Abstract
Two patterns from large-scale DNA sequence data have been put forward as evidence that speciation between humans and chimpanzees was complex, involving hybridization and strong selection: divergence between humans and chimpanzees varies considerably across the autosomes; and divergence between humans and chimpanzees, but not gorilla, is markedly lower on the X chromosome. Here, we describe how simple speciation and neutral molecular evolution explain both patterns. In particular, the wide range in autosomal divergence is consistent with stochastic variation in coalescence times in the ancestral population; and the lower human--chimpanzee divergence on the X chromosome is consistent with species differences in the strength of male-biased mutation caused by differences in mating system. We also highlight two further patterns of divergence that are problematic for the complex speciation model. Our conclusions thus raise doubts about complex speciation between humans and chimpanzees.
Signatures of complex speciation between humans and chimpanzees
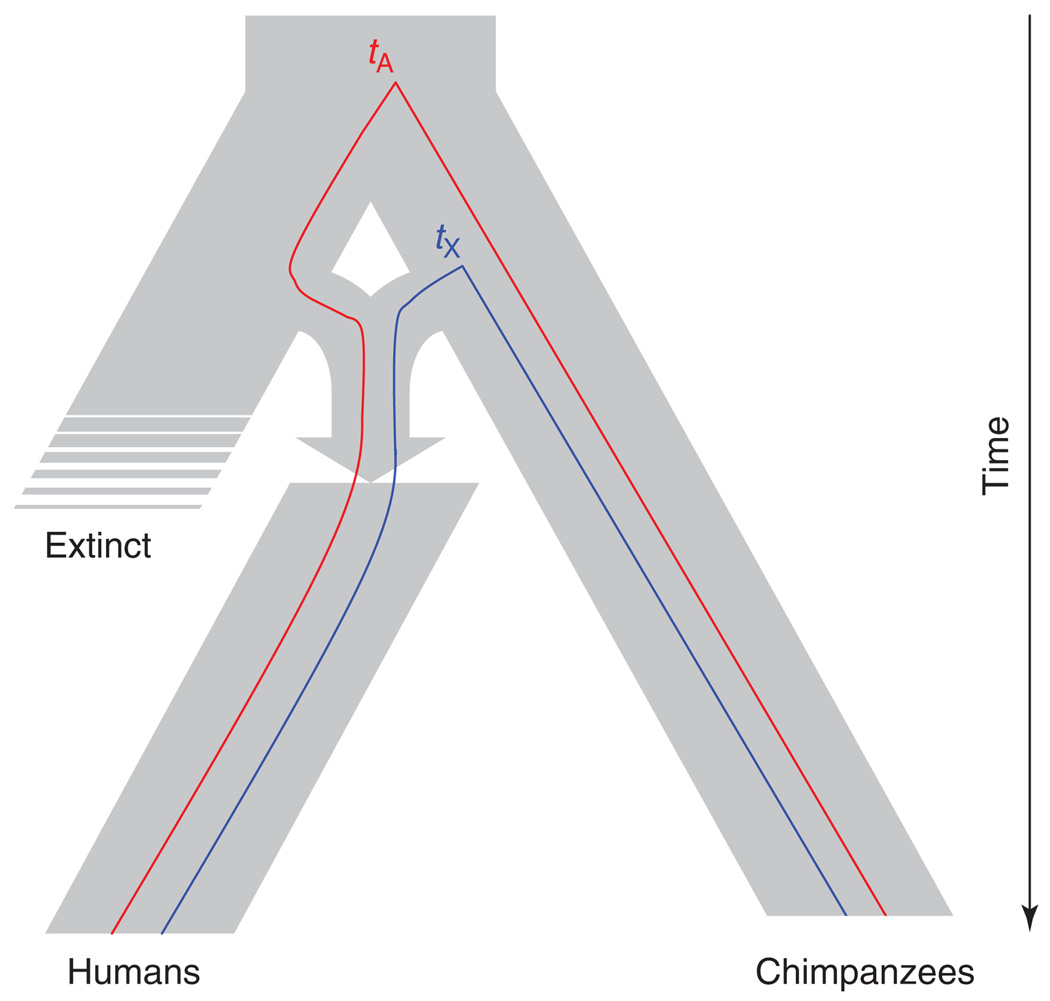
In 2006, Patterson et al. [1] made a surprising and provocative claim about speciation between humans and our closest living relatives, chimpanzees: one of the two species has hybrid origins. Two striking patterns emerged from their analyses of large-scale DNA sequence divergence data from four-species (HCGM, human–chimpanzee–gorilla–macaque) and five-species alignments (HCGOM, including orangutan). First, divergence between humans and chimpanzees varies widely across the autosomal genome. Second, divergence between humans and chimpanzees is markedly lower on the X chromosome than on the autosomes; there is no similar reduction of relative divergence on the X chromosome between humans and gorillas. After excluding several alternative hypotheses based on sex-biased mutation and demography, the authors concluded that these patterns provide genetic evidence for complex speciation during the split of the ancestral human and chimpanzee lineages [1]. In particular, after an initial period of separation and divergence, hominin and chimpanzee populations came into secondary contact and experienced a massive hybridization event, giving rise to a third population with mixed ancestry (Figure 1).
Figure 1.
Complex speciation between humans and chimpanzees. After an initial separation 5–7 Mya, the two lineages subsequently formed a third hybrid population. Strong selection against hybrid incompatibilities eliminated nearly the entire X chromosome from the hybrid population. In the case shown, the X chromosome from the ancestral chimpanzee lineage and autosomes mostly from a now extinct ancestral hominin lineage survived in the human descendants of the hybrid population. (The divergence data cannot distinguish if humans or chimpanzees descended from the hybrid population, but humans are arbitrarily shown as the descendants here, as in Ref. [1].) Under this scenario, the human-chimpanzee divergence time at autosomal loci is tA (red), whereas that for X chromosome loci is tX (blue).
The complex speciation scenario can explain the two patterns of divergence if the hybrid lineage and one of the two ancestral lineages survived, leading to extant humans and chimpanzees (Figure 1). First, under complex speciation, the wide range of autosomal divergence between humans and chimpanzees occurs because some loci coalesce in the hybrid population (recent divergence time) whereas others coalesce in the population of the common ancestors of the original separation event (older divergence time; Figure 1). Second, the low divergence on the X chromosome occurs because of the large X-effect [2–4]. Strong selection against hybrid sterility factors would disproportionately eliminate incompatible X-linked material from one of the ancestral lineages [1,5], leaving a hybrid lineage with autosomal contributions from both ancestors but X chromosome contributions from only one. If the X chromosome that survived in the hybrid population was contributed by the ancestor of the now extinct lineage, it would have an older divergence time; but if the X that survived was contributed by the ancestor of the extant lineage, it would have a younger divergence time [1,5]. The human–chimpanzee data are consistent with a younger divergence time on the X chromosome (Figure 1). The two genomic patterns of divergence could therefore be footprints of hybrid speciation for either chimpanzees or humans.
Here, we review why complex speciation is not necessary to explain the high variation in autosomal divergence between humans and chimpanzees. We then present a new explanation for why divergence is lower on the X chromosome than on the autosomes between humans and chimpanzees but not between humans and gorillas. Last, we note two other genomic patterns that are problematic for the complex speciation model. We conclude that all four genomic patterns of divergence between humans and chimpanzees are best explained by a simple splitting of lineages and neutral molecular evolutionary processes.
Explaining the wide range of autosomal divergence
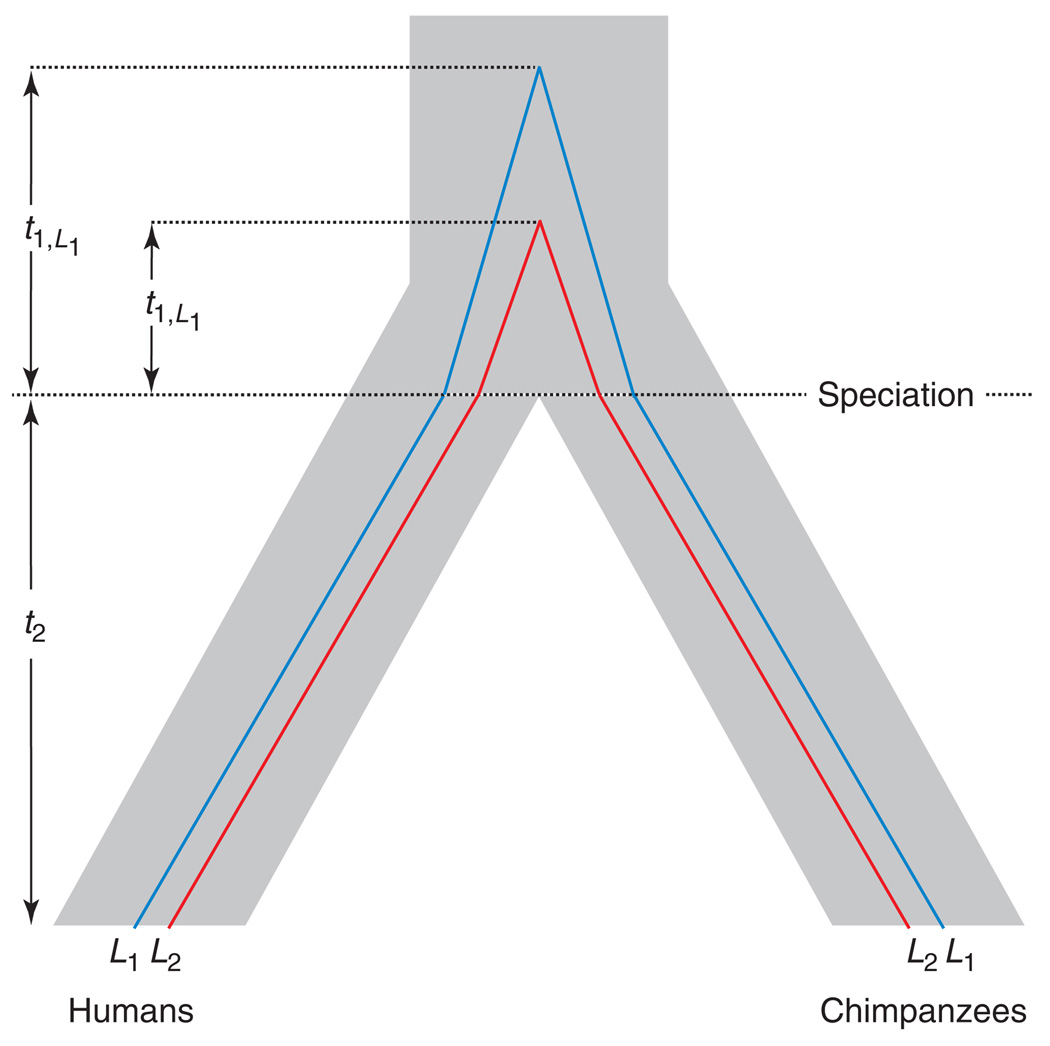
The number of differences between two DNA sequences depends on the mutation rate and the total time, going backwards into the past, until the two lineages coalesce in a common ancestor [6,7]. For two DNA sequences sampled from different species, the total time back to a common ancestor is the sum of two quantities: the split time between the species (t2) and the time to coalescence in the ancestral population (t1; Figure 2). Under simple speciation, all loci share the same species split time because there is no opportunity for two lineages separated by reproductive isolation to coalesce until they reach the common ancestral population (Figure 2). The distribution of neutral coalescence times in the ancestral population is exponential, with a mean of 2Na generations, where Na is the effective size of the ancestral population, and a variance of (2Na)2 generations [8,9]. As the histories of independent loci reflect different realizations of the genealogical process, the coalescence times among loci can vary enormously when Na is large. Therefore, to invoke complex speciation based on a wide range of divergence among loci requires statistical rejection of the simple null model (i.e. that the observed distribution of divergence times among loci can be explained by the variance in ancestral coalescence times). As noted by Barton [10] and Wakeley [11], this simple null model was not statistically tested by Patterson et al. [1].
Figure 2.
Divergence times vary among loci under simple speciation. The time separating two DNA sequences sampled from different species is the sum of the species split time (t2) and the time to coalescence in the ancestral population (t1). The divergence histories of two unlinked loci, L1 and L2, are shown. Both loci have the same species split time (t2) but different times to coalescence (t1,L1 and t1,L2, respectively), causing interlocus variation in divergence time.
Humans and chimpanzees differ at 1.23% of nucleotide sites genome-wide [12]. Because human–chimpanzee speciation occurred recently [5–7 million years ago (Mya)], a sizeable fraction of the observed differences between human and chimpanzee sequences accumulated during the coalescence history of the common ancestor: 0.17–0.55% accumulated during time t1 and 0.68–1.06% during time t2 [12–15]. Using a coalescent-based maximum likelihood approach, Innan and Watanabe [13] compared the fit of a simple speciation model to those of ones with differing levels of gene flow and found that the variance in autosomal divergence is best explained by a simple splitting of lineages with no subsequent gene flow (see also Refs [16,17]). There is now little dispute that this coalescent explanation accounts for the wide range of autosomal divergence between humans and chimpanzees. Indeed, setting autosomal divergence aside, Patterson et al. [5] now regard the reduced divergence on the X chromosome as the key observation supporting complex speciation: ‘Our argument for complex speciation rests on the difference in genetic divergence time that we observe between chromosome X and the autosomes, and not on the wide range of genetic divergence times observed within the autosomes (which can indeed be explained by a large ancestral population size). …To argue against the evidence for complex speciation, an alternative model is needed that explains the reduced chromosome X divergence in humans and chimpanzees with no similar reduction for humans and gorillas.’ Here, we present a simple alternative model.
Explaining X-autosome differences in divergence
The lower divergence on the X chromosome versus the autosomes could reflect its more recent species split time, its smaller ancestral population size, or its lower mutation rate. Whereas Patterson et al. [1] suggest that the X had a more recent split time resulting from its introgression between lineages (Figure 1), Burgess and Yang [16] and Hoboth et al.. [17] suggest that the X had a lower than expected effective population size in the human–chimpanzee ancestor. Rather than the expected NX/NA ≈ 0.75 (where NX and NA are the ancestral effective population sizes of the X and autosomes, respectively, and there are three X chromosomes for every four autosomes, assuming a population with an even sex ratio), they estimate that NX/NA = 0.51. Stronger, faster and more frequent selective sweeps on the X than on the autosomes might reduce X-linked polymorphism [18,19], but there are at least two difficulties with an ancestral sweeps model. First, the sweeps would have to be sufficient to reduce polymorphism across almost the entire X chromosome to explain its uniformly reduced divergence. Second, the ancestral sweeps model requires that sweeps reduced NX/NA in the human–chimpanzee ancestor more than in other ancestral populations [16] and more than in extant populations: NX/NA ≈ 1 in human populations in Africa [20] and NX/NA ≈ 0.76 in chimpanzees [21,22].
We believe that the reduced divergence on the X is best explained by its lower mutation rate, rather than by a more recent speciation time [1,5] or smaller ancestral population size [16,17]. In mammals, the number of germline stem cell divisions and, hence, the number of genome replications, is higher in males (which produce gametes continuously) than in females (which do not). The rate of replication-dependent errors (the source of most point mutations) is therefore higher in males than in females [23]. This male-biased mutation can cause the Y chromosome, the autosomes and the X chromosome to experience different mutational inputs. In each generation (assuming populations with equal sex ratios), male-biased mutation affects all Y-linked sequences (Y), half of autosomal sequences (A), and a third of X-linked sequences (X). Rates of neutral substitution therefore differ among chromosomes so that Y > A > X [24]. As male-driven evolution causes X to evolve slower than A, a reduced X/A ratio of divergence results. Indeed, ratios of substitution rates among the different chromosomes are commonly used to estimate α, the male:female ratio of mutation rates (Box 1).
Estimating α from DNA substitution data
Different male and female mutation rates (um and uf, respectively) cause neutral substitution rates to differ between Y-linked (Y), X-linked (X) and autosomal (A) sequences. (For simplicity, we consider only the case of male heterogametic X/Y chromosomal sex determination, but similar approaches can be used in the case of female heterogametic Z/W chromosomal sex determination.) The Y chromosome spends all of its time in males so that the neutral substitution rate for Y-linked sequences is Y = um. Similarly, assuming an equal sex ratio, the X chromosome spends one-third of its time in males and two-thirds in females, so that X = 1/3oum + 2/3ouf. Finally, autosomes spend half of their time in males and half in females, so that A = 1/2oum + 1/2ouf Taking ratios of neutral substitution rates on the sex chromosomes and the autosomes, we can estimate the underlying male:female ratio of mutation rates, α = um/uf. The X:autosome ratio of substitution rates is shown by:
Similarly, and . [24]. These can then be rearranged to estimate the male:female mutation rate from ratios of neutral divergence so that, for instance, .
These calculations do not account for ancestral polymorphism, which is important for estimating α with Y-linked sequences from closely related species for two reasons [32]. First, ancestral polymorphism makes a proportionally larger contribution to sequence differences between closely related species than between distantly related ones. Second, for the observed , where k is the post-speciation divergence in a single lineage and θ/2 is pre-speciation divergence in a single lineage, θY is typically smaller than θA, owing to the effects of recurrent hitchhiking [64], recurrent background selection [65] and the higher variance in male reproductive success [66] on the non-recombining Y chromosome. Because θY << θA, failing to correct for ancestral polymorphism in the Y/A case yields severe underestimates of Y/A and, hence, α. Failing to correct for ancestral polymorphism in the X/A case yields modest overestimates of α (Table 1, main text), but this bias tends to be weak as θX is typically only slightly smaller than, and often similar to, θA [20].
There is good evidence for male-driven molecular evolution from fish [25], birds [26] and mammals, including rodents, cats, perissodactyls, artiodactyls and primates [27,28]. The strength of male-biased mutation, however, appears to differ among taxa. In mammals, estimates of α range from ~2 in rodents [29,30] to ~4–7 in Old World primates [12,13,31–34]. These differences in α could mean that there are biologically meaningful differences in α among taxa or that there is one underlying ‘mammalian α ’ but its value is obscured by confounding factors. Noting that estimates of α from human genome repeats [35] and human–rat divergence are low (~1.9–2.1), whereas those from human–chimpanzee divergence are high (~4–7; Refs [12,34]), Patterson et al. [1] estimate α from human–macaque divergence and argue that ‘(t)he discrepancy in these estimates can be resolved if the low human-chimpanzee divergence on chromosome X reflects low divergence time. Correcting for this, we estimate that α ≈ 1.9, giving no evidence for an increase in α on the primate lineage.’
We suggest that there is no discrepancy that requires a lower divergence time (i.e. gene flow) for the X chromosome between humans and chimpanzees. Rather, the low estimates of α are inappropriate for the human–chimpanzee divergence, for two reasons. First, the difference between rodent and primate α is best explained by the generation-time effect. As males produce gametes continuously throughout adulthood, the number of germline cell divisions increases with paternal age; the number of female germline cell divisions is, by contrast, insensitive to age [36,37]. Species with longer generation times therefore experience greater discrepancies in the number of germline cell divisions between the sexes. Given the difference in generation times, we expect α to be higher in humans than in rats. Moreover, if α has evolved with generation time, then the human–rat α represents a long-term average over the deep branches connecting the two species (2 × 75 My [38]). Most of the divergence history between humanss and rat is undoubtedly characterized by short generation times and, hence, small α, as the long generation times of hominoids evolved recently. For the same reason, estimating α from human–macaque divergence might not accurately reflect α for recent human or chimpanzee molecular evolutionary history. Indeed, genome-scale data show that male-biased mutation is considerably lower in Old World monkeys than in hominoids [39–41].
Second, two notably low estimates of α from X and Y chromosome sequence data in humans [one based on interspersed repetitive DNAs (α ≈ 2.1; [35]) and another based on noncoding sequences (α ≈ 1.7; [42])] were previously shown to be problematic. Makova and Li [32] noted that the repeat-based analysis failed to correct for multiple substitutions and made the false assumption that repetitive elements in the same subfamily are the same age. They further showed that both estimates failed to account for ancestral polymorphism, which is essential in these cases as the effective population size (Ne) of non-recombining Y chromosomes differs qualitatively from other chromosomes: Y-linked sequences can have very small Ne and negligible sequence variation compared with X-linked or autosomal ones (Box 1). Correcting for these factors, virtually all recent estimates agree that α ≈ 4–7 in humans and chimpanzees [12,13,32,34].
We conclude that the difference in α between rodents and primates reflects a meaningful biological difference (the difference in generation times [31]) and that α has evolved. A high value of α in hominoids can explain the low X/A ratio of divergence between humans and chimpanzees [11]. However, if α ≈ 4–7, why is there no similarly low X/A ratio of divergence between humans and gorillas? We turn to this question next.
The strength of male-biased mutation evolves
We suggest that the strength of male-biased mutation can evolve to be lineage specific. The number of male germline cell divisions increases with paternal age at reproduction (i.e. the generation-time effect) [36,37] and, potentially, with the intensity of sperm competition [43]. Among hominoid species, generation times are comparable, with the possible exception of longer generation times in humans [31,44]. Hominoid mating systems, however, are markedly different. At one extreme, gorillas have a polygynous mating system (one male controls reproductive access to many females) and, at the other, chimpanzees have a promiscuous mating system (females often mate with as many as eight males during a periovulatory period). Thus, sperm competition is weak or absent in gorillas but intense in chimpanzees. Humans appear to be intermediate, having a largely monogamous mating system with weak sperm competition [45]. Not surprisingly, the different mating systems of gorillas, humans and chimpanzees have driven the evolution of pronounced differences in relative testis mass (relative to body mass) and the number of sperm per ejaculate: gorillas have low relative testis mass and produce few sperm per ejaculate; humans are intermediate; and chimpanzees have large relative testis mass and produce many sperm per ejaculate (Table 1 [46–48]).
Table 1.
Differences in the strength of male-biased mutation among hominoid lineages
| Human | Chimpanzee | Gorilla | Orangutan | |
|---|---|---|---|---|
| Body mass(kg)a | 65.7 | 44.3 | 169.0 | 74.64 |
| Testes mass(g)a | 40.5 | 118.8 | 29.6 | 35.3 |
| Relative testes massa | 0.616 | 2.682 | 0.175 | 0.473 |
| Sperm numbera | 175 × 106 | 603 × 106 | 65 × 106 | 91 × 106 |
| HCGM analyses | ||||
| X chromosome substitutionsb (747,260 bp) | 2008 | 1935 | 3138 | -- |
| Autosomal substitutionsb (17,552,410 bp) | 59175 | 59844 | 81671 | -- |
| kxc | 0.00269 | 0.00259 | 0.00420 | -- |
| kAc | 0.00337 | 0.00341 | 0.00465 | -- |
| X/A | 0.797 | 0.759 | 0.903 | -- |
| αd | 4.14 | 6.24 | 1.83 | -- |
| 95% C.I.e (lower,upper) | 3.03, 6.08 | 4.27, 10.56 | 1.49, 2.28 | -- |
| HCGOM analyses | ||||
| X chromosome substitutionsb (372,354 bp) | 936 | 944 | 1430 | 3086 |
| Autosomal substitutionsb (8,899,720 bp) | 28504 | 28495 | 38677 | 82670 |
| kx | 0.00251 | 0.00254 | 0.00384 | 0.00829 |
| kA | 0.00320 | 0.00320 | 0.00435 | 0.00929 |
| X/A | 0.785 | 0.792 | 0.884 | 0.892 |
| α | 4.71 | 4.39 | 2.08 | 1.96 |
| 95% C.I. (lower,upper) | 2.97, 9.16 | 2.80, 8.21 | 1.53, 2.93 | 1.59, 2.46 |
Data from four-species(HCGM) and five-species (HCGOM) alignments (see Table 1 of Ref. [1]); we excluded HG and CG changes as these probably reflect the sorting of ancestral polymorphisms rather than the fixation of lineage-specific mutations.
kx and kA are divergence (substitutions/site) on the X chromosome and the autosomes, respectively.
α calculated following Ref. [24], with no correction for ancestral polymorphism.
95% confidence intervals calculated by following Ref. [29]; bootstrap estimates (not shown) give nearly identical intervals.
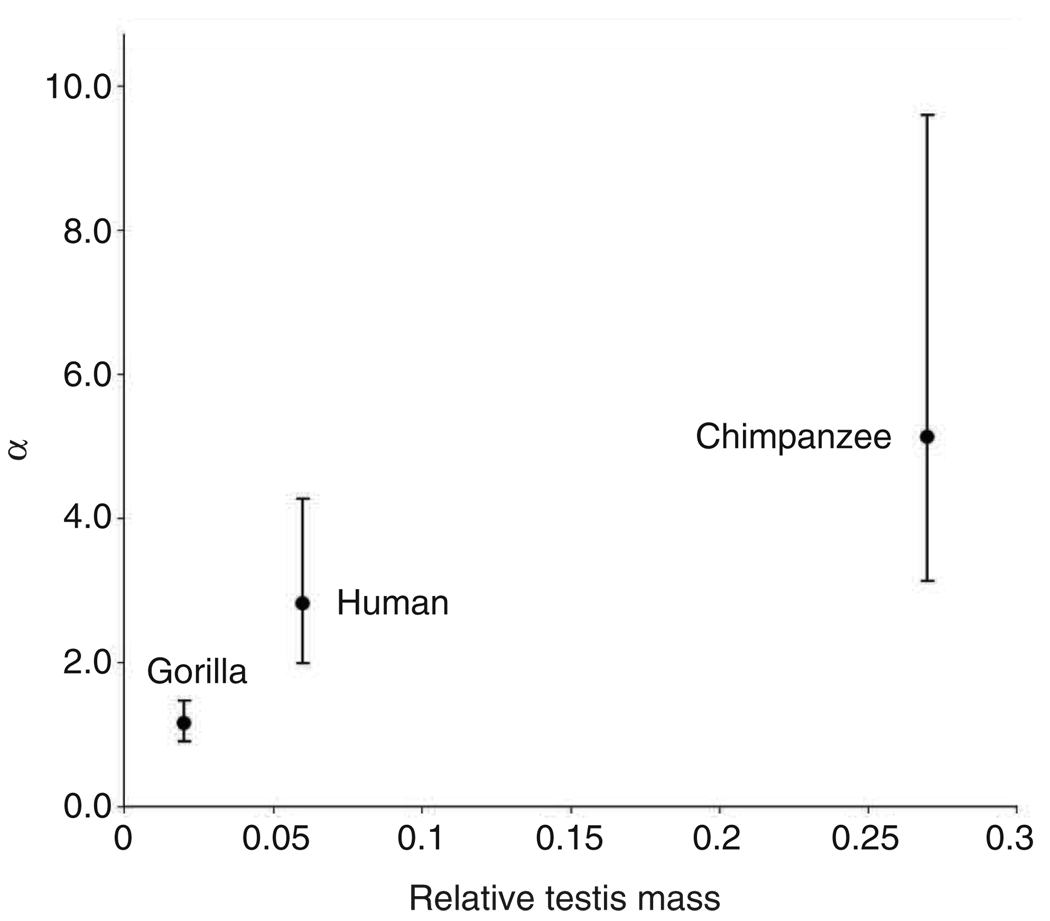
The increased demand on sperm production in promiscuous mating systems might have also driven the evolution of increased cell divisions in the male germline. If so, lineage-specific α should be lowest in gorillas, intermediate in humans, and highest in chimpanzees. To test this prediction, we estimated α using the X/A ratio of substitutions per site in the gorilla, human, and chimpanzee lineages separately using the data reported in Table 1 of Ref. [1]. We used the data from the four-species HCGM alignments spanning >0.74 Mb on the X and >17.55 Mb on the autosomes to estimate α in each lineage (Table 1; Box 1). The observed ratio of divergence is , where k = t2u (the divergence accumulated along a single lineage after speciation), and θ//2 = 2Nau (the divergence accumulated along a single lineage in the ancestral population before speciation; Box 1). We therefore corrected X/A for ancestral polymorphism by subtracting θX/2 and θA/2 from the numerator and denominator [12], respectively, and we assumed three possible ancestral diversities: chimpanzee-like diversity [21,22] and twofold and fourfold higher diversity than in human populations in Africa [49]. We used the corrected X/A ratios of divergence in each lineage to estimate lineage-specific α (Table 2 [21,22,32,34]). We found that α differs among the three lineages as predicted by the sperm competition hypothesis: α is lowest in gorillas (1.2–1.5, depending on the correction for ancestral polymorphism; Table 2), intermediate in humans (2.8–3.6), and highest in chimpanzees (5.1–5.8). Recent analyses of Y-linked sequence evolution further support higher rates of substitution in the chimpanzee lineage [50]. The correlation between α and the intensity of sperm competition (Figure 3) suggests that lineage-specific features of mating systems cause lineage-specific X/A ratios of substitution.
Table 2.
Estimates of α corrected for ancestral polymorphism
| Correction for ancestral polymorphism | Human | Chimpanzee | Gorilla | Orangutan |
|---|---|---|---|---|
| HCGM analyses | ||||
| Chimpanzee-like diversity, 1xa | 3.09 | 5.78 | 1.21 | -- |
| 2.17, 4.78d | 3.65, 11.52 | 0.95, 1.55 | -- | |
| Human-like diversity, 2xb | 3.56 | 5.79 | 1.51 | -- |
| 2.57, 5.31 | 3.86, 10.24 | 1.21, 1.90 | -- | |
| Human-like diversity, 4xc | 2.81 | 5.13 | 1.15 | -- |
| 1.99, 4.27 | 3.13, 9.59 | 0.90, 1.47 | -- | |
| HCGOM analyses | ||||
| Chimpanzee-like diversity, 1xa | 3.66 | 3.29 | 1.38 | 1.66 |
| 2.14, 8.06 | 1.96, 6.80 | 0.97, 2.02 | 1.33, 2.10 | |
| Human-like diversity, 2xb | 4.10 | 3.77 | 1.72 | 1.80 |
| 2.52,8.27 | 2.36, 7.27 | 1.24, 2.45 | 1.45, 2.26 | |
| Human-like diversity, 4xc | 3.28 | 2.96 | 1.31 | 1.63 |
| 1.94, 6.84 | 1.78, 5.86 | 0.91, 1.90 | 1.30, 2.06 | |
Assuming ancestral populations had chimpanzee-like diversity; estimates come from 10 kb on the X chromosome [22] and 22.4 kb on the autosomes [21]: θX = 0.0021 and θA = 0.0028, so that the X/A ration of diversity is 0.75. These estimates of diversity are relatively large as they include hypermutable CpG sites.
Assuming ancestral populations had 2x the level of human diversity in Africa [49]. Estimates based on non-CpG sites only for 31 kb on the X chromosome and 1.8 Mb on the autosomes [34, 49]. θx = 0.00053 and θA = 0.00071, so that the X/A ratio of diversity is 0.74.
Assuming ancestral populations had 4x the level of human diversity in Africa [49].
Lower, upper 95% confidence intervals [29].
Figure 3.
Correlation between the intensities of sperm competition in hominoid mating systems and lineage-specific strengths of male-biased mutation (α). For gorilla, human and chimpanzee lineages, α ± 95% confidence intervals from the HCGM analysis (four-fold human diversity correction for ancestral polymorphism; Table 2) are plotted against relative testis mass.
We also estimated α using data from the five-species HCGOM alignments spanning >0.37 Mb on the X and >8.89 Mb on the autosomes (Table 1). The HCGOM analysis has the advantage of including orangutans, which have a lower relative testis mass than do humans and little or no sperm competition (Table 1 [46,48]), but the drawback of estimating α from about half as much sequence data as the HCGM analysis. The HCGOM analysis nevertheless yields qualitatively similar results: α is relatively low in gorillas (1.3–1.7) and orangutans (1.6–1.8) and relatively high in humans (3.7–4.1) and chimpanzees (3.0–3.7; Table 2). The lack of separation between the αs of humans and chimpanzees in this analysis could reflect sampling error owing to the smaller dataset. Alternatively, the increased male germline divisions resulting from more sperm competition in chimpanzees might be offset by similar increases resulting from longer generation times in humans. In either case, the HCGM and HCGOM analyses show that the αs of humans and chimpanzees differ significantly from those of gorillas and orangutans (Table 2).
These results can explain the discrepancy between the X/A ratio of divergence for human–chimpanzee and that for human–gorilla: the low human–chimpanzee X/A ratio of divergence results from a higher average α during their 2 × ~5 My divergence history (α ≈ 2.8–5.8), whereas the higher human-gorilla X/A ratio of divergence results from a lower average α during their 2 × ~7 My divergence history (α ≈ 1.2–4.1). Put differently, the X chromosomes and autosomes of humans and chimpanzees do not have different divergence times [1]; they have different divergence rates. As α differs among branches, there is no need to invoke hybridization and strong selection against X-linked incompatibilities after the initial human–chimpanzee split. Instead, the data appear to be consistent with a simple splitting of lineages followed by the evolution of lineage-specific strengths of male-biased mutation.
Two problematic patterns for complex speciation
So far we have described how two genomic patterns taken as evidence for complex speciation are in fact consistent with simple speciation. We now consider two other genomic patterns that pose problems for complex, but not simple, speciation.
Different divergence patterns at CpG versus non-CpG sites
The first problem for complex speciation is the marked difference in the human-chimpanzee X/A ratio of divergence at CpG dinucleotide sites versus non-CpG sites [12,34]. Although mutation rates at CpG sites are higher than at non-CpG sites (uCpG > unon; [12]), the difference does not explain the discrepancy in X/A ratios of divergence: for CpG sites , and for non-CpG sites , where tX and tA are the divergence times on the X and autosomes, respectively (and where, for simplicity, we ignore ancestral polymorphism). Under complex speciation, the X chromosome has a more recent divergence time than do the autosomes (tX < tA), but the X/A ratios of divergence at CpG and non-CpG sites should be similarly reduced, so that XCpG/ACpG ≈ Xnon/Anon. Instead, human–chimpanzee X/A ratios of divergence at CpG (~0.84) and non-CpG (~0.76) sites differ significantly [34]. (CpG sites were excluded from the analyses in Ref. [1] as a precaution against possible multiple hits over the branches of the HCGM phylogeny.) It is difficult to imagine a historical scenario that would enable interspersed CpG and non-CpG sites on the X chromosome to have different tXs.
There is, however, a straightforward explanation under simple speciation with male-biased mutation. The cytosines of most CpG dinucleotides are methylated in vertebrates [51], and a higher CpG mutation rate results from a high rate of spontaneous deamination and repair of methylated cytosine, causing C → T (or G → A in the complementary strand) transitions [52,53]. As spontaneous deamination and repair is a clock time-dependent process that is largely decoupled from replication, CpG sites have predominantly clock time-dependent mutation rates, whereas non-CpG sites have predominantly replication-dependent mutation rates [54]. As a result, male-biased mutation will be weak or absent at CpG sites relative to non-CpG sites: for CpG sites , and for non-CpG sites , where unon,M and unon,F are the male and female mutation rates at non-CpG sites, respectively. Assuming no male-biased mutation at CpG sites, the CpG X/A ratio reflects relative divergence times, whereas the non-CpG X/A ratio reflects relative divergence times and the lower male mutational input on the X. Under simple speciation, tX/tA is the same for both CpG and non-CpG X/A ratios and the difference in ratios arises from the stronger male-biased mutation at non-CpG sites. Based on whole-genome data between humans and chimpanzees, αCpG ≈ 2–3, whereas αnon ≈ 6–7 [12,34]. A modest signature of male-biased is expected at CpG sites (αCpG > 1) because even though substitutions from CpG are largely clock time-dependent, substitutions to CpG and a small portion of substitutions from CpG are replication-dependent [34,54]. Thus, different mutational origins, and not different tXs, explain the different human-chimpanzee X/A ratios of divergence at CpG and non-CpG sites.
Genomic consequences of the large X-effect
The second problem for complex speciation is that human-chimpanzee divergence is reduced along most of the X chromosome (see Figure 3 of Ref. [1]). This surprisingly uniform pattern of lower divergence is difficult to explain under complex speciation, for two reasons. For one, the large X-effect predicts that the X should be the least likely chromosome to introgress between hybridizing species [10], as XY hybrids suffer especially low intrinsic fitness: not only do they experience the higher density of hybrid incompatibilities on the X [4,55,56], but, being hemizygous, they also experience the full effects of all recessive X-linked hybrid incompatibilities [57,58]. These two facts explain why, in most hybrid zones, the X (or Z) chromosome is distinguished by its general failure to introgress between species, leading to a relatively older, not younger, average divergence time [59,60]. For another, in the presence of recombination (assuming no fixed chromosomal inversion differences between species), it seems implausible that selection would eliminate most of the X chromosome (or even large, non-recombined ~50 Mbp segments) from one species (Figure 3b of Ref. [1]). Multi-locus studies of hybrid zones routinely find heterogeneous patterns of introgression along X (or Z) chromosomes: compatible regions introgress between species, but incompatible ones do not [61–63]. Thus, under complex speciation, we expect heterogeneous divergence times along the X between humans and chimpanzees. In particular, compatible regions should have a divergence time corresponding to the hybridization event and incompatible regions to the initial splitting of lineages (and the autosomes). The near uniformity of the reduced divergence on the X chromosome seems best explained by chromosome-wide mutational processes rather than introgression.
Conclusions
The original claim for complex speciation between humans and chimpanzees was based on the wide range of autosomal divergence and the lower than expected X/A ratio of divergence between humans and chimpanzees but not between humans and gorillas. Both signatures of complex speciation are consistent with a simple splitting of lineages and neutral substitution processes: the wide variance in autosomal divergence between humans and chimpanzees is consistent with a large variance in coalescence times in the ancestral population; and the reduced X/A ratio of divergence between humans and chimpanzees, but not humans and gorillas, is consistent with lineage-specific differences in the strength of male-biased mutation. Although these alternative explanations do not falsify the complex speciation hypothesis, two further observations raise additional difficulties: under complex speciation, the uniformity of the reduced divergence over most of the X chromosome is puzzling, and the disparity in the human–chimpanzee X/A ratio of divergence between CpG and non-CpG sites is especially difficult to explain.
Given the evidence outlined here, the notion that α has evolved with generation time and mating system seems more plausible than complex speciation between humans and chimpanzees. For now, support for the sperm competition hypothesis in primates rests on data from only a few lineages. Importantly, however, the sperm competition hypothesis predicted the rank order magnitude of ≥ among lineages a priori, with chimpanzee ≥ humans > gorillas, and, furthermore, has preliminary support in birds [43]. The sperm competition hypothesis thus warrants additional large-scale analyses to assess the separate effects of generation time and mating system on molecular evolution. Future support for complex speciation will require rejection of the null hypothesis that α has evolved among lineages.
Acknowledgements
We thank Danielle Jones, Colin Meiklejohn, Bret Payseur, three anonymous reviewers and the student participants of the University of Munich’s EES Summer School course on Sex Chromosome Evolution for comments and discussion. We are especially grateful to David Reich and Nick Patterson for sharing their thoughts on the manuscript. Eddie Loh provided computational support. D.C.P. is supported by funds from the David and Lucile Packard Foundation, the Radcliffe Institute for Advanced Study at Harvard University, the Alfred P. Sloan Foundation, the University of Rochester, and a grant from the National Institutes of Health (GM79543), and S.V.Y. is supported by funds from the Alfred P. Sloan Foundation, the National Science Foundation (BCS-0751508) and Georgia Tech.
Glossary
- Coalescence
the merging of two genealogical lineages, traced backwards in time, into a common ancestor.
- Complex speciation
the splitting of one species into two species in the face of hybridization and gene flow.
- Generation-time effect
the rate of DNA sequence evolution is generation-time dependent so that species with longer generation times tend to exhibit slower rates of molecular evolution.
- Hybrid zone
narrow geographic regions of overlap between species where hybridization occurs.
- Large X-effect
the disproportionately large effect of foreign X chromosomes on hybrid sterility resulting from both the hemizygous expression of recessive X-linked hybrid sterility factors in XY hybrids and the higher density of hybrid sterility factors on the X relative to the autosomes
- Reproductive isolation
the existence of intrinsic barriers to gene flow between species, including a reduced propensity for interspecific mating and reduced fitness of hybrid offspring.
- Sperm competition
in the reproductive tracts of females that have mated with more than one male, sperm from the different males compete for the opportunity to fertilize eggs.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Patterson N, et al. Genetic evidence for complex speciation of humans and chimpanzees. Nature. 2006;441:1103–1108. doi: 10.1038/nature04789. [DOI] [PubMed] [Google Scholar]
- 2.Coyne J. Genetics and speciation. Nature. 1992;355:511–515. doi: 10.1038/355511a0. [DOI] [PubMed] [Google Scholar]
- 3.Coyne JA, Orr HA. Two rules of speciation. In: Otte D, Endler J, editors. Speciation and Its Consequences. Sinauer Associates; 1989. pp. 180–207. [Google Scholar]
- 4.Presgraves DC. Sex chromosomes and speciation in Drosophila. Trends Genet. 2008;24:336–343. doi: 10.1016/j.tig.2008.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Patterson N, et al. Patterson et al. reply. Nature. 2008;452:E4. [Google Scholar]
- 6.Li WH. Distribution of nucleotide differences between two randomly chosen cistrons in a finite population. Genetics. 1977;85:331–337. doi: 10.1093/genetics/85.2.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takahata N. An attempt to estimate the effective size of the ancestral species common to two extant species from which homologous genes are sequenced. Genet. Res. 1986;48:187–190. doi: 10.1017/s001667230002499x. [DOI] [PubMed] [Google Scholar]
- 8.Hudson RR. Gene genealogies and the coalescent process. In: Futuyma D, Antonovics J, editors. Oxford Surveys In Evolutionary Biology. Oxford University Press; 1990. pp. 1–44. [Google Scholar]
- 9.Wakeley J. Coalescent Theory. Roberts & Company; 2008. [Google Scholar]
- 10.Barton NH. Evolutionary biology: how did the human species form? Curr. Biol. 2006;16:R647–R650. doi: 10.1016/j.cub.2006.07.032. [DOI] [PubMed] [Google Scholar]
- 11.Wakeley J. Complex speciation of humans and chimpanzees. Nature. 2008;452:E3–E4. doi: 10.1038/nature06805. [DOI] [PubMed] [Google Scholar]
- 12.Mikkelsen T, et al. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- 13.Innan H, Watanabe H. The effect of gene flow on the coalescent time in the human–chimpanzee ancestral population. Mol. Biol. Evol. 2006;23:1040–1047. doi: 10.1093/molbev/msj109. [DOI] [PubMed] [Google Scholar]
- 14.Takahasi K, Innan H. Encyclopedia of Life Sciences (ELS) John Wiley & Sons; 2008. Inferring the process of human-chimpanzee speciation. [Google Scholar]
- 15.Takahata N, et al. Divergence time and population size in the lineage leading to modern humans. Theor. Popul. Biol. 1995;48:198–221. doi: 10.1006/tpbi.1995.1026. [DOI] [PubMed] [Google Scholar]
- 16.Burgess R, Yang Z. Estimation of hominoid ancestral population sizes under Bayesian coalescent models incorporating mutation rate variation and sequencing errors. Mol. Biol. Evol. 2008;25:1979–1994. doi: 10.1093/molbev/msn148. [DOI] [PubMed] [Google Scholar]
- 17.Hobolth A, et al. Genomic relationships and speciation times of human, chimpanzee, and gorilla inferred from a coalescent hidden Markov model. PLoS Genet. 2007;3:294–304. doi: 10.1371/journal.pgen.0030007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aquadro CF, et al. Selection, recombination, and DNA polymorphism in Drosophila. In: Golding B, editor. Non-neutral Evolution: Theories and Molecular Data. Chapman & Hall; 1994. pp. 46–56. [Google Scholar]
- 19.Betancourt A, et al. A pseudohitchhiking model of X vs. autosomal diversity. Genetics. 2004;168:2261–2269. doi: 10.1534/genetics.104.030999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hammer MF, et al. Sex-biased evolutionary forces shape genomic patterns of human diversity. PLoS Genet. 2008;4:e1000202. doi: 10.1371/journal.pgen.1000202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fischer A, et al. Demographic history and genetic differentiation in apes. Curr. Biol. 2006;16:1133–1138. doi: 10.1016/j.cub.2006.04.033. [DOI] [PubMed] [Google Scholar]
- 22.Kaessmann H, et al. Great ape DNA sequences reveal a reduced diversity and an expansion in humans. Nat. Genet. 2001;27:155–156. doi: 10.1038/84773. [DOI] [PubMed] [Google Scholar]
- 23.Haldane J. The mutation rate of the gene for haemophilia, and its segregation ratios in males and females. Ann. Eugen. 1947;13:262–271. doi: 10.1111/j.1469-1809.1946.tb02367.x. [DOI] [PubMed] [Google Scholar]
- 24.Miyata T, et al. Male-driven molecular evolution: a model and nucleotide sequence analysis. Cold Spring Harb. Symp. Quant. Biol. 1987;52:863–867. doi: 10.1101/sqb.1987.052.01.094. [DOI] [PubMed] [Google Scholar]
- 25.Ellegren H, Fridolfsson A-K. Sex-specific mutation rates in salmonid fish. J. Mol. Evol. 2003;56:458–463. doi: 10.1007/s00239-002-2416-z. [DOI] [PubMed] [Google Scholar]
- 26.Ellegren H, Fridolfsson A-K. Male-driven evolution of DNA sequences in birds. Nat. Genet. 1997;17:182–184. doi: 10.1038/ng1097-182. [DOI] [PubMed] [Google Scholar]
- 27.Goetting-Minesky MP, Makova KD. Mammalian male mutation bias: Impacts of generation time and regional variation in substitution rates. J. Mol. Evol. 2006;63:537–544. doi: 10.1007/s00239-005-0308-8. [DOI] [PubMed] [Google Scholar]
- 28.Li WH, et al. Male-driven evolution. Curr.Opin. Genet. Dev. 2002;12:650–666. doi: 10.1016/s0959-437x(02)00354-4. [DOI] [PubMed] [Google Scholar]
- 29.Chang BHJ, et al. Weak male-driven molecular evolution in rodents. Proc. Natl. Acad. Sci. U. S. A. 1994;91:827–831. doi: 10.1073/pnas.91.2.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Makova KD, et al. Insertions and deletions are male biased too: a whole-genome analysis in rodents. Genome Res. 2004;14:567–573. doi: 10.1101/gr.1971104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li WH, et al. Rates of nucleotide substitution in primates and rodents and the generation-time effect hypothesis. Mol. Phylogenet. Evol. 1996;5:182–187. doi: 10.1006/mpev.1996.0012. [DOI] [PubMed] [Google Scholar]
- 32.Makova KD, Li WH. Strong male-driven evolution of DNA sequences in humans and apes. Nature. 2002;416:624–626. doi: 10.1038/416624a. [DOI] [PubMed] [Google Scholar]
- 33.Shimmin LC, et al. Male-driven evolution of DNA sequences. Nature. 1993;362:745–747. doi: 10.1038/362745a0. [DOI] [PubMed] [Google Scholar]
- 34.Taylor J, et al. Strong and weak male mutation bias at different sites in the primate genomes: insights from the human–chimpanzee comparison. Mol. Biol. Evol. 2006;23:565–573. doi: 10.1093/molbev/msj060. [DOI] [PubMed] [Google Scholar]
- 35.Lander E, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 36.Crow JF. The origins, patterns and implications of human spontaneous mutation. Nat. Rev. Genet. 2000;1:40–47. doi: 10.1038/35049558. [DOI] [PubMed] [Google Scholar]
- 37.Hurst LD, Ellegren H. Sex biases in the mutation rate. Trends Genet. 1998;14:446–452. doi: 10.1016/s0168-9525(98)01577-7. [DOI] [PubMed] [Google Scholar]
- 38.Waterston R, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 39.Elango N, et al. Mutations of different molecular origins exhibit contrasting patterns of regional substitution rate variation. PLOS Comput. Biol. 2008;4:e1000015. doi: 10.1371/journal.pcbi.1000015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gibbs R, et al. Evolutionary and biomedical insights from the rhesus macaque genome. Science. 2007;316:222–234. doi: 10.1126/science.1139247. [DOI] [PubMed] [Google Scholar]
- 41.Elango N, et al. Evolutionary rate variation in Old World monkeys. Biol. Lett. doi: 10.1098/rsbl.2008.0712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bohossian HB, et al. Unexpectedly similar rates of nucleotide substitution found in male and female hominids. Nature. 2000;406:622–625. doi: 10.1038/35020557. [DOI] [PubMed] [Google Scholar]
- 43.Bartosch-Harlid A, et al. Life history and the male mutation bias. Evolution Int. J. Org. Evolution. 2003;57:2398–2406. doi: 10.1554/03-036. [DOI] [PubMed] [Google Scholar]
- 44.Ruvolo M. Molecular phylogeny of the hominoids: Inferences from multiple independent DNA sequence data sets. Mol. Biol. Evol. 1997;14:248–265. doi: 10.1093/oxfordjournals.molbev.a025761. [DOI] [PubMed] [Google Scholar]
- 45.Martin R. The evolution of human reproduction: A primatological perspective. Am. J. Phys. Anthropol. 2007;45 Suppl.:59–84. doi: 10.1002/ajpa.20734. [DOI] [PubMed] [Google Scholar]
- 46.Harcourt AH, et al. Testis weight, body weight and breeding system in primates. Nature. 1981;293:55–57. doi: 10.1038/293055a0. [DOI] [PubMed] [Google Scholar]
- 47.Smith R. Human sperm competition. In: Smith R, editor. Sperm Competition and the Evolution of Animal Mating Systems. Academic Press; 1984. pp. 601–659. [Google Scholar]
- 48.Dixson A, Anderson M. Sexual selection and the comparative anatomy of reproduction in monkeys, apes, and human beings. Annu. Rev. Sex Res. 2001;12:121–144. [PubMed] [Google Scholar]
- 49.Ebersberger I, et al. Genome-wide comparison of DNA sequences between humans and chimpanzees. Am. J. Hum. Genet. 2002;70:1490–1497. doi: 10.1086/340787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goto H, et al. Evolution of X-degenerate Y chromosome genes in greater apes: conservation of gene content in human and gorilla, but not chimpanzee. J. Mol. Evol. 2009;68:134–144. doi: 10.1007/s00239-008-9189-y. [DOI] [PubMed] [Google Scholar]
- 51.Bird A. DNA methylation and the frequency of CpG in animal DNA. Nucleic Acids Res. 1980;8:1499–1504. doi: 10.1093/nar/8.7.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Duncan BK, Miller JH. Mutagenic deamination of cytosine residues in DNA. Nature. 1980;287:560–561. doi: 10.1038/287560a0. [DOI] [PubMed] [Google Scholar]
- 53.Ehrlich M, Wang RY. 5-Methylcytosine in eukaryotic DNA. Science. 1980;212:1350–1357. doi: 10.1126/science.6262918. [DOI] [PubMed] [Google Scholar]
- 54.Kim S-H, et al. Heterogeneous genomic molecular clocks in primates. PLoS Genet. 2006;2:1527–1534. doi: 10.1371/journal.pgen.0020163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Masly JP, Presgraves DC. High-resolution genome-wide dissection of the two rules of speciation in Drosophila. PLoS Biol. 2007;5:1890–1898. doi: 10.1371/journal.pbio.0050243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tao Y, et al. Genetic dissection of hybrid incompatibilities between Drosophila simulans and D. mauritiana. I. Differential accumulation of hybrid male sterility effects on the X and autosomes. Genetics. 2003;164:1383–1397. doi: 10.1093/genetics/164.4.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Turelli M, Orr HA. The dominance theory of Haldane’s rule. Genetics. 1995;140:389–402. doi: 10.1093/genetics/140.1.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Turelli M, Orr HA. Dominance, epistasis and the genetics of postzygotic isolation. Genetics. 2000;154:1663–1679. doi: 10.1093/genetics/154.4.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tucker PK, et al. Abrupt cline for sex chromosomes in a hybrid zone between two species of mice. Evolution Int. J. Org. Evolution. 1992;46:1146–1163. doi: 10.1111/j.1558-5646.1992.tb00625.x. [DOI] [PubMed] [Google Scholar]
- 60.Saetre G-P, et al. Sex chromosome evolution and speciation in Ficedula flycatchers. Proc. R. Soc. Lond. B. Biol. Sci. 2002;270:53–59. doi: 10.1098/rspb.2002.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Geraldes A, et al. Contrasting patterns of introgression at X-linked loci across the hybrid zone between subspecies of the European rabbit (Oryctolagus cuniculus) Genetics. 2006;173:919–933. doi: 10.1534/genetics.105.054106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Payseur BA, et al. Differential patterns of introgression across the X chromosome in a hybrid zone between two species of house mice. Evolution Int. J. Org. Evolution. 2004;58:2064–2078. doi: 10.1111/j.0014-3820.2004.tb00490.x. [DOI] [PubMed] [Google Scholar]
- 63.Putnam AS, et al. Discordand divergence times among Z-chromosome regions between two ecologically distinct swallowtail butterfly species. Evolution Int. J. Org. Evolution. 2007;61:912–927. doi: 10.1111/j.1558-5646.2007.00076.x. [DOI] [PubMed] [Google Scholar]
- 64.Maynard Smith J, Haigh J. The hitch-hiking effect of a favourable gene. Genet. Res. 1974;23:23–35. [PubMed] [Google Scholar]
- 65.Charlesworth B, et al. The effect of deleterious mutations on neutral molecular variation. Genetics. 1993;134:1289–1303. doi: 10.1093/genetics/134.4.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Charlesworth B. The effect of life-history and mode of inheritance on neutral genetic variability. Genet. Res. 2001;77:153–166. doi: 10.1017/s0016672301004979. [DOI] [PubMed] [Google Scholar]