Abstract
The 5′-cap structure that typifies all polymerase II-transcribed RNAs plays important roles in pre-mRNA processing as well as mRNA export, translation and quality control. Removal of the cap is a regulated process that is considered to be the first irreversible step in mRNA decay. An emerging view challenges this idea: mRNAs have been identified in mammalian cells that lack sequences from their 5′ ends but nevertheless appear to be modified with a cap or cap-like structure. Furthermore, a cytoplasmic form of capping enzyme was recently identified that, together with a novel kinase, generates capped ends from cleaved RNAs. These and other findings provide evidence for re-capping and its possible functions.
5′-cap structure and function
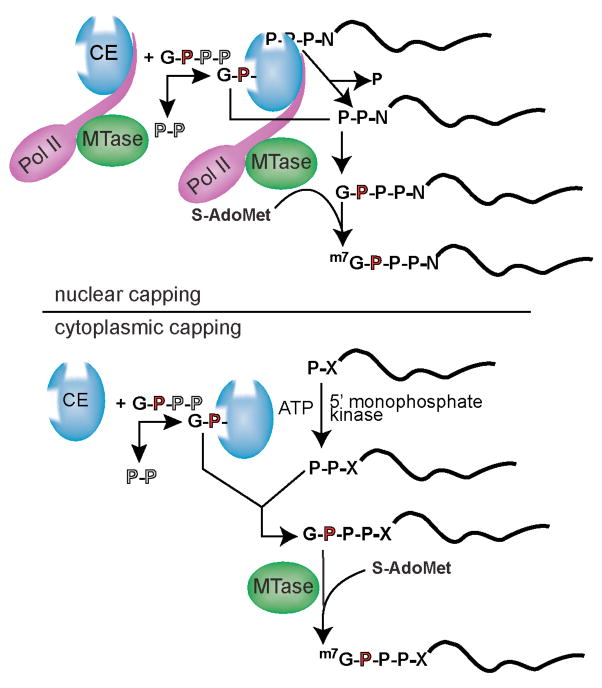
Nuclear capping occurs co-transcriptionally on all RNA polymerase II-synthesized RNAs in three steps (reviewed in [1–3]). The N-terminal triphosphatase domain of capping enzyme hydrolyzes the 5′ triphosphate of nascent pre-mRNA to a diphosphate (Fig. 1, upper panel). GMP that is covalently bound to the C-terminal guanylylation domain of capping enzyme is subsequently transferred onto this diphosphate to create a G(5′)ppp(5′)N terminus, where N denotes the first transcribed nucleotide. This terminus is then methylated by RNA (guanine-7)-methyltransferase to generate the m7G(5′)ppp(5′)N cap, also called cap 0, that typifies all pre-mRNAs and their product mRNAs. Subsequently, m7G(5′)ppp(5′)Nm, also called cap 1, or m7G(5′)ppp(5′)NmNm, also called cap 2, can be generated by methylation of the 2′-hydroxyl position of, respectively, the first or first and second transcribed riboses by cap-specific 2′-O nucleoside RNA methytransferases. The cap participates in many aspects of pre-mRNA and mRNA metabolism, including splicing, polyadenylation, nucleocytoplasmic transport, translation, quality control and stability [1–5]. Whereas cap removal is considered to be a requisite step in the mRNA degradation pathway, recent findings indicate that some portion of translationally inactive mRNAs might be stored in an uncapped state and subsequently returned to a translationally active state upon cytoplasmic re-capping.
Figure 1. Nuclear and cytoplasmic capping reactions.
Nuclear capping occurs co-transcriptionally on all RNA polymerase II-synthesized RNA, and recent evidence indicates that cytoplasmic capping can occur by a related mechanism. (i) The first steps of nuclear capping. Binding to the C-terminal domain of the large subunit of RNA polymerase II (Pol II; purple) positions the nuclear capping enzyme (CE; blue) near the 5′ end of newly synthesized transcripts. The N-terminal triphosphatase domain of capping enzyme hydrolyzes the 5′-triphosphate end of the first transcribed nucleotide (P-P-P-N, where N denotes the first transcribed nucleotide) to a diphosphate (P-P-N) capping substrate. Capping enzyme then transfers GMP that is bound covalently at its C-terminal guanylyltransferase domain to create the 5′-G(5′)-P-P-P-(5′)N terminus (G-P-P-P-N). RNA (guanine-7)-methytransferase (MTase; green) is also positioned near the transcript 5′ end via binding to Pol II. Transfer of a methyl group from S-adenosylmethionine (S-AdoMet) onto the N7 position of the terminal guanine generates 5′-m7G(5′)-P-P-P-(5′)N, thereby completing the synthesis of the cap structure referred to as cap 0. (ii) The steps of cytoplasmic capping. As in nuclear capping, GMP is bound covalently to the guanylyltransferase domain of capping enzyme. However, in contrast to nuclear capping, cytoplasmic capping enzyme forms a complex with a 5′-monophosphate kinase that converts uncapped RNA having a 5′-monophosphate (P-X) end to a diphosphate capping substrate (P-P-X). As in nuclear capping, GMP that is bound covalently to the guanylyltransferase domain of capping enzyme is then transferred to create the G(5′)-P-P-P-(5′)N end (G-P-P-P-X). Although it is present in the cytoplasm, cap methyltransferase (MTase; green) is not part of the cytoplasmic capping enzyme complex. The process by which the cap is methylated during cytoplasmic capping remains undetermined.
The various roles of the cap are mediated by associated proteins
In mammalian cells, the caps of pre-mRNAs and newly synthesized mRNAs are bound by the cap-binding protein (CBP) heterodimer CBP80–CBP20, which shuttles between the nucleus and the cytoplasm but is predominantly nuclear [1, 4, 5; Box 1]. CBP80–CBP20 not only promotes nuclear cap function during pre-mRNA processing, but it also is central to the cytoplasmic pioneer round of translation, the primary purpose of which is to monitor mRNA quality via the nonsense-mediated mRNA decay (NMD) pathway (Box 2). NMD degrades mRNAs that prematurely terminate translation during the pioneer round of translation, thereby eliminating the production of truncated proteins, which have the potential to be toxic to cellular metabolism [4, 5]. In contrast to the caps of pre-mRNAs and newly synthesized mRNAs, the caps of the majority of cytoplasmic mRNAs have lost CBP80–CBP20 and instead are associated with eukaryotic translation initiation factor (eIF)4E [6; Box 1]. eIF4E shuttles between the nucleus and the cytoplasm but predominantly localizes to the cytoplasm, where it supports the bulk of cellular protein synthesis. eIF4E is a prime target of translational regulation, and its activity is largely controlled by the phosphorylation status of eIF4E-binding repressor proteins in response to extracellular and intracellular effectors and by its interaction with other proteins [6].
Box 1. Cap binding proteins
CBP80 and CBP20 associate with caps of pre-mRNAs and newly synthesized mRNAs as a heterodimer rather than individually: CBP20, like eIF4E, sandwiches the 7-methylguanine of the cap structure between two amino acid side chains [51–54], and its binding to an mRNA cap requires CBP80 [55, 56]. eIF4E, which associates with the caps of the bulk of cytoplasmic mRNAs, has a lower affinity for caps than does CBP80–20 at least in vitro[57] and exists as a complex on mRNA with eIF4A, an ATPase and RNA helicase, and eIF4G, which connects the mRNA to initiating ribosomes via eIF3 [58].
Box 2. Nonsense-mediated mRNA decay (NMD)
This eukaryotic pathway eliminates the production of mRNAs which harbor either a frameshift or a nonsense mutation and, as a consequence, prematurely terminate translation [59]. The pathway appears to have evolved to protect cells from the potentially deleterious effects of routine inaccuracies in gene expression. Remarkably, approximately one-third of alternatively spliced transcripts are NMD targets that are generated by mistakes made during pre-mRNA splicing. NMD also silences genomic noise, such as non-functional transcripts that, e.g., have acquired transposons or retroviral sequences, and additionally serves regulatory roles by targeting physiologic transcripts that harbor an upstream open translational reading frame, or an intron within their 3′-untranslated region, or encode selenocysteine. NMD additionally functions in the homeostatic control of genes that encode serine-arginine-rich proteins and heterogeneous nuclear ribonucleoprotein splicing factors.
Identification of uncapped mRNAs
Until recently, there was little evidence for uncapped mRNAs in eukaryotic cells. The first suggestion that cells might contain populations of uncapped RNAs came in the 1970s from experiments examining the 5′-terminal nucleotides of what at the time were presumed to be primary transcripts in mouse L cells [7]. Compelling evidence for uncapped mRNAs appeared only recently through experiments in Arabidopsis thaliana directed at defining such populations. Gregory et al. [8] took advantage of the fact that uncapped RNAs have a 5′-monophosphate end to identify uncapped mRNAs in plants that were missing one subunit of the nuclear cap-binding protein ABH1 (CBP80) and the major 5′ exonuclease ethylene insensitive (EIN)5 (exoribonuclease (XRN)4)). After ligating a primer onto the 5′-monophosphate ends and generating cDNA libraries, uncapped RNAs were identified using deep sequencing. Jiao et al. [9] also used primer-ligation to tag uncapped mRNAs but with two important differences. They started with poly(A)-selected RNA, which might have removed deadenylated transcripts that were in the process of decay and, instead of using the primer for library synthesis and deep sequencing, the primer was employed to physically recover uncapped mRNAs that were subsequently identified on microarrays. Unexpectedly, >90% of protein-coding genes contributed RNAs that were recovered during floral bud development. Although the levels of uncapped mRNAs largely correlated with the abundance of the corresponding full-length mRNA, the finding that the levels of specific uncapped transcripts varied depending on the stage of floral development independently of full-length mRNA abundance provided evidence that decapping was regulated and of physiological significance. Also arguing for the physiological relevance of uncapped RNAs, regulatory-gene transcripts, e.g., those encoding kinases, nuclear and membrane-bound proteins, and proteins involved in signal transduction, as well as many known targets of microRNA-mediated silencing were uncapped to a greater extent than were transcripts encoding structural proteins. Remarkably, sequence-related transcripts usually shared similar levels of uncapping, offering additional evidence that decapping is to some extent an active, rather than a passive, process.
Roles for P bodies and the 5′ cap: transitioning mRNA between translationally active and inactive states
For simplicity, cytoplasmic mRNA can be viewed as existing in one of two states: translationally active, i.e., engaged by translating ribosomes, and transitionally inactive, i.e., silenced (e.g., by microRNAs), stored or undergoing decay. mRNA decay and most recognized forms of mRNA silencing begin with shortening of the poly(A) tail [10,11]. When a limit length – usually <30 As – is reached, the messenger ribonucleoprotein (mRNP) composition changes through a mechanism that is remains poorly understood (Fig. 2). Proteins that are involved in translation, including as poly(A)-binding protein and eIF4G, are lost; however, Lsm1 through 7, which is a complex of seven Sm-like proteins, and protein-associated with topoisomerase II (Pat1), which is both a translational repressor and an activator of decapping, are gained [12]. The transition to an inactive state (reviewed in [12]) is facilitated by the p54 (also called Rck) DEx[D/H]-box helicase and ultimately results in the formation of discrete cytoplasmic foci termed processing (P) bodies or, alternatively, Dcp or GW bodies [12]. P bodies are dynamic, and their size and number reflect the relative portion of mRNA within the cell that is not actively engaged in translation [12, 15]. The order of their assembly is not yet known, nor is it known whether all P bodies are the same or whether individual P bodies have different functions. In addition to Lsm1-7 and Pat1, the protein components of metazoan P-bodies include eIF4E, the decapping proteins Dcp1 and Dcp2, the 5′-3′ exonuclease Xrn1, several deadenylases, NMD factors Upf1, Smg5 and Smg7, human enhancer of decapping large subunit (Hedls, or Ge-1), and the Argonaute proteins that are central to microRNA-mediated silencing [16, 17]. In addition to Pat1, two other translational repressors are critical components of P bodies: the DEx[D/H]-box helicase discussed above and the eIF4E-transporter protein (eIF4E-T), which is a binding partner of the eIF4E cap-binding protein [18, 19].
Figure 2. A proposal that re-capping could rescue decapped mRNA from decay.

mRNA that is actively engaged in translation is bound by eIF4E (4E; beige) and eIF4G (4G; blue) at its 5′ end, poly(A)-binding protein (not shown for simplicity) at its 3′-poly(A) tail, and 80S ribosomes (green circles), among many other factors. The poly(A) tail is shortened by one or more deadenylases until a limit length of ~30 residues is achieved (Aoligo), at which point ribosomes no longer initiate translation, translationally active ribosomes presumably dissociate after directing translation termination, and the mRNA transits into a translationally inactive state. This transition, which can result in microscopically detectable P bodies, is likely facilitated by translational inhibitors, including Pat1 (red), the p54 (also called Rck) helicase (yellow) and, possibly, microRNAs. Little is known about the order of assembly, but a variety of proteins are recruited that include the decapping activators Lsm1-7 (blue ring), enhancer of mRNA decapping 3 homologue (Edc3; brown), Hedls (orange) and GW182 (blue oblong), the decapping enzyme Dcp2 (dark pink) and its activator Dcp1 (light pink), and the Xrn1 5′-3′ exonuclease (green). eIF4E is also present in P bodies, most likely bound to the 5′ cap on the silenced mRNA. The requirement of eIF4E-T for P-body assembly points to a central role for the cap plays a central role in the assembly process. The initial complex that contains silenced mRNA is submicroscopic, and the appearance of visible P bodies is thought to result from the aggregation of multiple silenced complexes. Little is known about the physical state of silenced mRNA in P bodies or what determines its fate. One possibility is that it is decapped and degraded (lower right). Alternatively, it might be stored in P bodies either with the cap intact or in a decapped state. We propose that the re-capping of decapped mRNA could facilitate its return to the translationally active pool. Given that poly(A) shortening precedes the translational silencing of most mRNAs, it seems likely that re-adenylation might also be a prerequisite for the re-activation of a stored or otherwise silenced mRNA.
A critical role of cap-binding proteins in translational silencing was appreciated well before the discovery of microRNAs [20]. eIF4E-T facilitates the incorporation of eIF4E, which is presumably associated with cap-bound mRNA, into P bodies [18, 19], and the disappearance of visible P bodies following knockdown of eIF4E-T (but not Dcp2) indicates a critical role for this protein and, by inference, eIF4E and the cap in the initial assembly of these silencing complexes. This concept is supported by results showing significantly greater silencing of an mRNA if it initiates translation in a cap-dependent manner compared to a cap-independent mechanism, such as through an internal ribosome entry site [21–23].
There is little doubt that at least some of the mRNAs that accumulate in P bodies can be degraded after they are decapped [24] (Fig. 2). Based on the loss of signal observed using Northern blotting, Xrn1 was originally thought to rapidly degrade the body of decapped mRNA with 5′-3′ polarity. However, a different picture emerged when the rate of decay as determined by Northern blotting was compared to the rate obtained using a fluorescence resonance energy transfer (FRET)-based assay that quantified loss of the 5′, middle and 3′ ends of a 600-nucleotide reporter mRNA in cells in which 3′-5′ decay was inactivated [25]. Sequences at the 5′ end disappeared at the rate that was determined by Northern blotting, but the middle portion decayed at half this rate and the 3′ end did not decay at all, indicating that Xrn1 is not sufficiently processive in vivo to degrade through the body of even this relatively small reporter mRNA.
Consistent with this finding, a growing body of literature indicates that P bodies function primarily as storage sites for non-translating mRNAs, from which they can be degraded or returned to the translating pool [12, 15, 26]. Remarkably little is known about the biochemical state of non-translating mRNAs in P bodies. As poly(A) shortening precedes the storage and/or silencing of most mRNAs, it is assumed that the mRNAs in P bodies have short poly(A) tails [27]. Nothing is known about the cap status of these mRNAs. Given that the mRNAs and the decapping proteins Dcp1 and Dcp2 are concentrated together in P bodies, it seems reasonable that a portion of these is intact or almost so except for lacking a cap. For these RNAs to be restored to the translating pool, they would have to be re-capped, and the poly(A) tail would have to be restored to a length that supports translation initiation (Fig. 2). Although some attempts have been made to identify uncapped mRNAs in mammalian cells [28], a systematic analysis of uncapped mRNAs as was done for A. thaliana has yet to be undertaken. Nevertheless, some evidence suggests that mammalian cells also contain populations of uncapped mRNAs.
Early evidence for re-capping: Stable NMD intermediates
The first direct evidence that mRNAs can undergo re-capping appeared in 1992 in work on the NMD of several human β°-thalassemic β-globin transcripts that harbor either a frameshift or a nonsense mutation and, as a consequence, contain a premature termination codon (PTC). Erythroid cells from mice that are transgenic for one of several of these alleles, and mouse erythroleukemia (MEL) cells that stably express one of several of these alleles, generate readily detectable β-globin mRNA decay intermediates that are polyadenylated but lack roughly the same regions from the mRNA 5′ end regardless of the position of the PTC [29–32]. The 5′ ends of these decay intermediates reside primarily at UG dinucleotides and are the result of a polysome-associated endonucleolytic activity that is similar to Xenopus laevis polysome ribonuclease (PMR)1 [32, 33]. Remarkably, these RNAs appear to be capped as evidenced by their in vivo stability, recovery with a monoclonal antibody to the methyl-cap structure, and sensitivity to decapping by tobacco acid pyrophosphatase (TAP) [30, 31].
Evidence for re-capping was next reported in 2001 in work studying the feasibility of using antisense oligonucleotides to inactivate hepatitis B virus by directing the RNase H-mediated cleavage of viral RNA [34]. Although antisense DNA oligonucleotide-directed RNase H cleavage had been thought to result in the complete degradation of the targeted transcript, the 3′-cleavage products of viral RNA were unexpectedly stable. Like the stable PTC-containing β-globin mRNA decay intermediates in erythroid cells, the viral 3′-cleavage products had an intact poly(A) tail. Unexpectedly, however, the viral 3′-cleavage products, like the 3′-cleavage product of a cellular mRNA, could be translated into N-terminally truncated proteins. Although no mechanism for re-capping was known at the time, it was speculated that the stability of the 3′-cleavage products and the appearance of a translated product were secondary to re-capping of the 3′-cleavage product.
Recent re-emergence of re-capping as a bone fide cellular mechanism
The endonucleolytic pathway responsible for cleaving PTC-containing β-globin mRNAs is a cytoplasmic process, and it appears to co-exist with the recently described Smg6-mediated endonucleolytic pathway of NMD [35], and with the exonucleolytic NMD pathways [36–38]. Otsuka et al. [39] re-examined the cap status of the β-globin mRNA decay intermediates and, in doing so, confirmed the 1992 results of Lim and Maquat [30] showing that the 5′-end modification of each decay intermediate was essentially indistinguishable from the cap on the parental mRNA. This finding posed questions about the origin of the cap. The 3′-cleavage products that derive from the endonucleolytic cleavage of PTC-containing β-globin mRNA, like the oligonucleotide-directed RNase H-mediated cleavage of viral transcripts [34], have a single phosphate on their 5′ ends. However, capping enzyme requires RNA with a 5′-diphosphate as substrate for transferring covalently bound GMP (Fig. 1, upper panel). Moreover, in mammals, only a single enzyme is known to catalyze this reaction, and it is generally thought to be restricted to the nucleus. Thus the issue of cytoplasmic capping faced two obstacles: identifying the enzyme that catalyzes capping, and defining its substrate.
One possibility for this enzyme was an activity that transfers GDP onto RNA having a 5′-monophosphate, which would be similar to the activity of the RNA-dependent RNA polymerase L protein of vesicular stomatitis virus [40]. However, the human genome lacks sequences even distantly related to RNA-dependent polymerase L-encoding sequence in the human genome. Another possibility for the enzyme was a so-called ‘cap snatching’ activity [41; Box 3], which catalyzes a process used by a number of viruses in which an endonuclease cleaves off the capped 5′ end of random cellular mRNAs and a viral RNA polymerase incorporates the resulting caps plus adjacent nucleotides onto the 5′-monophosphate ends of viral RNAs. Again, this was ruled out by the absence of both a detectable mammalian ortholog of the viral polymerase and heterologous sequences on the 5′ ends of the 3′-cleavage products. Therefore, two possibilities remained: a previously unrecognized cytoplasmic capping enzyme or some new type of cytoplasmic capping enzyme.
Box 3. P bodies
The term P body refers to focal concentrations of proteins that are generally involved in decapping and 5′-3′ decay [12, 15, 60]. P bodies were observed first in experiments examining the subcellular distribution of the Xrn1 5′-3′ exonuclease [61] and later in studies using autoimmune serum from a patient with motor and sensory neuropathy [62]. Evidence linking P bodies to mRNA decay initially derived from experiments studying the distribution of a battery of GFP-tagged proteins in Saccharomyces cerevisiae[14], and their focal concentration in mammalian cells was confirmed shortly thereafter [63]. To date, approximately 40 P-body proteins have been identified, and the number continues to increase. The best characterized of these proteins (Fig. 2) are the complex of seven cytoplasmic Lsm proteins (Lsm1-7), the Xrn1 5′-3′ exonuclease, the Dcp2 decapping activity with its activator(s) Dcp1a and/or Dcp1b, the Pat1 translational repressor, the Hedls and Edc3 enhancers of decapping [64], GW182 [65], the DEx[D/H]-box helicase p54 (also called Rck) [13], the various Argonaute proteins [17], and eIF4E and its binding partner eIF4E-T [18, 19]. Little is known about how P bodies assemble or disassemble. P bodies serve primarily as sites for storing non-translating mRNAs, and their size and number within cells change with fluctuations in this mRNA population. In metazoans, P bodies appear to play a critical role in microRNA-mediated mRNA silencing [15]. Related complexes include maternal P granules in Caenorhabditis elegans, neuronal granules and chromatoid bodies [12], all of which are sites for storing non-translating mRNAs.
Using biochemical fraction and immunofluorescence, Otsuka and co-workers [39] identified a cytoplasmic activity that, like nuclear-capping enzyme, forms a covalent active-site intermediate with GMP bound to its guanylylation domain. Strikingly, this cytoplasmic capping enzyme could transfer covalently bound GMP onto RNA with a 5′-monophosphate end to create a capped (GpppX) product, but it could not function on an RNA with a 5′-hydroxyl end. The transfer reaction requires ATP, and results implied that a kinase converted the 5′-monophosphate end to a 5′-diphosphate capping substrate (Fig. 1, lower panel). Indeed, immunoprecipitated cytoplasmic capping enzyme recovered a kinase activity that labeled 5′-monophosphate RNA with β-32P-ATP. The existence of such a kinase had been proposed more than 30 years ago [7], and contemporary evidence for its existence stemmed from vaccinia virus-infected cells [42]. Nevertheless, direct evidence for a mammalian polynucleotide 5′-monophosphate kinase was provided only this year: this kinase exists in a ~140-kDa complex together with cytoplasmic capping enzyme [39].
What function is served by re-capping the message?
Given the carefully choreographed steps that are used to remove the cap, one has to wonder why cells would have a process to restore a cap. On the face of it, cytoplasmic capping seems dangerous as it has the potential to generate N-terminally truncated proteins from mRNAs that have either undergone endonucleolytic cleavage, as was shown for a hepatitis B virus mRNA as well as a cellular mRNA [34], or have lost a portion of their 5′ end because of exonuclease decay. However, the translation of 5′-truncated mRNAs would be an infrequent event if these mRNAs were largely sequestered in P bodies, which are translationally inactive. As alluded to previously, it is possible that some portion of non-translating mRNAs is stored in an uncapped state and might be returned to the translating pool by re-capping (Fig. 2). In this situation, poly(A) tails that were shortened, for example during an initial stage of microRNA-mediated silencing, could be restored via cytoplasmic polyadenylation to a length that supports translation [43]. This scenario might reflect the changes observed in the population of uncapped yet polyadenylated mRNAs during flower development in A. thaliana [9], as noted above. Other possible physiologically relevant re-capping substrates include mRNAs in chromatoid bodies [44], mRNAs in neuronal granules, and perhaps populations of stem-cell mRNAs.
In addition to full-length mRNAs, recent work identified non-coding RNAs as likely substrates for cytoplasmic de novo capping. Accumulating evidence indicates that most of the genome is transcribed, with the majority of transcription producing non-coding RNAs (ncRNAs). Two classes of ncRNAs are promoter-associated short RNAs (PASRs), which map to regions immediately upstream or downstream of promoters, and longer non-promoter-associated RNAs termed non-PASRs, whose 5′ ends lie downstream of normal transcription initiation sites within the body of fully processed mRNAs [45]. Unexpectedly, the 5′ ends of both PASRs and non-PASRs map to CAGE tags, which are sequences that were identified on the basis of having a cap or cap-like structure [46]. The cap status of these RNAs was confirmed [46] using the same anti-cap monoclonal antibody that was used to recover β-globin mRNA decay intermediates [39].
Concluding remarks
The discoveries of uncapped RNAs and their cytoplasmic re-capping raise many questions (Box 5). What function is served by re-capping small or long RNAs, such as PASRs and non-PASRs [46]? Although a cap can protect against degradation by Xrn1 (or Xrn2 in the nucleus), it also can generate ligands for eIF4E binding. eIF4E is a proto-oncoprotein [47] whose expression is elevated in a number of cancers, and capped small RNAs could serve as ligands that impact eIF4E function. Is re-capping responsible for some of the dynamic changes observed in populations of uncapped mRNAs during flower development in A. thaliana [9]? Does a similar process occur in mammals? In human cells, sodium arsenite-induced oxidative stress can promote the movement of a microRNA-silenced mRNA in P bodies to the pool of translating mRNAs [48]. Consistent with the possibility that movement of an mRNA from P bodies to polysomes could involve mRNA re-capping, Otsuka et al. [39] showed that expressing a dominant-negative form of cytoplasmic capping enzyme in human cells reduces the recovery of cells from stress, suggesting that recovery may be promoted by the recapping of stored mRNAs. Can mRNAs be stored uncapped in a Lazarus-like state, to be reactivated later by cytoplasmic capping? Does cytoplasmic capping expand the proteome by enabling the translation of N-terminally truncated proteins from re-capped mRNAs? Is the cap on these RNAs the same as that on mRNA? Although this roughly appears to be the case, it has been observed that after TAP treatment to remove the cap, PTC-containing β-globin mRNA decay products, unlike the less abundant full-length PTC-containing β-globin mRNA, could not be amplified by cRT-PCR [49], raising the possibility that cytoplasmic capping may entail an additional unidentified modification. Although speculative, this modification might function to distinguish caps added in the cytoplasm from caps added in the nucleus. Other important questions pertain to the identity of the polynucleotide 5′-monophosphate kinase that generates the capping substrate from 5′-monophosphate RNA, and how its activity is regulated. Perhaps the most important issue that remains to be addressed is the degree to which uncapped mRNAs have physiological relevance. Although there are examples of their developmental regulation [14], it remains unclear whether the majority of uncapped mRNAs represent silenced or stored transcripts that can be returned to the functional pool, or if most have no physiological significance, as is the case for the bulk of alternatively spliced mRNAs that are targeted for NMD [50].
Existing studies of cytoplasmic capping have just scratched the surface of what could be a goldmine of new information about RNA metabolism and, possibly, gene regulation. Future work is likely to generate many more surprises about cellular RNomics.
Box 4. Outstanding questions
Most importantly, do uncapped mRNAs have physiological relevance? For example, are they involved in stem cell or embryonic development?
Are there populations of mRNAs that are stored in an uncapped state? Do they fall into specific groups? Do different cell types contain different populations of uncapped mRNAs?
What prevents uncapped RNAs from being immediately degraded? Does Xrn1 pause at specific sites to generate defined 5′ ends and/or is endonucleolytic cleavage responsible for generating 5′-truncated mRNAs?
How are the promoter-associated small RNAs (PASRS) created from primary transcripts? How do these become capped? What is their subcellular distribution? Do these transit between the nucleus and cytoplasm?
What is the purpose of capping small RNAs?
What is the cytoplasmic kinase that is responsible for creating a capping substrate from 5′-monophosphate RNA? How is this regulated?
Do re-capped mRNAs get translated? Do-recapped mRNAs undergo polyadenylation to restore the length of the poly(A) tail to one that promotes translation? Does recapping expand the proteome by facilitating translation of proteins from mRNAs that have lost sequences from their 5′ ends? What prevents re-capped mRNAs from being translated into what could be dominant-negative proteins?
Does eIF4E bind recapped RNAs in vivo? Does this impact translation? Is recapping related to the elevated expression of eIF4E in many cancers?
Acknowledgments
Work in the authors’ laboratories is supported by NIH R01 grants GM038277 and GM079707 (D.R.S) and GM074593 and GM059614 (L.E.M).
Glossary
- 5′ cap
A 5′m7GpppN that typifies the 5′ end of all RNA polymerase II-synthesized transcripts and consists of a 5′ N-methyl guanosine linked by an inverted 5′-5′ triphosphate bridge to the first gene-encoded nucleotide of the transcript. The 5′ cap promotes efficient pre-mRNA processing, mRNA export and mRNA translation. It also stabilizes pre-mRNA and mRNA against decapping and subsequent 5′-to-3′ exonucleolytic activities by virtue of its association with cap binding proteins
- Nuclear capping enzyme
A bifunctional enzyme that catalyzes the addition of an inverted GMP residue onto a diphosphate RNA 5′ end. Nuclear capping enzyme is bound to the C-terminal domain of the large subunit of RNA polymerase II. In nuclear capping, the N-terminal triphosphatase domain catalyzes the hydrolysis of the triphosphate end on the first transcribed nucleotide of newlysynthesized RNA to a diphosphate. The C-terminal portion of capping enzyme contains the guanylyltransferase domain. This reacts with GTP to form a covalent complex of GMP bound to lysine 294, which is then transferred to the diphosphate end to generate the basic GpppN cap structure (termed cap 0). The transfer of a methyl group from S-adenylmethionine to the N-7 of the guanine moiety by cap methyltransferase completes formation of the basic cap structure
- Cytoplasmic capping enzyme
A population of capping enzyme having the same guanylyltransferase activity as nuclear capping enzyme that is present in the cytoplasm in a ~140-kDa complex with a 5′-monophosphate kinase. The kinase converts uncapped RNA with a 5′-monophosphate end to a diphosphate capping substrate. GMP bound to the guanylyltransferase domain of capping enzyme is then transferred onto the diphosphate capping substrate to create the G(5′)-P-P-P-(5′)N end. Cap methyltransferase activity is not present in the cytoplasmic capping enzyme complex, and the process of cap methylation is currently unknown
- CAGE tags
A sequence tag used for identifying capped 5′ ends [46]. Tagging is accomplished by ligating a linker bearing an MmeI restriction site onto the 5′-ends of single-stranded cDNA. MmeI cleaves 20-bp downstream of its recognition site, and cleavage of double-stranded cDNA from this reaction generates a population of 20 bp 5′-end fragments. These are ligated together to form a concatamer that is subsequently sequenced to identify transcripts corresponding to each CAGE tag
- Cap snatching
A process used by a number of negative-strand RNA viruses (e.g., influenza) that allows viral transcripts to be translated by the host cell. In the case of influenza, one subunit of the viral polymerase has endonuclease activity that cleaves host-cell mRNAs to generate capped oligomers of 10–13 nucleotides [41]. These oligomers are used to prime the transcription of negative-strand viral RNAs to generate translationally active ‘capped’ mRNAs
- PASRs
Promoter-associated small RNAs. PASRs are short non-coding RNAs from both DNA strands that map to the regions surrounding transcription start sites and to first exons. The overlap between PASRs and CAGE tags led to the discovery that these short RNAs are capped [45]. Whereas some PASRs are likely capped during transcription, others are capped after being processed from longer precursors
- non-PASRs
Non-promoter-associated small RNAs. Non-PASTs are capped RNAs corresponding to CAGE tags whose 5′ ends map to regions downstream of the transcription start site of fully-processed mRNAs. Fejes-Toth et al. [45] determined that the capped 5′-ends of these RNAs lie within internal exons of 49% of all human genes
References
- 1.Lewis JD, Izaurralde E. The role of the cap structure in RNA processing and nuclear export. Eur J Biochem. 1997;247:461–469. doi: 10.1111/j.1432-1033.1997.00461.x. [DOI] [PubMed] [Google Scholar]
- 2.Furuichi Y, Shatkin AJ. Viral and cellular mRNA capping: past and prospects. Adv Virus Res. 2000;55:135–184. doi: 10.1016/S0065-3527(00)55003-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gu M, Lima CD. Processing the message: structural insights into capping and decapping mRNA. Curr Opin Struct Biol. 2005;15:99–106. doi: 10.1016/j.sbi.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 4.Maquat LE. Nonsense-mediated mRNA decay: splicing, translation and mRNP dynamics. Nat Rev Mol Cell Biol. 2004;5:89–99. doi: 10.1038/nrm1310. [DOI] [PubMed] [Google Scholar]
- 5.Isken O, Maquat LE. Quality control of eukaryotic mRNA: safeguarding cells from abnormal mRNA function. Genes Dev. 2007;21:1833–1856. doi: 10.1101/gad.1566807. [DOI] [PubMed] [Google Scholar]
- 6.Sonenberg N. eIF4E, the mRNA cap-binding protein: from basic discovery to translational research. Biochem Cell Biol. 2008;86:178–183. doi: 10.1139/O08-034. [DOI] [PubMed] [Google Scholar]
- 7.Schibler U, Perry RP. Characterization of the 5′ termini of hnRNA in mouse L cells: implications for processing and cap formation. Cell. 1976;9:121–130. doi: 10.1016/0092-8674(76)90058-1. [DOI] [PubMed] [Google Scholar]
- 8.Gregory BD, et al. A link between RNA metabolism and silencing affecting Arabidopsis development. Dev Cell. 2008;14:854–866. doi: 10.1016/j.devcel.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 9.Jiao Y, et al. Transcriptome-wide analysis of uncapped mRNAs in Arabidopsis reveals regulation of mRNA degradation. Plant Cell. 2008;20:2571–2585. doi: 10.1105/tpc.108.062786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zheng D, et al. Deadenylation is prerequisite for P-body formation and mRNA decay in mammalian cells. J Cell Biol. 2008;182:89–101. doi: 10.1083/jcb.200801196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu L, et al. MicroRNAs direct rapid deadenylation of mRNA. Proc Natl Acad Sci USA. 2006;103:4034–4039. doi: 10.1073/pnas.0510928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parker R, Sheth U. P bodies and the control of mRNA translation and degradation. Mol Cell. 2007;25:635–646. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 13.Chu CY, Rana TM. Translation repression in human cells by microRNA-Induced gene silencing requires RCK/p54. PLoS Biology. 2006;4:e210. doi: 10.1371/journal.pbio.0040210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sheth U, Parker R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 2003;300:805–808. doi: 10.1126/science.1082320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eulalio A, et al. P bodies: at the crossroads of post-transcriptional pathways. Nat Rev Mol Cell Biol. 2007;8:9–22. doi: 10.1038/nrm2080. [DOI] [PubMed] [Google Scholar]
- 16.Liu J, et al. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat Cell Biol. 2005;7:719–723. doi: 10.1038/ncb1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sen GL, Blau HM. Argonaute 2/RISC resides in sites of mammalian mRNA decay known as cytoplasmic bodies. Nat Cell Biol. 2005;7:633–636. doi: 10.1038/ncb1265. [DOI] [PubMed] [Google Scholar]
- 18.Ferraiuolo MA, et al. A role for the eIF4E-binding protein 4E-T in P-body formation and mRNA decay. J Cell Biol. 2005;170:913–924. doi: 10.1083/jcb.200504039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andrei MA, et al. A role for eIF4E and eIF4E-transporter in targeting mRNPs to mammalian processing bodies. RNA. 2005;11:717–727. doi: 10.1261/rna.2340405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Richter JD, Sonenberg N. Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature. 2005;433:477–480. doi: 10.1038/nature03205. [DOI] [PubMed] [Google Scholar]
- 21.Humphreys DT, et al. MicroRNAs control translation initiation by inhibiting eukaryotic initiation factor 4E/cap and poly(A) tail function. Proc Natl Acad Sci USA. 2005;102:16961–16966. doi: 10.1073/pnas.0506482102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thermann R, Hentze MW. Drosophila miR2 induces pseudo-polysomes and inhibits translation initiation. Nature. 2007;447:875–878. doi: 10.1038/nature05878. [DOI] [PubMed] [Google Scholar]
- 23.Mathonnet G, et al. MicroRNA inhibition of translation initiation in vitro by targeting the cap-binding complex eIF4F. Science. 2007;317:1764–1767. doi: 10.1126/science.1146067. [DOI] [PubMed] [Google Scholar]
- 24.Franks TM, Lykke-Andersen J. TTP and BRF proteins nucleate processing body formation to silence mRNAs with AU-rich elements. Genes Dev. 2007;21:719–735. doi: 10.1101/gad.1494707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murray EL, Schoenberg DR. A+U-rich instability elements differentially activate 5′-3′ and 3′-5′ mRNA decay. Mol Cell Biol. 2007;27:2791–2799. doi: 10.1128/MCB.01445-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Filipowicz W, et al. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 27.Wu L, Belasco JG. Let me count the ways: mechanisms of gene regulation by miRNAs and siRNAs. Mol Cell. 2008;29:1–7. doi: 10.1016/j.molcel.2007.12.010. [DOI] [PubMed] [Google Scholar]
- 28.Choi YH, Hagedorn CH. Purifying mRNAs with a high-affinity eIF4E mutant identifies the short 3′ poly(A) end phenotype. Proc Natl Acad Sci USA. 2003;100:7033–7038. doi: 10.1073/pnas.1232347100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lim S, et al. Novel metabolism of several β°-thalassemic β-globin mRNAs in the erythroid tissues of transgenic mice. EMBO J. 1989;8:2613–2619. doi: 10.1002/j.1460-2075.1989.tb08401.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lim SK, Maquat LE. Human beta-globin mRNAs that harbor a nonsense codon are degraded in murine erythroid tissues to intermediates lacking regions of exon I or exons I and II that have a cap-like structure at the 5′ termini. EMBO J. 1992;11:3271–3278. doi: 10.1002/j.1460-2075.1992.tb05405.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lim SK, et al. Nonsense codons in human beta-globin mRNA result in the production of mRNA degradation products. Mol Cell Biol. 1992;12:1149–1161. doi: 10.1128/mcb.12.3.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stevens A, et al. Beta-globin mRNA decay in erythroid cells: UG site-preferred endonucleolytic cleavage that is augmented by a premature termination codon. Proc Natl Acad Sci USA. 2002;99:12741–12746. doi: 10.1073/pnas.192442399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bremer KA, et al. An endonuclease activity similar to Xenopus PMR1 catalyzes the degradation of normal and nonsense-containing human beta-globin mRNA in erythroid cells. RNA. 2003;9:1157–1167. doi: 10.1261/rna.5720303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thoma C, et al. Generation of stable mRNA fragments and translation of N-truncated proteins induced by antisense oligodeoxynucleotides. Mol Cell. 2001;8:865–872. doi: 10.1016/s1097-2765(01)00364-1. [DOI] [PubMed] [Google Scholar]
- 35.Eberle AB, et al. SMG6 promotes endonucleolytic cleavage of nonsense mRNA in human cells. Nat Struct Mol Biol. 2009;16:49–55. doi: 10.1038/nsmb.1530. [DOI] [PubMed] [Google Scholar]
- 36.Lejeune F, et al. Nonsense-mediated mRNA decay in mammalian cells involves decapping, deadenylating, and exonucleolytic activities. Mol Cell. 2003;12:675–687. doi: 10.1016/s1097-2765(03)00349-6. [DOI] [PubMed] [Google Scholar]
- 37.Couttet P, Grange T. Premature termination codons enhance mRNA decapping in human cells. Nucleic Acids Res. 2004;32:488–494. doi: 10.1093/nar/gkh218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamashita A, et al. Concerted action of poly(A) nucleases and decapping enzyme in mammalian mRNA turnover. Nat Struct Mol Biol. 2005;12:1054–1063. doi: 10.1038/nsmb1016. [DOI] [PubMed] [Google Scholar]
- 39.Otsuka Y, et al. Identification of a cytoplasmic complex that adds a cap onto 5′-monophosphate RNA. Mol Cell Biol. 2009;29:2155–2167. doi: 10.1128/MCB.01325-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ogino T, Banerjee AK. Unconventional mechanism of mRNA capping by the RNA-dependent RNA polymerase of vesicular stomatitis virus. Mol Cell. 2007;25:85–97. doi: 10.1016/j.molcel.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 41.Dias A, et al. The cap-snatching endonuclease of influenza virus polymerase resides in the PA subunit. Nature. 2009;458:914–918. doi: 10.1038/nature07745. [DOI] [PubMed] [Google Scholar]
- 42.Spencer E, et al. Enzymatic conversion of 5′-phosphate-terminated RNA to 5′-di- and triphosphate-terminated RNA. Proc Natl Acad Sci USA. 1978;75:4793–4797. doi: 10.1073/pnas.75.10.4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kwak JE, et al. Mammalian GLD-2 homologs are poly(A) polymerases. Proc Natl Acad Sci USA. 2004;101:4407–4412. doi: 10.1073/pnas.0400779101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kotaja N, Sassone-Corsi P. The chromatoid body: a germ-cell-specific RNA-processing centre. Nat Rev Mol Cell Biol. 2007;8:85–90. doi: 10.1038/nrm2081. [DOI] [PubMed] [Google Scholar]
- 45.Fejes-Toth K, et al. Post-transcriptional processing generates a diversity of 5′-modified long and short RNAs. Nature. 2009;457:1028–1032. doi: 10.1038/nature07759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shiraki T, et al. Cap analysis gene expression for high-throughput analysis of transcriptional starting point and identification of promoter usage. Proc Natl Acad Sci USA. 2003;100:15776–15781. doi: 10.1073/pnas.2136655100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mamane Y, et al. eIF4E from translation to transformation. Oncogene. 2004;23:3172–3179. doi: 10.1038/sj.onc.1207549. [DOI] [PubMed] [Google Scholar]
- 48.Bhattacharyya SN, et al. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- 49.Grange T. Sensitive detection of mRNA decay products by use of reverse-ligation-mediated PCR (RL-PCR) Methods Enzymol. 2008;448:445–466. doi: 10.1016/S0076-6879(08)02622-0. [DOI] [PubMed] [Google Scholar]
- 50.Blencowe BJ. Alternative splicing: new insights from global analyses. Cell. 2006;136:37–4. doi: 10.1016/j.cell.2006.06.023. [DOI] [PubMed] [Google Scholar]
- 51.Marcotrigiano J, et al. Cocrystal structure of the messenger RNA 5′ cap-binding protein (eIF4E) bound to 7-methyl-GDP. Cell. 1997;89:951–961. doi: 10.1016/s0092-8674(00)80280-9. [DOI] [PubMed] [Google Scholar]
- 52.Matsuo H, et al. Structure of translation factor eIF4E bound to m7GDP and interaction with 4E-binding protein. Nat Struct Biol. 1997;4:717–724. doi: 10.1038/nsb0997-717. [DOI] [PubMed] [Google Scholar]
- 53.Niedzwiecka A, et al. Biophysical studies of eIF4E cap-binding protein: recognition of mRNA 5′ cap structure and synthetic fragments of eIF4G and 4E-BP1 proteins. J Mol Biol. 2002;319:615–635. doi: 10.1016/S0022-2836(02)00328-5. [DOI] [PubMed] [Google Scholar]
- 54.Tomoo K, et al. Crystal structures of 7-methylguanosine 5′-triphosphate (m(7)GTP)- and P(1)-7-methylguanosine-P(3)-adenosine-5′,5′-triphosphate (m(7)GpppA)-bound human full-length eukaryotic initiation factor 4E: biological importance of the C-terminal flexible region. Biochem J. 2002;362:539–544. doi: 10.1042/0264-6021:3620539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Izaurralde E, et al. A nuclear cap binding protein complex involved in pre-mRNA splicing. Cell. 1994;78:657–668. doi: 10.1016/0092-8674(94)90530-4. [DOI] [PubMed] [Google Scholar]
- 56.Mazza C, et al. Crystal structure of the human nuclear cap binding complex. Mol Cell. 2001;8:383–396. doi: 10.1016/s1097-2765(01)00299-4. [DOI] [PubMed] [Google Scholar]
- 57.Worch R, et al. Specificity of recognition of mRNA 5′ cap by human nuclear cap-binding complex. RNA. 2005;11:1355–1363. doi: 10.1261/rna.2850705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Martineau Y, et al. Poly(A)-binding protein-interacting protein 1 binds to eukaryotic translation initiation factor 3 to stimulate translation. Mol Cell Biol. 2008;28:6658–6667. doi: 10.1128/MCB.00738-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Isken O, Maquat LE. The multiple lives of NMD factors: balancing roles in gene and genome regulation. Nat Rev Genet. 2008;9:699–712. doi: 10.1038/nrg2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Franks TM, Lykke-Andersen J. The control of mRNA decapping and P-body formation. Mol Cell. 2008;32:605–615. doi: 10.1016/j.molcel.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bashkirov VI, et al. A mouse cytoplasmic exoribonuclease (mXRN1p) with preference for G4 tetraplex substrates. J Cell Biol. 1997;136:761–773. doi: 10.1083/jcb.136.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Eystathioy T, et al. A phosphorylated cytoplasmic autoantigen, GW182, associates with a unique population of human mRNAs within novel cytoplasmic speckles. Mol Biol Cell. 2002;13:1338–1351. doi: 10.1091/mbc.01-11-0544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.van DE, et al. Human Dcp2: a catalytically active mRNA decapping enzyme located in specific cytoplasmic structures. EMBO J. 2002;21:6915–6924. doi: 10.1093/emboj/cdf678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fenger-Gron M, et al. Multiple processing body factors and the ARE binding protein TTP activate mRNA decapping. Mol Cell. 2005;20:905–915. doi: 10.1016/j.molcel.2005.10.031. [DOI] [PubMed] [Google Scholar]
- 65.Eystathioy T, et al. The GW182 protein colocalizes with mRNA degradation associated proteins hDcp1 and hLSm4 in cytoplasmic GW bodies. RNA. 2003;9:1171–1173. doi: 10.1261/rna.5810203. [DOI] [PMC free article] [PubMed] [Google Scholar]