Abstract
The ability to examine real time reaction kinetics for multimeric enzymes in their native state may offer unique insights toward understanding the catalytic mechanism and its interplay with three-dimensional structure. In this study, we have utilized a time-resolved electrospray mass spectrometry approach to probe the kinetic mechanism of 4-hydroxybenzoyl-coenzyme A (4-HBA-CoA) thioesterase from Arthrobacter sp. Strain SU in a millisecond time domain. Intact tetrameric complexes of 4-HBA-CoA thioesterase with up to four natural substrate (4-HBA-CoA) molecules bound were detected at times as early as 6 milliseconds using an online rapid-mixing device directly coupled to an electrospray ionization time-of-flight mass spectrometer. Species corresponding to the formation of a folded tetramer of the thioesterase at charge states 16+, 17+, 18+ and 19+, around m/z 3800, were observed and assigned as individual tetramers of thioesterase and noncovalent complexes of the tetramers with up to four substrate and /or product molecules. Real time evaluation of the reaction kinetics was accomplished by monitoring change in peak intensity corresponding to the substrate and product complexes of the tetrameric protein. The mass spectral data suggest that product 4-HBA is released from the active site of the enzyme prior to the release of product CoA following catalytic turnover. This study demonstrates the utility of this technique to provide additional molecular details for an understanding of the individual enzyme states during the thioesterase catalysis and ability to observe real-time interactions between enzyme and substrates and /or products in millisecond time range.
Keywords: time-resolved electrospray ionization mass spectrometry, thioesterase, non-covalent enzyme ligand complex, mechanistic enzymology
Mass spectrometry has proven to be a powerful tool for the detection and study of protein-protein and protein-ligand complexes and enzyme reaction kinetics (1, 2). A common approach for characterizing the reaction kinetics for enzyme reaction pathways involves determining pre-steady-state and steady-state kinetic constants using spectrophotometric techniques (fluorescence and UV) and rapid-mixing methodologies such as stopped-flow and rapid chemical quench. However, these techniques measure the average properties of biomacromolecules or small molecules such as substrates and products. Electrospray ionization mass spectrometry (ESI-MS) is an increasingly important methodology that has grown in recent years to become a well-accepted and complementary technique in structural biology. Unlike spectrophotometric techniques, ESI-MS can provide detailed information about the enzymatic mechanism and reaction kinetics at the level of the quaternary protein structure (3–8). This technique is well suited for the detection of non-covalent protein complexes (5, 9–19) and their interactions with DNA (20), RNA (21) and ligands (22).
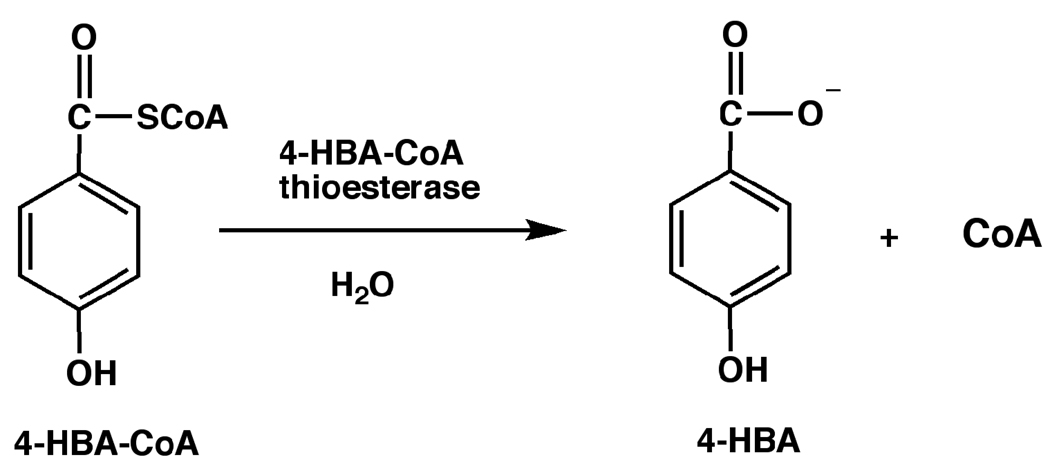
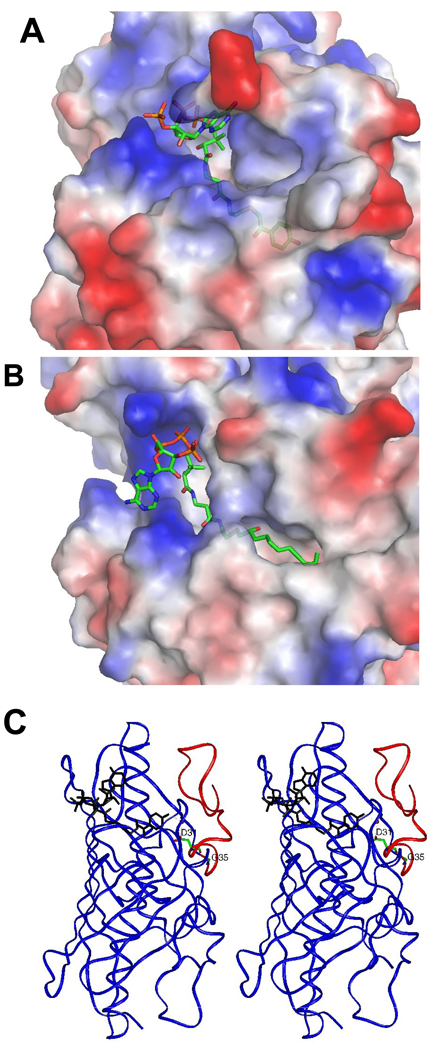
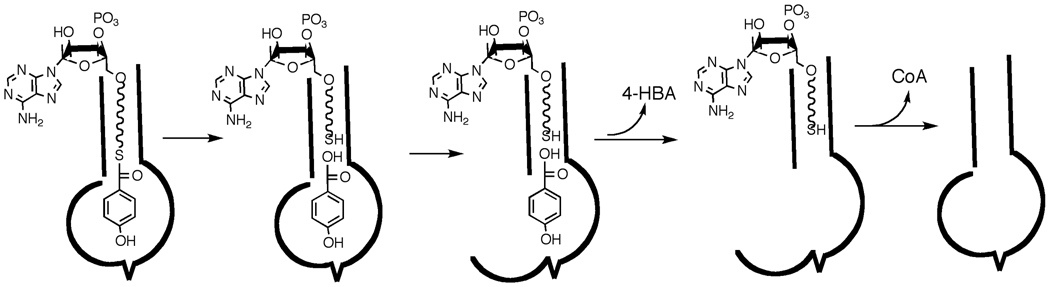
In the present investigation of the 4-hydroxybenzoyl-coenzyme A (4-HBA-CoA) thioesterase from Arthrobacter sp. strain SU, we apply quantitative time-resolved electrospray ionization mass spectrometry with time of flight detection (ESI-TOF) in the millisecond time range to monitor substrate and product occupancy of the four active sites of the native tetramer during the course of a single turnover reaction. 4-HBA-CoA thioesterase catalyzes the hydrolysis of 4-HBA-CoA to 4-HBA and CoA (Scheme 1) as the final step of the bacterial 4-chlorobenzoate degradation pathway (23). The structure of the ligated enzyme defined the location of the four substrate binding sites on the native tetramer (24). The binding site extends from a cupped shaped depression at the protein surface that binds the nucleotide moiety through a narrow tunnel that binds the pathetheine arm into an enclosed pocket that binds the hydroxybenzoyl group (Figure 1A). Structures of 4-HBA-CoA thioesterase homologs that hydrolyze long chain fatty acyl-CoA thioesters differ in that the acyl binding pocket is open to solvent in order to accommodate the acyl group (Figure 1B) (25, 26). Whereas the substrate specificity of the long chain fatty acyl-CoA thioesterases is low (23, 26), the substrate range of 4-HBA-CoA thioesterase is limited and does not include fatty acyl-CoAs (27). The key structural unit unique to the 4-HBA-CoA thioesterase is a highly extended loop-like N-terminus that forms the back wall of the hydroxybenzoyl binding pocket (Figure 1C). If the loop is fixed in position, then product release must be ordered with CoA departure required to open the tunnel through which the 4-HBA can diffuse to solvent. On the other hand, if the N-terminus segment is flexible and acts simply as a flap, the 4-HBA might be able to diffuse “out the back way” when the flap is open. In this case, 4-HBA release need not follow CoA release.
Scheme 1.
The reaction catalyzed by Arthrobacter thioesterase.
Figure 1.
(A) The Grasp generated representation of the steric and electrostatic topological features of the 4-HBA-CoA binding site of Arthrobacter 4-HBA-CoA thioesterase bound with 4-hydroxyphenacyl-CoA (PDB code 1Q4T). The ligand carbon atoms are green, oxygen atoms red, nitrogen atoms blue and phosphorus atoms orange.
(B) The Grasp generated representation of the steric and electrostatic topological features of the long chain fatty acyl-CoA binding site of hHTEM2 bound with undecan-2-one-CoA (PDB code 35FO) (same coloring scheme as in (A)). (C) Stereorepresentation of the backbone trace (blue) of the Arthrobacter 4-HBA-CoA thioesterase dimer unit (PDB code 1Q4T). One of the two active sites is identified by the 4-hydroxyphenacyl-CoA ligand shown in black along with the N-terminal loop shown in red.
The present work was carried out to determine if time-resolved ESI-MS TOF could be used to determine the timing release of release of CoA and 4-HBA-CoA from 4-HBA-CoA thioesterase active site following catalytic turnover of substrate. In addition, it might be feasible to examine the multimeric state of the enzyme concurrent with catalysis. For this purpose we used an online rapid-mixing device directly coupled to an electrospray ionization time-of-flight mass spectrometer to monitor the substrate and product ligation state of the tetramer during the single turnover reaction (4, 5). Herein, we present our experimental findings that demonstrate that product release is indeed ordered, but that 4-HBA rather than CoA is the first to dissociate. This result suggests that 4-HBA dissociates directly from the hydroxybenzoyl binding pocket more quickly than CoA can move out of the connecting tunnel. Moreover, we have defined the impact of the multimeric state of the enzyme in various states of ligand occupancy upon catalytic turnover.
Materials and methods
4-HBA-CoA thioesterase preparation
Arthrobacter sp. strain SU 4-HBA-CoA thioesterase was purified as previously described (23). The enzyme solution for analysis was prepared by overnight dialysis against 2 × 4 L 10 mM ammonium acetate (pH 7.5) at 4°C. The concentration of the enzyme subunits was determined using an extinction coefficient of 23.4 mM−1cm−1 at 280 nm. The concentrated thioesterase was diluted to 80 µM with 10 mM ammonium acetate (pH 7.5) prior to the mass spectrometric experiments.
4-HBA-CoA preparation
4-HBA-CoA was prepared according to the published procedure (29). It was dissolved in 10 mM ammonium acetate (pH 7.5) and then metal ions such as Li, Na and K in the solution of 4-HBA-CoA were removed using IC-H Maxi-Clean Cartridge (Part # 30256, Alltech Corp.). The concentration of 4-HBA-CoA was determined using an extinction coefficient of 11.8 mM−1cm−1 at 300 nm. The concentrated substrate was separately diluted to 10, 20, 30, 40, 50 and 60 µM with 10 mM ammonium acetate (pH 7.5) prior to the mass spectrometric experiments.
Electrospray ionization time-of-flight mass spectrometry (ESI-ToF MS)
ESI mass spectral experiments were carried out by two electrospray ionization time-of-flight mass spectrometers (Agilent Technologies, Inc., and Analytica of Branford, Inc., USA). Each mass spectrum represents the average of 10–15 full scans recorded in the positive ion mode under the following conditions: drying gas temperature and flow-rate: 180 °C and 15L/min, respectively; nebulized gas flow-rate, 10 L/min; the scan time for one spectrum, 1s; resolution at m/z 1500, 7000 FWHM. Mass spectra are without smoothing and background subtraction. The deconvoluted mass spectra were obtained by BioAnalyst1.15 (Applied Biosystems). The method is based on a novel rapid-mixing setup as previously described (4, 5). Two syringes were advanced simultaneously by a syringe pump (model PHD 2000 infusion, Harvard Apparatus, South Natick, MA) at several different total flow rates of 260, 240, 200, 160, 80, 40, 20 and 10 µL/min. Syringe 1 (1 mL volume) contained the solution of 80 µM thioesterase in 10 mM ammonium acetate (pH 7.5); syringe 2 (1 mL volume) contained 4-HBA-CoA in 10 mM ammonium acetate (pH 7.5). Both syringes were connected to a ‘reaction’ fused silica capillary (TSP030150, i.d. of 30 ± 3 µm, Polymicro Technologies, Phoenix, AZ, USA) by a zero dead volume-mixing tee (MY1XCS6, Valco Instrument Co. Inc., Houston, TX, USA). The reaction time is calculated according to the total flow rate for the same length of the ‘reaction’ capillary between the mixing point and other end of the fused silica capillary located in electrospray source. The length of the ‘reaction’ fused silica capillary and flow rates allowed reaction time points 6, 7, 8, 10, 20, 40, 80 and 160 ms, corresponding to the total flow rates 260, 240, 200, 160, 80, 40, 20 and 10 µL/min, respectively. The length for electrospray ionization probe is 3.7 cm. Our previous experiments have established that these flow rates and reaction conditions can provide adequate mixing of reactants to collect reaction times as short as 6 milliseconds and longer times greater than 30 seconds (4,5).
Results
Examination of the native conformation of Arthrobacter thioesterase using ESI-TOF MS in the absence of ligands
As a first step in exploring enzyme catalysis in the multimeric state of the protein, the quaternary state of Arthrobacter thioesterase was examined by ESI-TOF MS using a buffer both suitable for electrospray ionization and compatible with the thioesterase native structure (see Methods). The native tetramer (theoretical mass Mr: 65,577.64 Da) was observed from m/z 3400 to 4200 with charge states 16+, 17+, 18+ and 19+ (see Fig.2). Mass spectral data were measured as a function of flow rate (10 µl/min to 260 µl/min) and protein concentration. The flow rates employed allow sampling times from 6 milliseconds to multiple seconds which is an ideal time frame for monitoring enzyme catalysis (4, 5). The charge state distribution of the tetramer was not affected by the flow rate or the protein concentration (Figure S1 and Figure S2, see the supplementary material).
Figure 2.
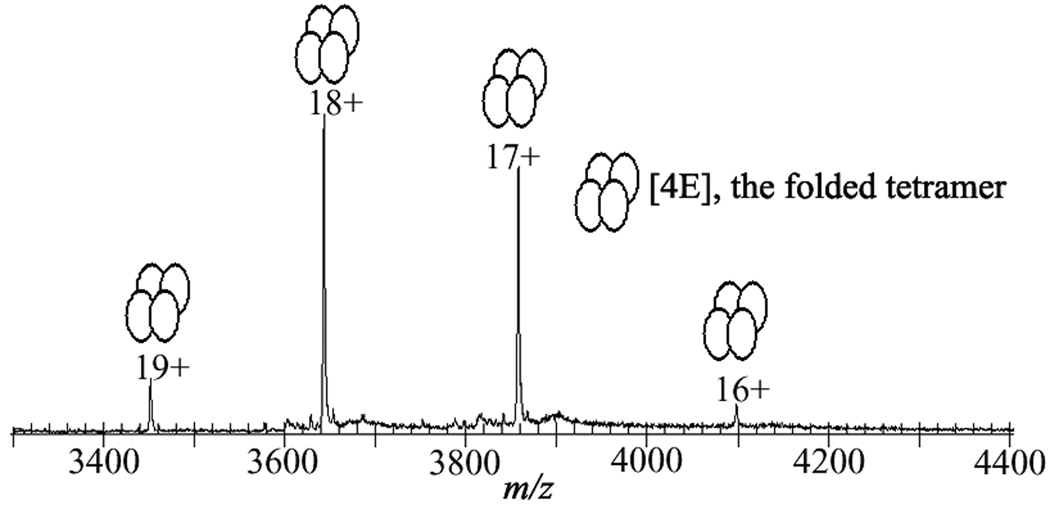
Observation of the native conformation of Arthrobacter 4-hydroxybenzoyl-Coenzyme A thioesterase as a tetramer. ESI-TOF mass spectrum of the thioesterase with a solution of 40 µM concentration in 10 mM ammonium acetate buffer, pH 7.5.
The observation of the tetrameric state of the enzyme by ESI-TOF MS is in accord with the X-ray structure and additionally shows that the tetramer is the predominant form of the enzyme in solution (24). Together this set of initial results established the feasibility of using electrospray mass spectrometry to directly monitor the native enzyme in the gas phase, and in a time dependent fashion.
Detection of multimeric, non-covalent substrate and product complexes of Arthrobacter thioesterase in a substrate titration experiment
Having established that the native tetramer can be readily observed by ESI-TOF MS, the next step was to demonstrate that non-covalent complexes of protein with substrate and/or product ligands could be detected. Accordingly, the native tetramer bound with substrate (4-HBA-CoA) and/or products (4-HBA and CoA) can be monitored in a substrate titration experiment. Because the binding of one molecule of 4-HBA-CoA, CoA, or 4-HBA increases the mass of the enzyme (67,577.94) by 887.62, 767.53, or 138.12 Da, respectively, the peaks derived from the apo enzyme and the complexes of the enzyme with its substrate (4-HBA-CoA) and/or products (CoA, HBA) can be easily distinguished (see Table 1).
Table 1.
Experimental and theoretical mass of the complexes of thioesterase with its substrate and/or products during the catalysis.
| Complexes of enzyme with its substrate and/or products |
Experimental mass (Da)* |
Theoretical mass (Da) |
|---|---|---|
| 4(E+S) | 69123 ± 6 | 69127.70 |
| (3(E+S)+(E+CoA)) | 68001 ± 4 | 69008.01 |
| (2(E+S)+2(E+CoA)) | 68883 ± 5 | 68887.74 |
| ((E+S)+3(E+CoA)) | 68762 ± 6 | 68767.65 |
| (4(E+CoA)) | 68641 ± 4 | 68647.76 |
| (3(E+S)+E) | 68238 ± 3 | 68240.50 |
| (2(E+S)+(E+CoA)+E) | 68116 ± 5 | 68120.41 |
| ((E+S)+2(E+CoA)+E) | 67996 ± 6 | 68000.32 |
| (3(E+CoA)+E) | 67874 ± 6 | 67880.23 |
| (2(E+S)+2E) | 67347 ± 3 | 67352.88 |
| ((E+S)+(E+CoA)+2E | 67231 ± 4 | 67232.79 |
| (2(E+CoA)+2E) | 67106 ± 4 | 67112.70 |
| ((E+CoA)+3E) | 66461 ± 4 | 66465.26 |
| ((E+S)+3E | 66338 ± 5 | 66345.17 |
| 4E | 65572 ± 6 | 65577.64 |
Note: theoretical mass of one subunit E, substrate S, CoA, and 4-HBA are 16394.38, 887.62, 767.53 and 138.12 Da, respectively,
represents the average data of three experiments.
To proceed with the investigation, solutions of the thioesterase and 4-HBA-CoA were mixed using an online rapid-mixing device coupled to a conventional ESI-TOF MS for analysis of the mixture as a function of incubation time under non-denaturing conditions using ammonium acetate at pH 7.5 as a buffer. The ligation state of the tetramer was examined as a function of 4-HBA-CoA concentration (5–30 µM) at a fixed tetramer concentration (40 µM) and fixed reaction time (7 ms). The single turnover rate for chemical catalysis measured under similar conditions (HEPES buffer at pH 7.5 instead of ammonium acetate at pH 7.5) using stopped-flow/absorption or rapid quench techniques is ~ 50 s−1 (t1/2 ~ 14 ms) (28). Thus, we anticipated that both products (CoA and 4-HBA) and substrate (4-HBA-CoA) would be present in the MS sample at 7 ms. The deconvoluted mass spectrum shown in Figure 3A was collected at 7 ms reaction of 40 µM tetramer with 30 µM 4-HBA-CoA. The dissociation constant for 4-HBA-CoA is estimated at ~ 1 µM and thus, ~ 70% of thioesterase is expected to be bound with substrate at the start of the single turnover reaction (23, 24). The unligated tetramer is observed as well as complexes with 1, 2, 3 or 4, 4-HBA-CoA substrate ligands bound and no product (CoA or 4-HBA) ligands bound, and with 1, 2 or 3 substrate ligands bound and 0–2 CoA product ligands bound. Notably, no tetramers contain 4-HBA ligands, the other product of the enzymatic reaction (Scheme 1). The Ki values of 4-HBA and CoA are 240 µM and 16 µM, respectively (28). Thus, once the reaction has completed, ~10 % of the total subunits present are expected to be bound with 4-HBA and ~44 % of the total subunits present are expected to be bound with CoA. We note that the spectrum of the reaction of 40 µM tetramer with 30 µM 4-HBA-CoA at 160 ms incubation time (see Figure 4G) reveals ~50% occupancy with CoA and no 4-HBA.
Figure 3.
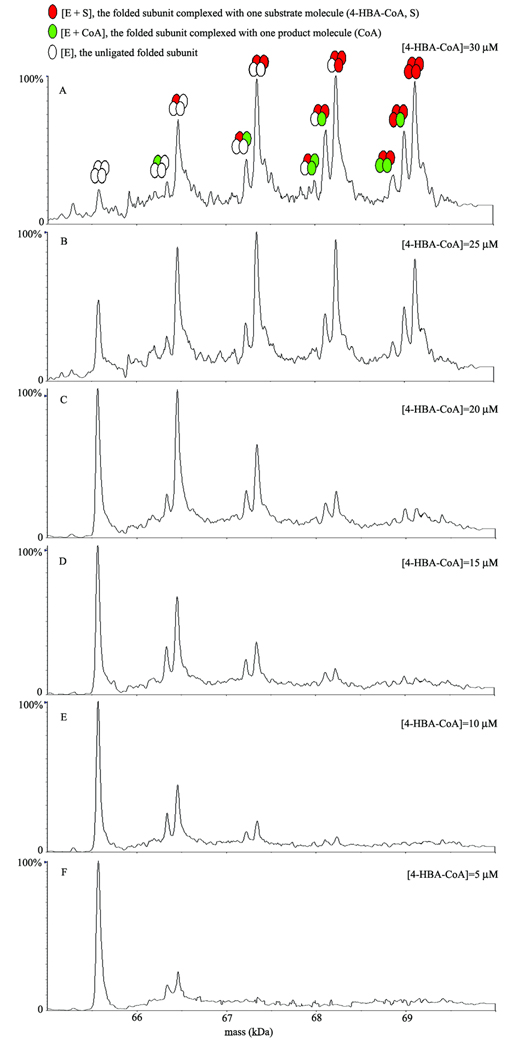
Detection of multimeric, non-covalent substrate and product complexes of Arthrobacter thioesterase. ESI-TOF mass spectra were obtained after rapidly mixing Arthrobacter thioesterase (40 µM) with varying concentrations (5–30 µM) of 4-HBA-CoA substrate (final concentrations). Representative spectra at 30 µM (A), 25 µM (B), 20 µM (C), 15 µM (D), 10 µM (E) and 5 µM (F) of 4-HBA-CoA were recorded at the reaction time point 7 ms in the positive ion mode under the same conditions.
Figure 4.
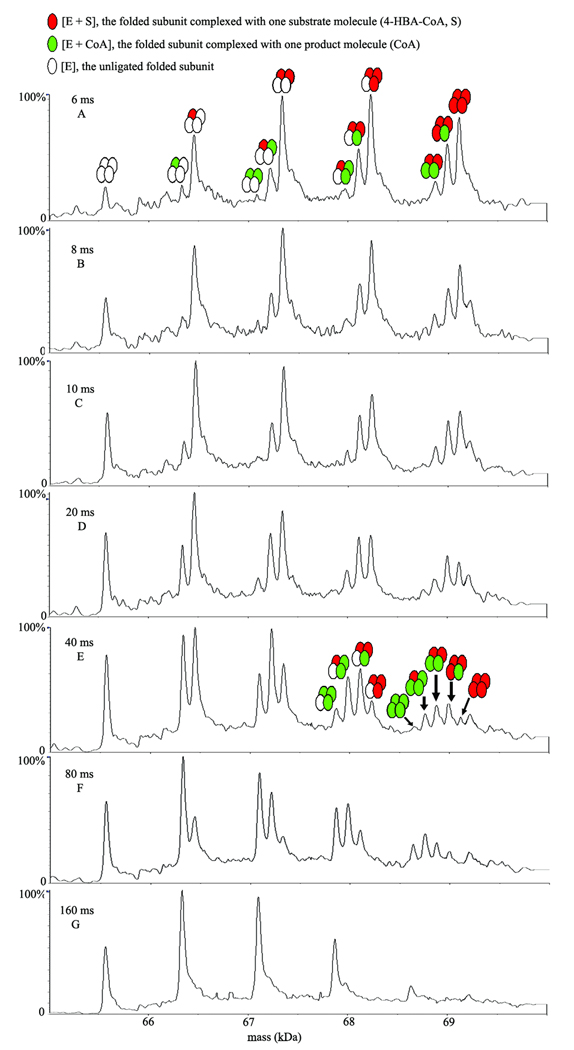
A reaction time course of Arthrobacter thioesterase catalysis under single turnover conditions. ESI-TOF mass spectra after online rapid mixing of 40 µM Arthrobacter thioesterase with the natural substrate (4-HBA-CoA) at 30 µM concentration (final concentrations) in 10 mM ammonium acetate buffer at pH 7.5 were recorded at the different reaction time points: 6, 8, 10, 20, 40, 80, and 160 ms.
Comparison of the deconvoluted mass spectra shown in Figures 3A–3F shows that as the concentration of 4-HBA-CoA employed in the 7 ms reaction is increased from 5 to 30 µM the amount of apo tetramer decreases as the amount of ligand bound tetramer increases. Starting at 5 µM 4-HBA-CoA (only 12 % of the thioesterase active sites are predicted to be occupied) the peaks corresponding to a tetramer with one CoA ligand bound and a tetramer with one 4-HBA-CoA ligand bound are clearly observable in a ~1:2 ratio. At a substrate concentration of 10 µM 4-HBA-CoA, the peaks corresponding to a tetramer with one CoA ligand and one 4-HBA-CoA ligand bound, and a tetramer with two 4-HBA-CoA ligands bound are clearly observable in a ~1:2 ratio. With increasing substrate concentration, tetramers with triple and quadruple occupancy increase and at 30 µM substrate they dominate the mixture (see discussion in the following section).
It is also evident that the ratio of peak intensities for a tetramer with one CoA ligand bound and a tetramer with one 4-HBA-CoA ligand bound decreases with increasing substrate concentration (~1:2 at 5 µM substrate to ~1:10 at 20 µM). Detection of multimeric, non-covalent substrate and product complexes of Arthrobacter thioesterase in a time course experiment
To proceed with the investigation, solutions of the thioesterase (40 µM final concentration) and 4-HBA-CoA (30 µM final concentration) in 10 mM ammonium acetate buffer (pH 7.5) were rapidly mixed online with ESI-TOF MS detection for analysis of the reaction kinetics as a function of time. Shown in Fig. 4 are the deconvoluted mass spectra collected at 6–160 ms reaction times after mixing enzyme and substrate solutions of 40 µM thioesterase and 30 µM 4-HBA-CoA in 10 mM ammonium acetate buffer (pH 7.5)(final concentrations). At the 6 ms time point (Fig. 4A), a distribution of tetrameric enzyme-substrate complexes with 1, 2, or 3 substrate molecules bound are detected along with a small amount of unligated tetrameric enzyme. Also evident at this time point are several tetrameric enzyme-product complexes containing 1 or 2 CoA molecules. For example, an enzyme-substrate complex (4(E+S)) of the tetrameric enzyme with four substrate (S) molecules (mass = 69,123 Da, Table 1) is observed at 6 ms along with two ligand bound complexes each containing four ligands: product (CoA) in addition to substrate (i.e., (3(E+S) + (E+CoA)) and (2(E+S)+2(E+CoA)) where E and S represent one subunit of the tetrameric enzyme and one substrate molecule, respectively, via the loss, respectively, of one and two 4-HBA molecules. These resulting complexes, (3(E+S)+(E+CoA)), (2(E+S)+2(E+CoA)), ((E+S)+3(E+CoA)), and (4(E+CoA)), can be generated from the complex (4(E+S)) via the loss of respectively, one to four molecules of product (4-HBA), as the reaction time progresses from 6 to 40 ms (Fig. 4A–E, see Table 1). In a competing pathway, the complex (4(E+S)) can also simultaneously release the multiple substrate(s) or products CoA and 4-HBA after the turnover occurred to produce ligand bound enzyme substrate(s) and product (s) complexes with three ligands bound. Similar catalysis is observed for the ligand bound complexes with one or two ligands.
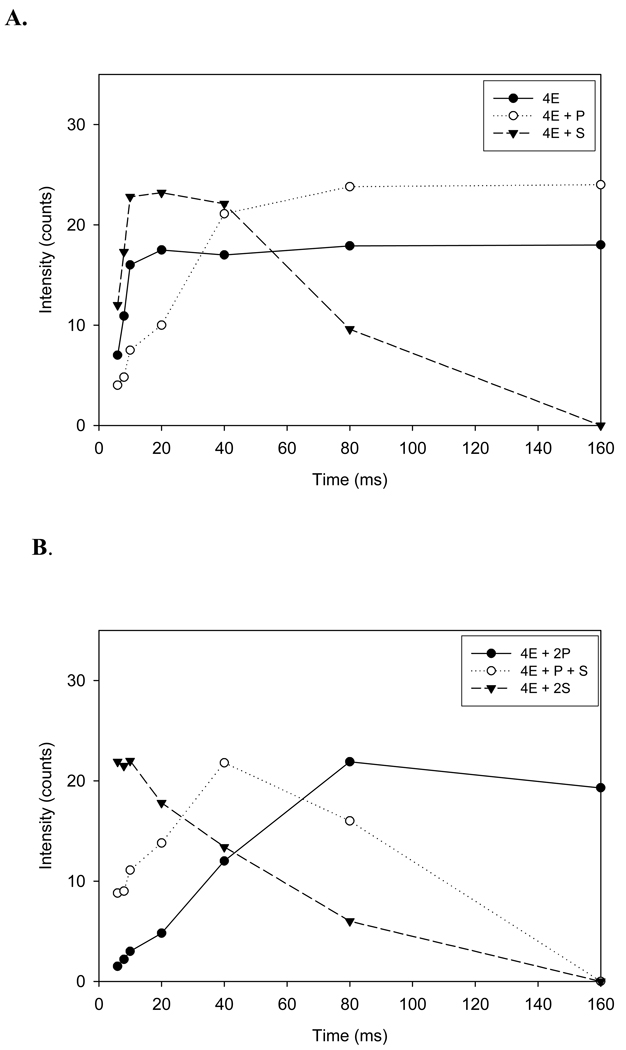
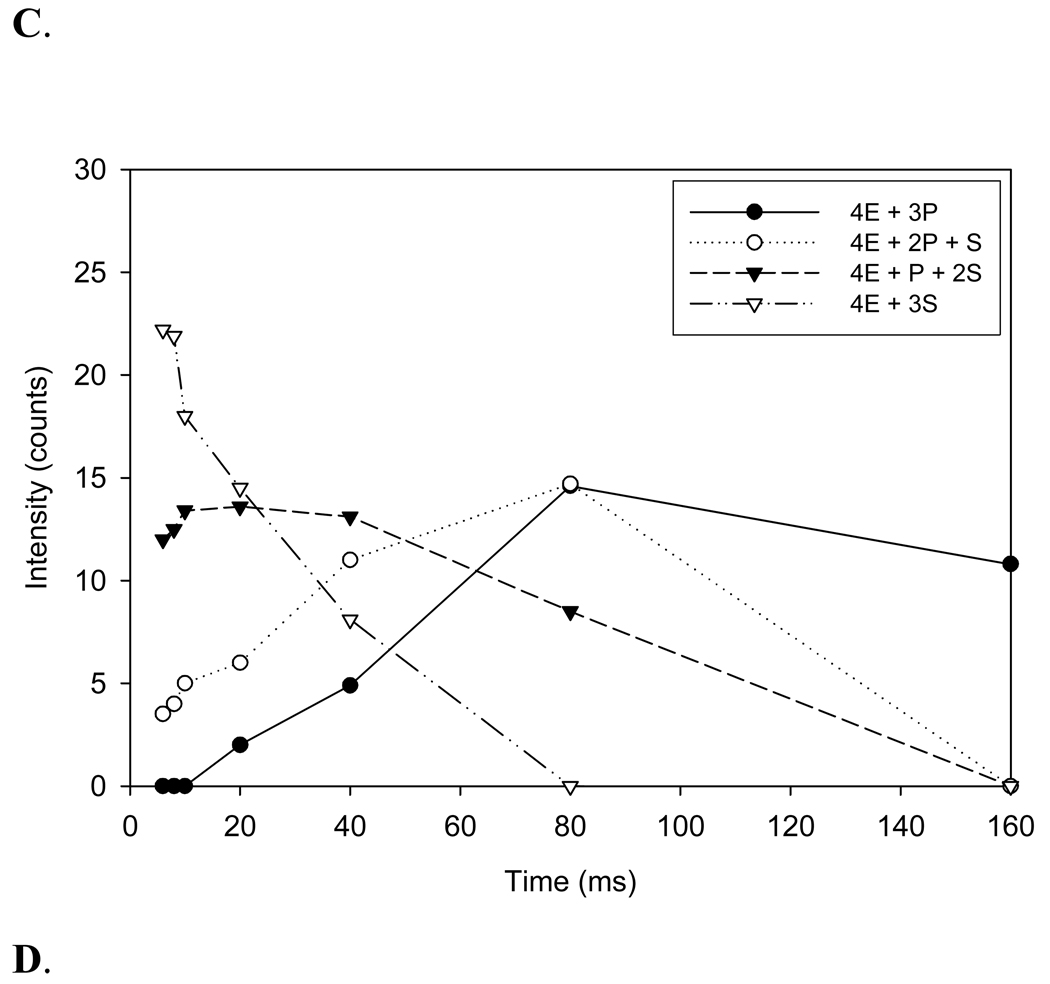
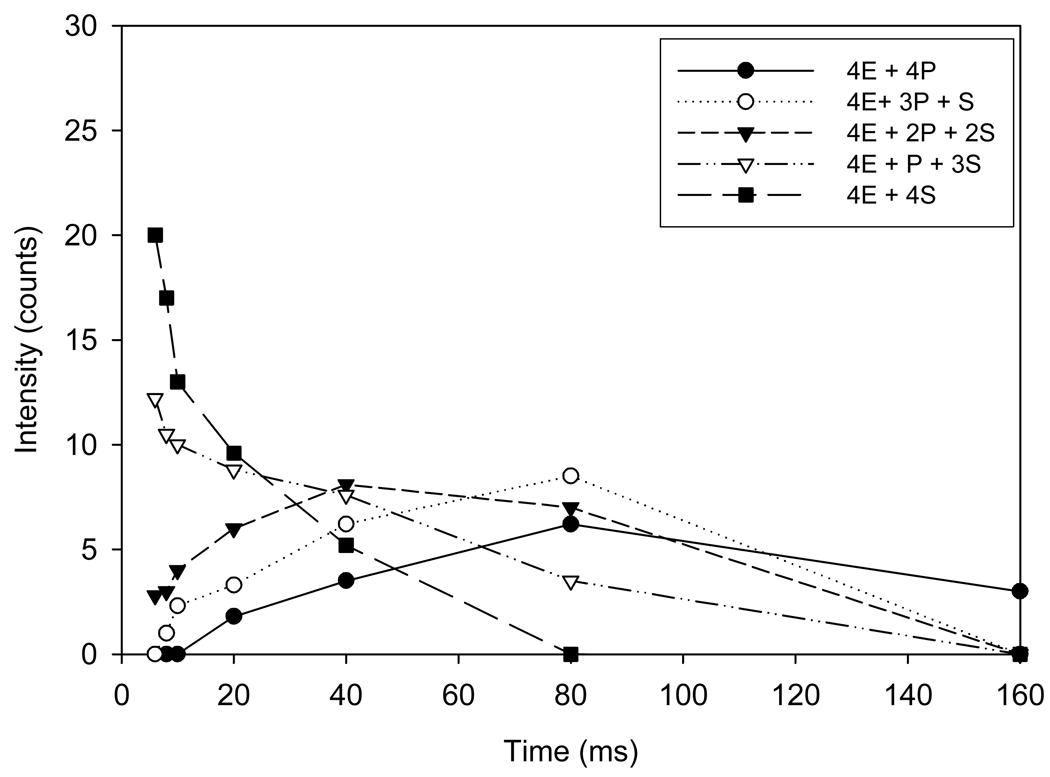
A summary of the changes in intensities of each enzyme-substrate and product species for a reaction time course from 6 to 160 ms is shown in Figure 5 A–D. A comparison of relative intensities for the tetrameric enzyme species (4E) containing one to four substrate molecules: (4E+1S, 4E+2S, 4E+3S, and 4E+4S) over a reaction time course from 6 to 160 ms is shown in Figure 5A. The data suggest that the tetrameric enzyme-substrate complex containing 4 substrates (4E+4S) disappears more rapidly than the complexes containing 3 or 2 substrate molecules while the tetrameric enzyme complex containing only one substrate increases up to 20 ms, plateaus slightly, and then slowly disappears up to 160 ms. The corresponding intensities for tetrameric enzyme species (4E) containing 1 to 4 product (CoA) molecules (4E +1P; 4E+2P; 4E +3P; and 4E+4P) are shown in Figure 5B. In this case, the tetrameric enzyme complex with a single ligand bound containing one product is formed faster than 2, 3, or 4 liganded species. In each case, the time course suggests that the peak for reaction is approximately 80 ms and CoA appears to stay bound to the enzyme and dissociate from the active site very slowly. Additional evidence for slow CoA release is illustrated in Fig. 4G in which the mass spectra show that a substantial amount of CoA product remains even after 160 ms. The time course for 3 and 4 ligand bound enzyme complexes are shown in Figures 5 C and D, respectively. Collectively, the data show that catalysis occurs more rapidly when 3 or 4 substrate molecules are bound as compared with 1 or 2 and that 4-HBA is rapidly released following catalytic turnover whereas CoA is slowly released.
Figure 5.
The time dependent intensity change of substrate species and product species (A–D) bound to the tetrameric enzyme. Intensity change of species, tetrameric enzyme (4E) with one substrate (S), [4E+1S] tetrameric enzyme (4E) with two substrates, [4E+2S]; tetrameric enzyme (4E) with three substrates, [4E+3S]; tetrameric enzyme (4E) with four substrates, [4E+4S]; with an increase in reaction time. Intensity change of species, tetrameric enzyme (4E) with one product (CoA) (P), [4E+1P]; tetrameric enzyme (4E) with two products (CoA), [4E+2P]; tetrameric enzyme (4E) with three products (CoA), [4E+3P]; tetrameric enzyme (4E) with four products (CoA), [4E+4P]; with an increase in reaction time.
Discussion
This study has demonstrated the utility of time-resolved ESI-MS technique to examine the details of the reaction kinetics of HBA-CoA thioesterase in the multimeric state. These data offer information on the reaction pathway, in particular, release of reaction products that allow further interpretation in the context of structure and reveal new insights for how catalysis is governed by the oligomeric state of the enzyme and ligand occupancy.
Our experimental findings provide evidence that product release is predominately ordered with 4-HBA dissociating before CoA as illustrated in Scheme 2. These results are in accord with previous kinetic studies showing that that HBA has an affinity 15-fold weaker relative to that of CoA (28). Structural studies with Arthrobacter HBA-CoA thioesterase complexed with the 4-hydroxyphenacyl-CoA inhibitor (see Fig. 1) shows the CoA pantothenic acid moiety is inserted in the outer part of the active site tunnel and the nucleotide moiety is anchored to the protein surface while the hydroxybenzoyl group binds in an enclosed pocket. The back wall of the hydroxybenzoyl pocket is formed by the N-terminal segment in an extended loop-like conformation. Based upon the kinetic data, this suggests that the 4-HBA product dissociates directly from the hydroxybenzoyl binding pocket more quickly than CoA can move out of the connecting tunnel. Accordingly, the N-terminal loop must act as a flexible flap that allows 4-HBA release as illustrated in Scheme 2.
Scheme 2.
Illustration of kinetic mechanism of thioesterase catalysis and order of product release
The time-resolved ESI-MS TOF technique also allows us to directly investigate catalysis of the tetramer in various states of ligand occupancy. These data reveal that the tetrameric form of the enzyme with no ligands bound differs in catalytic efficiency from a tetramer with bound ligands.. An analysis of the reaction kinetics establishes that catalytic turnover occurs more rapidly in the tetramer at full active site occupancy than at partial active site occupancy, suggesting some form of positive cooperativity.
In summary, the present study illustrates the ability of time resolved ESI-MS TOF to monitor multimeric states of an enzyme, as the reaction is occurring, by effectively capturing a properly folded tetrameric enzyme complexed with its substrate and/or product in the gas phase of the mass spectrometer in the millisecond time range. It allows the direct observation of distinct enzymatic complexes with ligands during the catalysis. Moreover, this technique allows us to directly study the individual enzyme states that are unique in terms of catalysis. The ability to observe the tetramer complexed with substrates and/or products permits the order of products released from the thioesterase enzyme to be established and furthermore offers insight for how product dissociation may occur at a structural level.
Supplementary Material
Acknowledgements
This work was supported by NIH GM71805 to K.S. A. and NIH GM28688 to D.D.M.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Zhili Li, Email: lizhili@ibms.pumc.edu.cn.
Debra Dunaway-Mariano, Email: dd39@unm.edu.
Karen S. Anderson, Email: karen.anderson@yale.edu.
References
- 1.Sobott F, Robinson CV. Protein complexes gain momentum. Curr Opin Struct Biol. 2002;12:729–734. doi: 10.1016/s0959-440x(02)00400-1. [DOI] [PubMed] [Google Scholar]
- 2.Hernandez H, Robinson CV. Dynamic protein complexes: insights from mass spectrometry. J Biol Chem. 2001;276:46685–46688. doi: 10.1074/jbc.R100024200. [DOI] [PubMed] [Google Scholar]
- 3.Gross JW, Frey PA. Rapid mix-quench MALDI-TOF mass spectrometry for analysis of enzymatic systems. Methods Enzymol. 2002;354:27–49. doi: 10.1016/s0076-6879(02)54004-0. [DOI] [PubMed] [Google Scholar]
- 4.Li Z, Sau A, Shen S, Whitehouse C, Baasov T, Anderson KSA. Snapshot of Enzyme Catalysis Using Electrospray Ionization Mass Spectrometry. J. Am. Chem. Soc. 2003;125:9938–9939. doi: 10.1021/ja0354768. [DOI] [PubMed] [Google Scholar]
- 5.Li Z, Sau AK, Furdui CM, Anderson KS. Probing the role of tightly bound phosphoenolpyruvate in Escherichia coli 3-deoxy-d-manno-octulosonate 8-phosphate synthase catalysis using quantitative time-resolved electrospray ionization mass spectrometry in the millisecond time range. Anal Biochem. 2005;343:35–47. doi: 10.1016/j.ab.2005.04.021. [DOI] [PubMed] [Google Scholar]
- 6.Pi N, Meyers CL, Pacholec M, Walsh CT, Leary JA. Mass spectrometric characterization of a three-enzyme tandem reaction for assembly and modification of the novobiocin skeleton. Proc Natl Acad Sci U S A. 2004;101:10036–10041. doi: 10.1073/pnas.0403526101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rhomberg AJ, Shriver Z, Biemann K, Sasisekharan R. Mass spectrometric evidence for the enzymatic mechanism of the depolymerization of heparin-like glycosaminoglycans by heparinase II. Proc Natl Acad Sci U S A. 1998;95:12232–12237. doi: 10.1073/pnas.95.21.12232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zechel DL, Konermann L, Withers SG, Douglas DJ. Pre-steady state kinetic analysis of an enzymatic reaction monitored by time-resolved electrospray ionization mass spectrometry. Biochemistry. 1998;37:7664–7669. doi: 10.1021/bi980445o. [DOI] [PubMed] [Google Scholar]
- 9.Benesch JL, Robinson CV. Mass spectrometry of macromolecular assemblies: preservation and dissociation. Curr Opin Struct Biol. 2006;16:245–251. doi: 10.1016/j.sbi.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 10.Benesch JL, Sobott F, Robinson CV. Thermal dissociation of multimeric protein complexes by using nanoelectrospray mass spectrometry. Anal Chem. 2003;75:2208–2214. doi: 10.1021/ac034132x. [DOI] [PubMed] [Google Scholar]
- 11.Hanson CL, Fucini P, Ilag LL, Nierhaus KH, Robinson CV. Dissociation of intact Escherichia coli ribosomes in a mass spectrometer. Evidence for conformational change in a ribosome elongation factor G complex. J Biol Chem. 2003;278:1259–1267. doi: 10.1074/jbc.M208966200. [DOI] [PubMed] [Google Scholar]
- 12.Ilag LL, Videler H, McKay AR, Sobott F, Fucini P, Nierhaus KH, Robinson CV. Heptameric (L12)6/L10 rather than canonical pentameric complexes are found by tandem MS of intact ribosomes from thermophilic bacteria. Proc Natl Acad Sci U S A. 2005;102:8192–8197. doi: 10.1073/pnas.0502193102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Y, Aubert SD, Maes EG, Raushel FM. Enzymatic resolution of chiral phosphinate esters. J Am Chem Soc. 2004;126:8888–8889. doi: 10.1021/ja048457m. [DOI] [PubMed] [Google Scholar]
- 14.McCammon MG, Hernandez H, Sobott F, Robinson CV. Tandem mass spectrometry defines the stoichiometry and quaternary structural arrangement of tryptophan molecules in the multiprotein complex TRAP. J Am Chem Soc. 2004;126:5950–5951. doi: 10.1021/ja0317170. [DOI] [PubMed] [Google Scholar]
- 15.McCammon MG, Robinson CV. Structural change in response to ligand binding. Curr Opin Chem Biol. 2004;8:60–65. doi: 10.1016/j.cbpa.2003.11.005. [DOI] [PubMed] [Google Scholar]
- 16.McLaughlin SH, Sobott F, Yao ZP, Zhang W, Nielsen PR, Grossmann JG, Laue ED, Robinson CV, Jackson SE. The co-chaperone p23 arrests the Hsp90 ATPase cycle to trap client proteins. J Mol Biol. 2006;356:746–758. doi: 10.1016/j.jmb.2005.11.085. [DOI] [PubMed] [Google Scholar]
- 17.Ruotolo BT, Giles K, Campuzano I, Sandercock AM, Bateman RH, Robinson CV. Evidence for macromolecular protein rings in the absence of bulk water. Science. 2005;310:1658–1661. doi: 10.1126/science.1120177. [DOI] [PubMed] [Google Scholar]
- 18.Sobott F, Benesch JL, Vierling E, Robinson CV. Subunit exchange of multimeric protein complexes. Real-time monitoring of subunit exchange between small heat shock proteins by using electrospray mass spectrometry. J Biol Chem. 2002;277:38921–38929. doi: 10.1074/jbc.M206060200. [DOI] [PubMed] [Google Scholar]
- 19.Westblade LF, Ilag LL, Powell AK, Kolb A, Robinson CV, Busby SJ. Studies of the Escherichia coli Rsd-sigma70 complex. J Mol Biol. 2004;335:685–692. doi: 10.1016/j.jmb.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 20.McDonald WH, Pavlova Y, Yates JR, 3rd, Boddy MN. Novel essential DNA repair proteins Nse1 and Nse2 are subunits of the fission yeast Smc5-Smc6 complex. J Biol Chem. 2003;278:45460–45467. doi: 10.1074/jbc.M308828200. [DOI] [PubMed] [Google Scholar]
- 21.Ilag LL, Westblade LF, Deshayes C, Kolb A, Busby SJ, Robinson CV. Mass spectrometry of Escherichia coli RNA polymerase: interactions of the core enzyme with sigma70 and Rsd protein. Structure. 2004;12:269–275. doi: 10.1016/j.str.2004.01.007. [DOI] [PubMed] [Google Scholar]
- 22.Benning MM, Hong SB, Raushel FM, Holden HM. The binding of substrate analogs to phosphotriesterase. J Biol Chem. 2000;275:30556–30560. doi: 10.1074/jbc.M003852200. [DOI] [PubMed] [Google Scholar]
- 23.Zhuang Z, Gartemann KH, Eichenlaub R, Dunaway-Mariano D. Characterization of the 4-hydroxybenzoyl-coenzyme A thioesterase from Arthrobacter sp. strain SU. Appl Environ Microbiol. 2003;69:2707–2711. doi: 10.1128/AEM.69.5.2707-2711.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thoden JB, Zhuang Z, Dunaway-Mariano D, Holden HM. The structure of 4-hydroxybenzoyl-CoA thioesterase from arthrobacter sp. strain SU. J Biol Chem. 2003;278:43709–43716. doi: 10.1074/jbc.M308198200. [DOI] [PubMed] [Google Scholar]
- 25.Willis MA, Zhuang Z, Song F, Howard A, Dunaway-Mariano D, Herzberg O. Structure of YciA from Haemophilus influenzae (HI0827), a hexameric broad specificity acyl-coenzyme A thioesterase. Biochemistry. 2008;47:2797–2805. doi: 10.1021/bi702336d. [DOI] [PubMed] [Google Scholar]
- 26.Cao J, Xu H, Zhao H, Gong W, Dunaway-Mariano D. The Mechanisms of Human Hotdog-fold Thioesterase 2 (hTHEM2) Substrate Recognition and Catalysis Illuminated by a Structure and Function Based Analysis (dagger) (,) (double dagger) Biochemistry. 2009 doi: 10.1021/bi801879z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhuang Z, Song F, Zhao H, Li L, Cao J, Eisenstein E, Herzberg O, Dunaway-Mariano D. Divergence of function in the hot dog fold enzyme superfamily: the bacterial thioesterase YciA. Biochemistry. 2008;47:2789–2796. doi: 10.1021/bi702334h. [DOI] [PubMed] [Google Scholar]
- 28.Song F. Department of Chemistry and Chemical Biology Ph.D. Thesis. Albuquerque NM: University of New Mexico; 2005. Structure, function and mechanism of hotdog-fold enzyme superfamily thioesterases; pp. 38–81. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.