (De Rycke et al., 1999), have also been reported in a very small percentage of human faecal E. coli isolates (Kadhum et al., 2008). The objective of this study was to evaluate the presence of specific virulence genes and toxin activity in several β-haemolytic E. coli isolates cultured from a macaque colony used for neurobiology research.
METHODS
Animals.
Macaques (Macaca mulatta and Macaca fascicularis) received from three US-based vendors in 2005–2006 received physical examinations and routine diagnostic evaluations during quarantine and at quarterly intervals. Animals were routinely pair-housed and maintained in an animal facility accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International. They were fed specified amounts of primate chow (Purina Lab Diet 5038) twice a day and provided with water ad libitum. Housing conditions were maintained at 20–22 °C, 30–70 % humidity, 10–15 non-recirculated air changes h−1 and a light cycle of 12 h light : 12 h dark.
Microbiological analysis.
Faecal samples were collected over a 2-year period from 2005 to 2006, either rectally or from cage pans, from a cohort of 92 clinically normal macaques (those received in quarantine as well as the established cohort of macaques actively being used in research). For those animals that were pair-housed and not isolated prior to sample collection, faeces were taken from both cage pans but it is not definitively known which animal the faeces came from, although it is likely that cage mates would both be affected. E. coli colonies were isolated on MacConkey lactose agar (Remel). For each isolate, a lactose-positive colony was streaked onto a sheep blood agar plate (Remel) and β-haemolysis was visualized directly on the blood agar plate. Several β-haemolytic colonies were then used to identify each isolate using API 20E strips (bioMérieux). Six isolates representing two macaque species, two vendors and four rooms in two animal facilities were chosen for further characterization.
Antibiotic susceptibility testing was performed for all six E. coli isolates using antimicrobial susceptibility discs (Remel for all discs except for the tetracycline disc, which was from Becton Dickinson) for the following antibiotics: ampicillin 10 μg, amoxicillin/clavulanic acid 20/10 μg, cephalothin 30 μg, gentamicin 10 μg, trimethoprim/sulfamethoxazole 1.25/23.75 μg, tetracycline 30 μg and enrofloxacin 5 μg. A few bacterial colonies from a pure culture isolated on blood agar were transferred to tryptic soy broth and incubated for 1 h at 37 °C. The bacterial suspension was then plated on Mueller–Hinton agar (Remel) and incubated overnight at 37 °C. Following incubation, the diameters of inhibition zones were measured with a standard ruler.
Serotyping and PCR.
Isolates were submitted on trypticase soy agar (Remel) to the E. coli Reference Center at Pennsylvania State University, PA, USA, for serotyping. Isolates underwent O- and H-typing as well as virulence testing for the selected toxins and adhesins heat-labile enterotoxin (LT), heat-stable enterotoxin a and b (STa and STb), Shiga-like toxin types I and II (stx I and stx II), intimin-γ (eae), and CNF1 and CNF2 (cnf1 and cnf2) by PCR following the procedures outlined on the Reference Center's website (http://ecoli.cas.psu.edu). We performed additional PCRs for α-haemolysin (hlyA) and pyelonephritis-associated pilus (papG I, II and III) using previously published protocols and designated primers, as these genes share the same pathogenicity island as cnf1 (Kadhum et al., 2008). In addition, there are E. coli strains that produce both CNF and cytolethal distending toxin (CDT); therefore, a multiplex PCR using two pairs of cdtB-specific primers was performed (Tóth et al., 2003).
CNF activity assays.
HeLa cell culture experiments were conducted to correlate CNF activity with the presence of cnf1. After overnight incubation at 37 °C in Difco LB broth (Becton Dickinson), E. coli cultures were centrifuged for 3 min at 17 900 g at room temperature. Supernatants were filtered using a 0.2 μm Acrodisc syringe filter (Pall Gelman Laboratory) and stored at −80 °C until use. Bacterial pellets were resuspended in 1 ml PBS and stored at −80 °C for 15 min before sonication. Bacteria were sonicated for three 30 s pulses on ice with a 30 s cooling time in between using an Artek Sonic Dismembrator (model 150; Artek Systems). Sonicates were centrifuged for 3 min at 17 900 g at room temperature and syringe-filtered before storage at −80 °C until use. E. coli sonicates or supernatants from the six macaque isolates and a non-pathogenic laboratory E. coli strain (XL-1 Blue), as well as controls of PBS and Eagle's minimal essential medium (Sigma-Aldrich) supplemented with 10 % fetal calf serum, were added to HeLa cell monolayers and incubated for 72 h. HeLa cells (5.5×103) were plated in a Lab-Tek II Chamber Slide System and incubated at 37 °C, 6 % CO2 for 3 h. Twelve microlitres of sonicate or supernatant was added to each chamber of HeLa cells and incubated at 37 °C, 6 % CO2 for 72 h. Media chambers were removed from the slide system and the slides were stained using an eosin Y/azure A Diff-Quick procedure. Cytopathic effect (CPE), specifically multinucleation and cell enlargement, was visualized by light microscopy.
Genotyping by repetitive element sequence-based PCR (rep-PCR).
High-resolution genotyping using a DiversiLab Escherichia DNA Fingerprinting kit based on the rep-PCR method of Versalovic et al. (1994) was carried out. DiversiLab software was used to compare the amplification patterns obtained and to analyse genetic similarity among the six tested isolates.
RESULTS
Microbiology results
Twenty-five (27 %) of 92 faecal samples were culture-positive for β-haemolytic E. coli on sheep blood agar plates. Serotyping and virulence factor testing results for six of these isolates are summarized in Table 1. Five serotypes were identified: O1 : H7, O88w : H7, O6 : H7, O6 : H1 and O2 : H1 (two isolates). All isolates were β-haemolytic and hlyA- and papG-positive (all isolates were papG I-negative and papG II-positive, and three of the six isolates were papG III-positive), and five of the six isolates were cnf1-positive. All other assays used to screen for virulence determinants were negative for all six isolates (LT, STa, STb, stx1, stx2, eae, cnf2 and cdtB). Five of the isolates showed multiple antibiotic resistance (Table 2). The antibiotic to which the greatest number of isolates had resistance was cephalothin, a first-generation cephalosporin. Five of the isolates were resistant and one isolate had an intermediate zone of susceptibility. Five of the isolates were susceptible to enrofloxacin, and one isolate had an intermediate zone of susceptibility.
Table 1.
Serotyping and virulence testing results
All isolates were β-haemolytic and negative for LT, STa, STb, stx1, stx2, eae, cnf2 and cdtB.
| Isolate no. | O type | H type | cnf1 | hlyA | papG |
|---|---|---|---|---|---|
| 06-2830 | 2 | 1 | + | + | + |
| 06-2832 | 2 | 1 | + | + | + |
| 06-2725 | 6 | 1 | + | + | + |
| 06-3320 | 6 | 7 | + | + | + |
| 06-2828 | 88w | 7 | + | + | + |
| 06-2934 | 1 | 7 | − | + | + |
Table 2.
Antibiotic susceptibility testing results
AMP, ampicillin; AMC, amoxicillin/clavulanic acid; CEF, cephalothin; GEN, gentamicin; SXT, trimethoprim/sulfamethoxazole; TET, tetracycline; ENRO, enrofloxacin; S, susceptible; I, intermediate; R, resistant.
| Isolate no. | AMP | AMC | CEF | GEN | SXT | TET | ENRO |
|---|---|---|---|---|---|---|---|
| 06-2830 | S | S | R | S | S | R | S |
| 06-2832 | S | S | R | S | S | S | S |
| 06-2725 | S | S | R | S | S | R | S |
| 06-3320 | S | S | R | S | S | R | I |
| 06-2828 | R | S | R | S | R | I | S |
| 06-2934 | S | S | I | R | R | R | S |
CNF1 activity shown in vitro
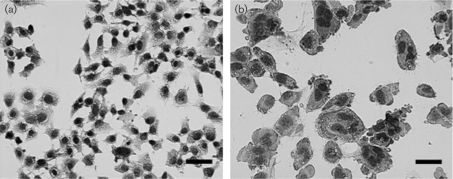
The five cnf1-positive E. coli isolates produced CPE (Fig. 1), except for 06-2725, which caused cell death at the 72 h time point. The cnf1-negative isolate, 06-2934, showed no CPE.
Fig. 1.
CPE representative of CNF1, most notably multinucleation and cell enlargement, in HeLa cell monolayers after 72 h. (a) An untreated HeLa cell monolayer. (b) HeLa cell monolayer treated with a sonicate from a cnf1+ isolate, 06-3320. Monolayers were stained with Diff-Quick and pictures were taken at the same magnification. Bars, 40 μm.
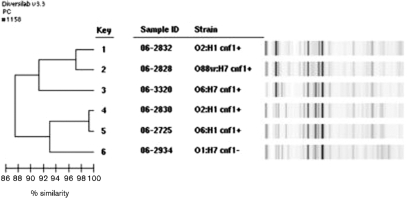
Genotyping by rep-PCR quantifies genetic similarity
Isolates 06-2828 and 06-2832 showed 97.2 % similarity by rep-PCR, whilst isolates 06-2725 and 06-2830 were indistinguishable by rep-PCR (Fig. 2). Isolates 06-2934 and 06-3320 were clearly distinguishable by rep-PCR and showed no evidence of a genetic relationship.
Fig. 2.
Analysis of the six E. coli macaque isolates using a DiversiLab Escherichia DNA Fingerprinting kit and DiversiLab software. Isolates 06-2828 and 06-2832 showed 97.2 % similarity and may represent the same clone. Isolates 06-2725 and 06-2830 were indistinguishable and probably represent a second clone. Isolates 06-2934 and 06-3320 were clearly distinguishable and showed no evidence of a genetic relationship.
DISCUSSION
To our knowledge, this is the first reported isolation of cnf1-positive E. coli in macaques. The cnf1-positive isolates had confirmed CNF1 activity in vitro, whilst the cnf1-negative isolate did not, showing that the cnf1 gene was functional. Some E. coli strains also produce CDTs, although CDT typically causes mononucleation in vitro in contrast to the multinucleation seen with CNF (Kadhum et al., 2008). Although our in vitro cell culture assays showed multinucleated giant cells typical of CNF, a cdt PCR was run to verify that these isolates were negative for the presence of cdt. The macaque isolates represent a heterogeneous population of CNF-1-producing E. coli, most likely due to the historical acceptance of animals positive for β-haemolytic E. coli into the research colony from quarantine. Our rep-PCR results suggest a common point source for several of the strains, as two of the rep-PCR patterns were represented by two isolates each. 06-2828 and 06-2832 were highly similar comparing rep-PCR results but had different antibiotic profiles and serotypes. 06-2725 and 06-2830 were indistinguishable by rep-PCR and had identical antibiotic susceptibility patterns, but differed in their serotype, having different O groups. Five of the six monkeys colonized with these E. coli strains came from the same vendor, were captive-bred of Chinese origin and entered our quarantine facility on the same day, although only four of these strains were cnf1+. One monkey was acquired the following year from a different vendor, was wild-caught of Chinese origin and the E. coli strain isolated was cnf1+. Their exact age was unknown but they were all young adult male rhesus macaques. They most likely entered our facility harbouring these E. coli strains, as these animals were housed in different holding rooms and were used by different investigators. Antibiotic susceptibility testing revealed that the E. coli isolates had several multidrug-resistance patterns. Despite this, a high percentage of isolates were susceptible to enrofloxacin. Enrofloxacin is a fluoroquinolone antibiotic commonly used to treat various diseases in non-human primates, for example as a prophylactic treatment for subclinical Shigella infections while in quarantine before shipment to institutions (Fox et al., 2001). The usage of antibiotics as prophylaxis or in the absence of culture and sensitivity results may have exacerbated the antibiotic resistance noted in these E. coli isolates. Indeed, Shigella flexneri strains isolated from monkeys housed in the Massachusetts Institute of Technology (MIT) colony have multiple antibiotic resistance, including enrofloxacin, which is plasmid-mediated (Martin et al., 2008). The method of acquisition of antibiotic resistance in the E. coli isolates is unknown and requires further study. Irrespective of the mechanism of antibiotic resistance in the E. coli strains isolated and characterized in this study, in the absence of clinical signs and disease, our recommendation is to avoid antibiotic treatment, thereby limiting the potential for promoting antibiotic resistance. If an animal is clinically compromised, antibiotic therapy is recommended based on culture and sensitivity results.
CNF1-producing E. coli was isolated previously from ferrets housed at MIT (Marini et al., 2004). Extra-intestinal E. coli-associated diseases are a clinically significant cause of morbidity and mortality in post-parturient jills and neonates. Similarly, these ferret E. coli isolates were all β-haemolytic, and α-haemolysin- and P fimbriae-positive, and were negative for other virulence factors tested. Another common feature between the ferret and macaque isolates was the somatic O-antigens, which were types 2 and 6. E. coli is a normal inhabitant of the gastrointestinal tract of macaques and several different serotypes of E. coli have been isolated from asymptomatic rhesus macaques (Schiff et al., 1972). There are reports of E. coli-associated diarrhoeal disease in young chimpanzees and an orang-utan, with some of the chimpanzees developing peracute disease and dying within hours (McClure et al., 1972). Tamarins also have mortality attributed to enteropathogenic E. coli infection (Potkay, 1992). Enteropathogenic E. coli was also reported as a common opportunistic pathogen causing diarrhoea and wasting in rhesus macaques infected with simian immunodeficiency virus (Mansfield et al., 2001). In a preliminary report, clinically inapparent E. coli infections in squirrel monkey infants were attributed as the cause of peracute deaths, although these organisms were non-motile and did not produce enterotoxin. Colitis and septic meningitis were seen histopathologically (Scimeca & Brady, 1990). Although gastrointestinal disease has been attributed to pathogenic E. coli infections in these non-human primate species, toxin production was either not found or not evaluated, and the serotypes identified differed from those of the six E. coli isolates described in our study. As NTEC has been isolated from human and animal populations, and these isolates can be highly related on a molecular basis, there is concern about potential zoonotic spread (Mainil et al., 1999). The presence of CNF1-producing E. coli strains in primates used in neurobiology emphasizes the need to use appropriate personnel protection and hygienic practices when handling these primates to minimize this zoonotic risk. In cases where zoonotic spread is suspected, molecular as well as epidemiological evidence are needed to substantiate animal-to-human transmission.
Acknowledgments
This study was supported by NIH grants T32 RR07036 and P30 ES02109. We would like to thank Dr Eric Oswald (Ecole Nationale Vétérinaire de Toulouse, France) for supplying a CDT-positive E. coli strain, and Dr James Versalovic's Laboratory at Baylor College of Medicine for performing the rep-PCR.
Abbreviations
CNF, cytotoxic necrotizing factor
CPE, cytopathic effect
NTEC, necrotoxigenic Escherichia coli
rep-PCR, repetitive element sequence-based PCR
References
- De Rycke, J., Milon, A. & Oswald, E. (1999). Necrotoxic Escherichia coli (NTEC): two emerging categories of human and animal pathogens. Vet Res 30, 221–233. [PubMed] [Google Scholar]
- Falbo, V., Pace, T., Picci, L., Pizzi, E. & Caprioli, A. (1993). Isolation and nucleotide sequence of the gene encoding cytotoxic necrotizing factor 1 of Escherichia coli. Infect Immun 61, 4909–4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Féria, C., Machado, J., Correia, J. D., Gonçalves, J. & Gaastra, W. (2001). Virulence genes and P fimbriae PapA subunit diversity in canine and feline uropathogenic Escherichia coli. Vet Microbiol 82, 81–89. [DOI] [PubMed] [Google Scholar]
- Fox, J. G., Handt, L., Xu, S., Shen, Z., Dewhirst, F. E., Paster, B. J., Dangler, C. A., Lodge, K., Motzel, S. & Klein, H. (2001). Novel Helicobacter species isolated from rhesus monkeys with chronic idiopathic colitis. J Med Microbiol 50, 421–429. [DOI] [PubMed] [Google Scholar]
- Johnson, J. R., Kaster, N., Kuskowski, M. A. & Ling, G. V. (2003). Identification of urovirulence traits in Escherichia coli by comparison of urinary and rectal E. coli isolates from dogs with urinary tract infection. J Clin Microbiol 41, 337–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadhum, H. J., Finlay, D., Rowe, M. T., Wilson, I. G. & Ball, H. J. (2008). Occurrence and characteristics of cytotoxic necrotizing factors, cytolethal distending toxins and other virulence factors in Escherichia coli from human blood and faecal samples. Epidemiol Infect 136, 752–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaper, J. B., Nataro, J. P. & Mobley, H. L. (2004). Pathogenic Escherichia coli. Nat Rev Microbiol 2, 123–140. [DOI] [PubMed] [Google Scholar]
- Mainil, J. G., Jacquemin, E., Pohl, P., Fairbrother, J. M., Ansuini, A., Le Bouguénec, C., Ball, H. J., De Rycke, J. & Oswald, E. (1999). Comparison of necrotoxigenic Escherichia coli isolates from farm animals and from humans. Vet Microbiol 70, 123–135. [DOI] [PubMed] [Google Scholar]
- Mansfield, K. G., Lin, K. C., Newman, J., Schauer, D., MacKey, J., Lackner, A. A. & Carville, A. (2001). Identification of enteropathogenic Escherichia coli in simian immunodeficiency virus-infected infant and adult rhesus macaques. J Clin Microbiol 39, 971–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marini, R. P., Taylor, N. S., Liang, A. Y., Knox, K. A., Peña, J. A., Schauer, D. B. & Fox, J. G. (2004). Characterization of hemolytic Escherichia coli strains in ferrets: recognition of candidate virulence factor CNF1. J Clin Microbiol 42, 5904–5908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, H. R., Shen, Z., Buckley, E. M. & Fox, J. G. (2008). Plasmid-mediated quinolone resistance in Shigella flexneri isolated from macaques (Macaca mulatta). In Abstracts of the 58th Annual AALAS National Meeting, Indianapolis, IN, USA, Abstract PS53, p. 13.
- McClure, H. M., Strozier, L. M. & Keeling, M. E. (1972). Enteropathogenic Escherichia coli infection in anthropoid apes. J Am Vet Med Assoc 161, 687–689. [PubMed] [Google Scholar]
- Orden, J. A., Domínguez-Bernal, G., Martínez-Pulgarín, S., Blanco, M., Blanco, J. E., Mora, A., Blanco, J. & de la Fuente, R. (2007). Necrotoxigenic Escherichia coli from sheep and goats produce a new type of cytotoxic necrotizing factor (CNF3) associated with the eae and ehxA genes. Int Microbiol 10, 47–55. [PubMed] [Google Scholar]
- Oswald, E., De Rycke, J., Guillot, J. F. & Boivin, R. (1989). Cytotoxic effect of multinucleation in HeLa cell cultures associated with the presence of Vir plasmid in Escherichia coli strains. FEMS Microbiol Lett 49, 95–99. [DOI] [PubMed] [Google Scholar]
- Potkay, S. (1992). Diseases of the Callitrichidae: a review. J Med Primatol 21, 189–236. [PubMed] [Google Scholar]
- Rodriguez-Siek, K. E., Giddings, C. W., Doetkott, C., Johnson, T. J., Fakhr, M. K. & Nolan, L. K. (2005). Comparison of Escherichia coli isolates implicated in human urinary tract infection and avian colibacillosis. Microbiology 151, 2097–2110. [DOI] [PubMed] [Google Scholar]
- Schiff, L. J., Barbera, P. W., Port, C. D., Yamashiroya, H. M., Shefner, A. M. & Poiley, S. M. (1972). Enteropathogenic Escherichia coli infections: increasing awareness of a problem in laboratory animals. Lab Anim Sci 22, 705–708. [PubMed] [Google Scholar]
- Scimeca, J. M. & Brady, A. G. (1990). Neonatal mortality in a captive breeding squirrel monkey colony associated with an invasive Escherichia coli. Lab Anim Sci 40, 546–547. [Google Scholar]
- Siqueira, A. K., Ribeiro, M. G., Leite, D. S., Tiba, M. R., Moura, C. D., Lopes, M. D., Prestes, N. C., Salerno, T. & Silva, A. V. (2009). Virulence factors in Escherichia coli strains isolated from urinary tract infection and pyometra cases and from feces of healthy dogs. Res Vet Sci 86, 206–210. [DOI] [PubMed] [Google Scholar]
- Starcic, M., Johnson, J. R., Stell, A. L., van der Goot, J., Hendriks, H. G., van Vorstenbosch, C., van Dijk, L. & Gaastra, W. (2002). Haemolytic Escherichia coli isolated from dogs with diarrhea have characteristics of both uropathogenic and necrotoxigenic strains. Vet Microbiol 85, 361–377. [DOI] [PubMed] [Google Scholar]
- Tibbetts, R. J., White, D. G., Dyer, N. W., Giddings, C. W. & Nolan, L. K. (2003). Characterization of Escherichia coli isolates incriminated in colisepticaemia in mink. Vet Res Commun 27, 341–357. [DOI] [PubMed] [Google Scholar]
- Tóth, I., Oswald, E., Mainil, J. G., Awad-Masalmeh, M. & Nagy, B. (2000). Characterization of intestinal cnf1+ Escherichia coli from weaned pigs. Int J Med Microbiol 290, 539–542. [DOI] [PubMed] [Google Scholar]
- Tóth, I., Hérault, F., Beutin, L. & Oswald, E. (2003). Production of cytolethal distending toxins by pathogenic Escherichia coli strains isolated from human and animal sources: establishment of the existence of a new cdt variant (type IV). J Clin Microbiol 41, 4285–4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versalovic, J., Schneider, M., de Bruijn, F. J. & Lupski, J. R. (1994). Genomic fingerprinting of bacteria using repetitive sequence-based polymerase chain reaction. Methods Mol Cell Biol 5, 25–40. [Google Scholar]
- Yuri, K., Nakata, K., Katae, H., Yamamoto, S. & Hasegawa, A. (1998). Distribution of uropathogenic virulence factors among Escherichia coli strains isolated from dogs and cats. J Vet Med Sci 60, 287–290. [DOI] [PubMed] [Google Scholar]